Summary
Background
miRNAs are an abundant class of small, endogenous regulatory RNAs. Although it is now appreciated that miRNAs are involved in a broad range of biological processes, relatively little is known about the actual mechanism by which miRNAs down-regulate target gene expression. An exploration of what protein co-factors are necessary for a miRNA to down-regulate a target gene should reveal more fully the molecular mechanisms by which miRNAs are processed, trafficked, and regulate their target genes.
Results
A weak allele of the C. elegans miRNA gene let-7 was used as a sensitized genetic background for a whole-genome RNAi screen to detect miRNA pathway genes, and 213 candidate miRNA pathway genes were identified. About 2/3 of the 61 candidates with the strongest phenotype were validated through genetic tests examining the dependence of the let-7 phenotype on target genes known to function in the let-7 pathway. Biochemical tests for let-7 miRNA production place the function of nearly all of these new miRNA pathway genes downstream of let-7 expression and processing. By monitoring the down-regulation of the protein product of the lin-14 mRNA, which is the target of the lin-4 miRNA, we have identified 19 general miRNA pathway genes.
Conclusions
The 213 candidate miRNA pathway genes identified could act at steps that produce and traffic miRNAs or in downstream steps that detect miRNA::mRNA duplexes to regulate mRNA translation. The 19 validated general miRNA pathway genes are good candidates for genes that may define protein cofactors for sorting or targeting miRNA::mRNA duplexes, or recognizing the miRNA basepaired to the target mRNA to down-regulate translation.
Introduction
miRNAs are a class of small (~22 nt), non-coding RNAs that regulate gene expression through complementary base-pairing to target mRNAs, typically at sites located in the 3’UTR region [1]. In animals, discontinuous base-pairing between a miRNA and its target mRNA yields a secondary structure containing extensive loops and bulges that generally trigger potent inhibition of translation and/or a less robust degradation of the mRNA. Since the discovery of the first two miRNAs in C. elegans, hundreds of miRNAs have been identified in organisms from plants to flies to humans. miRNAs play roles in a wide variety of processes, including developmental events, programmed cell death, stem cell differentiation, neuronal specification and more [2]. Although our understanding of the basic biology of miRNA biogenesis and function has grown, there is still much we do not understand about the mechanisms by which miRNAs act to modulate message stability and translation.
As miRNAs play diverse roles in development, loss of general miRNA pathway factors would be expected to yield devastating phenotypes. Indeed, complete loss of known miRNA pathway factors, such as the ribonuclease III enzyme Dicer, cause early embryonic lethality [3–5]. Genetic screens aimed at uncovering miRNA pathway factors are hampered by the fact that the desired mutants would likely be lethal. This problem may be solved through the application of whole-genome RNAi screening in a background sensitized for defects in a specific miRNA pathway.
The let-7 miRNA was the second miRNA discovered and the first demonstrated to be conserved in other organisms [6, 7]. Genetic analysis of suppressors of let-7 loss-of-function phenotypes in C. elegans has revealed several mRNAs targeted by let-7, including lin-41, which encodes a Ring finger protein and hbl-1, which encodes a Hunchback-like protein [8–10]. A null mutation in let-7 causes retarded heterochronic phenotypes during larval development and lethality at the larval to adult transition when the worms are unable to properly form a vulva and consequently burst [7]. The crucial mRNA target of let-7 with regard to the heterochronic bursting phenotype is lin-41; bursting is suppressed in lin-41;let-7 double mutants [10]. Furthermore, down-regulation of dcr-1 (the worm homolog of Dicer) by dsRNA injection, which impairs the processing of miRNAs, causes phenotypes strikingly similar to those of the let-7 mutant, including vulval bursting [3]. Thus let-7 is sensitive to changes in the activity of the general miRNA pathway.
A weak allele of let-7, mg279, shows slightly reduced levels of the mature let-7 miRNA due to defects in processing of the primary transcript, but is viable and does not show the let-7 null phenotype of lethality due to bursting [7, 11]. We reasoned that this weak allele of let-7 would provide a sensitized background in which to detect defects in the miRNA pathway, by producing a specific and scorable miRNA-associated phenotype rather than lethality. We carried out a whole-genome feeding RNAi screen and identified 213 gene inactivations that give rise to a bursting phenotype in a let-7(mg279) background. Examination of other phenotypes in the let-7 temporal patterning pathway confirmed a role in the heterochronic pathway for many of the candidates. Northern blotting indicated that the new miRNA pathway genes function downstream of let-7 expression and processing. Some of the gene inactivations also affected lin-4 miRNA regulation of the lin-14 transcript, implicating this subset of genes in processes generally required for miRNA function.
Results and Discussion
let-7(mg279) is sensitized for defects in the miRNA pathway
As in other organisms, the ribonuclease III enzyme DCR-1 is required for the processing of miRNA precursors to mature miRNAs in C. elegans [3, 12, 13]. Complete loss of DCR-1 function causes early embryonic lethality [3]. Probably because of partial inactivation of dcr-1, dsRNA targeting dcr-1 causes no obvious phenotype in the first or second generation of feeding to wild-type C. elegans (Figure S1A). However, an accumulation of the let-7 miRNA precursor can be detected by Northern blot, indicating that DCR-1 activity is decreased by dcr-1 RNAi feeding(Figure 2 and [12]). We hypothesized that in the let-7(mg279) worms, where mature levels of let-7 are already lower, dcr-1 RNAi feeding might produce a genetic enhancement to the bursting let-7 null phenotype. Indeed, let-7(mg279) worms fed dcr-1 dsRNA show an increased incidence of bursting at the larval to adult transition as compared to let-7(mg279) worms fed a control RNAi clone or wild-type worms fed either dcr-1 or control dsRNA (Figure S1A). Thus, let-7(mg279) worms are sensitized to detect subtle defects in the miRNA pathway and provide a useful genetic background to perform an enhancer screen to identify new components of this pathway.
Figure 2. let-7 Northern blot.
