Abstract
For the first time, 15N solid-state NMR experiments were conducted on wild-type phospholamban (WT-PLB) embedded inside mechanically oriented phospholipid bilayers to investigate the topology of its cytoplasmic and transmembrane domains. 15N solid-state NMR spectra of site-specific 15N-labeled WT-PLB indicate that the transmembrane domain has a tilt angle of 13° ± 6° with respect to the POPC (1-palmitoyl-2-oleoyl-sn-glycero-phosphocholine) bilayer normal and that the cytoplasmic domain of WT-PLB lies on the surface of the phospholipid bilayers. Comparable results were obtained from site-specific 15N-labeled WT-PLB embedded inside DOPC/DOPE (1,2-dioleoyl-sn-glycero-3-phosphocholine/1,2-dioleoyl-sn-glycero-3-phosphoethanolamine) mechanically oriented phospholipids' bilayers. The new NMR data support a pinwheel geometry of WT-PLB, but disagree with a bellflower structure in micelles, and indicate that the orientation of the cytoplasmic domain of the WT-PLB is similar to that reported for the monomeric AFA-PLB mutant.
Keywords: wild-type phospholamban, oriented phospholipids, structural topology, 15N solid-state NMR spectroscopy
Wild-type phospholamban (WT-PLB) is a 52-amino-acid transmembrane protein that interacts with the Ca-ATPase pump and lowers its affinity for Ca2+. WT-PLB plays a major role in the regulation process of the cardiac cycle (contraction and relaxation), which controls the heartbeat (Simmerman and Jones 1998). Unphosphorylated WT-PLB inhibits sacroplasmic reticulum ATPase activity and stops the flow of Ca2+ ions, and this inhibition can be relieved by the cyclic AMP- and calmodulin-dependent phosphorylation of WT-PLB (Simmerman and Jones 1998). Previously, the Smith group demonstrated using 2H solid-state NMR spectroscopy that WT-PLB forms a pentamer in phosphatidylcholine (PC) membranes (www.avantilipids.com/SyntheticPhosphatidylcholine.asp) (Ying et al. 2000). Several theoretical and biophysical experimental studies have been conducted to investigate its structure embedded into a membrane (Arkin et al. 1995; Tatulian et al. 1995; Mascioni et al. 2002; Zamoon et al. 2003; Clayton et al. 2005; Oxenoid and Chou 2005; Robia et al. 2005; Abu-Baker and Lorigan 2006).
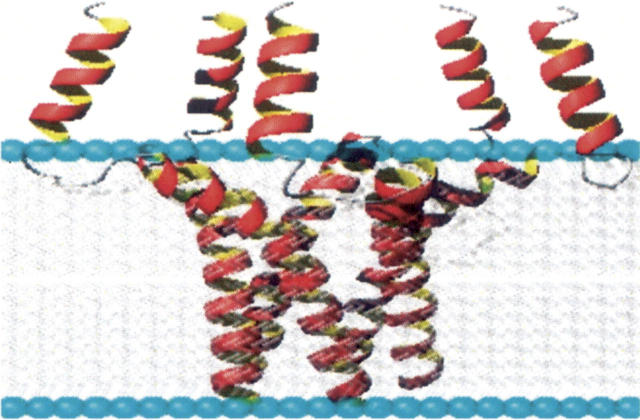
One early structural model was proposed by the Arkin group, in which WT-PLB is a continuous α-helix with a tilt angle of ∼28° with respect to the bilayer normal (Arkin et al. 1995). In another structural model suggested by Tatulian and coworkers, WT-PLB is composed of two α-helices connected by an unstructured/β-sheet region with a cytoplasmic domain tilted in a range of 50°–60° with respect to the bilayer normal of 1,2-Dimyristoyl-sn-Glycero-3-phosphocholine (DMPC) and 1-palmitoyl-2-oleoyl-sn-glycero-phosphocholine (POPC) phospholipid bilayers (Tatulian et al. 1995). A third structural model suggested by the Thomas group, using fluorescence resonance energy transfer (FRET), indicates that the cytoplasmic domain of each subunit of WT-PLB is in direct contact with the membrane surface, forming an L-shaped pinwheel geometry (Robia et al. 2005). In agreement with this model, NMR studies by the Lorigan and Middleton groups suggest that a direct interaction takes place between the cytoplasmic domain of WT-PLB and the phospholipid head groups of the bilayers (Clayton et al. 2005; Abu-Baker and Lorigan 2006). Conversely, using solution NMR spectroscopy, the Chou group recently revealed an unusual bellflower-like structural assembly for pentameric WT-PLB in micelles that indicates an α-helical cytoplasmic domain of the pentamer that on average points away from the membrane surface (Fig. 1; Oxenoid and Chou 2005). This new high-resolution NMR solution structure suggests minimal interaction between the cytoplasmic domain and the membrane surface. This structure represents the most highly constrained WT-PLB structure of the pentamer currently available in the literature.
