Abstract
The phenomena of protein reconstitution and three-dimensional domain swapping reveal that highly similar structures can be obtained whether a protein is comprised of one or more polypeptide chains. In this review, we use protein reconstitution as a lens through which to examine the range of protein tolerance to chain interruptions and the roles of the primary structure in related features of protein structure and folding, including circular permutation, natively unfolded proteins, allostery, and amyloid fibril formation. The results imply that noncovalent interactions in a protein are sufficient to specify its structure under the constraints imposed by the covalent backbone.
Keywords: stability, metastability, steric constraints, cooperativity, ligand binding
Protein structure and folding reflect the large number of noncovalent contacts that form under the very substantial constraints imposed by the covalent chain. Due to steric overlap, roughly 90% of the combinations of backbone torsion angles phi and psi are inaccessible. Nevertheless, the accessible ∼10% of the Ramachandran map allows for a remarkable variation in protein folds through combinations of extended or helical segments and loops. Although chain connectivity restricts protein conformational space through steric hindrance, it also significantly limits losses due to configurational entropy as the protein folds. Thus, chain connectivity is at once both restrictive and permissive, and is commonly viewed as essential for protein folding and structure. Yet the widespread phenomenon of protein reconstitution (Hirs and Timasheff 1986; Fisher and Taniuchi 1992; Håkansson and Linse 2002) demonstrates that many native structures tolerate breaks in the polypeptide chain, and native contacts reform from separated fragments in an assembly reaction akin to intramolecular folding (Fig. 1). Like reconstitution, the process of three-dimensional domain swapping (Fig. 2) relies on native contacts, and, in fact, often uses chain segments that are also observed to reconstitute (Fig. 3). Swapping has enjoyed several excellent influential reviews (Heringa and Taylor 1997; Schlunegger et al. 1997; Liu and Eisenberg 2002; Newcomer 2002; Rousseau et al. 2003). The present review assimilates with swapping the closely related process of fragment reconstitution to consider a diverse group of protein phenomena with the aim of shedding light on common underlying features of protein structure and folding. The ideas developed here use examples from the literature and from our own work that are chosen for their conceptual value rather than to make an exhaustive review of the literature. The discussion relates to in vitro folding, acknowledging that in vivo folding can be governed by additional factors.
Figure 1.
Protein reconstitution and intramolecular folding. Monellin (Mn, 4MON.pdb) is an example of a naturally complemented protein. As isolated from the serendipity berry Mn occurs as two chains that form a single globular domain (Kohmura et al. 1990). The separated chains reform the native structure upon mixing (Kohmura et al. 1991). An artificial single-chain variant (scMn, 1MOL.pdb) with the two chains linked folds to the same native structure (Kim et al. 1989; Somoza et al. 1993). The equilibrium constants for the two folding processes reflect the difference in their molecularity, with consequent need to define the standard state for the intermolecular reconstitution reaction. In the example given, a standard state of 1 M is used.
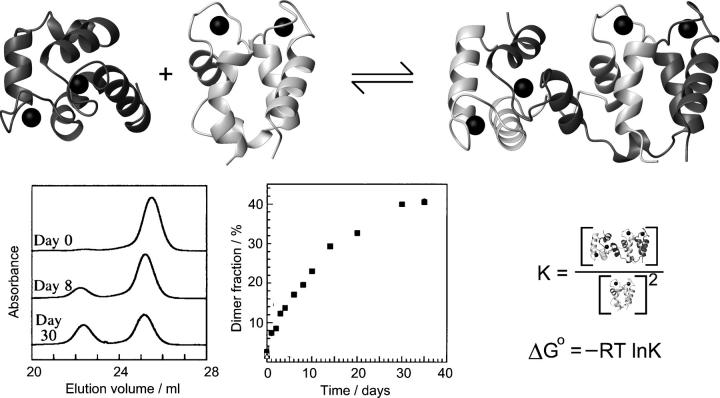
Figure 2.
Domain swapping. The example shown is calbindin D9k (Håkansson et al. 2001). Formation of intertwined dimers occurs in which the monomer structure is restored twice through reciprocal exchange of structural elements between the chains. Each monomer-like unit thus contains secondary structure elements from both chains. The swapping process can be observed experimentally by gel filtration and is typically characterized by slow kinetics of both forward and reverse reactions.
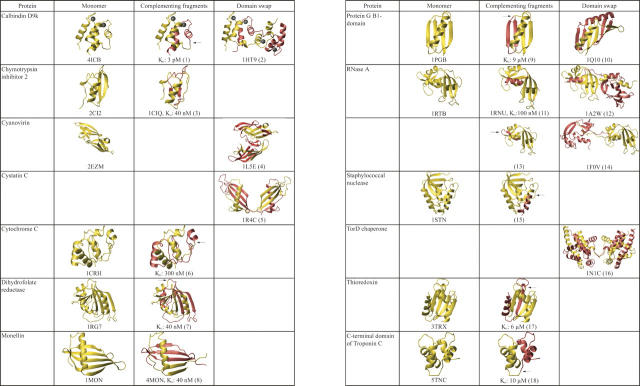
Figure 3.
Structural similarity of fragment-complemented and domain-swapped proteins. Structures of each monomeric protein and its corresponding complementing fragments and domain-swapped states were prepared from the indicated pdb files. For reconstituted proteins where the structure of the complex has not been determined, the monomeric structure is colored to indicate the complementing fragments, and an arrow marks the break in the covalent chain. Dissociation constants are given for complexes where these have been determined experimentally. References: (1) Berggard et al. (2001); (2) Håkansson et al. (2001); (3) de Prat Gay et al. (1994); (4) Barrientos et al. (2002); (5) Janowski et al. (2001); (6) Hantgan and Taniuchi (1977); (7) Smith and Matthews (2001); (8) Xue et al. (2004); (9) Kobayashi et al. (1995); (10) Byeon et al. (2003), (11) Richards (1958); (12) Liu et al. (1998); (13) Gutte et al. (1972); (14) Liu et al. (2001); (15) Taniuchi and Anfinsen (1969); (16) Tranier et al. (2003); (17) Holmgren (1972); (18) Shaw et al. (1994).
It has long been known that proteolytic fragments of some enzymes recombine noncovalently to regenerate activity (Kalman et al. 1955; Richards 1958). Protein reconstitution and fragment complementation are used as interchangeable terms to describe this phenomenon. Regain of activity upon mixing separated fragments implies that they interact spontaneously in the correct molar ratio to reform native-like tertiary structure in the complex. More recent direct structural analyses have demonstrated that reconstituted proteins adopt structures very similar to those of their intact progenitors despite the presence of one or more nicks in the reconstituted assembly, even when the isolated fragments display little or no residual structure. The degree of folded structure that persists in isolated fragments prior to reconstitution is quite variable, ranging from completely disordered to nearly native-like (Kobayashi et al. 1993; de Prat-Gay et al. 1994; Neira et al. 1996; Julenius et al. 2002). This finding indicates that pre-existing folded structure is not required for reconstitution, but rather that folding can occur concomitantly with association of fragments. The range of documented complementation reactions (Hirs and Timasheff 1986; de Prat-Gay 1996; Peng and Wu 2000; Håkansson and Linse 2002) includes proteins of many structural classes that reconstitute from diverse sizes and numbers of fragments (Fig. 3). Both monomeric and multimeric proteins have been reconstituted, and chain redundancies or deletions are sometimes accommodated. This versatility strongly suggests that protein reconstitution is not an aberrant feature of a few unusual proteins, but rather derives from common properties of proteins.
Interdependence of protein structural levels
The remarkable ability of even disordered protein fragments to associate into native-like folded structures demonstrates that chain connectivity cannot be considered the defining feature of a protein's structure. This interpretation is reinforced and extended by the related phenomenon of circular permutation (Fig. 4), in which the native termini of a protein are joined and new ends are created in another position (for review, see Lindqvist and Schneider 1997). As for fragment complementation, a wide range of proteins of different structural classes tolerate circular permutation with retention of structure and/or activity. Systematic variation of the positions of the ends (Shiba and Schimmel 1992; Graf and Schachman 1996) reveals that in some proteins many different locations of the termini are tolerated. Like fragment reconstitution, circular permutation presents an example of formation of native tertiary structure from nonnative primary structure (Smith and Matthews 2001). In addition, permutation demonstrates that native structures are maintained even when the linear order of secondary structure elements, i.e., the chain topology, is altered. Thus, neither chain connectivity nor chain topology is the defining feature of a protein's fold.
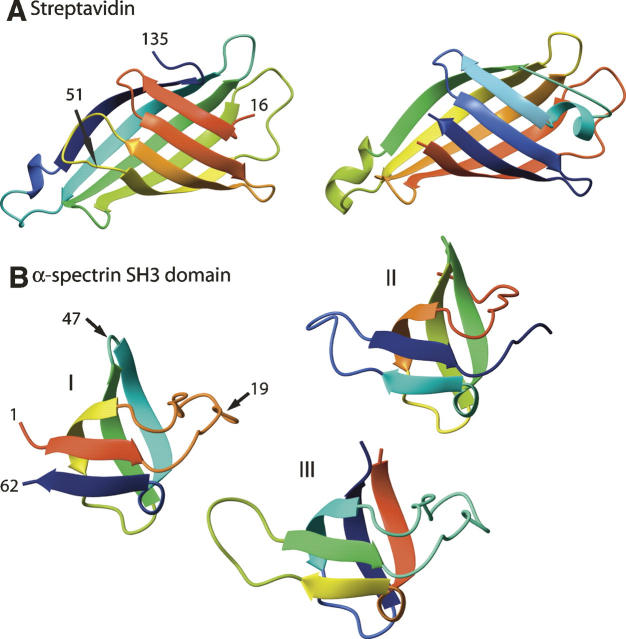
Figure 4.
Circular permutation rearranges the linear order of secondary structure elements. Rainbow colors trace each chain from red N to blue C terminus. (A) Native streptavidin (left, 1SWB.pdb) permuted (right) with new N terminus at original position 51 (arrow). The two structures are similar with some differences in details, but biotin affinity is reduced by six orders of magnitude in the permutant (Chu et al. 1998). (B) Native α-spectrin (I; 1SHG.pdb) and permutants with new N termini at original position 19 (II, 1TUC.pdb) or 47 (III, 1TUD.pdb). Fold stability is only slightly reduced in the permutants with little change in structure, but radical changes occurred in folding kinetics (Viguera et al. 1995).