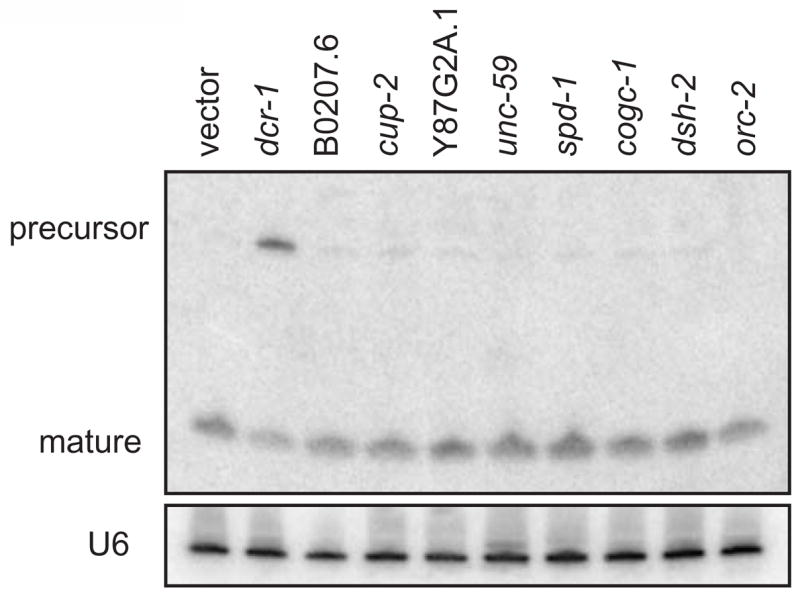
Total RNA from young adult worms raised for two generations on feeding RNAi probed for let-7. dcr-1 (RNAi) results in an accumulation of precursor and loss of mature let-7 miRNA. Other RNAi treatments do not affect let-7 processing or expression.
A genome-wide feeding RNAi screen uncovers 213 gene inactivations that enhance eri-1(mg366); let-7(mg279)
The eri-1(mg366) mutation renders worms hypersensitive to RNAi[14]. The eri-1(mg366); let-7(mg279) double mutant exhibits significant bursting when fed dcr-1 dsRNA (Figure S1A). The eri-1(mg366) single mutant also shows significant bursting when fed dcr-1 RNAi, most likely due to the increased efficiency of gene knockdown, but the presence of the let-7(mg279) allele increased the bursting response significantly (Figure S1A). We used this double mutant to perform a whole-genome feeding RNAi screen. The screen utilized the Ahringer feeding RNAi library supplemented with clones from the Vidal feeding RNAi library for a total of ~17,900 dsRNA clones, corresponding to 94% of the protein-coding genes in the C. elegans genome [15–17]. RNAi feeding was initiated with L1 larvae and their progeny were scored for bursting at the L4 to young adult transition. This two-generation screen gives RNA interference time to reach maximal gene inactivation. In addition, a cherry-picked library of approximately 2700 clones from the whole-genome library known to cause developmental arrest, lethality, and sterility in the first generation of an eri-1 mutant strain [15] was screened by initiating feeding with L1 larvae and scoring for bursting at the larval to adult transition in the first generation of RNAi. Individual wells were scored on a 4 point scale, with 1 corresponding to ~1% burst, 2 corresponding to up to ~15% burst, 3 corresponding to up to ~50% burst and 4 corresponding to greater than ~50% burst. The bursting phenotype is highly penetrant in a let-7 null mutant [10].
As an important set of controls, empty vector clones and dcr-1 clones were added to the library and screened blindly. A total of 110 empty vector controls were scored and in each case caused no bursting, suggesting that the false positive rate of the screen should be very low (Figure S1B). Of the 112 dcr-1 RNAi clones scored, the majority were assigned the strong bursting score of 3 or 4. However, 19 of the dcr-1 clones were scored as generating lower levels of bursting and even, in 5 examples, no bursting at all. That dcr-1 RNAi occasionally fails to induce bursting indicates that some real positives may be missed in the initial screen, possibly due to the variability of RNAi.
The initial screen identified 350 RNAi feeding clones, targeting 332 individual genes, that induce let-7-mutant-like bursting in the eri-1(mg366); let-7(mg279) background (Figure S1C). Upon re-testing in triplicate, 231 clones, targeting 213 individual genes, reproducibly generated this bursting phenotype (Table S1). It is interesting to note that the lethal/sterile sub-library was significantly enriched for clones that scored positive. Of these 213 genes, most are conserved across phylogeny: for example 172 of these genes are included among the KOG classification, which identifies genes having clear orthologs in plants, animals or fungi [18]. The animal and plant orthologs identified by the KOG classification of the miRNA pathway genes revealed in our screen are good candidates to mediate miRNA function in those phyla as well.
After eight individual trials, an average burst score was calculated for each positive RNAi clone. While all clones that reproducibly enhanced bursting may be considered positives, a cut-off was set at a score of 2 for initial more detailed analysis. In addition, due to the variability of RNAi, those clones that had an average score of less than 2, but at least two individual scores of 3 or 4 were also included. This strict positive category included 69 clones, targeting 61 genes (Table S1).
Genetic tests place many of the candidate genes in the let-7 miRNA pathway
A trivial explanation for the bursting induced by the RNAi clones is that they target genes crucial for vulval development, but in pathways parallel to that of the let-7 miRNA pathway. To address this we compared the bursting phenotype of a dsRNA clone on the eri-1(mg366); let-7(mg279) strain to that induced by feeding eri-1(mg366). Given our hypothesis that the let-7(mg279) allele provides a sensitized background due to lower levels of mature let-7, we reasoned that those clones targeting genes required for let-7 activity would be more likely to enhance the bursting phenotype in the presence of that allele. To measure the dependence of the bursting on let-7(mg279) we subtracted the bursting score of eri-1(mg366) from that of eri-1(mg366); let-7(mg279). Bursting was considered dependent on let-7(mg279) if the difference was greater than 1.5. Inactivation of 20 of the 61 genes in the strict positive class showed a dependence on let-7(mg279) for the bursting phenotype (Figure 1B). These gene inactivations also failed to enhance the weak bursting phenotype of the unrelated multi-vulva mutant lin-1(e1777) (Figure S2), further arguing against general defects in vulval development.