Figure 1.
Side view of the high-resolution solution NMR structure for WT-PLB by the Chou group (bellflower-like assembly) obtained from the Protein Data Bank (1ZLL) (Oxenoid and Chou 2005). The WT-PLB structure embedded inside membrane bilayers was generated using MOLMOL software and a Macintosh G5 Apple computer.
Clearly, the structural topology of pentameric WT-PLB and its interaction with the membrane are under serious debate. Determining the correct structural topology of WT-PLB and its interaction with the membrane is important to understanding its function. Similarly, a disagreement has been reported on the physiologically active monomeric AFA-PLB mutant. The Veglia (Mascioni et al. 2002; Zamoon et al. 2003) and Baldus (Andronesi et al. 2005) groups, respectively, debate whether the transmembrane domain of the monomeric AFA-PLB mutant structure is connected to an α-helical cytosolic segment that lies on and interacts with the membrane surface (L-shaped) or a non-α-helical disordered cytosolic domain that has minimal interaction with the 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC)/1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE) membrane surface.
15N solid-state NMR spectroscopy is a powerful tool to ascertain direct information regarding the structural topology of membrane proteins in oriented phospholipid bilayers (Cross and Opella 1994). In this approach, specific 15N-labeled amide sites can be used to probe the alignment of the helix with respect to the bilayer normal. In this communication, the orientation of both the transmembrane and cytoplasmic domains of WT-PLB embedded inside mechanically oriented phospholipids will be probed with solid-state NMR spectroscopy.
Results and Discussion
Uniformly 15N-labeled WT-PLB was overexpressed and purified as described previously (Buck et al. 2003). Site-specific 15N-labeled WT-PLB was synthesized using a modified Fmoc-based solid-phase method as reported previously (Abu-Baker and Lorigan 2006). 15N-labeled amide residues were chosen at Ala11 (in the cytoplasmic domain) and Leu42 and Leu51 (both in the transmembrane helix). As indicated in the Electronic supplemental material (Supplemental Fig. S1), all three of these residues are consistent with an α-helical structure according to the 15N cross-polarization magic angle spinning (CPMAS) chemical-shift values (Wildman et al. 2003). CD-spectroscopy has indicated that WT-PLB is 79% α-helical (Arkin et al. 1995). Previously, we showed using 13C CPMAS solid-state NMR experiments that Leu39 and Ala15 representing the transmembrane and the cytoplasmic domains, respectively, are both parts of an α-helical structure (Abu-Baker and Lorigan 2006).
Mechanically oriented samples containing 3–4 mg of WT-PLB in 60 mg of either POPC lipids or a mixture of DOPC and DOPE lipids (4:1 ratio) were prepared as described previously (Tiburu et al. 2004). This DOPC/DOPE combination can be used for functional reconstitution of phospholamban into lipid bilayers as determined by Ca+2 ATPase activity (Karim et al. 2004). A 500-MHz WB Bruker Avance solid-state NMR spectrometer and a flat-coil 1H-X Low-E SS NMR probe (designed and built at the National Magnetic Field Laboratory, Tallahassee, Florida) with coil dimensions 7.6 × 5.6 × 11 mm were used to collect the 15N solid-state NMR spectra (Gor′kov et al. 2007). The magnetic field was parallel to the membrane bilayer normal for all the oriented samples.
Figure 2 shows the one-dimensional 15N solid-state NMR spectra of 15N-labeled WT-PLB embedded in unoriented (Fig. 2A) and oriented (Fig. 2B–E) POPC phospholipids as well as oriented DOPC/DOPE (4:1) phospholipids (Fig. 2G–J). To verify the formation of uniformly oriented lipid bilayers, one-dimensional 31P NMR spectra were conduced for all the samples (two representative examples are shown in Fig. 2L,M). The 31P peaks at 23 ppm and 26 ppm indicate that the POPC and DOPC/DOPE bilayer normals are aligned parallel to the static magnetic field.
Figure 2.