Examination of sequence context effects reveals that the primary, secondary, and tertiary levels of protein structure are interdependent. A given residue substitution in different positions typically has very different effects on protein conformational stability. Point mutations are known that convert β-strands into α-helices (Cordes et al. 1999). The secondary structures of chameleon sequences depend on their location in the primary structure (Minor and Kim 1996), and chameleon proteins can adopt an unrelated fold with only a limited number of residue substitutions (Dalal et al. 1997). The interdependence of levels in protein structural organization reflects the inherent metastability of secondary structures that is rooted in steric constraints. All residue types can occupy both α and β regions of φ, ψ space, with slightly different energies in the two regions and relatively low energy barriers between them. The inherent metastability of secondary structures, and the reciprocity between secondary and tertiary structures that is implied by their interdependence, together account for the fact that structural specificity is highly dependent on primary structure context (Lattman and Rose 1993). This view suggests that the interdependence of secondary and tertiary structures is both the molecular origin of global cooperativity of folding and the defining feature of a protein's structure.
Given that the native structure of a protein represents a free-energy minimum that is strongly sequence dependent, it may seem surprising that in many cases the same protein sequence can adopt structures representing alternative minima. The most dramatic example appears to be that of amyloid fibrils, which form a highly similar multimeric state with a characteristic cross-β structure regardless of the native fold or sequence of the parent protein. The secondary, tertiary, and quaternary structure common to fibrils thus has very wide tolerance for primary structure. Fibril formation is documented for so many proteins that it may well be universal, and it has been proposed to reflect universal properties of proteins (Dobson 1999). Despite intensive and widespread effort, these underlying properties have not been elucidated.
A less dramatic example of an alternative free-energy minimum is the three-dimensional domain-swapped assembly (Fig. 2), in which part of the tertiary structure of one chain is replaced by the corresponding part of another chain (Bennett et al. 1994). Swaps can be reciprocated, resulting in a dimer, or nonreciprocated, propagating a multimeric assembly. Swapping has in common with fragment reconstitution that the tertiary structure begins in one chain and is completed using segments that originate from another chain (Håkansson and Linse 2002). Indeed, several examples are known in which swapped regions correspond closely to fragments for which reconstitution is observed (Fig. 3). Antiparallel coiled coils may be considered a special case of domain swapping. Both three-dimensional domain swapping and amyloid formation illustrate that protein quaternary structures are neither uniquely encoded by the primary structure nor independent of secondary and tertiary structural context.
Finally, allostery and folding coupled to ligand binding (Tsai et al. 1999; Dyson and Wright 2002) focus attention on the very important distinction between the stability and specificity of protein structures (Lattman and Rose 1993). Ligand binding to proteins can be accompanied by a very wide range of structural consequences, including even global folding in the growing class of natively unfolded proteins (Dyson and Wright 2005). In this class, the role of the primary structure in specifying the fold appears to be ambiguous because it is required to encode the fold even if it is not sufficient by itself to express the structure. On the contrary, such proteins reinforce the view that a protein's structure is specified by its sequence, because they show that specific conformations can be achieved even if they are not the lowest free-energy state of the isolated protein. Whereas natively folded proteins are marginally stable, natively unfolded proteins may be viewed as marginally unstable, and part of the free-energy change contributed by the ligand-binding event completes the folding process. Thus, the free energy required to order a partly ordered protein lowers the ligand affinity compared with a protein that is already in the bound conformation. Ligands ranging from simple small ions to partners of comparable size including nucleic acids or other proteins are known to induce such structural changes. Recent reports of allosteric behavior in seemingly nonallosteric proteins (Wand 2001; Gunasekaran et al. 2004; Clarkson et al. 2006) support the idea that proteins are inherently able to transduce local binding events into global responses, implying that allostery may be a property of all proteins. The molecular basis of this coupling has long been appreciated (Weber 1972), even if details are only now being revealed through NMR and other analyses.
The above phenomena collectively illustrate that native secondary, tertiary, and quaternary structures can be formed from nonnative primary structures; that primary structures in slightly different contexts can yield quite different secondary, tertiary, and/or quaternary structures; and that the primary, secondary, tertiary, and quaternary levels of the protein structural hierarchy are intimately interdependent. Protein reconstitution offers a unique perspective from which to re-examine how protein structures are encoded in their sequences.
Nature and limits of reconstitution
Before the discovery of natively unfolded proteins, domain swaps, and single-domain proteins composed of more than one chain, there appeared to be a distinction between reconstitution and other processes leading to folded structures. However, already in the very first example of reconstitution, RNase A, Richards suggested that the distinction between proteins and their cofactors was blurred by the fact that the 20-residue RNase S-peptide restores the enzymatic activity of S-protein (residues 21–124) in the RNase S complex (Richards 1958). Richards' (1958) suggestion leads to the question of how reconstitution is related to ligand binding on the one hand and intramolecular folding on the other. Where is the boundary between covalent and noncovalent processes in proteins? What is the lower limit of fragment size for reconstitution? The examples introduced here to address these questions suggest that the processes of reconstitution, folding, swapping, and ligand binding share common underlying structural and energetic features.
Truncated S-peptides of only 13 residues bind to S-protein with essentially the same affinity as does the 20-residue S-peptide (Shoemaker et al. 1987). These short peptides are among the very few in this size range from any source that display residual native-like structure as monomers in the absence of the rest of the protein. Preformed native-like structure in separated fragments enhances reconstitution affinity by lowering the configurational entropy loss on association, but it is not a prerequisite for reconstitution, as many other examples show. The residual helical structure of RNase peptides offered a rare opportunity to study in parallel the effects of sequence alterations on peptide helix stability and on protein reconstitution affinity (Mitchinson and Baldwin 1986). The results showed a clear correlation, providing one of the earliest indications that folding and binding are indeed closely related. Thus, RNase A could be considered a covalent analog of the RNase S complex.
The smallest unit that can be considered to complement is a single amino acid or even just a side chain, as exemplified by several cases. The helix-turn-helix (hth) motif sequence of TrpR (Fig. 5A,B) differs from other bacterial hth repressors in presenting a glycine instead of an aromatic residue at a key position in the second helix. The TrpR hth motif is relatively dynamic and becomes more ordered upon binding of the corepressing ligand L-tryptophan (for review, see Jardetzky 1996), which occupies a crevice afforded by the glycine. Replacement of this glycine by a tryptophan residue yields a constitutively activated repressor (Komeiji et al. 1994), presumably by increasing local order in the hth, as do other bulky substitutions in this region (Gryk and Jardetzky 1996). This example further suggests that evolution can take advantage of spontaneous destabilizing mutations that bring folding under the control of desirable exogenous ligands, in this case, the end-product of the pathway regulated by TrpR. An example where only a side chain binds in a void within the structure is provided by a phenylalanine-to-alanine mutant of bacteriophage T4 lysozyme, which binds benzene in place of the phenyl ring with concomitant gain of stability (Eriksson et al. 1992).
Figure 5.
Tryptophan repressor, TrpR. (A) One intact TrpR dimer (1P6Z.pdb), with helix-turn-helix regions colored to identify the two chain segments of each monomer that reconstitute to reform a native-like dimer. One intact monomer consists of the pink N-terminal segment (residues 8–71; residues 1–7 are disordered) and the green C-terminal segment (residues 72–108), and the other monomer is blue and gray. The domain swap of the intact native dimer is evident from the pink and blue segments, which extend across the subunit interface in opposite directions. (B) 1H-NMR spectra of (top to bottom) intact TrpR, fragment 8–71, fragment 72–108, and equimolar mixture of 8–71 and 72–108. (C) The intertwined multimeric crystalline array formed by intact TrpR in aqueous alcohol. Two nodes of the extended array are shown. Colors correspond to A to depict the structural roles and chain origins of each segment, but all chains in the array are intact: Each node contains segments of four chains, and each chain participates in two nodes. A single blue chain spans both of the nodes shown; the central long helical segment spans ∼50 residues. Each node differs in structure from ordinary dimeric TrpR by only ∼1.0 Å. RSMD. (D) Section of the hexagonal crystal lattice (AB plane view) showing all protein atoms (gray) of one unit cell c repeat (parallelogram). The model shown is refined at 2.5 Å for one intact chain representing one asymmetric unit. The pores within the lattice have diameter ∼50 Å. A, C, and D are from Lawson et al. (2004) (reprinted with permission from Elsevier © 2004); B is from Tasayco and Carey (1992) (reprinted with permission from the American Association for the Advancement of Science © 1992).
Single-monomer complementation is also known for RNA, as in the ATP-binding aptamer that forms a well-defined tertiary structure in which the nucleotide ligand plays a key role (Dieckmann et al. 1996; Jiang et al. 1996). Other examples include the binding of natural protein ligands like fatty acids, which sometimes form an integral part of a protein's hydrophobic core (Prinsen and Veerkamp 1996; Zimmerman et al. 2001). Many other kinds of ligands and cofactors often cause dramatic increases in protein thermal stability, indicating that the ligand-binding equilibrium is coupled to the unfolding equilibrium and implying that the ligand may be integral to the structure (for review, see Higgins et al. 2005). These examples make it clear that reconstitution cannot be distinguished from ligand binding, whether or not folding accompanies the event.
There is at least one well-known example of macromolecular self-assembly that proceeds entirely from constituent monomers with no covalent bonds between them. Lipid monomers spontaneously assemble into supramolecular structures whose size, composition, and physical properties can nevertheless be well-defined (Gennis 1989; Lafleur et al. 1996). In order for amino acids to self-assemble into a unique three-dimensional structure, each one would have to adopt a unique location. Thus, the entropy difference between free and assembled states is much larger in the protein case than for lipids. Similarly, the ability of monomeric nucleobases to self-associate through stacking interactions is well-known, and is often invoked in “RNA world” scenarios. In this light, proteins appear unique in the inability of their constituent monomers to self-associate, another evidence of the interdependence of protein structural levels. Chain connectivity plays a proportionately more restrictive role for proteins than for RNAs. RNA monomers display remarkable steric restriction consistent with the three-dimensional conformational preferences of RNA polymers (Murthy et al. 1999). For proteins, the main steric constraints on the backbone come from steric clash between peptide groups and side chains.
Given that short peptides and even single residues exhibit high-affinity reconstitution, a logical corollary question is, How far can a native protein be dissected and still undergo reconstitution? Even proteins of rather small size are known to reassemble noncovalently from multiple fragments. For cytochrome c, a complex of three fragments has electron acceptor activity (Fisher and Taniuchi 1992). Cytochrome c might be considered a special case because the heme remains covalently attached via thioether links to one fragment, and its iron ion provides coordination with both fragments. Heme covalency cannot be the only factor, however, because two apocytochrome c peptides plus heme form a structured ternary complex (Kang and Carey 1999). Other small proteins that reconstitute from multiple fragments also exploit facilitating ligands. For example, calbindin D28k reassembles from six fragments of 33 residues each (Fig. 6), but only in the presence of calcium (Linse et al. 1997); some of its isolated individual peptides also acquire secondary structure in the presence of calcium.
Figure 6.

