Abstract
A key step in activation of the p53 tumor suppressor is its transport into the nucleus; however, despite intensive study of p53, the regulation of its subcellular localization is still poorly understood. Here we examined the p53 nuclear importation using a series of mutant cell lines that were resistant to the growth inhibitory effects of temperature-sensitive murine p53 (tsp53). Examination of the p53 subcellular localization in these cell lines showed that the protein was cytoplasmic in most of them. Using a digitonin-permeabilized cell in vitro nuclear import system, we showthat cytosols from these cell lines do not support nuclear translocation of a p53 nuclear localization signal (NLS)-containing substrate protein, but promote nuclear localization of a SV40TAgNLS-containing substrate. Complementation assays and use of the mutant cells themselves in the in vitro assays demonstrate that both soluble and insoluble protein components are involved in p53 nuclear import. Collectively, our results suggest that there is a p53 NLS-selective nuclear import pathway and that both soluble and insoluble proteins are involved in its function.
Keywords: p53, temperature sensitive, mutant, nuclear localization
Introduction
The p53 tumor suppressor is a nuclear transcription factor involved in regulation of different cellular processes such as cell cycle control, apoptosis, differentiation and DNA repair in respond to a wide variety of stress signals (Levine, 1997). The inactivation of this tumor suppressor is a key factor in the development of human cancer. Although genetic mutations in the p53 gene are the most frequent mechanism by which it is inactivated, it is now clear that mislocalization of the p53 protein within the cell can also nullify p53 tumor suppressor activity (Goldman et al., 1996).
Nuclear–cytoplasmic shuttling of p53 is a fast, energy-dependent pathway and depends on nuclear export signals (NESs) and nuclear localization signals (NLSs) located in the N and C termini of the protein (Middeler et al., 1997). Three putative NLSs residing in the C terminus of p53 were first identified, and NLSI (PQPKKKP) is the most important one for directing p53 nuclear import (Shaulsky et al., 1990). However, two additional amino acids located 5′ of the original NLSI were showed to be required to form a fully functional NLS, indicating that the p53 NLS was bipartite in nature (Liang and Clarke, 1999). Nuclear import is initialized by binding of importin-α/β complex, which mediates the docking of proteins at the nuclear pore complex (Gorlich and Mattaj, 1996). However, the mechanism that regulates the importin-mediated nuclear import is still poorly understood.
To study the regulation of p53 nuclear import, A1–5 cells were adopted as the model system in this lab because it express a temperature-sensitive murine p53 (tsp53), which can be induced to concentrate either in the cytoplasm or nucleus in response to the incubation temperature (Martinez et al., 1991). At 37°C, the tsp53 in A1–5 cells is cytoplasmically sequestered; however, when temperature is reduced to 32°C the tsp53 accumulates in the nucleus and induces p21, which results in suppression of cell proliferation. In this work, 20 ALTR lines (A1–5 Low Temperature Resistant) were generated from A1–5 cells by chemical mutagenesis with EMS and their p53 subcellular localization was characterized using indirect immunofluorescence. By assaying for nuclear transport of an Escherichia coli expressed GST-p53NLS-EGFP fusion protein using cytosols from different ALTR cells, we showed that both soluble and insoluble components are required for p53 nuclear importation.
Results
Cell lines defective for p53 nuclear localization
A1–5 cells were mutagenized and mutants that could proliferate at 32°C were selected as described in Materials and methods. Twenty ALTR lines were stably expanded and propagated. Using indirect immunofluorescence, the tsp53 was found to distribute primarily in the cytoplasm of all the ALTR lines at 37°C, similar to that observed for A1–5 cells at this temperature. However, at 32°C we found that in the majority of the ALTR lines (15 out of 20 or 75%), tsp53 was concentrated in the cytoplasm. In the remaining five ALTR lines (25%), tsp53 was found concentrated in the nucleus (Table 1).
Table 1.
Categorization of ALTR cells
| P53val135 mutational statusa | Subcellular localization of p53val135 |
|
|---|---|---|
| Nuclear at 32°C | Cytoplasmic at 32°C | |
| Wild-type | ALTR10, ALTR17, ALTR24 | ALTR1, ALTR7, ALTR8, ALTR11, ALTR12, ALTR16, ALTR25 |
| Mutant | ALTR9, ALTR18 | ALTR3, ALTR4, ALTR13, ALTR14, ALTR15, ALTR19, ALTR23, ALTR26 |
Abbreviation: ALTR, A1–5 Low Temperature Resistant.
Wild-type indicates that the protein is identical to the p53val135 in A1–5 cells. Mutant indicates that p53val135 has an additional mutation.