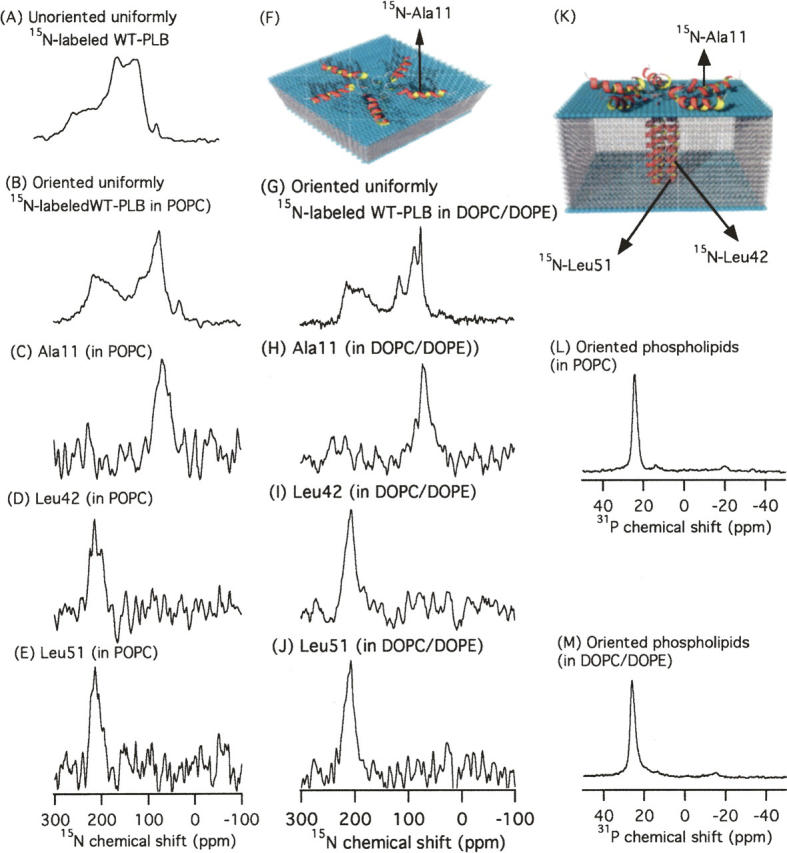
(A,B) One-dimensional 15N solid-state NMR spectra of uniformly 15N-labeled WT-PLB spectrum in randomly dispersed POPC MLVs (A) and oriented POPC and DOPC/DOPE (4:1) bilayers (B). (C–E) NMR spectra of site-specific 15N-labeled WT-PLB at Ala11 (in the cytoplasmic domain) (C), Leu42 (D), and Leu51 (E) (both in the transmembrane helix) in POPC oriented bilayers. (H–J) NMR spectra of site-specific 15N-labeled WT-PLB at Ala11 (H), Leu42 (I), and Leu51 (J) in DOPC/DOPE oriented bilayers. (L) One-dimensional 31P NMR spectrum of oriented POPC bilayers (one example is shown). (M) One-dimensional 31P NMR spectrum of oriented DOPC/DOPE bilayers (one example is shown). (K) Side view and (F) top view of a proposed structural model of WT-PLB embedded in POPC phospholipid bilayers.
The 15N powder pattern spectrum of uniformly 15N-labeled WT-PLB in unoriented POPC multilamellar vesicles (MLVs) is shown in Figure 2A. The isotropic component in this spectrum most likely results from the side-chain dynamics of 15N-labeled arginine and lysine residues. When uniform 15N-labeled WT-PLB is embedded inside mechanically aligned POPC or DOPC/DOPE phospholipids using glass plates, both spectra (Fig. 2B,G) show two distinct components at ∼73 ppm and at ∼211 ppm. The component at 211 ppm (near σ∥) indicates that a segment of WT-PLB is transmembrane (nearly parallel with B0), whereas the component at 73 ppm (near σ⊥) indicates that part of WT-PLB lies on the membrane surface.