High-affinity reconstitution from multiple fragments. Intact calbindin D28k (2G9B.pdb) shown as a space-filling model (top) and ribbon diagram (bottom) with the same orientation and color coding of the six individual EF-hands that reconstitute the protein (Linse et al. 1997).
These examples demonstrate that the boundary between covalent and noncovalent features of polymeric macromolecules extends at its lower limit to single individual monomers. For proteins, it is difficult to understand why there are so few examples of naturally reconstituting proteins, considering that the covalent/noncovalent boundary is demonstrably a moving target. The scarcity of such forms in vivo when they are so amply demonstrated in vitro implies the existence of negative selection factors that operate at a level other than folding itself. Although there are relatively few documented natural examples of fragment complementation, some of these unify fragment complementation with protein folding.
Relationship of reconstitution to folding
The major histocompatibility complex (MHC) proteins of the immune response present a variation on the theme of interdependency among protein organizational levels. A relatively small repertoire of MHCs is responsible for presenting to T-cells all possible peptides processed from a lifetime of antigen exposure. Thus, MHCs must tolerate essentially unlimited peptide variation while maintaining relatively high affinity. This combination of high affinity and low specificity is achieved using a coupled folding and binding mechanism. Free MHCs display properties of molten globules in solution (Bouvier and Wiley 1998), and binding of a peptide ligand confers dramatic stability toward thermal and chemical denaturation. The original high-resolution structures of isolated natural MHCs (Björkman et al. 1987) revealed bound peptides of heterogeneous sequence that nevertheless shared similar binding features: an extended structure with generic functional groups (backbone H-bond donors and acceptors; hydrophobic groups of similar volume) that contact MHC in a network of interactions like those found within folded proteins. Thus, peptide binding organizes the MHC tertiary fold through quaternary interactions, analogously to protein reconstitution and ligand-coupled folding. Peptide affinity is high despite its low specificity, because dissociation cannot be faster than the slow global unfolding of the complex. The low specificity of MHC-peptide binding reveals an inherent plasticity that allows folding by mutual adaptation of the partners.
Calmodulin-target protein recognition is a slight variation on the theme. Calmodulin binds to and regulates a number of enzymes through calcium-induced protein–protein interaction (for review, see Ikura and Ames 2006). The calmodulin regulatory network thus enables coordinated control of a wide range of cellular processes through binding to many unrelated proteins. However, similar regulatory events also occur through intramolecular conformational change in proteins with covalently attached calmodulin-like domains like the enzyme calpain. Such proteins can be regulated independently of the calmodulin regulatory network, offering a biological rationale for the existence of both independent calmodulin and covalently connected calmodulin-like domains. However, the underlying molecular events are similar in both cases. Structures of calmodulin–peptide complexes (Fig. 7) reveal that diverse peptides form an integral part of the protein's fold, similar to the MHC-peptide case. Consistent with its plasticity and global adaptation, calmodulin reassembles with high affinity from two fragments if Ca2+ and a target peptide are present (Shuman et al. 2006).
Figure 7.

Reconstitution of calmodulin facilitated by target peptide. (Top) Calmodulin in complex with a target peptide from smooth muscle mysosin light chain kinase (1CDL.pdb). (Middle) The N- and C-terminal domains of calmodulin are pulled apart and rotated 90 degrees to show the bound peptide and the contact surfaces. (Bottom) The complex and the separated lobes as a ribbon diagram.
Domain swapping and higher-order oligomerization
Domain swapping has been reviewed recently by Liu and Eisenberg (2002), and also by Håkansson and Linse (2002) who pointed out its relationship to reconstitution. Numerous proteins are found to undergo both processes using similar chain segments (Fig. 3). In all documented cases, there is a remarkable structural similarity between the native, reconstituted, and domain-swapped protein, implying a key role for noncovalent interactions. Most of the original native contacts are reformed in a reconstituted protein or a domain-swapped oligomer. The swapping process can be described as a transition between states that have similar balances of favorable and unfavorable free-energy contributions, and that are separated by a transition state in which the protein has opened up to permit exchange of segments between chains. Whereas the monomeric and multimeric states are nearly isoenergetic (i.e., the value of the equilibrium association constant is low), the barrier between them is often very high (i.e., the kinetics of conversion are slow). Because domain swapping is an intermolecular reaction, the fraction of multimer at equilibrium is concentration dependent (Fig. 8), and the free-energy difference between the two states depends on the choice of standard state. Merely raising the protein concentration thus shifts the equilibrium toward domain-swapped states due to the law of mass action. For example, if the equilibrium association constant for dimerization is 105 M−1 (KD = 10 μM), monomers dominate (∼90%) at 1 μM total protein concentration, and dimers dominate (∼90%) at 100 μM. At typical crystallization conditions of 1 mM, dimers dominate even for very weakly dimerizing proteins (>50% if dimerization constant is 103 M−1 or higher [KD ≤ 1 mM]).
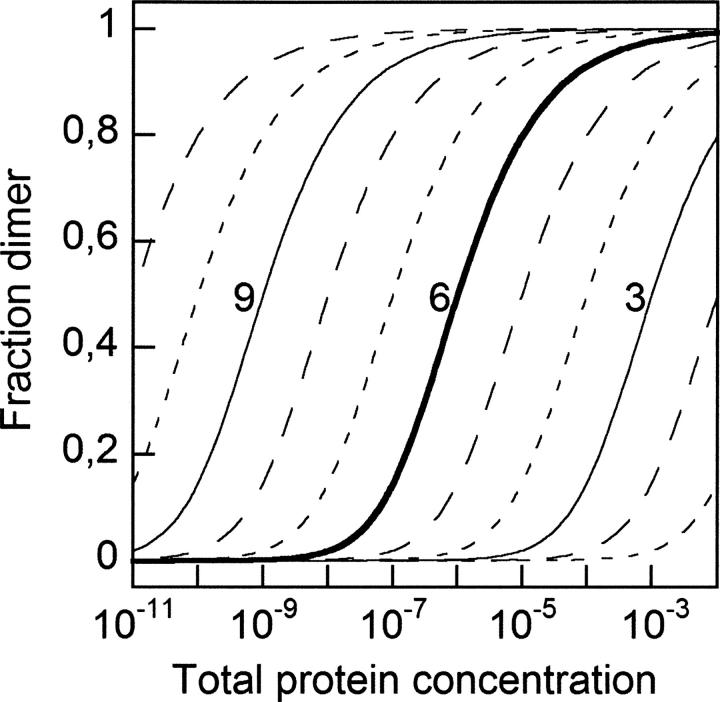
Figure 8.
Association governed by concentration and equilibrium constant. For the association of two monomers to form a dimer, the fraction of protein present as dimer is shown as a function of total protein concentration for equilibrium association constants (K) ranging from 1011 to 101. Binding curves for K = 109, 106, and 103 are labeled by their values of log K.
Although hinge-like and/or strained regions are often invoked as controlling the swapping process, their energetics are not always clear. For example, a stiff hinge can favor domain swapping by disfavoring a compact monomeric structure, but a flexible hinge can also promote swapping by favoring monomer opening. Thus, the domain-swapped state can be favored by low stability of the monomer, by flexibility of a linker region favoring opening of the monomer, and/or by stiffness of a linker region disfavoring a monomeric fold. An apparently straightforward example is the hinge loop of p13suc1, where mutations that modulate strain change the equilibrium between monomer and dimer (Rousseau et al. 2001). TrpR seems to present an example in which the monomer is so unstable that only the domain-swapped dimer is significantly populated. Domain-swapped proteins that have no apparent monomeric counterpart under a given set of conditions indicate an equilibrium that strongly favors the oligomeric state.
This equilibrium view of swapping defines a broad class of domain-swapped proteins, which in turn suggests that swapping is a common property of proteins, consistent with the diversity of protein structures known to swap (Fig. 3). A survey of the structure database (April, 2007) revealed about 70 nonredundant cases for which domain-swapped structures have been reported, representing all-α, all-β, and mixed proteins of widely varying tertiary structures. A single example that shows the great variance of swapping is RNaseA, which can swap either the N-terminal helical peptide segment, a C-terminal β-strand, or both segments at once with two other chains. It seems possible that most proteins might be able to domain swap given the right combination of conditions that destabilize the monomeric form and favor the domain-swapped state. These conditions may require nothing more than slightly reduced solubility of folded monomers at very high protein concentration, as suggested by the frequency with which domain-swapped forms are identified in crystalline proteins.
The biological relevance of domain swapping has been questioned, mainly because direct evidence is often lacking that domain-swapped species form at physiological protein concentrations and solution conditions. Because so many domain-swapped proteins are known only from crystal structures, it is reasonable to be concerned that swapping may be induced by the extreme conditions typically used, particularly the very high concentrations of not only proteins but also solution components, some of which have unknown effects on proteins. On the other hand, clear evidence of biological function is available for some domain-swapped forms. TrpR, for example, exists only as domain-swapped dimers both in crystals and in solution. X-ray, NMR, and biochemical analyses confirm that the domain-swapped dimer binds to DNA (Otwinowski et al. 1988; Zhang et al. 1994).
Not only the biological relevance but also the biological rationale for domain swapping seems to be clear in the TrpR case. A domain-swapped dimer permits TrpR subunits to remain stably associated without compromising flexibility of the hth; other ways of evolving a stable dimer, e.g., by selecting for greater stability of the monomers or the dimer interface, might be imagined to reduce hth dynamics. The functional role of hth dynamics and dimerization is known from quantitative analysis of DNA affinity, specificity, and cooperativity (Yang et al. 1996). A stable dynamic dimer enables TrpR to bind relatively poor palindromes through mutual fit with DNA. Adjacent dimers also mutually adapt to allow cooperative binding to even poorer adjacent palindromes. Thus, poor DNA affinity is compensated for by protein–protein cooperativity, both within and between dimers, ensuring specific recognition of trp operator sequences even with low symmetry. As bacterial operators are embedded within the promoters they control, it may be impossible to evolve better trp operators without altering trp promoter function. Stable dynamic TrpR dimers thus apparently co-evolved with poor trp operators to modulate affinity and specificity of DNA recognition through cooperative protein–DNA and protein–protein binding that uses mutual adaptation and responds to the ligand L-tryptophan.
Other plausible biological roles for domain swapping can be envisioned even if evidence is presently lacking. Domain swapping confers kinetic stabilization because simultaneous events are required in each native-like domain to reach a fully open state. This could increase the lifetime of proteins that need to be long-lived, like viral coat proteins and eye lens proteins, two groups of proteins with known domain swaps (Lapatto et al. 1991; Chen et al. 2006). It may be no coincidence that both of these groups of proteins include enzymes that have been co-opted for structural roles, and domain swapping could be relevant to this adaptation. In the case of one lens crystallin, circular permutation alters the hierarchy of domain assembly involving domain swapping, again reflecting the interdependency of levels in protein structural organization (Wright et al. 1998). Other potential biological benefits of domain swapping may include the possibility to use it as a switch for inactivating proteins. Human cystatin C provides a probable example, although biological relevance is not yet directly demonstrated. The monomer uses two sides of the protein to inhibit two different proteases. The domain-swapped dimer obscures one of the two sides and eliminates the corresponding inhibitory activity (Ekiel and Abrahamson 1996). Among the more speculative potential advantages of domain swapping, it may be a means to permit storage of proteins at high concentration without the risk of amorphous aggregation.