Because mutational inactivation of p53 could also result in a nonfunctional p53 and/or cytoplasmic sequestration, we examined the sequence of the tsp53 transcript in each of the ALTR lines using reverse transcription (RT)–PCR. Sequence analysis data determined that 10 out of the 20 ALTR cells (50%) expressed the same tsp53 as that in parental A1–5 cells. In the remaining 10 ALTR lines (50%), tsp53 had acquired an additional mutation. On the basis of their p53 mutation status and subcellular localization, the ALTR lines were segregated into four categories: wild-type p53/nuclear, wild-type p53/cytoplasmic, mutant p53/nuclear and mutant p53/cytoplasmic (Table 1). This analysis showed that the ratio of ALTR lines with wild-type tsp53 to mutated tsp53 and in which the protein was cytoplasmic was 50:50. Similarly, the ratio of ALTR lines with wild-type tsp53 to mutated tsp53 and in which the protein was nuclear was also 50:50. Hence, wild-type and mutant tsp53s are equally likely to be sequestered in the cytoplasm.
p53 can be excluded from the nucleus by virtue of an active nuclear export pathway mediated by Crm1 whose activity can be inhibited by leptomycin B (Nishi et al., 1994). Hence, we treated ALTR lines with cytoplasmic tsp53 with leptomycin B and then stained for p53 protein using indirect immunofluorescence to ascertain whether inhibition of nuclear export resulted in tsp53 nuclear localization (Table 2). We found that 7 out of 14 (50%) showed a marked redistribution of tsp53 to the nucleus when exposed to leptomycin B, suggesting that the tsp53 in these lines was actively exported from the nucleus. However, the other six (43%) lines showed no change in cytoplasmic p53 localization. One line, ALTR3, showed only a minor response to leptomycin B. Our results indicate that in approximately half of the ALTR lines, tsp53 is sequestered in the cytoplasm because it is excluded from entering the nucleus.
Table 2.
Leptomycin B-induced p53 nuclear localization in ALTR cells
| Cell line | Leptomycin B (24 h, 4 μl/ml) |
|---|---|
| A1–5 | 93% (++++)a |
| ALTR1 | 37% (++) |
| ALTR3 | 8.8% |
| ALTR4 | 59% (++) |
| ALTR7 | 38.5% (++) |
| ALTR8 | 70% (+++) |
| ALTR11 | ND |
| ALTR12 | ND |
| ALTR14 | 54% (++) |
| ALTR15 | ND |
| ALTR16 | ND |
| ALTR19 | ND |
| ALTR23 | ND |
| ALTR25 | 50% (++) |
| ALTR26 | 23% (+) |
Abbreviations: ALTR, A1–5 Low Temperature Resistant; ND, not detectable nuclear localization.
Values indicate the % of cells with nuclear p53 after leptomycin B treatment. Qualitative ranking is given in parentheses.
Full-length bipartite NLS is required for p53 nuclear translocation
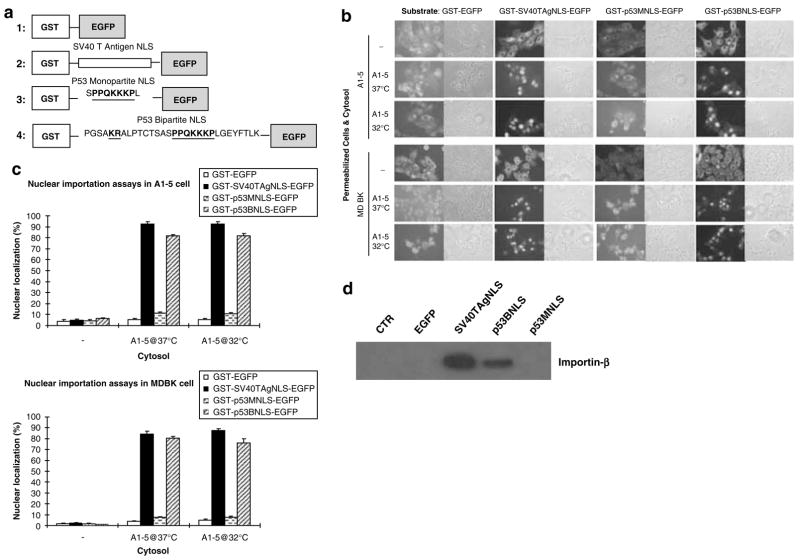
We next decided to characterize further the ALTR lines, in which p53 was excluded from entering the nucleus using an in vitro nuclear transport system. To accomplish this, we first tested for the in vitro requirement for nuclear importation of an EGFP-tagged p53 NLS substrate. Two substrate proteins, which contained either a monopartite p53 NLS (GST-p53MNLS-GFP) or a bipartite p53 NLS (GST-p53BNLS-EGFP), were produced by expression in E. coli. In addition, the GST-EGFP fusion served as a negative control, and the GST-SV40TAgNLS-EGFP fusion served as a positive control (Figure 1a).
Figure 1.
Full bipartite NLS is required for p53 nuclear import. (a) Construction of nuclear localization signal (NLS)-bearing substrates. (b) In vitro nuclear import assays. Digitonin-permeabilized A1–5 or MDBK cells were incubated with different substrates as indicated in a reaction with or without cytoplasm from A1–5 cells grown at 32 or 37°C. (c) The graphs show the percentage of cells exhibiting nuclear-localized substrates calculated from ~500 cells in random microscope fields. Bars represent the average ±s.e. from three separate experiments. (d) Analysis of importin-β binding with different substrates. A1–5 cytoplasm was subjected to GST-pull-down assays with unbound (lane 1) or substrates bound (lanes 2–5) glutathione-Sepharose 4B. The pull-down products were analysed by immunoblotting using anti-importin-β.