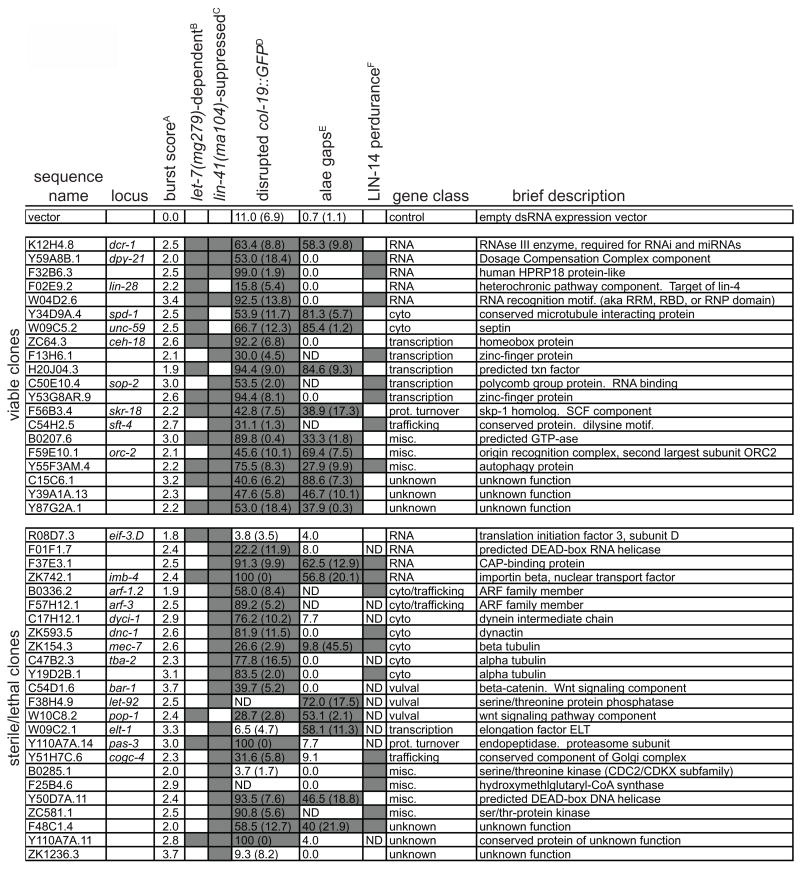
Figure 1. let-7 miRNA pathway candidate genes.
A. The average burst score from all trials (7–11 trials for each).
B. Gene inactivations for which bursting depends on the presence of the let-7(mg279) allele. Positives are defined as a difference of at least 1.5 in average bursting score over 3 trials comparing eri-1(mg366); let-7(mg279) to eri-1(mg366).
C. Gene inactivations where bursting is suppressed by lin-41(ma104). Positives which are shaded are defined as a difference of at least 1.5 in average bursting score over 3 trials comparing eri-1(mg366); let-7(mg279) to lin-41(ma104); eri-1(mg366); let-7(mg279).
D. Percentage of col-19::GFP;let-7(mg279) worms with disrupted patterns of expression. Standard deviation of three trials is shown in parentheses. Shading indicates a p value ≤ 0.05 determined by Chi square test comparing RNAi feeding to empty vector control.
E. Percentage of eri-1(mg366) worms with gaps in adult alae after RNAi feeding. Standard deviation of three trials is shown in parentheses, when appropriate. Shading indicates a p value ≤ 0.05 determined by Chi square test comparing RNAi feeding to empty vector control.
F. Shading indicates a perdurance of LIN-14 protein in L2 worms as determined by a comparison on a LIN-14 Western blot to the empty vector-fed control (see Figure 3).
dcr-1 RNAi by injection induces bursting even in the wild-type background [3], suggesting that let-7 phenotypes can result from a down-regulation of the general miRNA pathway machinery even in the absence of a sensitized background. By this view, some of the RNAi clones that induce a bursting phenotype independently of the let-7(mg279) allele may still target genes in the miRNA pathway. To identify this class of gene targets, we asked if the induced bursting could be suppressed in the lin-41(ma104) background, which suppresses bursting of a let-7 genetic mutant [7, 10]. We used a similar approach to that described for examining let-7(mg279) dependence, comparing the bursting induced in the eri-1(mg366); let-7(mg279) strain with a triple mutant, lin-41(ma104); eri-1(mg366); let-7(mg279). The bursting induced by inactivation of 39 of the 61 strict positive genes was suppressed by lin-41(ma104) (Figure 1C). Taken together, these genetic tests pl ace 44 genes, targeted by 49 of the 69 strict positive RNAi clones, in the developmental pathway controlled by let-7.
Classes of candidate miRNA pathway genes identified
RNA binding/processing
Genes in one large class of candidate miRNA pathway genes encode proteins with known or predicted roles in RNA binding or processing. Included in this class is dcr-1, with its well-established role in miRNA processing [1]. Also included in this category is imb-4, which encodes the worm ortholog of exportin-1, an importin-β-like protein involved in nuclear trafficking. Although most similar to exportin-1, by BLAST imb-4 also represents the closest worm homolog of exportin-5, which plays a role in the export of pre-miRNAs from the nucleus [1]. The known heterochronic pathway gene and miRNA target lin-28 [19] also was identified in the screen. LIN-28 interacts with target mRNAs to regulate their translation efficiency and has been localized to stress-granules [20, 21], which are related to P-bodies, and may be sites of miRNA regulation [21].
Several of the genes identified encode proteins predicted to act in mRNA processing. F32B6.3 encodes a protein with similarity to human HPRP18, which interacts with the U5 snRNP involved in splicing [22]. F37E3.1 encodes a conserved nuclear cap-binding protein, which has been found to interact with the m(7)G cap and also with translation initiation factors [23, 24]. A translation inititation factor itself, eif-3.D, was also identified, as was a DEAD-box RNA helicase, F01F1.7, which is predicted to interact with F37E3.1 [25]. Kiriakidou et al. identified a m(7)G cap-binding domain in human Ago2 that is required for translational inhibition and is conserved in C. elegans alg-1 and alg-2, suggesting that the m(7)G cap plays a crucial role in miRNA regulation [26]. One of the strongest positives in the screen that was completely dependent on let-7(mg279) and suppressed by lin-41(ma104) was W04D2.6, which encodes a conserved protein with an RNP-1 RNA binding domain and a PWI domain. PWI domains have been shown to bind both single-stranded and double-stranded nucleic acids [27], making this protein an intriguing candidate for a miRNA pathway component.