One much-disputed area in which domain swapping has been proposed to play a role is amyloid fibril formation (for a recent review, see Bennett et al. 2006). Despite extensive efforts to understand the pathology of deposition diseases in general and the formation of protein fibrils in particular, there are still many gaps in our knowledge of the molecular mechanisms of fibril formation for any native protein, and reservations about the role of domain swapping in fibril formation. Circumstantial evidence for a proposed connection is that several amyloidogenic proteins have domain-swapped crystal structures including prion protein (Knaus et al. 2001) and cystatin C (Janowski et al. 2001), and that proteins can be engineered to form fibrils by fusing amyloidogenic segments with swapping domains (Sambashivan et al. 2005; Guo and Eisenberg 2006). The single-crystal X-ray structure for a peptide model of a cross-β fibril was solved recently (Nelson et al. 2005). Its general features are consistent with protein fibrils, but unlike native globular proteins or their domain-swapped relatives, it presents tightly interlaced and dehydrated β-strands.
By introducing a disulfide bond in the monomer of cystatin C, Nilsson et al. (2004) aimed to prevent opening of the swapping interface and thus prevent domain swapping and amyloid formation. Formation of domain-swapped dimers was abolished, and fibril formation was diminished by 80%. This incomplete inhibition was attributed to the very harsh conditions used to promote fibril formation. On the other hand, the fact that fibrils accumulate when domain-swapped dimers do not implies that the dimers might not be on the pathway for fibril formation. Furthermore, Sanders et al. (2004) showed that in the closely related chicken cystatin, tetrameric states are intermediates in fibril formation, and the rate-limiting step for tetramer formation is intramolecular rearrangement within dimers. Thus, it appears to still be an open question whether domain-swapped forms of native proteins are a dead end in fibrillation or if they proceed to larger complexes via domain swapping and are essential in the growth of fibers. Indeed, the only structure, to our knowledge, of an extended three-dimensional intermolecular network formed by domain swapping of a native protein is not fibrillar. This example is again for TrpR, which in aqueous alcohol forms an all-helical crystalline network (Fig. 5C,D) that is gel-like in its topological entanglement and very high (70%) solvent content (Lawson et al. 2004). Many other domain swaps propagate intermolecularly to form head-to-tail polymers of native-like units, but these are not fibrillar either.
Thus, in spite of several indications of a linkage, there is yet no solid proof for domain swapping in amyloid fibrils of pathogenic proteins, and it might still turn out that fibril formation via domain swapping is a property of engineered proteins rather than a pathologically relevant phenomenon. A distinction between domain swapping and fibril formation that seems to be significant in the frame of this review is that domain swapping, like fragment reconstitution, uses native interactions and fibrils use nonnative interactions. Indeed, native interactions are expected to compete with formation of fibrillar structures by favoring globular folds that oppose opening of short extended regions. Additional factors, such as dehydrating conditions, may be required to tip the balance away from native interactions. It is also unclear whether destabilization of native interactions or stabilization of multimers is the dominating influence, though they could both be important.
Energetics of protein folding and fragment reconstitution
In many cases, fragment reconstitution results in essentially the same native structure as intramolecular folding, sometimes with quite high fragment affinities (Fig. 3). Thus, it is instructive to compare energetic contributions in the two cases. Which determinants govern affinity among fragments, and how does this inform us about the nature of protein folding? The viewpoint of fragment reconstitution sharpens our focus on the energetic tradeoffs involved in protein structure and folding, and suggests the utility of reconstitution for studies of folding and stability.
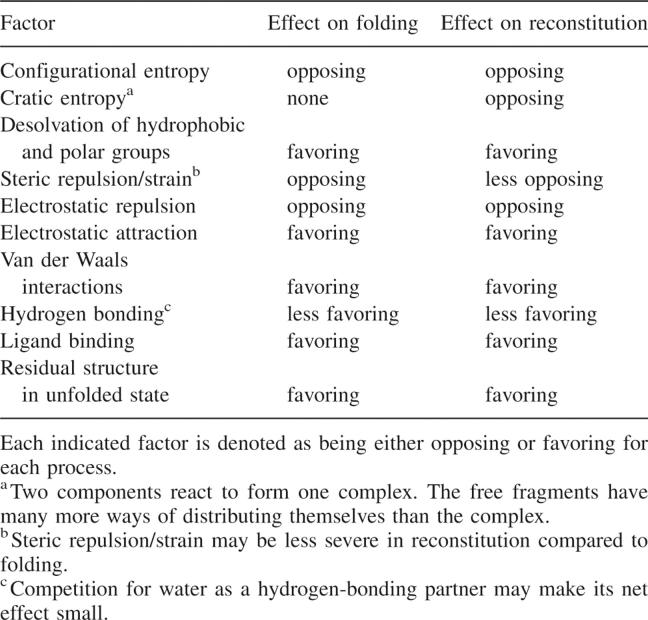
For a protein-folding equilibrium, the difference in free energy between the native and unfolded states defines the structural stability (ΔGNU). The free energies of the native and unfolded states are each determined by a net balance of favorable and unfavorable contributions. Table 1 represents an attempt to list favorable and unfavorable contributions to the processes of intramolecular folding and fragment reconstitution and shows that they are governed largely by the same factors. Although magnitudes of the listed effects are not given, it is generally agreed that compaction of the chain is a large unfavorable entropic contribution that is offset by the large enthalpically and entropically favorable burial of hydrophobic groups. For each particular protein, the exact offset is slightly different, as is the net effect of other contributions, resulting in a characteristic folding free energy under a given set of conditions. The presence of very large terms of similar and offsetting magnitudes, and the small magnitudes of all other effects, accounts for two well-known facts about protein-folding free energies: that the net stability of most native states is rather small, only about 20–40 kJ/mol under typical conditions, and it is easily altered by minor changes in conditions or sequence.
Table 1.
Determinants of protein folding and reconstitution
In both intramolecular folding and intermolecular reconstitution, the fact that stability is due to offsetting contributions from numerous factors implies that stability cannot be attributed to specific interactions formed within the structure. Of course, individual interactions are important to global stability, but it is strictly correct to consider that stability is due only to the net difference between all favorable and unfavorable contributions. The fact that stabilities can be greatly altered by certain point mutations derives from the inherently small magnitude of the net free energy, which therefore is hypersensitive to minor changes in any of its contributing factors. Mutational changes in free energy cannot be associated with specific interactions gained or lost upon mutation, as has been carefully argued by Mark and van Gunsteren (1994). However, even if energies cannot be assigned to individual interactions, protein structures are defined largely by noncovalent interactions that have been optimized by evolution. As discussed above, these interactions typically involve many residues from distant parts of the linear chain, coupling secondary structure elements to the tertiary structure with resulting mutual stabilization. The same long-range coupling of secondary and tertiary structural levels that promotes a protein's fold also promotes its reconstitution from fragments.
The principal energetic differences between intra- and intermolecular folding reactions lie in the configurational entropy factors that come into play in the reconstitution process due to bringing together two or more separate fragments. The free energy change upon reconstitution will contain an entropic contribution that stems from the fact that two components react to form one complex. The free fragments have many more ways of distributing themselves than the complex. Many researchers relate this term (“cratic entropy”) to the loss in translational entropy upon association (Amzel 1997). Most other contributions to free energy are likely very similar for intramolecular folding and intermolecular reconstitution. Energetic differences arising from the presence of a chain break typically depend on its location and are quite variable, reflecting the role of the interrupted segment in the fold and its effect on the balance of forces. At one extreme, TrpR reconstituted from fragments produced by proteolytic cleavage in the hth region (Fig. 5) is virtually indistinguishable from the native protein (Tasayco and Carey 1992), presumably due to the highly dynamic nature of the hth region. At the other extreme are the many cases where fragments are not observed to recombine. If there are no side reactions like aggregation of isolated fragments, then the low affinity of recombination indicates that the net balance of free energy change is shifted by the chain break, although, again, no single cause can be assigned.
Even when the net free energy of folding becomes unfavorable due to destabilizing mutations or chain breaks, proteins do not adopt structures of unrelated fold-classes. As Lattman and Rose (1993) have pointed out, these considerations help to focus the distinction between affinity and specificity in protein folding. This distinction is clear in the case of ligand binding, where each property can be related to free energy: Affinity is defined as the free-energy difference between bound and free states, and specificity can be defined as the difference in affinity between any pair of ligands. Both properties are readily quantified by measuring the equilibrium constants. If protein reconstitution is essentially equivalent to protein folding, then reconstitution offers a means to understand stability and specificity of protein structures, analogously as in ligand-binding reactions.
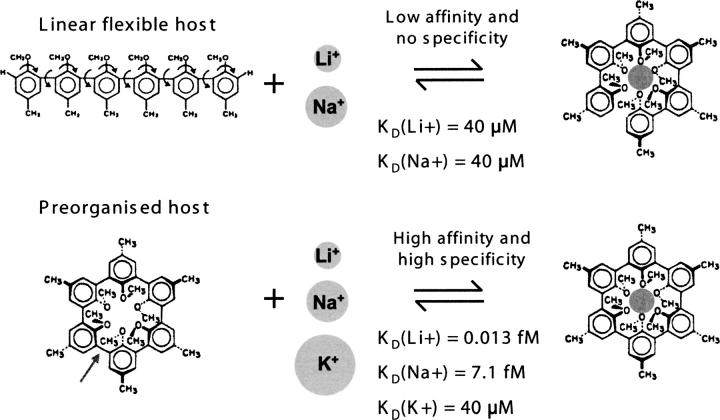
The energetic tradeoffs are also easier to understand and quantify for intermolecular interactions than for intramolecular processes like folding. For example, the roles of noncovalent forces in lipid assembly seem particularly clear (Gennis 1989). Another well-known and very simple case of molecular recognition clarifies the tradeoffs in terms of affinity and specificity. Host-guest chemistry (for review, see Cram 1987) had among its practical aims the design of industrial chelators to selectively remove one ion type among many. Early hosts were linear, flexible oligomers that became organized around the guest ion upon binding, with loss of host configurational entropy upon binding. Their affinities were relatively weak and approximately the same for different guests, i.e., hosts were not very specific. Later designs incorporated the knowledge that in the bound state the host was organized around the guest, leading to the idea of circularizing the oligomer by creating a bond between its ends (Fig. 9). Such preorganized hosts conferred much higher affinities and very high specificities for guests, because configurational entropic losses are minimized and the cavity size and shape are constrained. Significantly, in the bound state, bonding between host and guest is identical for preorganized or linear hosts, clearly revealing that the difference between the two cases is energetic rather than structural.