The purified fusion proteins were then used as substrates assaying for their ability to transport into the nucleus of digitonin-permeabilized cells. No nuclear localization of GST-EGFP was observed in either A1–5 or MDBK cells demonstrating that the NLS is indispensable for nuclear importation (Figure 1b). In contrast, both GST-SV40TAgNLS-EGFP and GST-p53BNLS-EGFP substrates were translocated into the nucleus of A1–5 and MDBK cells, with 80–90% of the cells showing nuclear-localized substrate (Figure 1c). Importantly, nuclear localization of GST-p53MNLS-EGFP was much reduced (~10%) compared to either GST-SV40TAgNLS-EGFP or GST-p53BNLS-EGFP. This suggested that the full bipartite p53 NLS is required for nuclear import in this system and is consistent with previous reports, which demonstrated that the p53 NLS has a bipartite structure (Liang and Clarke, 1999). In no case was nuclear localization of the substrate proteins observed in the absence of cytosol. Hence, soluble cytoplasmic factors are essential for the nuclear import of NLS-bearing substrates.
Finally, we tested the substrate proteins for their binding to importin-β, which is a key transporter that promotes transportation of cargo proteins across the nuclear membrane (Gorlich and Mattaj, 1996). GST-pull-down showed that importin-β coprecipitated with the GST-p53BNLS-EGFP protein, but not GST-p53MNLS-EGFP (Figure 1d), suggesting that the lack of nuclear import activity of the monopartite p53 NLS is due to its inability to bind importin-β. Interestingly, importin-β binding of GST-SV40TAgNLS-EGFP was notably more robust than that of GST-p53BNLS-EGFP, suggesting that the p53 bipartite NLS is a less potent NLS than the SV40 T-antigen NLS. Collectively, these results indicated that the biochemical requirements for nuclear importation in our in vitro system were similar to that seen for p53 in vivo.
Cytosols from several ALTR lines are defective in supporting p53 NLS nuclear import
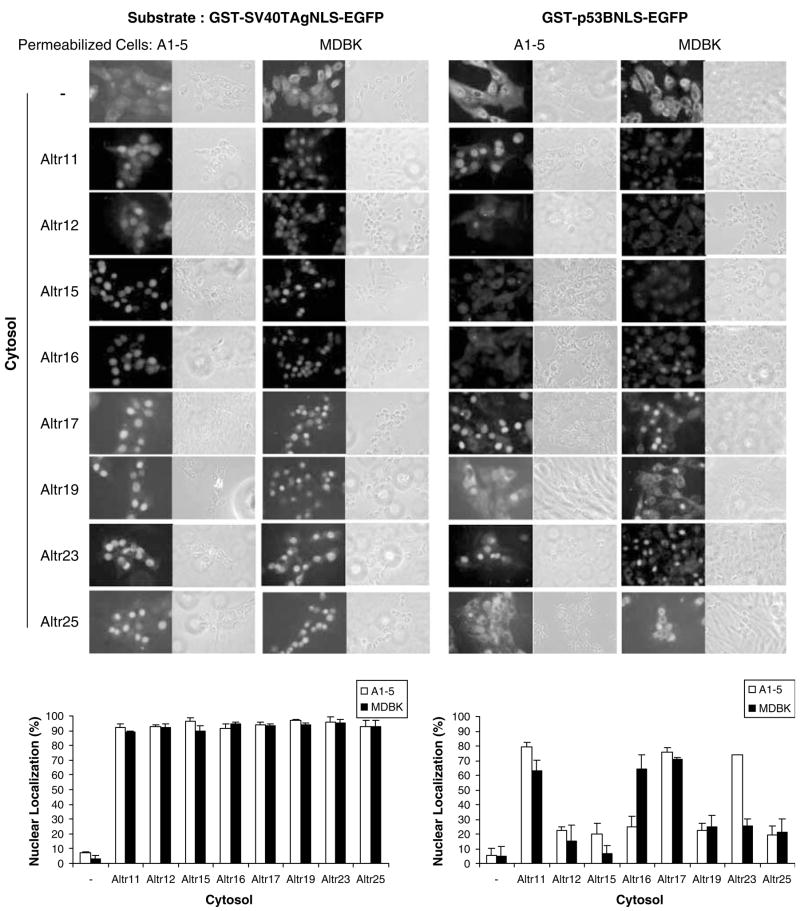
We next used our in vitro system to investigate further the failure of p53 nuclear localization in ALTR cells. Cytosols from the ALTR lines, which showed no response to leptomycin B, were isolated and assayed for their activities in supporting the GST-p53BNLS-EGFP nuclear import. ALTR17 was included as a positive control since tsp53 is nuclear localized in this line. Cytosols from all tested ALTR lines were found to support robust nuclear importation of the GST-SV40TAgNLS-EGFP with ~90% of cells showing nuclear-localized substrate in A1–5 or MDBK cells. In contrast, only some of the ALTR cytosols could support nuclear import of GST-p53BNLS-EGFP substrate. Only ~20% or lower nuclear localization was observed when the supplied cytosol was from ALTR12, ALTR15, ALTR19 or ALTR25 cells (Figure 2), suggesting a defect in cytoplasmic factor(s) specific for p53 NLS nuclear translocation in these four ALTR lines. Interestingly, cytosol from ALTR23 and ALTR16 could only support the nuclear import of GST-p53BNLS-EGFP substrate in A1–5 and MDBK cells, respectively, indicating that the origin of the insoluble components provided by the permeabilized cells could, in some cases, influence nuclear import.