In order to distinguish between the different domains of WT-PLB, site-specific 15N-labeled amide residues located on both the cytosolic domain (Ala11) and the transmembrane domain (Leu42 and Leu51) of WT-PLB were incorporated into oriented POPC and DOPC/DOPE bilayers and analyzed via 15N solid-state NMR spectroscopy. The 15N-labeled Ala11 sample shows resonance peaks at ∼69 ppm (Fig. 2C) and 71 ppm (Fig. 2H). Both resonant peaks are close to the σ⊥ component of the 15N CSA tensor of the corresponding powder spectrum, indicating that this residue located in the cytoplasmic domain is oriented approximately perpendicular to the bilayer normal. This implies that the structural topology of WT-PLB embedded inside oriented POPC and DOPC/DOPE bilayers is not similar to the bellflower-like structural assembly in DPC micelles reported previously (Fig. 1; Oxenoid and Chou 2005). According to the bellflower-like structural assembly, Ala11 should be oriented approximately parallel to the bilayer normal (Oxenoid and Chou 2005).
15N resonance peaks corresponding to the hydrophobic part of WT-PLB were observed at ∼211 ppm (Fig. 2D, Leu42 in POPC), 207 ppm (Fig. 2I, Leu42 in DOPC/DOPE), 212 ppm (Fig. 2E, Leu51 in POPC), and 209 ppm (Fig. 2J, Leu 51 in DOPC/DOPE) in oriented membrane samples. All of these peaks are close to the σ∥ component, indicating that the amide backbone vector of both residues is nearly parallel to the bilayer normal.
For the POPC oriented samples (Fig. 2C,D), the resonant peaks were broad, indicating some conformational heterogeneity and/or dynamics. Interestingly, using DOPC/DOPE phospholipids, the 15N spectral resolution of uniform 15N-labeled WT-PLB improved (Fig. 2G) when compared with the WT-PLB/POPC sample (Fig. 2B).
The cytoplasmic segment of the bellflower-like structure reported for WT-PLB in DPC micelles points away from the micelle surface (Oxenoid and Chou 2005). Micelles have a smaller surface area than POPC phospholipids bilayers; therefore, the cytoplasmic domain of the WT-PLB may not have sufficient area to remain bound to the micelle surface. Additionally, taking together both our POPC and DOPC/DOPE data as well as the continuous α-helical structure reported in DMPC by the Smith group (Arkin et al. 1995), it can be concluded that the WT-PLB might adapt different unique structural conformations in different membrane systems. Additionally, it is important to emphasize that the spacing between the phospholipid bilayer of interest and its water content plays an important role in determining the structural topology of the protein in the aligned samples (Rand and Parseian 1989).
The tilt angle of the transmembrane helix can be calculated using the method provided previously by the Cross group (Kovacs et al. 2000). Using a similar approach, a tilt angle of 13° ± 7° was calculated using the chemical shift tensor values of site-specific 15N-labeled WT-PLB at Leu51and Leu42 inserted into unoriented POPC phospholipid bilayers (Supplemental Table S1) and the 15N chemical shift from the oriented samples of the same residues (Fig. 2D,E). Previously, tilt angles of 28° ± 6°, 23°, and 19° were reported using FTIR spectroscopy (Arkin et al. 1995), computer modeling studies (Adams et al. 1995), and solution NMR methods (Oxenoid and Chou 2005), respectively, on the transmembrane domain of the WT-PLB. In the current study, WT-PLB was embedded inside POPC lipids (two long acyl chains of 16 and 18 carbons). Conversely, Arkin and coworkers conducted their WT-PLB studies in DMPC phospholipid bilayers (the acyl chain length is 14 carbons). It is quite common for peptides to adjust their corresponding tilt angle due to hydrophobic mismatch (de Planque et al. 1998; Harzer and Bechinger 2000). For the monomeric form of AFA-PLB, a tilt angle of 10° was reported in DOPC/DOPE bilayers (Mascioni et al. 2002). Previously, we have shown indirectly using 31P NMR spectroscopy that WT-PLB in MLVs interacts with the phospholipid head groups (Abu-Baker and Lorigan 2006). That work coupled with the new 15N solid-state NMR spectra presented in this article clearly indicate that the cytoplasmic domain of WT-PLB interacts with and lays on the surface of the membrane (Fig. 2C,H). However, this conclusion disagrees with the recent solution NMR structure of PLB in a micelle complex (Oxenoid and Chou 2005). The Chou PLB structure indicates that the cytoplasmic domain of PLB does not interact with the membrane surface. Understanding the structural topology of unbound PLB with respect to the membrane is essential to completely describe the function and binding mechanism of PLB to sarcoplasmic reticulum calcium ATPase (SERCA). Several models have been proposed that describe this interaction for function (Simmerman and Jones 1998; Oxenoid and Chou 2005; Zamoon et al. 2005). Zamoon and coworkers have indicated that when AFA-PLB is in its free form (not interacting with SERCA), the monomer interconverts between a predominant bent form (T-state, highly ordered L-shape, cytoplasmic domain facing the lipids) and an extended form (R-state, with higher affinity to SERCA) (Zamoon et al. 2005). In agreement with these observations, our data suggest that pentameric WT-PLB in POPC and DOPC/DOPE lipids exists mainly in a bent/pinwheel form in the absence of SERCA. Alternatively, the Chou group suggests that the cytoplasmic domain of pentameric WT-PLB, which on average points away from the membrane surface, can potentially fit into a groove in the SERCA cytoplasmic domain (Oxenoid and Chou 2005). The Chou PLB samples were prepared in the absence of SERCA.