Figure 9.
Host-guest chemistry. Introduction of one bond (arrow at bottom, left) preorganizes the host in the bound conformation and dramatically alters its ligand affinity (KD) and specificity, as discussed in the text. (Adapted from Cram 1987 © The Nobel Foundation 1987.)
One feature of this kind of preorganized system is that affinity and specificity accrue approximately in parallel. This feature is not adaptive for biological systems that may require unusual combinations of affinity and specificity to carry out their physiological purposes, as in the MHC example. Thus, biological systems are expected to use molecular mechanisms that allow independent modulation of affinity and specificity. One such mechanism is protein folding coupled to ligand binding, and its effect on ligand-binding affinity and specificity can be understood in direct analogy with the host-guest case (Szwajkajzer and Carey 1997). Conformational adjustment upon ligand binding means that different ligands can be accommodated with similar free-energy change, leading to relatively low specificity. In fact, because it is energetic tradeoffs and not structural details that determine affinity and specificity, the structural details of accommodation can differ among ligands with similar affinities, and affinities can differ among structurally similar complexes.
The stabilizing contacts that develop during folding and association must overcome the additional losses in configurational entropy upon bringing fragments together. This balance thus determines the number and locations of breaks in a peptide chain that can be tolerated in a reconstitution reaction. Several examples show that even when different cleavage positions are compatible with reconstitution, the structure and/or activity of the assembly can differ with cleavage position, consistent with variations in the net balance of all factors (Andria et al. 1971; Kobayashi et al. 1995; Dutta et al. 2005). Nicks can often be accommodated in flexible loops, where entropic effects may be smallest. Active sites often contain such loops, as in the hth of TrpR. Consistent with their domain-swapped structure, apoTrpR dimers are readily proteolyzed in each hth, and the resulting four fragments associate and yield an NMR spectrum essentially indistinguishable from that of native dimers (Fig. 5B). This result implies that the reconstituted assembly represents the native dimer more accurately than does the crystal structure of intact TrpR, in which the hth is seen as a well-folded subdomain. This example led to development of nonspecific proteolysis as a systematic tool to probe native-state structures and dynamics and to identify sites for protein dissection (Carey 2000).
Not all fragment combinations of a given protein reconstitute with equal affinity. Similarly, only some end positions are tolerated well in circular permutation. Such results indicate that a chain break at some sites increases the cost of assembly more than a break at other sites, altering the balance of energetic tradeoffs. A systematic method for generating and selecting functional circular permutants (Graf and Schachman 1996) reveals that termini can be tolerated even within secondary structures. In contrast, for reconstitution, binding free energy may be inadequate to overcome the effects of a break in such locations. However, other factors may be at work in both cases. One such factor can be the presence of strain in the structure. The development of strain in folded proteins requires that the primary structure acts in concert with the secondary and tertiary structure, thus offering another example of the interdependency of levels in the protein structural hierarchy. This interdependence is energetic: Development of strain requires that the structure be sufficiently stable to overcome the unfavorable energetic contribution due to strain. It seems worth emphasizing that strain is a global structural and free-energy property of the structure, and not a property that can be assigned to a specific location, not even to a bond whose cleavage may relieve it. In this sense of being unassignable, strain is similar to the internal interactions that cannot be regarded as individually stabilizing to a structure. Strain represents stored free energy that can be used to do work on the system, as, for example, in enzyme reactions where it may be harnessed during catalysis. The fact that strain can modulate the stability of a structure, do enzymatic work, and be reduced by mutation (Hodel et al. 1993; Stites et al. 1994; Karplus 1996) implies that strain may be an evolutionarily selectable property of protein sequences.
The inference that primary structures may sometimes allow development of strain has also been suggested in studies of the growing class of self-cleaving proteins (Blair and Semler 1991; Brannigan et al. 1995; Paulus 2000; Macao et al. 2006), which naturally should be compared with artificial fragment reconstitution. Many different folds and sequences are now known in which an internal peptide bond in a preprotein is apparently cleaved by autocatalysis without any classical catalytic triad. Typically, the catalytic center is nothing more than an acidic serine or threonine hydroxyl or cysteine sulfhydryl immediately C-terminal to the cleaved bond, occasionally with assistance from a nearby residue that may facilitate deprotonation by stabilizing the anion and/or accepting the proton. A role for strain in facilitating self-cleavage seems likely, particularly considering that serine and threonine are among the most common Ramachandran outliers (Gunasekaran et al. 1996), but the relationships between strain and other factors in autocleavage (Håkansson 2002) are largely unknown. Many self-cleaving proteins also autocatalyze chemical transformation of the newly exposed internal N terminus (Oinonen and Rouvinen 2000; Saarela et al. 2004), or carry out peptide transfer by attack on another internal bond as in the case of the inteins (Perler 2006).
In some self-cleaved proteins the separated fragments do not reassemble with high affinity. This is surprising, given that so many intact native proteins can be artificially dissected into two or more reassembling pieces. Presumably, covalent connection prohibits conformations that become available after cleavage. Some ordinary fragment reconstitution reactions also depend upon co-dialysis of fragments from denaturing conditions, suggesting that kinetic traps may occur and unfolding must precede assembly; this could also be true for the separated fragments of some self-cleaving proteins, making their dissociation essentially irreversible.
A final example that should be considered along with reconstitution is the serpins (for a recent review, see Gettins 2002). These proteins inhibit serine proteases by presenting a reactive loop that is cleaved by the enzyme. The cleaved serpin undergoes a dramatic rearrangement in which the loop segment is inserted into the body of the protein's globular structure, intruding into a seemingly ordinary pre-existing sheet to form an extra strand. The strand-inserted forms generally have higher thermal stability to denaturation than the uncleaved native forms. The strand exchange reaction can also occur intermolecularly, leading to an indefinite multimer formed by domain swapping. Although the duality of serpin structures recalls the alternative free-energy minima of domain swapping and fibril formation, the free energies of serpin forms cannot be compared directly, due to the cleavage in one state. However, some uncleaved serpins have a more stable state in which partial strand insertion occurs without cleavage, representing a true alternative minimum and revealing the metastability of the native state. Serpins, like circular permutation, domain swaps, and fibrils, reveal the great plasticity of protein structures, reflecting both the mutual adaptation of structural levels and the delicate balance of interactions that characterize protein folding and structure.
Folding and binding; stability and affinity
The fact that reconstituted proteins typically adopt native-like structures strongly suggests that the folding of intact proteins can also be thought of as an association reaction, but one in which the binding partners are imprisoned in the same chain. Consequently, chain segments enjoy extremely high effective concentrations that favor mutual interactions among residues, including those that may be distant in the primary structure. This apparent affinity gain is analogous to the chelate effect, in which higher affinity for a ligand can be achieved when multiple coordinating groups are covalently connected.
The analogy between folding and reconstitution leads to the suggestion that reconstitution may be an alternative way to study protein folding. Although reconstitution reactions lose the advantage of high effective concentration compared with intramolecular folding, they gain a very significant practical advantage by bringing protein folding under the control of mass action. This feature enables direct experimental approaches to studies of structural stability under physiologically relevant solution conditions (Fig. 10). The population of the bound (folded) state depends only on the concentrations of the interacting partners and on the magnitude of the equilibrium constant under the chosen conditions (Fig. 8). In contrast, in intramolecular folding, the stability of the native state often can be determined only by extrapolation from high denaturant concentrations where the denatured protein is populated sufficiently to permit estimation of the equilibrium constant.
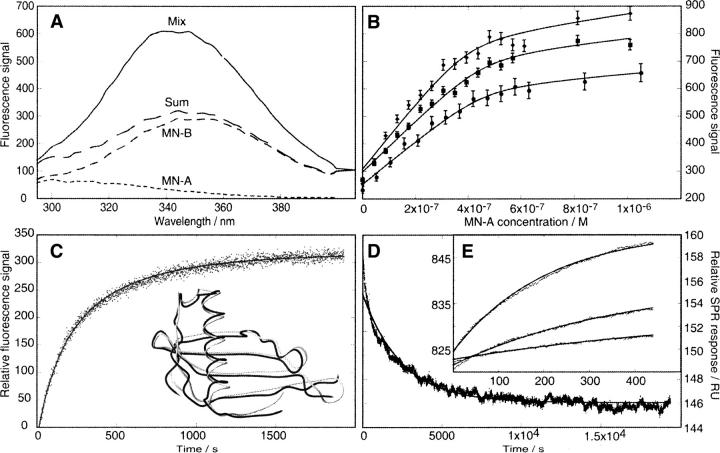
Figure 10.
Experimental approaches to protein reconstitution. Monellin has been studied using complementary methods that yield both kinetic and equilibrium data. (A) Fluorescence spectra of individual Mn-A and -B chains, the arithmetric sum of individual spectra, and the spectrum of the A and B chains mixed in 1:1 proportion. (B) Titration of Mn-B at three concentrations with Mn-A, monitored in equilibrium mixtures using the fluorescence change illustrated in A. (C) Kinetics of association of Mn-A and -B chains monitored by the change in fluorescence upon mixing equal amounts of Mn-A and Mn-B. (Inset) The superimposed structures of scMn (gray, 1MOL.pdb) and natural two-chain Mn (black, 4MON.pdb) show high identity despite the break between A and B chains. (D) Kinetics monitored by surface plasmon resonance. Main figure, dissociation of monellin A and B chains; (inset) association to Mn-B at three concentrations of Mn-A. (Adapted from Xue et al. 2004 © John Wiley & Sons, Inc. 2004.)
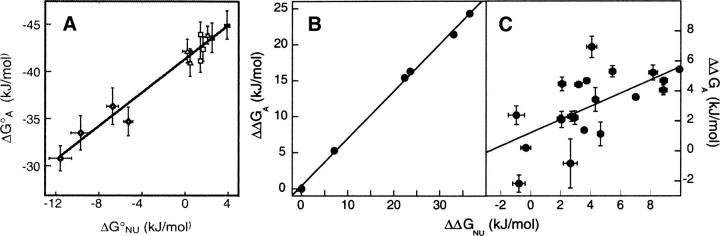
The analogy between folding and binding makes it tempting to compare the affinity of fragments with the stability of the corresponding intact protein chain. The results of mutational studies in several reconstitution systems suggest that a correlation may exist (Fig. 11). Barley chymotrypsin inhibitor 2 shows changes in equilibrium dissociation constant of reconstituting fragments that parallel changes in the free energy of unfolding of the intact chain (Ruiz-Sanz et al. 1995). The rate constants for fragment association also correlate with the rate constants for folding of the intact protein. The fact that several of the mutations are away from the interaction surface is consistent with globally cooperative folding. Mutations in the hydrophobic core of calbindin D9k change the affinity between fragments in parallel with the stability of the intact chain (Berggard et al. 2001). The rate of fragment dissociation is also correlated with stability of the intact protein, implying a common association rate for all mutated fragments. In the naturally reconstituted monellin, mutations altering Coulombic interactions on the surface affect fragment affinity in parallel with the stability of the single-chain protein (Xue et al. 2006). Charge mutations in monellin alter the association rate constant in concert with the stability of the single-chain protein, suggesting a role for charged residues in the association mechanism.