Figure 2.
Cytosols from several A1–5 Low Temperature Resistant (ALTR) lines are defective in supporting p53 nuclear localization signal (NLS) nuclear importation. GST-SV40TAgNLS-EGFP and GST-p53BNLS-EGFP substrates were assayed for nuclear import in digitonin-permeabilized A1–5 and MDBK cells with or without ALTR cytoplasm as indicated. The graphs show the percentage of cells exhibiting nuclear-localized substrates calculated from ~500 cells in random microscope fields. Bars represent the average ±s.e. from three separate experiments.
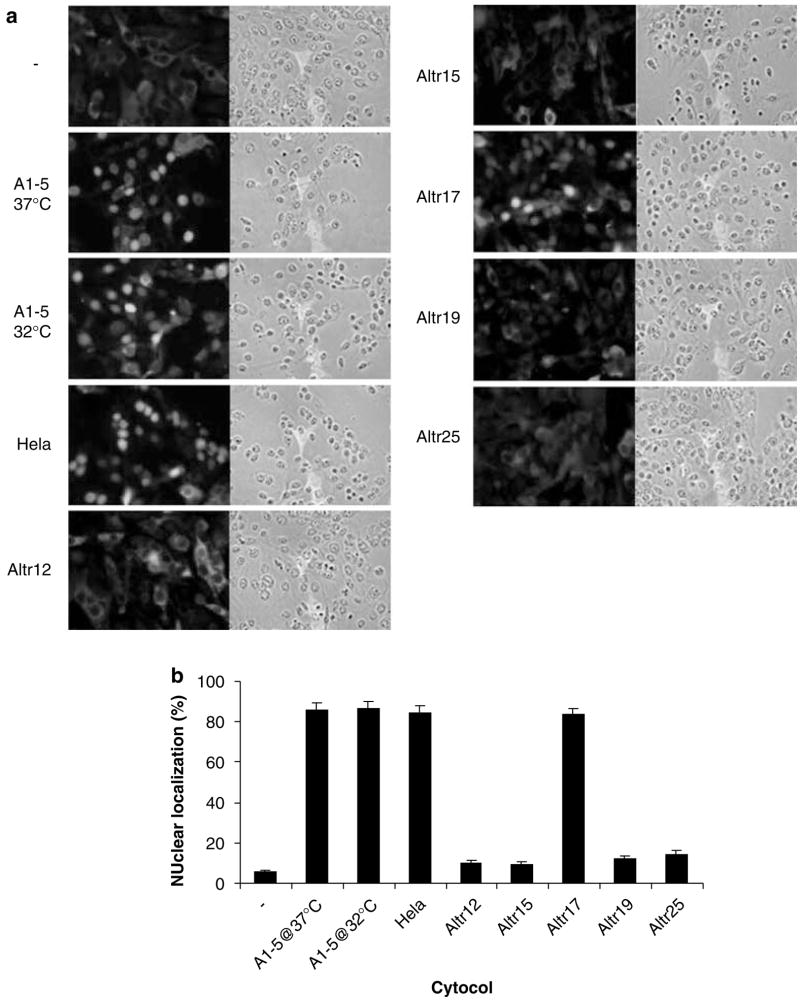
To confirm the above results, GST-tagged wild-type p53 was used as a substrate and assayed for in vitro nuclear importation using different cytosols. The subcellular localization of GST-p53 was then determined by indirect immunofluorescence using anti-GST antibody (Figure 3a). Again, while cytosols from A1–5, HeLa and ALTR17 cells supported robust nuclear localization of the GST-p53 substrate with ~90% of cells showing nuclear-localized substrate in permeabilized A1–5 cells, only ~15% or lower nuclear localization was observed when the supplied cytosol was from ALTR12, ALTR15, ALTR19 or ALTR25 cells (Figure 3b). Taken together, these results indicate that four of the ALTR lines (ALTR12, ALTR15, ALTR19 and ALTR25) show no p53 nuclear importation either in vivo or in vitro.
Figure 3.
ALTR (A1–5 Low Temperature Resistant) cytosols defective for nuclear importation of full-length p53. (a) Digitonin-permeabilized A1–5 cells were incubated with GST-p53 substrates with cytosol as indicated. The subcellular localization of GST-P53 was then determined by indirect immunofluorescence using anti-GST antibody. (b) The graphs show the percentage of cells exhibiting nuclear-localized GST-P53 calculated from ~500 cells in random microscope fields. Bars represent the average ±s.e. from three separate experiments.