Cytoskeleton
A number of the candidate genes encode proteins that play a structural or regulatory role in the cytoskeleton. Y19D2B.1 and tba-2 both encode α-tubulins and mec-7 encodes a β-tubulin. spd-1 encodes a microtubule interacting protein and dyci-1, which encodes an intermediate chain of the motor protein dynein, and dnc-1, which encodes the dynein interactor dynactin, also interact with microtubules. Two genes encoding members of the ADP-ribosylation factor family, arf-1.2 and arf-3, were also identified. Members of this protein family play roles in controlling microtubule dynamics as well as intracellular trafficking [28]. unc-59, which encodes a septin, a filament-forming protein with known roles in cytokinesis [29], was also identified. Given the requirements for cell division and morphogenesis during vulval development, it would seem reasonable that these candidates may be required for cellular events downstream of the activity of let-7. Indeed, most of these gene inactivations cause bursting even in the absence of the let-7(mg279) allele (Figure 1B). However, this bursting is suppressed in the lin-41(ma104) background, raising the possibility that the connection between miRNAs and the cytoskeleton may be more tightly linked. In addition, inactivation of several of these cytoskeleton factors also abrogated miRNA down-regulation of lin-14 mRNA translation (see below), arguing for a more general role in miRNA function. Evidence for a connection between small RNAs and the cytoskeleton does exist. In sea urchin eggs, Seawi, a homolog of the small-RNA factor Piwi, is a major component of the microtubule ribonucleoprotein complex that also includes ribosomes and mRNAs [30]. The discovery that RNAs interact with and play a regulatory role in mitotic spindle formation [31] provides a precedent for this potential miRNA/cytoskeleton regulatory connection. Although we have no direct evidence, it is tantalizing to imagine localized zones of miRNA regulation guided by association with the cytoskeleton.
Vulval induction signaling
Two signaling pathways that play well-characterized roles in induction of the vulva are the Wnt and the Ras-mediated pathways [32]. pop-1, which encodes the downstream transcription factor of the Wnt pathway was identified as a let-7 enhancing gene inactivation, as was bar-1, which encodes the beta-catenin homolog that interacts with POP-1. We also identified let-92, which encodes a homolog of the catalytic subunit of protein phosphatase 2A that plays a positive role in Ras-mediated vulval induction [33]. let-92 is thought to play a role downstream of let-60, the worm homolog of RAS and a possible target of the let-7 family of miRNAs [34]. lin-41(ma104) suppresses the bursting caused by let-92 gene inactivation, suggesting crosstalk between the pathways regulated by different let-7 targets. These candidates represent a connection between the let-7 miRNA pathway and the established vulval induction pathways.
Transcriptional regulation
Of most interest in this class is the Polycomb Group gene sop-2, which represses Hox gene expression and binds RNA [35]. sop-2 was also identified in an RNAi screen for disruption of RNAi-mediated transcriptional gene silencing [36]. The identification of the sop-2 inactivation as an enhancer of let-7(mg279) raises the possibility of a similar transcriptional silencing mechanism involving miRNAs. Interestingly, SOP-2 and another Hox gene regulator, SOR-1, are localized to a subnuclear compartment that may be where they function, for example, in epigenetic silencing [37]. sor-1 was also detected as a strong let-7 enhancer in our genome screen. The involvement of such polycomb-like factors in miRNA function suggests a possible transcriptional silencing aspect to miRNA function. Alternatively, these polycomb-like factors could positively regulate the transcription of miRNAs, though we do not see any change in let-7 expression levels when these genes are inactivated (see below).
Several known or predicted transcription factors are also let-7 enhancing gene inactivations, including ceh-18, elt-1, H20J04.3, Y53G8AR.9 and F13H6.1. Many miRNAs are expressed at higher levels in differentiated cells than tumor cells [38], suggesting that as cells differentiate during development, the expression of miRNAs is under significant transcriptional control. The transcription factors identified in our screen could be the direct regulators of miRNA expression that couple their activation to the differentiation of particular cell types during development. There is also evidence of post-transcriptional regulation of miRNA expression in tumors [39]; in these tumors, the cofactors for miRNA maturation or function could be under transcriptional control. Similarly, the transcription factors identified in our screen could control the expression of general miRNA pathway components, including other genes identified in this screen. In addition, dpy-21, which encodes a component of the dosage compensation complex was identified. It is also possible that miRNAs associate with these chromatin factors in the nucleus, in analogy to the siRNA regulation of heterochromatin formation in fungi.
Protein turnover
Inactivation of a proteasome subunit encoding gene, pas-3, or skr-18, which encodes a homolog of the SCF ubiquitin ligase component Skp-1, each induce let-7(mg279)-dependent bursting. These gene identifications raise the intriguing possibility that active turnover of the proteins whose mRNAs are targeted by a miRNA may contribute to target down-regulation.
Protein traffickings
In addition to the two ADP-ribosylation factor family members mentioned above, arf-2.1 and arf-3, two other genes encoding proteins involved in trafficking were uncovered. cogc-4 encodes a conserved member of the oligomeric golgi complex which is required for proper golgi function and plays a role in vesicle trafficking [40]. sft-4 encodes a conserved protein whose homologs are localized to transport vesicles [41]. With the exception of arf-3, for which the LIN-14 assay was not performed, each of these gene inactivations also inhibited LIN-14 down-regulation (see below), suggesting a general connection between protein trafficking and miRNA regulation.
Miscellaneous and unknown function
A number of candidate genes were identified that did not cluster into any obvious functional classification. These include Y50D7A.11, encoding another DEAD-box helicase that is predicted to act on DNA instead of RNA, two serine/threonine kinases encoded by B0285.1 and ZC581.1, a predicted GTPase encoded by B0207.6 and others. Five genes encoding proteins with unknown functions were also identified.
A common worry about all RNAi screens is specificity of the gene inactivation, most especially whether the dsRNA targets more than the intended mRNA. In general, RNAi screens in C. elegans have been validated to target the intended gene: for example dsRNAs targeting genes of known loss of function phenotypes from classical genetics induced just those phenotypes in most cases [17]. However, there are some indications that genes targeting some of the members of highly conserved protein families may inactivate more than the exactly homologous mRNA. For example, the mec-7 dsRNA strongly enhances let-7 and strongly affects LIN-14 down-regulation, but a mec-7 null mutation is viable and is only known to be mechanosensory defective, and the mec-7 tubulin is only expressed in mechanosensory neurons [42]. Blastn analysis of mec-7 shows that there are regions of 20 to 30 nt conserved between beta tubulin paralogs that may be broadly targeted by these dsRNAs. However, most of the hits from the RNAi screen do not target members of conserved families of paralogs. In addition, several of the genes identified (ceh-18, sop-2, dyci-1 and Y53G8AR.9) were targeted by multiple non-overlapping RNAi clones, all of which scored as positives, arguing against off-target effects of specific clones.