Conclusion
In conclusion, this report clearly shows for the first time using 15N solid-state NMR of mechanically aligned bilayers that the orientation of the cytoplasmic domain of WT-PLB lies on the membrane surface, suggesting that the WT-PLB pentamer could function and be regulated in a similar way as the AFA-PLB monomeric mutant (Mascioni et al. 2002; Zamoon et al. 2003, 2005). Further experiments are needed to understand the effect of phosphorylation as well as SERCA binding on the orientation of the cytoplasmic domain of WT-PLB embedded inside phospholipids bilayers.
Electronic supplemental material
The supplemental material includes additional information about the sample conditions, 15N CPMAS spectra of site-specific 15N-labeled amide WT-PLB incorporated into POPC bilayers (Supplemental Fig. S1), and 15N CSA tensor values of unoriented POPC samples (Supplemental Table S1).
Acknowledgments
We thank Dr. Veglia and Dr. Thomas at the University of Minnesota for the help with the preparation of uniformly 15N-labeled WT-PLB. This work was supported by NIH grant GM080542 and AHA grant 0755602B. The 500-MHz wide-bore NMR spectrometer was obtained from NSF Grant (10116333).
Footnotes
Supplemental material: see www.proteinscience.org
Reprint requests to: Gary A. Lorigan, Miami University, Department of Chemistry and Biochemistry, Oxford, OH 45056, USA; e-mail: garylorigan@muohio.edu; fax: (513) 529-5715.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.072977707.
References
- Abu-Baker S. and Lorigan, G.A. 2006. Phospholamban and its phosphorylated form interact differently with lipid bilayers: A 31P, 2H, and 13C solid-state NMR spectroscopic study. Biochemistry 45: 13312–13322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams P.D., Arkin, I.T., Engelman, D.M., and Brunger, A.T. 1995. Computational searching and mutagenesis suggest a structure for the pentameric transmembrane domain of phospholamban. Nat. Struct. Biol. 2: 145–162. [DOI] [PubMed] [Google Scholar]
- Andronesi C.A., Becker, S., Seidel, K., Heise, H., Young, H.S., and Baldus, M. 2005. Detemination of membrane protein structure and dynamics by magic-angle-spininng solid-state NMR spectroscopy. J. Am. Chem. Soc. 127: 12965–12974. [DOI] [PubMed] [Google Scholar]
- Arkin I.T., Rothman, M., Ludlam, C.F.C., Aimoto, S., Engelman, D.M., Rothschild, K.J., and Smith, S.O. 1995. Structural model of the phospholamban ion-channel complex in phospholipid-membranes. J. Mol. Biol. 248: 824–834. [DOI] [PubMed] [Google Scholar]
- Buck B., Zamoon, J., Kirby, T.L., DeSilva, T.M., Karim, C., Thomas, D., and Veglia, G. 2003. Overexpression, purification, and characterization of recombinant Ca-ATPase regulators for high-resolution solution and solid-state NMR studies. Protein Expr. Purif. 30: 253–261. [DOI] [PubMed] [Google Scholar]
- Clayton J.C., Hughes, E., and Middleton, D.A. 2005. The cytoplasmic domains of phospholamban and phospholemman associate with phospholipid membrane surfaces. Biochemistry 44: 17016–17026. [DOI] [PubMed] [Google Scholar]
- Cross T.A. and Opella, S.J. 1994. Solid-state NMR structural studies of peptides and proteins in membranes. Curr. Opin. Struct. Biol. 4: 574–581. [Google Scholar]
- de Planque M.R.R., Greathouse, D.V., Koeppe, R.E., Schafer, H., Marsh, D., and Killian, J.A. 1998. Influence of lipid/peptide hydrophobic mismatch on the thickness of diacylphosphatidylcholine bilayers. A H-2 NMR and ESR study using designed transmembrane α-helical peptides and gramicidin A. Biochemistry 37: 9333–9345. [DOI] [PubMed] [Google Scholar]