Insoluble component is involved in p53 NLS nuclear importation
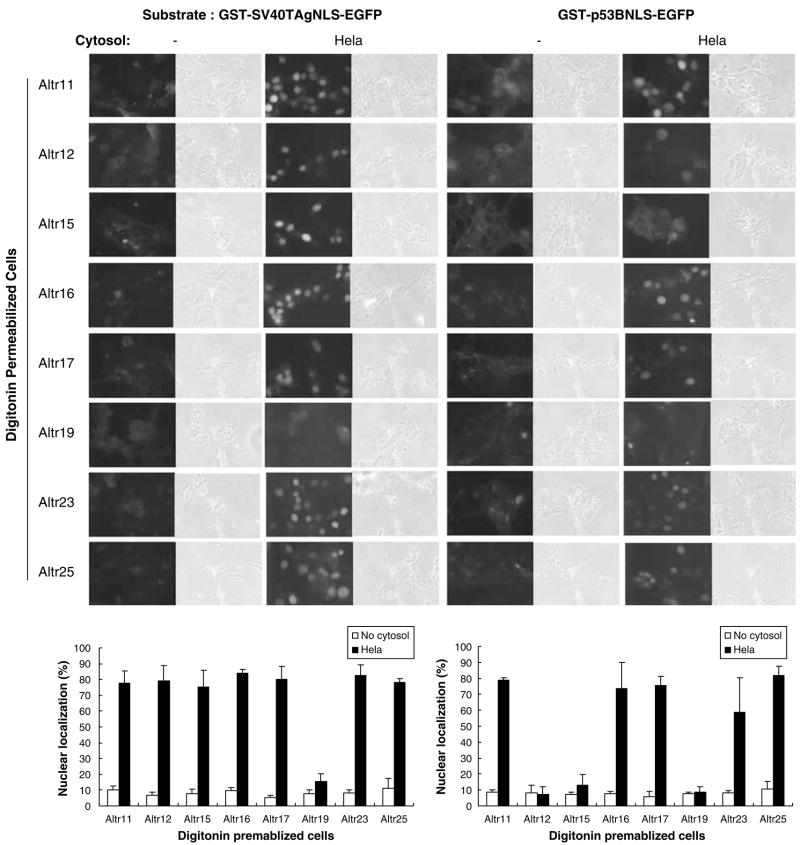
In addition to the soluble cytoplasmic factors, insoluble proteins that are a part of the nuclear pore complex are also essential for nuclear import. Hence, ALTR cells were permeabilized with digitonin and assayed for nuclear import to test whether insoluble proteins might have been affected (Figure 4). In the presence of HeLa cytosol, nuclear localization of GST-SV40TAgNLS-EGFP substrate was uniformly robust with ~80% or higher in all tested ALTR cells, except for ALTR19s. In contrast, nuclear localization of GST-p53BNLS-EGFP was more variable. Nuclear localization of the p53 bipartite NLS substrate was nearly equal to that seen for the SV40 NLS in five of the ALTR lines (ALTR11, ALTR16, ALTR17, ALTR23 and ALTR25). However, in ALTR12s and ALTR15s nuclear localization of GST-p53BNLS-EGFP was reduced to 10% or lower (Figure 4), suggesting that an insoluble factor involved in p53 NLS nuclear import may be defective in these two ALTR lines. Moreover, neither the GST-SV40TAgNLS-EGFP nor the GST-p53BNLS-EGFP substrates could enter into the nucleus of ALTR19 cells, suggesting that an insoluble factor required for nuclear import of both substrates may be defective in this line.
Figure 4.
Insoluble factor(s) is involved in p53 nuclear localization signal (NLS) nuclear importation. Digitonin-permeabilized ALTR (A1–5 Low Temperature Resistant) cells as indicated were incubated with GST-SV40TAgNLS-EGFP or GST-p53BNLS-EGFP substrates with or without HeLa cytosol. The graphs show the percentage of cells exhibiting nuclear-localized substrates calculated from 500–600 cells in random microscope fields. Bars represent the average ±s.e. from three separate experiments.
Complementation of ALTR cytosols in p53 NLS nuclear import
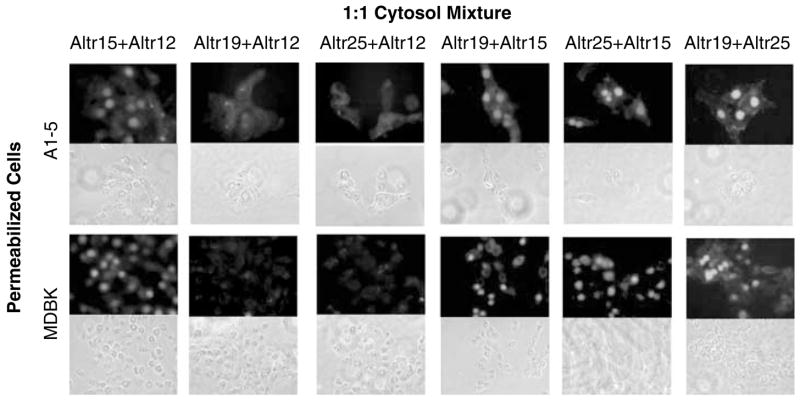
The similarity in p53 nuclear localization phenotypes of some of the ALTR lines prompted us to determine whether the ALTR lines could be segregated into complementation groups. To examine this, the ALTR12, ALTR15, ALTR19 and ALTR25 lines were selected for testing because of the inability of their cytosol to support nuclear import of the p53 bipartite NLS substrate. To test for complementation, the cytosol of each of these ALTR lines were mixed in a 1:1 ratio and then assayed for their ability to support GST-p53BNLS-EGFP nuclear import (Figure 5). Individually, the cytosols from ALTR15, ALTR19 and ALTR25 cells could not support nuclear importation of GST-p53BNLS-EGFP; they did support nuclear importation when mixed together with ~70% nuclear localization observed in both A1–5 and MDBK cells (Table 3). Interestingly, the cytosol of ALTR12 could not complement the cytosol of either ALTR19 or ALTR25 although it did complement the cytosol from ALTR15. Hence, ALTR12 and ALTR15 appear to constitute two different complementation groups whereas ALTR12, ALTR19 and ALTR25s may be in the same complementation group.