Figure 11.
Structural stability correlated with fragment affinity. Mutations affect stability of the intact chain and affinity of reconstituting fragments. Each point represents a mutant for which free energies (A) or free-energy differences from wild type (B,C) have been determined for both the stability of the intact chain (ΔGNU, ΔΔGNU) and the affinity of reconstituting fragments (ΔGA, ΔΔGA). (A) Charge mutations in monellin (Xue et al. 2006). Symbols refer to different salt concentrations; slope, 0.9; correlation coefficient, 0.98. (B) Hydrophobic core mutations in calbindin D9k (Berggard et al. 2001); slope, 0.7; correlation coefficient, 0.99. (C) Various mutations in chymotrypsin inhibitor 2 (Ruiz-Sanz et al. 1995); slope, 0.4; correlation coefficient, 0.63; this value for the 22 measurements shown implies only ∼0.2% probability that the correlation is due to chance.
The slope of affinity versus stability should reflect the degree to which reconstitution and refolding report on the same structural transition. A slope of one is expected when both the folded and unfolded states are similarly structured in the two cases. Deviations from this slope may sometimes be interpreted to gain insight about these states. The structures of reconstituted assemblies and intact proteins are often similar, and this can be relatively easy to discover experimentally. In such cases, deviations from a slope of one may point to differences between the unfolded intact chain and the dissociated fragments. This is potentially useful because it is difficult to evaluate the degree of residual structure in unfolded states of proteins (Shortle 1996). Residual structure in the intact chain that is not present in the dissociated fragments will yield a slope greater than one when affinity is plotted as the independent variable. However, this method reports only on differences between intact and fragment assemblies, and not on the presence or absence of residual structure in the intact chain itself: If residual structure is similar in the dissociated fragments and in the unfolded intact chain, the correlation plot will still have a slope of one. A slope of less than one, when structures are similar in the reconstituted assembly and intact native protein, reveals that fragment association has a more favorable free-energy change than refolding of the intact chain. Provided that standard states are adequately treated and conditions are similar, such a slope could point to unfavorable energetic contributions to stability of the intact native chain that are relieved by fragmentation, such as strain or steric clash.
Phage display has been applied to RNase S-peptide and S-protein to select variants with increased affinity in reconstitution (Schultz et al. 1998; Chakravarty et al. 2000; Dwyer et al. 2001; Dutta et al. 2005). If stability and affinity are related as suggested here, then mutations selected to increase fragment affinity should also stabilize the corresponding intact protein. Analysis of folding and reconstitution by harnessing the combinatorial power of phage display may eventually provide the systematic data required to evaluate the correlation that is implied by the analogy between folding and binding.
Conclusions
Reconstitution and domain swapping are common processes for proteins. Like folding of intact proteins, reconstitution and domain swapping follow ordinary thermodynamic laws and are governed by the search for free-energy minima. Both phenomena thus reflect general, universal properties of proteins, and lead to restoration of native interactions in a concentration-dependent manner. Considering both processes together offers insights into native protein structures and folding that neither one provides alone. This synergistic viewpoint suggests that novel approaches using reconstitution and three-dimensional domain swapping may deepen our understanding of proteins.
Dedication
This paper is dedicated to the memory of Zheng-yu Peng, a champion of the utility of protein reconstitution as a route to better understanding of proteins.
Acknowledgments
We thank Robert Baldwin, Pamela Björkman, Bengt Jönsson, Wei-Feng Xue, and Charles Yanofsky for valuable discussions or sharing of data. This work was supported by the Swedish Research Council, VR (S. Linse), the Tage Erlander Foundation (J.C.) and NSF 01-36094 (J.C.).
Footnotes
Reprint requests to: Jannette Carey, Chemistry Department, Princeton University, Princeton, New Jersey, 08544-1009 USA; e-mail: jcarey@princeton.edu; fax: (609) 258-6746.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.072985007.
References
- Amzel L.M. 1997. Loss of translational entropy in binding, folding, and catalysis. Proteins 28: 144–149. [PubMed] [Google Scholar]
- Andria G., Taniuchi, H., and Cone, J.L. 1971. Specific binding of 3 fragments of staphylococcal nuclease. J. Biol. Chem. 246: 7421–7428. [PubMed] [Google Scholar]
- Barrientos L.G., Louis, J.M., Botos, I., Mori, T., Han, Z.Z., O'Keefe, B.R., Boyd, M.R., Wlodawer, A., and Gronenborn, A.M. 2002. The domain-swapped dimer of cyanovirin-N is in a metastable folded state: Reconciliation of X-ray and NMR structures. Structure 10: 673–686. [DOI] [PubMed] [Google Scholar]
- Bennett M.J., Choe, S., and Eisenberg, D. 1994. Domain swapping - entangling alliances between proteins. Proc. Natl. Acad. Sci. 91: 3127–3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett M.J., Sawaya, M.R., and Eisenberg, D. 2006. Deposition diseases and 3D domain swapping. Structure 14: 811–824. [DOI] [PubMed] [Google Scholar]
- Berggard T., Julenius, K., Ogard, A., Drakenberg, T., and Linse, S. 2001. Fragment complementation studies of protein stabilization by hydrophobic core residues. Biochemistry 40: 1257–1264. [DOI] [PubMed] [Google Scholar]
- Björkman P.J., Saper, M.A., Samraoui, B., Bennett, W.S., Strominger, J.L., and Wiley, D.C. 1987. Structure of the human class-I histocompatibility antigen, Hla-A2. Nature 329: 506–512. [DOI] [PubMed] [Google Scholar]
- Blair W. and Semler, B. 1991. Self-cleaving proteases. Curr. Opin. Cell Biol. 3: 1039–1045. [DOI] [PubMed] [Google Scholar]
- Bouvier M. and Wiley, D.C. 1998. Structural characterization of a soluble and partially folded class I major histocompatibility heavy chain/β(2)m heterodimer. Nat. Struct. Biol. 5: 377–384. [DOI] [PubMed] [Google Scholar]
- Brannigan J.A., Dodson, G., Duggleby, H.J., Moody, P.C.E., Smith, J.L., Tomchick, D.R., and Murzin, A.G. 1995. A protein catalytic framework with an N-terminal nucleophile is capable of self-activation. Nature 378: 416–419. [DOI] [PubMed] [Google Scholar]
- Byeon I.J.L., Louis, J.M., and Gronenborn, A.M. 2003. A protein contortionist: Core mutations of GBI that induce dimerization and domain swapping. J. Mol. Biol. 334: 605. [DOI] [PubMed] [Google Scholar]
- Carey J. 2000. A systematic and general proteolytic method for defining structural and functional domains of proteins. Methods Enzymol. 328: 499–514. [DOI] [PubMed] [Google Scholar]
- Chakravarty S., Mitra, N., Queitsch, I., Surolia, A., Varadarajan, R., and Dubel, S. 2000. Protein stabilization through phage display. FEBS Lett. 476: 296–300. [DOI] [PubMed] [Google Scholar]
- Chen R., Neill, J.D., Estes, M.K., and Prasad, B.V.V. 2006. X-ray structure of a native calicivirus: Structural insights into antigenic diversity and host specificity. Proc. Natl. Acad. Sci. 103: 8048–8053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu V., Freitag, S., Le Trong, I., Stenkamp, R.E., and Stayton, P.S. 1998. Thermodynamic and structural consequences of flexible loop deletion by circular permutation in the streptavidin-biotin system. Protein Sci. 7: 848–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson M.W., Gilmore, S.A., Edgell, M.H., and Lee, A.L. 2006. Dynamic coupling and allosteric behavior in a nonallosteric protein. Biochemistry 45: 7693–7699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes M.H.J., Walsh, N.P., McKnight, C.J., and Sauer, R.T. 1999. Evolution of a protein fold in vitro. Science 284: 325–327. [DOI] [PubMed] [Google Scholar]
- Cram D.J. 1987. The design of molecular hosts, guests, and their complexes. Nobel lecture. The Nobel Foundation, Stockholm, Sweden, http://nobelprize.org/nobel_prizes/chemistry/laureates/1987/cram-lecture.html.
- Dalal S., Balasubramanian, S., and Regan, L. 1997. Protein alchemy: Changing β-sheet into α-helix. Nat. Struct. Biol. 4: 548–552. [DOI] [PubMed] [Google Scholar]
- de Prat-Gay G. 1996. Association of complementary fragments and the elucidation of protein folding pathways. Protein Eng. 9: 843–847. [DOI] [PubMed] [Google Scholar]
- de Prat-Gay G., Ruiz-Sanz, J., Davis, B., and Fersht, A.R. 1994. The structure of the transition state for the association of two fragments of the barley chymotrypsin inhibitor 2 to generate native-like protein: Implications for mechanisms of protein folding. Proc. Natl. Acad. Sci. 91: 10943–10946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieckmann T., Suzuki, E., Nakamura, G.K., and Feigon, J. 1996. Solution structure of an ATP-binding RNA aptamer reveals a novel fold. RNA 2: 628–640. [PMC free article] [PubMed] [Google Scholar]
- Dobson C.M. 1999. Protein misfolding, evolution and disease. Trends Biochem. Sci. 24: 329–332. [DOI] [PubMed] [Google Scholar]
- Dutta S., Batori, V., Koide, A., and Koide, S. 2005. High-affinity fragment complementation of a fibronectin type III domain and its application to stability enhancement. Protein Sci. 14: 2838–2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwyer J.J., Dwyer, M.A., and Kossiakoff, A.A. 2001. High affinity RNase S-peptide variants obtained by phage display have a novel “hot-spot” of binding energy. Biochemistry 40: 13491–13500. [DOI] [PubMed] [Google Scholar]
- Dyson H.J. and Wright, P.E. 2002. Coupling of folding and binding for unstructured proteins. Curr. Opin. Struct. Biol. 12: 54–60. [DOI] [PubMed] [Google Scholar]
- Dyson H.J. and Wright, P.E. 2005. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 6: 197–208. [DOI] [PubMed] [Google Scholar]
- Ekiel I. and Abrahamson, M. 1996. Folding-related dimerization of human cystatin C. J. Biol. Chem. 271: 1314–1321. [DOI] [PubMed] [Google Scholar]
- Eriksson A.E., Baase, W.A., Wozniak, J.A., and Matthews, B.W. 1992. A cavity-containing mutant of T4 lysozyme is stabilized by buried benzene. Nature 355: 371–373. [DOI] [PubMed] [Google Scholar]
- Fisher A. and Taniuchi, H. 1992. A study of core domains, and the core domain domain interaction of cytochrome-C fragment-complex. Arch. Biochem. Biophys. 296: 1–16. [DOI] [PubMed] [Google Scholar]
- Gennis R.B. 1989. Biomembranes: Molecular structure and function. Springer-Verlag, New York.