- Gor′kov P.L., Chekmenev, E.Y., Li, C., Cotten, M., Buffy, J.J., Traaseth, N.J., Veglia, G., and Brey, W.W. 2007. Using low-E resonators to reduce RF heat in biological samples for static solid-state NMR up to 900 MHz. J. Magn. Reson. 185: 77–93. [DOI] [PubMed] [Google Scholar]
- Harzer U. and Bechinger, B. 2000. Alignment of lysine-anchored membrane peptides under conditions of hydrophobic mismatch: A CD, 15N, and 31P solid-state NMR spectroscopy investigation. Biochemistry 39: 13106–13114. [DOI] [PubMed] [Google Scholar]
- Karim C.B., Kirby, T.L., Zhang, Z.W., Nesmelov, Y., and Thomas, D.D. 2004. Phospholamban structural dynamics in lipid bilayers probed by a spin label rigidly coupled to the peptide backbone. Proc. Natl. Acad. Sci. 101: 14437–14442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs F.A., Denny, J.K., Song, Z., Quine, J.R., and Cross, T.A. 2000. Helix tilt of the M2 transmembrane peptide from Influenza A virus. J. Mol. Biol. 295: 117–125. [DOI] [PubMed] [Google Scholar]
- Mascioni A., Karim, C., Zamoon, J., Thomas, D.D., and Veglia, G. 2002. Solid-state NMR and rigid body molecular dynamics to determine domain orientations of monomeric phospholamban. J. Am. Chem. Soc. 124: 9392–9393. [DOI] [PubMed] [Google Scholar]
- Oxenoid K. and Chou, J.J. 2005. The structure of phospholamban pentamer reveals a channel-like architecture in membrane. Proc. Natl. Acad. Sci. 102: 10870–10875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand R.P. and Parseian, V.A. 1989. Hydration forces between phospholipid bilayers. Biochim. Biophys. Acta 988: 351–376. [Google Scholar]
- Robia S.L., Flohr, N.C., and Thomas, D.D. 2005. Phospholamban pentamer quaternary conformation determined by in-gel fluorescence anisotropy. Biochemistry 44: 4302–4311. [DOI] [PubMed] [Google Scholar]
- Simmerman H.K.B. and Jones, L.R. 1998. Phospholamban: Protein structure, mechanism of action, and role in cardiac function. Physiol. Rev. 78: 921–947. [DOI] [PubMed] [Google Scholar]
- Tatulian S.A., Jones, L.R., Reddy, L.G., Stokes, D.L., and Tamm, L.K. 1995. Secondary structure and orientation of phospholamban reconstituted in supported bilayers from polarized attenuated total-reflection FTIR spectroscopy. Biochemistry 34: 4448–4456. [DOI] [PubMed] [Google Scholar]
- Tiburu E.K., Dave, P.C., Damodaran, K., and Lorigan, G.A. 2004. Investigating leucine side-chain dynamics and backbone conformations of phospholamban incorporated in phospholipid bilayers utilizing 2H and 15N solid-state NMR spectroscopy. Biochemistry 43: 13899–13909. [DOI] [PubMed] [Google Scholar]
- Wildman K.A.H., Lee, D.K., and Ramamoorthy, A. 2003. Mechanism of lipid bilayer disruption by the human antimicrobial peptide, LL-37. Biochemistry 42: 6545–6558. [DOI] [PubMed] [Google Scholar]
- Ying W., Irvine, S.E., Beekman, R.A., Siminovitch, D.J., and Smith, S.O. 2000. Deuterium NMR reveals helix packing interactions in phospholamban. J. Am. Chem. Soc. 122: 11125–11128. [Google Scholar]
- Zamoon J., Mascioni, A., Thomas, D.D., and Veglia, G. 2003. NMR solution structure and topological orientation of monomeric phospholamban in dodecylphosphocholine micelles. Biophys. J. 85: 2589–2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamoon J., Nitu, F., Karim, C., Thomas, D.D., and Veglia, G. 2005. Mapping the interaction surface of a membrane protein: Unveiling the conformational switch of phospholamban in calcium pump regulation. Proc. Natl. Acad. Sci. 102: 4747–4752. [DOI] [PMC free article] [PubMed] [Google Scholar]