Figure 5.
Complementation of ALTR (A1–5 Low Temperature Resistant) cytosols in p53 nuclear localization signal (NLS) nuclear import. GST-p53BNLS-EGFP substrates were assayed for nuclear import in digitonin-permeabilized A1–5 or MDBK cells with cytoplasm mixture (1:1) of different ALTR lines as indicated.
Table 3.
Complementation of ALTR cytosols in supporting p53NLS nuclear import
| Altr12 | Altr15 | Altr19 | Altr25 | |
|---|---|---|---|---|
| Altr12 | ||||
| A1–5 | — | 61.3±3.9a | 15.2±2.3 | 7.0±3.5 |
| MDBK | — | 58.4±10.7 | 8.2±3.1 | 2.8±4.2 |
| Altr15 | ||||
| A1–5 | — | — | 66.9±4.3 | 69.8±5.4 |
| MDBK | — | — | 68.6±11.3 | 68.5±6.3 |
| Altr19 | ||||
| A1–5 | — | — | — | 72.5±5.7 |
| MDBK | — | — | — | 62.8±6.8 |
Abbreviation: ALTR, A1–5 Low Temperature Resistant.
Values indicate the % of cells with nuclear-localized GST-p53BNLS-EGFP ±s.e. from three separate experiments.
The binding of p53 with importin-β remains intact in ALTR cells
One trivial explanation is that the lack of nuclear import capacity of ALTR cells is due to the absence of importin-α/β, or that these importation factors cannot bind with the substrate. To test this we examined importin-β binding to GST-p53BNLS-EGFP or p53 using either GST-pull-down or co-immunoprecipitation. As shown in Figure 6a, the binding of GST-p53BNLS-EGFP with importin-β in cytosol from all tested ALTR cells was detected and remained normal when compared with A1–5 cells, suggesting that the importin-β binding with substrate is intact in all of these ALTR cells. The same result was also observed in the p53 co-immunoprecipitation assays (Figure 6b). Hence, the defect in p53 nuclear importation in these ALTR cells cannot be attributed to the lack of importin-β binding.
Figure 6.
The binding of p53 with importin-β remains intact in ALTR (A1–5 Low Temperature Resistant) cells. (a) Analysis of importin-β binding by GST-pull-down assays. TEN buffer (lane 1) or cytosols from different cells (lanes 2–11) were subjected to GST-pull- down with GST-p53BNLS-EGFP bound glutathione-Sepharose 4B. The pull-down products were analysed by immunoblotting using anti-importin-β. (b) Analysis of importin-β binding by co-immunoprecipitation. The lysis buffer (lane 1) or cell lysate from different cells (lanes 2–11) were subjected to co-immunoprecipitation with p53 antibody PAb 421. The bound proteins were analysed by immunoblotting using anti-importin-β. NLS, nuclear localization signal.
Discussion
The p53 nuclear import process is crucial for activation of its tumor suppressor functions. Here we probed the p53 nuclear importation by deriving a set of mutant cell lines in which the capacity to import p53 into the nucleus was disrupted. Mutagenesis of A1–5 cells and subsequent characterization of the resulting ALTR lines demonstrated that in the majority of these lines p53 was sequestered in the cytoplasm. In approximately half of these lines, p53 could be induced to enter the nucleus to varying degrees by leptomycin B treatment, suggesting that in some instances, cytoplasmic sequestration might be due to an accelerated export process. However, in the remaining ALTR lines, no p53 nuclear accumulation could be induced to by either leptomycin B or genotoxic stress (unpublished data), indicating that in these lines, the p53 nuclear import process had been compromised.
Nuclear localization of p53 is mediated by a bipartite NLS in the C terminus of the protein (Liang and Clarke, 1999). In our in vitro nuclear importation system, the full bipartite p53 NLS is also required for nuclear import of a p53 NLS-containing protein. Moreover, the bipartite p53 NLS, but not the monopartite motif, could bind with importin-β, indicating that p53 nuclear localization is importin-β dependent. Importin-β is expressed in each of the ALTR lines and interacted with the p53 NLS in vitro and with the tsp53 protein in vivo. Hence, the cytoplasmic sequestration of p53 in ALTR cells could not be attributed to the absence of importin-β or to the inability of p53 to interact with it. Clearly, interaction between a cargo protein and importin-β is only one step in the complete nuclear import process. Nuclear pore composition must favor protein transit, cytoplasmic cargo proteins must be physically transported to the nuclear membrane, and many of transportation steps are energy-dependent. Hence, ALTR cell mutations appear to have preferentially affected these other steps instead of cargo/importin interaction, which is likely to be essential for cell viability.