Known miRNA pathway components
The screen identified the known miRNA pathway components dcr-1 and imb-4, the closest homolog of Exportin-5. This confirms the utility of the sensitized eri-1(mg366);let-7(mg279) background for screening. There are, however, known miRNA pathway components that were not identified in the screen, either because they induced a more severe lethal phenotype or because they did not induce any detectable phenotype in the strain screened. The Argonaute family member alg-1 has been shown to play a role in miRNA function in C. elegans and was not identified in our screen. More direct tests using known miRNA pathway genes showed that, in the eri-1(mg366) enhanced RNAi background, alg-1 RNAi causes a larval lethal phenotype in the parental generation (data not shown); these arrested animals could not be scored for thelet-7 phenotype at the larval/adult transition. Two other known miRNA pathway components, drsh-1 and pash-1, which mediate the processing of pri-miRNAs, were also not identified in the screen, but not due to earlier lethality. Interestingly, feeding RNAi targeting these genes does lead to bursting in some genetic backgrounds. We found that let-7(mg279) worms carrying a GFP transgene expressed in the hypodermis were enhanced to bursting upon feeding of dsRNA targeting drsh-1 or pash-1, although feeding to let-7(mg279) or the GFP transgene strains alone did not induce bursting (data not shown). It is unclear why this transgene sensitizes let-7(mg279) to drsh-1 and pash-1 knock-down while adding eri-1(mg366) in the genetic background did not. These observations illustrate that while our sensitized RNAi screen approach will identify many miRNA factors, some bona fide miRNA pathway components may be missed.
Many of the candidate miRNA pathway gene inactivations induce phenotypes consistent with defects in the miRNA function
For the 44 candidate genes, we carried out further secondary tests to confirm a role in the let-7 miRNA pathway. let-7 has a well-characterized role in the heterochronic developmental pathway in C. elegans that determines the timing of specific cellular events during development. One of the final readouts of the heterochronic pathway is the expression of the cuticle collagen gene col-19 in the hypodermis and seam of adults. The let-7(mg279) allele causes modest defects in the expression of a col-19::GFP fusion gene, with 11% of worms showing either no GFP signal or GFP in the seam, but not in the hypodermis (Figure 1D and [43]). dsRNA treatment led to a statistically significant increase in worms displaying disrupted col-19::GFP expression for 38 of the 42 candidate genes for which the assay could be performed (Figure 1D).
Another assay for the mis-coordination of developmental events is the presence of defects in adult alae, the small ridges that run along both sides of the adult. Loss or knockdown of many heterochronic pathway gene products and also of known miRNA pathway components causes defects in these alae, including the presence of gaps in the ridges [3, 7, 10, 12]. The candidate RNAi clones were fed to eri-1(mg366) worms and the state of the adult alae were assessed visually for any gaps. Inactivation of 19 of the 44 genes caused significant defects in the continuity of adult alae (Figure 1E). It is significant that each of these assays examine heterochronic phenotypes in a tissue that is distinct from the vulval cells scored in the primary screen.
Most of the genes act downstream of let-7 biogenesis
To determine if any of the targeted genes are required for let-7 biogenesis, Northern blots with let-7 probes were performed on samples from young adult animals, when let-7 is normally highly expressed, after inactivation of each of the candidate miRNA pathway genes. The fact that the RNAi clones examined produce a bursting phenotype in the eri-1(mg366); let-7(mg279) background presents a hurdle for collecting staged young adult animals on which to perform let-7 Northerns. We knew that feeding RNAi targeting dcr-1 to wild-type worms leads to a noticeable increase in let-7 precursor accumulation, although there is no bursting phenotype associated with this molecular phenotype. Given this observation, we expected gene inactivations affecting let-7 miRNA processing may produce a molecular phenotype in the absence of bursting in the wild-type background. dcr-1 RNAi demonstrated a clear, although incomplete, defect in processing of the let-7 precursor to the mature form in young adults, as expected (Figure 2 and Figure S3). In contrast, no other dsRNA treatment induced a dramatic increase in precursor or decrease in mature let-7 (Figure 2 and Figure S3). A subset of RNAi clones were also fed to lin-41(ma104);eri-1(mg366);let-7(mg279) worms to determine if miRNA processing defects could be detected in this genetic background, but again no defects in let-7 maturation were observed (data not shown). These data suggest that these miRNA debilitating gene inactivations abrogate function downstream of the expression and processing of let-7.
Many of the miRNA pathway genes are necessary for the down-regulation of target mRNA translation
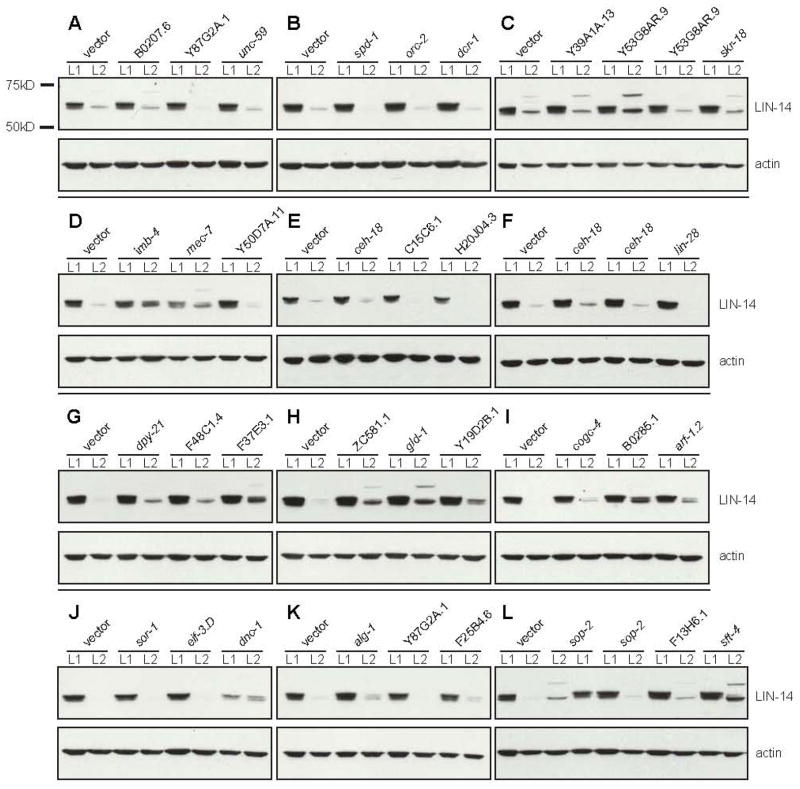