- Gettins P.G.W. 2002. Serpin structure, mechanism, and function. Chem. Rev. 102: 4751–4803. [DOI] [PubMed] [Google Scholar]
- Graf R. and Schachman, H.K. 1996. Random circular permutation of genes and expressed polypeptide chains: Application of the method to the catalytic chains of aspartate transcarbamoylase. Proc. Natl. Acad. Sci. 93: 11591–11596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryk M.R. and Jardetzky, O. 1996. AV77 hinge mutation stabilizes the helix-turn-helix domain of trp repressor. J. Mol. Biol. 255: 204–214. [DOI] [PubMed] [Google Scholar]
- Gunasekaran K., Ramakrishnan, C., and Balaram, P. 1996. Disallowed Ramachandran conformations of amino acid residues in protein structures. J. Mol. Biol. 264: 191–198. [DOI] [PubMed] [Google Scholar]
- Gunasekaran K., Ma, B.Y., and Nussinov, R. 2004. Is allostery an intrinsic property of all dynamic proteins? Proteins 57: 433–443. [DOI] [PubMed] [Google Scholar]
- Guo Z.F. and Eisenberg, D. 2006. Runaway domain swapping in amyloid-like fibrils of T7 endonuclease I. Proc. Natl. Acad. Sci. 103: 8042–8047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutte B., Lin, M.C., Caldi, D.G., and Merrifield, R.B. 1972. Reactivation of des (119-124, 120-124, or 121-124) ribonuclease-a by mixture with synthetic COOH-terminal peptides of varying lengths. J. Biol. Chem. 247: 4763–4767. [PubMed] [Google Scholar]
- Håkansson K. 2002. The strand-helix motif is a recurring theme in biological hydrolysis. Does the conformation of the Ramachandran outlier enhance its electrophilicity? Int. J. Biol. Macromol. 30: 273–277. [DOI] [PubMed] [Google Scholar]
- Hantgan R.R. and Taniuchi, H. 1977. Formation of a biologically active, ordered complex from 2 overlapping fragments of cytochrome c. J. Biol. Chem. 252: 1367–1374. [PubMed] [Google Scholar]
- Heringa J. and Taylor, W.R. 1997. Three-dimensional domain duplication, swapping and stealing. Curr. Opin. Struct. Biol. 7: 416–421. [DOI] [PubMed] [Google Scholar]
- Higgins C.L., Muralidhara, B.K., and Wittung-Stafshede, P. 2005. How do cofactors modulate protein folding? Protein Pept. Lett. 12: 165–170. [DOI] [PubMed] [Google Scholar]
- Hirs C.H.W. and Timasheff, S.N. 1986. Complementation in folding and fragment exchange. In Methods of enzymology, enzyme structure Part L, pp. 185–217. Academic Press, New York. [DOI] [PubMed]
- Hodel A., Kautz, R.A., Jacobs, M.D., and Fox, R.O. 1993. Stress and strain in staphylococcal nuclease. Protein Sci. 2: 838–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmgren A. 1972. Thioredoxin-C'—reconstitution of an active form of Escherichia coli thioredoxin from 2 noncovalently linked cyanogen bromide peptide fragments. FEBS Lett. 24: 351–354. [DOI] [PubMed] [Google Scholar]
- Håkansson M. and Linse, S. 2002. Protein reconstitution and 3D domain swapping. Curr. Protein Pept. Sci. 3: 629–642. [DOI] [PubMed] [Google Scholar]
- Håkansson M., Svensson, A., Fast, J., and Linse, S. 2001. An extended hydrophobic core induces EF-hand swapping. Protein Sci. 10: 927–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikura M. and Ames, J.B. 2006. Genetic polymorphism and protein conformational plasticity in the calmodulin superfamily: Two ways to promote multifunctionality. Proc. Natl. Acad. Sci. 103: 1159–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janowski R., Kozak, M., Jankowska, E., Grzonka, Z., Grubb, A., Abrahamson, M., and Jaskolski, M. 2001. Human cystatin C, an amyloidogenic protein, dimerizes through three-dimensional domain swapping. Nat. Struct. Biol. 8: 316–320. [DOI] [PubMed] [Google Scholar]
- Jardetzky O. 1996. Protein dynamics and conformational transitions in allosteric proteins. Prog. Biophys. Mol. Biol. 65: 171–219. [DOI] [PubMed] [Google Scholar]
- Jiang F., Kumar, R.A., Jones, R.A., and Patel, D.J. 1996. Structural basis of RNA folding and recognition in an AMP-RNA aptamer complex. Nature 382: 183–186. [DOI] [PubMed] [Google Scholar]
- Julenius K., Robblee, J., Thulin, E., Finn, B.E., Fairman, R., and Linse, S. 2002. Coupling of ligand binding and dimerization of helix-loop-helix peptides: Spectroscopic and sedimentation analyses of calbindin D9k EF-hands. Proteins 47: 323–333. [DOI] [PubMed] [Google Scholar]
- Kalman S.M., Linderstrom-Lang, K., Ottesen, M., and Richards, F.M. 1955. Degradation of ribonuclease by subtilisin. Biochim. Biophys. Acta 16: 297–299. [DOI] [PubMed] [Google Scholar]
- Kang X.S. and Carey, J. 1999. Structural organization in peptide fragments of cytochrome c by heme binding. J. Mol. Biol. 285: 463–468. [DOI] [PubMed] [Google Scholar]
- Karplus P.A. 1996. Experimentally observed conformation-dependent geometry and hidden strain in proteins. Protein Sci. 5: 1406–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.H., Kang, C.H., Kim, R., Cho, J.M., Lee, Y.B., and Lee, T.K. 1989. Redesigning a sweet protein: Increased stability and renaturability. Protein Eng. 2: 571–575. [DOI] [PubMed] [Google Scholar]
- Knaus K.J., Morillas, M., Swietnicki, W., Malone, M., Surewicz, W.K., and Yee, V.C. 2001. Crystal structure of the human prion protein reveals a mechanism for oligomerization. Nat. Struct. Biol. 8: 770–774. [DOI] [PubMed] [Google Scholar]
- Kobayashi N., Endo, S., and Munekata, E. 1993. Conformational study on the IgG binding domain of protein G. Peptide Chem. 1992: 278–280. [Google Scholar]
- Kobayashi N., Honda, S., Yoshii, H., Uedaira, H., and Munekata, E. 1995. Complement assembly of 2 fragments of the streptococcal protein-G B1 domain in aqueous-solution. FEBS Lett. 366: 99–103. [DOI] [PubMed] [Google Scholar]
- Kohmura M., Nio, N., and Ariyoshi, Y. 1990. Complete amino acid sequence of the sweet protein monellin. Agric. Biol. Chem. 54: 2219–2224. [PubMed] [Google Scholar]
- Kohmura M., Nio, N., and Ariyoshi, Y. 1991. Solid-phase synthesis of crystalline monellin, a sweet protein. Agric. Biol. Chem. 55: 539–545. [PubMed] [Google Scholar]
- Komeiji Y., Fujita, I., Honda, N., Tsutsui, M., Tamura, T., and Yamato, I. 1994. Glycine-85 of the Trp-repressor of Escherichia coli is important in forming the hydrophobic tryptophan binding pocket—Experimental and computational approaches. Protein Eng. 7: 1239–1247. [DOI] [PubMed] [Google Scholar]
- Lafleur M., Bloom, M., Eikenberry, E.F., Gruner, S.M., Han, Y.Q., and Cullis, P.R. 1996. Correlation between lipid plane curvature and lipid chain order. Biophys. J. 70: 2747–2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapatto R., Nalini, V., Bax, B., Driessen, H., Lindley, P.F., Blundell, T.L., and Slingsby, C. 1991. High-resolution structure of an oligomeric eye lens β-crystallin—loops, arches, linkers and interfaces in β-B2 dimer compared to a monomeric γ-crystallin. J. Mol. Biol. 222: 1067–1083. [DOI] [PubMed] [Google Scholar]
- Lattman E.E. and Rose, G.D. 1993. Protein folding—Whats the question? Proc. Natl. Acad. Sci. 90: 439–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson C.L., Benoff, B., Berger, T., Berman, H.M., and Carey, J. 2004. E. coli trp forms a domain-swapped array in aqueous alcohol. Structure 12: 1099–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindqvist Y. and Schneider, G. 1997. Circular permutations of natural protein sequences: Structural evidence. Curr. Opin. Struct. Biol. 7: 422–427. [DOI] [PubMed] [Google Scholar]
- Linse S., Thulin, E., Gifford, L.K., Radzewsky, D., Hagan, J., Wilk, R.R., and Akerfeldt, K.S. 1997. Domain organization of calbindin D(28k) as determined from the association of six synthetic EF-hand fragments. Protein Sci. 6: 2385–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. and Eisenberg, D. 2002. 3D domain swapping: As domains continue to swap. Protein Sci. 11: 1285–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.S., Gotte, G., Libonati, M., and Eisenberg, D. 2001. A domain-swapped RNase A dimer with implications for amyloid formation. Nat. Struct. Biol. 8: 211–214. [DOI] [PubMed] [Google Scholar]
- Liu Y.S., Hart, P.J., Schlunegger, M.P., and Eisenberg, D. 1998. The crystal structure of a 3D domain-swapped dimer of RNase A at a 2.1 angstrom resolution. Proc. Natl. Acad. Sci. 95: 3437–3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macao B., Johansson, D.G.A., Hansson, G.C., and Hard, T. 2006. Autoproteolysis coupled to protein folding in the SEA domain of the membrane-bound MUC1 mucin. Nat. Struct. Mol. Biol. 13: 71–76. [DOI] [PubMed] [Google Scholar]
- Mark A.E. and van Gunsteren, W.F. 1994. Decomposition of the free energy of a system in terms of specific interactions: Implications for theoretical and experimental studies. J. Mol. Biol. 240: 167–176. [DOI] [PubMed] [Google Scholar]
- Minor D.L. and Kim, P.S. 1996. Context-dependent secondary structure formation of a designed protein sequence. Nature 380: 730–734. [DOI] [PubMed] [Google Scholar]
- Mitchinson C. and Baldwin, R.L. 1986. The design and production of semisynthetic ribonucleases with increased thermostability by incorporation of S-peptide analogues with enhanced helical stability. Proteins 1: 23–33. [DOI] [PubMed] [Google Scholar]