Testing the SV40TAgNLS for in vitro nuclear importation demonstrated that this motif exhibited strong activity in all of the reactions using ALTR cytoplasm. However, when the same assays were conducted using the p53 NLS, we found that only the cytoplasm from ALTR11s showed nuclear localization that was equivalent to that seen with the cytoplasm from A1–5 cells. Moreover, cytosols from four ALTR lines (ALTR12, ALTR15, ALTR19 and ALTR25) showed no ability to promote nuclear import of the bipartite p53 NLS. Hence, although the ALTR cytosols supported SV40TAgNLS nuclear importation the p53 NLS could not be imported in the nucleus under the same conditions. This suggested that the nuclear-trafficking mechanism that acts on p53 NLS is defective in ALTR cells, but that other nuclear-trafficking pathways that may act on housekeeping remained intact. We think it unlikely that the p53 NLS nuclear importation pathway is dedicated to p53 alone. Instead, because p53 activation is stimulated by cell stress, we speculate that this pathway may serve to promote nuclear localization of stress response proteins.
These studies also provide insight into the nature of the p53 nuclear localization pathway. Previously, we showed that ALTR cells with cytoplasmically sequestered tsp53 at 32°C also exhibited a predominantly mutant conformation of tsp53 and only in cells with nuclear-localized p53 was the tsp53 present in a wild-type conformation, suggesting that there is a correlation between the localization of p53 protein and its conformation (Gaitonde et al., 2000). This correlation suggests that there is a link between protein conformation and nuclear localization. Hence, proper p53 nuclear localization may involve a pathway that also ensures correct folding of the protein.
In this work, cytosols from ALTR12, ALTR15, ALTR19 and ALTR25 cells showed little capacity to promote nuclear localization of the p53 NLS in vitro, suggesting that one or several soluble factors necessary for p53 nuclear trafficking were either absent or defective. Complementation assays suggest that ALTR12, ALTR19 and ALTR25s belong to the same complementation group. This is surprising considering that the cytoplasm from ALTR19 and ALTR25 do complement each other. Nevertheless, the inability of ALTR12 cytoplasm to complement either ALTR19 or ALTR25 cytoplasm suggests that mutation in these two cell lines has affected functionally related components. One potential explanation is that the affected component in ALTR12s is required for the production or activation of the components in ALTR19 and ALTR25 cells.
The in vitro nuclear importation assay using permeabilized ALTR cells demonstrated that insoluble factors that could not diffuse out of the cell when permeabilized with digitonin are also involved in the p53 nuclear transport. One potential explanation is that components of the nuclear pore also participate in regulation of p53 nuclear localization. Recent evidence indicates that the nuclear pore is itself a dynamic protein complex that can regulate transport of cargo proteins across the nuclear membrane (Tran and Wente, 2006). Hence, mutations in some nuclear pore components might also be expected to affect p53 nuclear localization. Collectively, our results suggest that p53 nuclear localization is a complex process involving many proteins which, when mutated, can lead to exclusion of p53 and perhaps other stress-responsible proteins from the nucleus.
Materials and methods
Cell culture, reagents and irradiation
Cells were maintained in DMEM medium with 10% fetal bovine serum, 100 U/ml penicillin/streptomycin (Gibco BRL, Gaithersburg, MD, USA). HeLa, MDBK and A1–5 cells were incubated at 37°C with 5% CO2 unless otherwise noted and ALTR lines at 32°C.
Antibody for p53 was PAb 421. Anti-importin-β was from BD Biosciences (San Diego, CA, USA). Anti-GST was from Santa Cruz Biotechnology (Santa Cruz, CA, USA). GST-tagged human p53 was from SignalChem (Richmond, BC, Canada). Cells were exposed to 5Gy ionizing radiation using 60Co source at 47 cGy/min.
Selection for ALTR cell lines
ALTR lines were derived by mutagenesis with ethyl methane sulfonate (EMS; Sigma, St Louis, MO, USA). Briefly, two 150 cm2 flasks containing A1–5 cells at 90% confluence were incubated with 500 μg/ml EMS for 13 h at 37°C. After mutagenesis, cells were rinsed 3× with DMEM and incubated for 24 h. Subsequently, the cells were trypsinized and replated onto twenty 10 cm dishes. After incubation for 16 h at 37°C, the cultures were transferred to 32°C until colonies began to appear (~3 weeks). Forty resistant colonies were selected, 20 of which were successfully expanded into stable cell lines.
RT–PCR and sequencing of p53val135
Reverse transcription of 2 μg total RNA from A1–5 and ALTR cells was carried out using Superscript II RT (Gibco BRL). Full-length p53 was divided into three sections and amplified by PCR using different primers: 5′-ATGGAGGAGTCA CAGTCGGATA-3′ and 5′-ATAACAGACTTGGCTGTCC CAG-3′ were used to amplify the 366 bp N terminus; 5′-GCC AAGTCTGTTATGTGCA-3′ and 5′-GAGGCGCTTGTGC AGGTGG-3′ were used to amplify the 590 bp DNA-binding domain; 5′-AGAAAATTTCCGCAAAAAGGAA-3′ and 5′-CCCCACTTTCTTGACCATTGTT-3′ were used to amplify the 330 bp C terminus. The PCR products were sequenced on an automated sequencer.