To classify the functions of candidate genes as either acting generally in the miRNA pathway or specifically to the let-7 pathway, we assayed whether these gene inactivations also interfere with the lin-4 miRNA pathway. lin-4 was the first miRNA identified and it is well established that lin-4 down-regulates lin-14 mRNA translation during larval development [44, 45]. Western blots of LIN-14 protein comparing levels in L1 and L2 larval stage worms show a clear down-regulation of protein in wild-type (Figure 3). We asked if RNAi inactivation of the miRNA pathway candidate genes abrogates this down-regulation of LIN-14 protein levels. LIN-14 runs as a doublet at ~67 kD. During normal development, the top ~67 kD band disappears first, followed by the lower band. For many of the let-7-enhancing gene inactivations, the down-regulation of LIN-14 at late stages was not as pronounced as in wild-type. For a significant subset of the miRNA pathway gene inactivations, in addition to the slower disappearance of the ~67 kD LIN-14 bands, a new LIN-14 band appears at ~75 kD. This ~75 kD band appears transiently in a wild-type time course of LIN-14 down-regulation (data not shown), suggesting that it is a short-lived intermediate in the wild-type pathway that becomes more stable when steps in the miRNA pathway are inactivated. The appearance of the ~75 kD band coincides in wild-type with loss of the upper band of the ~67 kD doublet, raising the possibility of a protein modification, such as sumoylation or ubiquination, during turnover (data not shown). Inactivation of 19 of 34 genes in the eri-1(mg366) background caused an increase in LIN-14 abundance at ~67 kD or ~75 kD as compared to comparably staged worms fed vector alone controls (Figure 1F, Figure 3). This is consistent with a role for these genes in the general miRNA pathway upstream of miRNA-mediated down-regulation of protein synthesis. The enhanced let-7 miRNA phenotype resulting from knockdown of these genes in the whole-genome screen cannot be explained by a decrement in lin-4 activity only, as the lin-4(e912);let-7(mg279) double mutant does not show enhanced bursting (data not shown). In order to collect L1 and L2 larval samples that had been exposed to RNAi for the lethal/sterile clones, we started RNAi treatment in the first generation as late L4s. Even this late treatment led to lethality or sterility for many of the sterile/lethal clones, so only a subset of these clones was tested. With this caveat in mind, it is notable that almost all of the sterile/lethal RNAi clones for which we were able to perform the assay showed altered LIN-14 levels.
Figure 3. LIN-14 Western blots.
Protein samples from synchronized L1 and L2 (24 hours at 20 °C) worms are shown for each RNAi treatment. LIN-14 levels drop dramatically from L1 to L2 when fed empty vector control. Due to variations in LIN-14 intensity between blots all comparisons were made to the empty vector control lanes for each blot to determine if LIN-14 down-regulation was affected.
Taken together with the let-7 Northern blot data, these LIN-14 results suggest that a major subset of the genes identified in our screen function between the point of miRNA biogenesis and the point of target mRNA down-regulation. Given how little is known about how miRNAs actually induce down-regulation of target mRNA translation and/or stability, the genes identified in this screen are promising leads towards discerning the molecular mechanisms by which miRNAs down-regulate their targets. In this set are several genes from the RNA binding/processing class, including the RNA binding W04D2.6, the nuclear cap-binding F37E3.1 and the predicted U5 snRNP interactor F32B6.3. It may be that these factors facilitate the interaction of the target mRNA with its corresponding miRNA or detect that interaction to in-turn recruit the mRNA::miRNA complex to P bodies and down-regulate translation. It is also notable that all of the sterile/lethal cytoskeleton factors for which we were able to perform LIN-14 Westerns abrogated LIN-14 down-regulation, including the ARF family member arf-1.2, the dynactin dnc-1 and both α-(tba-2) and β-(mec-7) tubulins. This suggests a more general role for the cytoskeleton in miRNA regulation. Perhaps subcellular sites of miRNA regulation, such as the P-bodies, stress granules, or other unknown sites, are organized by the cytoskeleton.
The miRNA pathway screen has identified genes distinct from those identified in a screen for RNAi factors
DCR-1 is an example of a protein that functions in both the miRNA and RNAi pathways [3, 12, 13]. Kim et al. performed a whole-genome RNAi screen designed to identify genes required for RNAi. They identified only three genes (dcr-1, pop-1 and kin-10) from their list of 90 candidates that when inactivated caused significant enhancement of let-7(mg279), suggestive of a role in the miRNA pathway as well [15]. Consistent with this, only two of our 44 confirmed let-7 pathway genes (dcr-1 and pop-1) appear on the RNAi candidate list. It appears that, by these assays, the RNAi and miRNA pathways are largely molecularly distinct.
Experimental Procedures
Strains
Standard C. elegans cultivation techniques were used for all worm handling. Strains and mutant alleles used in this study: wild-type Bristol N2, let-7(mg279)X, eri-1(mg366)IV, eri-1(mg366)IV; let-7(mg279), lin-41(ma104)I; eri-1(mg366)IV; let-7(mg279)X, veIs13[col-19::GFP;rol-6(su1006)]V; let-7(mg279)X, Is: [col-10p::lac-Z-lin-41 3’UTR] (integrated transgene strain kindly provided by the Plasterk lab).
RNAi screen for enhancement to bursting
Handling of the Ahringer RNAi library supplemented with the unique clones from the Vidal RNAi library and the cherry-picked sterile/lethal sub-library was done largely as previously described [15–17]. Synchronized L1 worms of the eri-1(mg366);let-7(mg279) genotype were placed onto 6-well RNAi plates; ~6 (for second generation screening) or ~100 (for parental generation screening). These RNAi plates were then kept at 20 °C and either scored for bursting 3 days later (for parental generation screening) or shifted to 25 °C 3 days later and then scored for bursting 2 days after that (for second generation screening). All scoring was done using a Nikon SMZ645 dissection microscope. Subsequent re-testing and bursting assays in other genetic backgrounds were performed as above.