- Murthy V.L., Srinivasan, R., Draper, D.E., and Rose, G.D. 1999. A complete conformational map for RNA. J. Mol. Biol. 291: 313–327. [DOI] [PubMed] [Google Scholar]
- Neira J.L., Davis, B., Ladurner, A.G., Buckle, A.M., Gay, G.D., and Fersht, A.R. 1996. Toward the complete structural characterization of a protein folding pathway: The structures of the denatured, transition and native states for the association/folding of two complementary fragments of cleaved chymotrypsin inhibitor 2. Direct evidence for a nucleation-condensation mechanism. Fold. Des. 1: 189–208. [DOI] [PubMed] [Google Scholar]
- Nelson R., Sawaya, M.R., Balbirnie, M., Madsen, A.O., Riekel, C., Grothe, R., and Eisenberg, D. 2005. Structure of the cross-β spine of amyloid-like fibrils. Nature 435: 773–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newcomer M.E. 2002. Protein folding and three-dimensional domain swapping: A strained relationship? Curr. Opin. Struct. Biol. 12: 48–53. [DOI] [PubMed] [Google Scholar]
- Nilsson M., Wang, X., Rodziewicz-Motowidlo, S., Janowski, R., Lindstrom, V., Onnerfjord, P., Westermark, G., Grzonka, Z., Jaskolski, M., and Grubb, A. 2004. Prevention of domain swapping inhibits dimerization and amyloid fibril formation of cystatin C—Use of engineered disulfide bridges, antibodies, and carboxymethylpapain to stabilize the monomeric form of cystatin C. J. Biol. Chem. 279: 24236–24245. [DOI] [PubMed] [Google Scholar]
- Oinonen C. and Rouvinen, J. 2000. Structural comparison of Ntn-hydrolases. Protein Sci. 9: 2329–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z., Schevitz, R.W., Zhang, R.G., Lawson, C.L., Joachimiak, A., Marmorstein, R.Q., Luisi, B.F., and Sigler, P.B. 1988. Crystal-structure of Trp repressor operator complex at atomic resolution. Nature 335: 321–329. [DOI] [PubMed] [Google Scholar]
- Paulus H. 2000. Protein splicing and related forms of protein autoprocessing. Annu. Rev. Biochem. 69: 447–496. [DOI] [PubMed] [Google Scholar]
- Peng Z.Y. and Wu, L.C. 2000. Autonomous protein folding units. Adv. Protein Chem. 53: 1–46. [DOI] [PubMed] [Google Scholar]
- Perler F.B. 2006. Protein splicing mechanisms and applications. IUBMB Life 58: 63. [DOI] [PubMed] [Google Scholar]
- Prinsen C.F.M. and Veerkamp, J.H. 1996. Fatty acid binding and conformational stability of mutants of human muscle fatty acid-binding protein. Biochem. J. 314: 253–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards F.M. 1958. On the enzymatic activity of subtilisin-modified ribonuclease. Proc. Natl. Acad. Sci. 44: 162–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseau F., Schymkowitz, J.W.H., Wilkinson, H.R., and Itzhaki, L.S. 2001. Three-dimensional domain swapping in p13suc1 occurs in the unfolded state and is controlled by conserved proline residues. Proc. Natl. Acad. Sci. 98: 5596–5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseau F., Schymkowitz, J.W., and Itzhaki, L.S. 2003. The unfolding story of three-dimensional domain swapping. Structure 11: 243–251. [DOI] [PubMed] [Google Scholar]
- Ruiz-Sanz J., Prat-Gay, G.D., Otzen, D.E., and Fersht, A.R. 1995. Protein-fragments as models for events in protein-folding pathways—protein engineering analysis of the association of 2 complementary fragments of the barley chymotrypsin inhibitor-2 (Ci-2). Biochemistry 34: 1695–1701. [DOI] [PubMed] [Google Scholar]
- Saarela J., Oinonen, C., Jalanko, A., Rouvinen, J., and Peltonen, L. 2004. Autoproteolytic activation of human aspartylglucosaminidase. Biochem. J. 378: 363–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambashivan S., Liu, Y.S., Sawaya, M.R., Gingery, M., and Eisenberg, D. 2005. Amyloid-like fibrils of ribonuclease A with three-dimensional domain-swapped and native-like structure. Nature 437: 266–269. [DOI] [PubMed] [Google Scholar]
- Sanders A., Craven, C.J., Higgins, L.D., Giannini, S., Conroy, M.J., Hounslow, A.M., Waltho, J.P., and Staniforth, R.A. 2004. Cystatin forms a tetramer through structural rearrangement of domain-swapped dimers prior to amyloidogenesis. J. Mol. Biol. 336: 165–178. [DOI] [PubMed] [Google Scholar]
- Schlunegger M.P., Bennett, M.J., and Eisenberg, D. 1997. Oligomer formation by 3D domain swapping: A model for protein assembly and misassembly. Adv. Protein Chem. 50: 61–122. [DOI] [PubMed] [Google Scholar]
- Schultz D.R., Ladbury, J.E., Smith, G.P., and Fox, R.O. 1998. Interactions of ribonuclease S with ligands from random peptide libraries. In Applications of calorimetry in the biological sciences (eds. J.E.a.C. B.Z. Ladbury et al.), pp. 123–138. John Wiley and Sons, New York.
- Shaw G.S., Hodges, R.S., Kay, C.M., and Sykes, B.D. 1994. Relative stabilities of synthetic peptide homodimeric and heterodimeric troponin-c domains. Protein Sci. 3: 1010–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba K. and Schimmel, P. 1992. Functional assembly of a randomly cleaved protein. Proc. Natl. Acad. Sci. 89: 1880–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker K.R., Fairman, R., Kim, P.S., York, E.J., Stewart, J.M., and Baldwin, R.L. 1987. The C-peptide helix from ribonuclease-a considered as an autonomous folding unit. Cold Spring Harb. Symp. Quant. Biol. 52: 391–398. [DOI] [PubMed] [Google Scholar]
- Shortle D.R. 1996. Structural analysis of nonnative states of proteins by NMR methods. Curr. Opin. Struct. Biol. 6: 24–30. [DOI] [PubMed] [Google Scholar]
- Shuman C.F., Jiji, R., Akerfeldt, K.S., and Linse, S. 2006. Reconstitution of calmodulin from domains and subdomains: Influence of target peptide. J. Mol. Biol. 358: 870–881. [DOI] [PubMed] [Google Scholar]
- Smith V.F. and Matthews, C.R. 2001. Testing the role of chain connectivity on the stability and structure of dihydrofolate reductase from E. coli: Fragment complementation and circular permutation reveal stable, alternatively folded forms. Protein Sci. 10: 116–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somoza J.R., Jiang, F., Tong, L., Kang, C.H., Cho, J.M., and Kim, S.H. 1993. Two crystal structures of a potently sweet protein. Natural monellin at 2.75 A resolution and single-chain monellin at 1.7 A resolution. J. Mol. Biol. 234: 390–404. [DOI] [PubMed] [Google Scholar]
- Stites W.E., Meeker, A.K., and Shortle, D. 1994. Evidence for strained interactions between side-chains and the polypeptide backbone. J. Mol. Biol. 235: 27–32. [DOI] [PubMed] [Google Scholar]
- Szwajkajzer D. and Carey, J. 1997. Molecular and biological constraints on ligand-binding affinity and specificity. Biopolymers 44: 181–198. [DOI] [PubMed] [Google Scholar]
- Taniuchi H. and Anfinsen, C.B. 1969. An experimental approach to study of folding of staphylococcal nuclease. J. Biol. Chem. 244: 3864–3875. [PubMed] [Google Scholar]
- Tasayco M.L. and Carey, J. 1992. Ordered self-assembly of polypeptide fragments to form native-like dimeric trp repressor. Science. www.sciencemag.org. [DOI] [PubMed]
- Tranier S., Iobbi-Nivol, C., Birck, C., Ilbert, M., Mortier-Barriere, I., Mejean, V., and Samama, J.P. 2003. A novel protein fold and extreme domain swapping in the dimeric TorD chaperone from Shewanella massilia. Structure 11: 165–174. [DOI] [PubMed] [Google Scholar]
- Tsai C.J., Ma, B.Y., and Nussinov, R. 1999. Folding and binding cascades: Shifts in energy landscapes. Proc. Natl. Acad. Sci. 96: 9970–9972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viguera A.R., Blanco, F.J., and Serrano, L. 1995. The order of secondary structure elements does not determine the structure of a protein but does affect its folding kinetics. J. Mol. Biol. 247: 670–681. [DOI] [PubMed] [Google Scholar]
- Wand A.J. 2001. Dynamic activation of protein function: A view emerging from NMR spectroscopy. Nat. Struct. Biol. 8: 926–931. [DOI] [PubMed] [Google Scholar]
- Weber G. 1972. Ligand binding and internal equilibria in proteins. Biochemistry 11: 864–878. [DOI] [PubMed] [Google Scholar]
- Wright G., Basak, A.K., Wieligmann, K., Mayr, E.M., and Slingsby, C. 1998. Circular permutation of β B2-crystallin changes the hierarchy of domain assembly. Protein Sci. 7: 1280–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W.-F., Carey, J., and Linse, S. 2004. Multi-method global analysis of thermodynamics and kinetics in reconstitution of monellin. Bioinformatics 57: 586–595. [DOI] [PubMed] [Google Scholar]
- Xue W.F., Szczepankiewicz, O., Bauer, M.C., Thulin, E., and Linse, S. 2006. Intra- versus intermolecular interactions in monellin: Contribution of surface charges to protein assembly. J. Mol. Biol. 358: 1244–1255. [DOI] [PubMed] [Google Scholar]
- Yang J., Gunasekera, A., Lavoie, T.A., Jin, L.H., Lewis, D.E.A., and Carey, J. 1996. In vivo and in vitro studies of TrpR-DNA interactions. J. Mol. Biol. 258: 37–52. [DOI] [PubMed] [Google Scholar]
- Zhang H., Zhao, D.Q., Revington, M., Lee, W.T., Jia, X., Arrowsmith, C., and Jardetzky, O. 1994. The solution structures of the Trp repressor-operator DNA complex. J. Mol. Biol. 238: 592–614. [DOI] [PubMed] [Google Scholar]
- Zimmerman A.W., van Moerkerk, H.T.B., and Veerkamp, J.H. 2001. Ligand specificity and conformational stability of human fatty acid-binding proteins. Int. J. Biochem. Cell Biol. 33: 865–876. [DOI] [PubMed] [Google Scholar]