Proteins expression and purification
The p53 monopartite NLS (p53MNLS) was obtained by annealing of two oligonucleotides, 5′-GATCCTCTCCCCCG CAAAAGAAAAAACCACTTG-3′ and 5′-AATTCAAGTG GTTTTTTCTTTTGCGGGGGAGAG-3′. The p53 bipartite NLS were amplified by PCR using 5′-CCGCGGATCCCCA GGGAGCGCAAAGAGAGC-3′ and 5′-GCCGGAATTCCT TGAGGGTGAAATACTCTC-3′ as primers from pP53-A1 plasmid. These two NLSs were inserted in frame to pGEX-2T vector. An EGFP-coding sequence amplified by PCR using 5′-GCCGGAATTCATGGTGAGCAAGGGCGAGGA-3′ and 5′-CCGCGAATTCTTACTTGTACAGCTCGTCCA-3′ as primers from pEGFP-C1 was then inserted to the resulting plasmids obtaining pGST-p53MNLS-EGFP and pGST-p53BNLS-EGFP, respectively. The EGFP-coding sequence was inserted into pGEX-2T to obtain pGST-EGFP. All plasmids were confirmed by DNA sequencing.
The GST-tagged proteins were expressed in E. coli BL21(DE3) and purified according to the manufacturer’s directions (Amersham Biosciences, Piscataway, NJ, USA). All proteins were analysed for purity by SDS–PAGE and Coomassie blue staining (not shown).
Preparation of cytosols and in vitro nuclear import
The preparation of cytosols and in vitro nuclear import were performed as described previously (Adam et al., 1990). Assays in the absence of cytosol were conducted as a negative control.
GST-pull-down, immunoprecipitation and indirect immunofluorescence
GST-tagged proteins (100 μg) bound glutathione-Sepharose 4B were incubated with cytosol (1 mg) in TEN buffer (20mM Tris-HCl, pH 7.4, 0.1mM EDTA and 100mM NaCl) at 4°C for 3 h. After three washes with NTEN (20mM Tris-HCl, pH 7.4, 1mM EDTA, 0.5% NP40 and 100mM NaCl), pull-down products were analysed by immunoblotting.
The immunoprecipitation and indirect immunofluorescence were performed as described previously (Gaitonde et al., 2000).
Acknowledgments
We thank Dr Minoru Yoshida for providing leptomycin B and Dr Stephen A Adam for the pGST-SV40TAgNLS-EGFP plasmid. This work was supported by the National Institutes of Health Grant CA090776 to JDM.
References
- Adam SA, Marr RS, Gerace L. Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J Cell Biol. 1990;111:807–816. doi: 10.1083/jcb.111.3.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaitonde SV, Riley JR, Qiao D, Martinez JD. Conformational phenotype of p53 is linked to nuclear translocation. Oncogene. 2000;19:4042–4049. doi: 10.1038/sj.onc.1203756. [DOI] [PubMed] [Google Scholar]
- Goldman SC, Chen CY, Lansing TJ, Gilmer TM, Kastan MB. The p53 signal transduction pathway is intact in human neuroblastoma despite cytoplasmic localization. Am J Pathol. 1996;148:1381–1385. [PMC free article] [PubMed] [Google Scholar]
- Gorlich D, Mattaj IW. Nucleocytoplasmic transport. Science. 1996;271:1513–1518. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- Liang SH, Clarke MF. A bipartite nuclear localization signal is required for p53 nuclear import regulated by a carboxyl-terminal domain. J Biol Chem. 1999;274:32699–32703. doi: 10.1074/jbc.274.46.32699. [DOI] [PubMed] [Google Scholar]
- Martinez J, Georgoff I, Martinez J, Levine AJ. Cellular localization and cell cycle regulation by a temperature-sensitive p53 protein. Genes Dev. 1991;5:151–159. doi: 10.1101/gad.5.2.151. [DOI] [PubMed] [Google Scholar]
- Middeler G, Zerf K, Jenovai S, Thulig A, Tschodrich-Rotter M, Kubitscheck U, et al. The tumor suppressor p53 is subject to both nuclear import and export, and both are fast, energy-dependent and lectin-inhibited. Oncogene. 1997;14:1407–1417. doi: 10.1038/sj.onc.1200949. [DOI] [PubMed] [Google Scholar]
- Nishi K, Yoshida M, Fujiwara D, Nishikawa M, Horinouchi S, Beppu T. Leptomycin B targets a regulatory cascade of crm1, a fission yeast nuclear protein, involved in control of higher order chromosome structure and gene expression. J Biol Chem. 1994;269:6320–6324. [PubMed] [Google Scholar]
- Shaulsky G, Goldfinger N, Ben-Ze’ev A, Rotter V. Nuclear accumulation of p53 protein is mediated by several nuclear localization signals and plays a role in tumorigenesis. Mol Cell Biol. 1990;10:6565–6577. doi: 10.1128/mcb.10.12.6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran EJ, Wente SR. Dynamic nuclear pore complexes: life on the edge. Cell. 2006;125:1041–1053. doi: 10.1016/j.cell.2006.05.027. [DOI] [PubMed] [Google Scholar]