RNAi controls
RNAi clones targeting dcr-1 and the empty vector feeding clone pPD129.36 [46] were used as positive and negative controls, respectively. For the initial screening, these clones were added to approximately half of the 96-well O/N cultures in the place of clones known to give no growth. They were then scored blindly.
col-19::GFP assays
Gravid adult let-7(mg279) worms carrying an integrated transgene expressing GFP under the col-19 promoter and fed RNAi as above were examined for their GFP expression pattern with a fluorescence dissection microscope. Worms were scored as either have normal expression (in the hypodermis and seam) or disrupted expression (absent or in the seam alone). Chi squared analysis was performed to determine significance, with the cut-off at a p-value of 0.05.
Alae assays
Young adult eri-1(mg366) worms fed RNAi as above were mounted for imaging and the alae examined for gaps with DIC microscopy using a 40X lens. It should be noted that it was not possible to score alae in worms that had burst, so for those gene inactivations that induce some bursting in the eri-1(mg366) background, only worms that had survived past the larval/adult transition were scored. This could lead to an underestimate of the impact of the gene inactivation on alae formation. Chi squared analysis was performed as for the col-19::GFP assays.
let-7 Northern blots
RNAi feeding in a worm strain carrying a reporter transgene were performed largely as for the bursting screen, with a few exceptions. In order to prepare RNA from synchronized worms in the second generation an egg prep was performed on the gravid adults from the RNAi-fed parental generation and the resulting synchronized L1s were then plated onto the same RNAi clone and grown to adulthood. Total RNA preparation and Northern blots for the let-7 precursor and mature miRNA were performed as previously described [7], with the exception that Invitrogen 15% TBE-Urea gels were used and ULTRAHyb (Ambion) was used for the hybridization solution. 20 μg total RNA was loaded into each lane, except for the last 7 lanes for blot H, where 10 μg was loaded. Blots were reprobed for the U6 snRNA as a loading control.
LIN-14 Western blots
RNAi feeding was performed as described for the let-7 Northern blots for RNAi clones scored in the second generation. For the sterile/lethal RNAi clones, worms were initially fed on the empty vector control and then transferred to RNAi plates as L4s before subsequent egg prepping of gravid adults. In both cases, a fraction of the synchronized L1s were flash-frozen in liquid nitrogen and others were placed onto the appropriate RNAi clone and fed for 24 hours at 20 °C before flash-freezing as L2s. Before L2 harvesting, worms were visually inspected to verify they were all at the same developmental stage. Worm lysate preparation and LIN-14 Western blotting was performed as previously described [47]. 50 μg of total protein (as assayed by Bio-Rad DC protein assay) was loaded into each lane of an Invitrogen NuPAGE 4–12% Bis-Tris gel. Blots were re-probed with an actin antibody (MP Biomedicals 691001) as a loading control.
Supplementary Material
Figure S1. let-7 enhancement screen details
A. let-7(mg279) is sensitized for defects in miRNA processing. Feeding of dsRNA targeting dcr-1 induces significant bursting in the let-7(mg279) background as compared to wild-type N2. Dcr-1 RNAi also induces bursting in eri-1(mg366) and this bursting is significantly increased in the eri-1(mg366); let-7(mg279) background (by chi square test, p value = 9.15 × 10-9). Each of these worm strains fed empty vector control showed negligible bursting.
B. Positive and negative dsRNA feeding controls. Empty vector negative controls were consistently scored 0 in the let-7 enhancement screen. dcr-1 positive controls were mostly scored as strong inducers of bursting (3 or 4), but a minority of these control clones were scored as inducing weak or no bursting. All controls were scored blindly.
C. let-7 enhancement screen summary. The whole-genome library consists of the Ahringer library supplemented with clones from the Vidal Orfeome library for a total of ~17,900 clones and was screened in the second generation. The ~2,700 sterile/lethal sub-library clones have been previously described [15] and were screened in the first generation. The primary screen was done in a single pass, the re-testing was done in triplicate. A total of 8 tests (for most clones) were used to calculate an average burst score (see Table S1) and those clones with an average of 2 or greater or that were scored as 3 or 4 on at least 2 separate occasions make up the strict positive class.
Figure S2. Bursting enhancement test with the general vulval development mutant lin-1(e1777).
RNAi clones that induced let-7(mg279)-dependent bursting were fed to the lin-1(e1777) mutant, which has a multiple vulva phenotype and bursts at a low level on its own. The percent burst after dsRNA treatment was normalized to that seen for lin-1(e1117) fed empty vector control. None of the dsRNA treatments significantly enhanced the bursting of this vulval development mutant.
Figure S3. let-7 Northern blots.
Total RNA was isolated from young adults fed RNAi. Each blot contains an empty vector-fed negative control and a dcr-1 (RNAi) positive control. Blots were re-probed with a U6 snRNA probe as a loading control. Only dcr-1 (RNAi) shows a significant increase in precursor accumulation indicating a defect in let-7 miRNA processing.
Table S1. Re-tested positives from the eri-1(mg366); let-7(mg279) whole-genome RNAi screen for bursting.
Listed are all genes identified from the primary screen whose inactivation resulted in an average burst score >0 when re-tested in triplicate. The burst score shown is a more complete average score encompassing 8 separate burst tests (for most genes). Genes classified as strict positives are indicated with a “+”. These include all those with an average score of 2 or greater and those that scored a 3 or 4 in at least 2 separate tests. Gene descriptions are from WormBase version WS170 and were acquired using WormMart.
Acknowledgments
Some strains were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources. We are grateful to John Kim, Harrison Gabel and Ravi Kamath for assembling the sterile/lethal RNAi sub-library. We thank Ronald Plasterk for worm strains. Chris Carr provided much help with the chi square statistical test. We thank Justine Melo and Gabe Hayes for critical reading of the manuscript and Harrison Gabel and members of the Ruvkun, Kaplan and Ausubel labs for engaging discussions and suggestions. DHP is a Damon Runyon Fellow supported by the Damon Runyon Cancer Research Foundation (DRG-1816-04). This work was supported by a grant from the NIH to GR (R01-GM44619).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 3.Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 4.Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 5.Wienholds E, Koudijs MJ, van Eeden FJ, Cuppen E, Plasterk RH. The microRNA-producing enzyme Dicer1 is essential for zebrafish development. Nat Genet. 2003;35:217–218. doi: 10.1038/ng1251. [DOI] [PubMed] [Google Scholar]
- 6.Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Muller P, Spring J, Srinivasan A, Fishman M, Finnerty J, Corbo J, Levine M, Leahy P, Davidson E, Ruvkun G. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 7.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 8.Lin SY, Johnson SM, Abraham M, Vella MC, Pasquinelli A, Gamberi C, Gottlieb E, Slack FJ. The C elegans hunchback homolog, hbl-1, controls temporal patterning and is a probable microRNA target. Dev Cell. 2003;4:639–650. doi: 10.1016/s1534-5807(03)00124-2. [DOI] [PubMed] [Google Scholar]
- 9.Abrahante JE, Daul AL, Li M, Volk ML, Tennessen JM, Miller EA, Rougvie AE. The Caenorhabditis elegans hunchback-like gene lin-57/hbl-1 controls developmental time and is regulated by microRNAs. Dev Cell. 2003;4:625–637. doi: 10.1016/s1534-5807(03)00127-8. [DOI] [PubMed] [Google Scholar]
- 10.Slack FJ, Basson M, Liu Z, Ambros V, Horvitz HR, Ruvkun G. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol Cell. 2000;5:659–669. doi: 10.1016/s1097-2765(00)80245-2. [DOI] [PubMed] [Google Scholar]
- 11.Bracht J, Hunter S, Eachus R, Weeks P, Pasquinelli AE. Trans-splicing and polyadenylation of let-7 microRNA primary transcripts. RNA. 2004;10:1586–1594. doi: 10.1261/rna.7122604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 14.Kennedy S, Wang D, Ruvkun G. A conserved siRNA-degrading RNase negatively regulates RNA interference in C. elegans. Nature. 2004;427:645–649. doi: 10.1038/nature02302. [DOI] [PubMed] [Google Scholar]
- 15.Kim JK, Gabel HW, Kamath RS, Tewari M, Pasquinelli A, Rual JF, Kennedy S, Dybbs M, Bertin N, Kaplan JM, Vidal M, Ruvkun G. Functional genomic analysis of RNA interference in C. elegans. Science. 2005;308:1164–1167. doi: 10.1126/science.1109267. [DOI] [PubMed] [Google Scholar]
- 16.Rual JF, Ceron J, Koreth J, Hao T, Nicot AS, Hirozane-Kishikawa T, Vandenhaute J, Orkin SH, Hill DE, van den Heuvel S, Vidal M. Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res. 2004;14:2162–2168. doi: 10.1101/gr.2505604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, Welchman DP, Zipperlen P, Ahringer J. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- 18.Koonin E, Fedorova N, Jackson J, Jacobs A, Krylov D, Makarova K, Mazumder R, Mekhedov S, Nikolskaya A, Rao B, Rogozin I, Smirnov S, Sorokin A, Sverdlov A, Vasudevan S, Wolf Y, Yin J, Natale D. A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biology. 2004;5:R7. doi: 10.1186/gb-2004-5-2-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moss EG, Lee RC, Ambros V. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell. 1997;88:637–646. doi: 10.1016/s0092-8674(00)81906-6. [DOI] [PubMed] [Google Scholar]
- 20.Polesskaya A, Cuvellier S, Naguibneva I, Duquet A, Moss EG, Harel-Bellan A. Lin-28 binds IGF-2 mRNA and participates in skeletal myogenesis by increasing translation efficiency. Genes Dev. 2007;21:1125–1138. doi: 10.1101/gad.415007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leung AK, Calabrese JM, Sharp PA. Quantitative analysis of Argonaute protein reveals microRNA-dependent localization to stress granules. Proc Natl Acad Sci U S A. 2006;103:18125–18130. doi: 10.1073/pnas.0608845103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horowitz DS, Abelson J. Stages in the second reaction of pre-mRNA splicing: the final step is ATP independent. Genes Dev. 1993;7:320–329. doi: 10.1101/gad.7.2.320. [DOI] [PubMed] [Google Scholar]
- 23.McKendrick L, Thompson E, Ferreira J, Morley SJ, Lewis JD. Interaction of eukaryotic translation initiation factor 4G with the nuclear cap-binding complex provides a link between nuclear and cytoplasmic functions of the m(7) guanosine cap. Mol Cell Biol. 2001;21:3632–3641. doi: 10.1128/MCB.21.11.3632-3641.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Izaurralde E, Lewis J, McGuigan C, Jankowska M, Darzynkiewicz E, Mattaj IW. A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell. 1994;78:657–668. doi: 10.1016/0092-8674(94)90530-4. [DOI] [PubMed] [Google Scholar]
- 25.Zhong W, Sternberg PW. Genome-wide prediction of C. elegans genetic interactions. Science. 2006;311:1481–1484. doi: 10.1126/science.1123287. [DOI] [PubMed] [Google Scholar]
- 26.Kiriakidou M, Tan GS, Lamprinaki S, De Planell-Saguer M, Nelson PT, Mourelatos Z. An mRNA m(7)G Cap Binding-like Motif within Human Ago2 Represses Translation. Cell. 2007 doi: 10.1016/j.cell.2007.05.016. [DOI] [PubMed] [Google Scholar]
- 27.Szymczyna BR, Bowman J, McCracken S, Pineda-Lucena A, Lu Y, Cox B, Lambermon M, Graveley BR, Arrowsmith CH, Blencowe BJ. Structure and function of the PWI motif: a novel nucleic acid-binding domain that facilitates pre-mRNA processing. Genes Dev. 2003;17:461–475. doi: 10.1101/gad.1060403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.D'Souza-Schorey C, Chavrier P. ARF proteins: roles in membrane traffic and beyond. Nat Rev Mol Cell Biol. 2006;7:347–358. doi: 10.1038/nrm1910. [DOI] [PubMed] [Google Scholar]
- 29.Spiliotis ET, Nelson WJ. Here come the septins: novel polymers that coordinate intracellular functions and organization. J Cell Sci. 2006;119:4–10. doi: 10.1242/jcs.02746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodriguez AJ, Seipel SA, Hamill DR, Romancino DP, Di Carlo M, Suprenant KA, Bonder EM. Seawi--a sea urchin piwi/argonaute family member is a component of MT-RNP complexes. RNA. 2005;11:646–656. doi: 10.1261/rna.7198205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blower MD, Nachury M, Heald R, Weis K. A Rae1-containing ribonucleoprotein complex is required for mitotic spindle assembly. Cell. 2005;121:223–234. doi: 10.1016/j.cell.2005.02.016. [DOI] [PubMed] [Google Scholar]
- 32.Sternberg PW. Vulval development. In: Meyer B, editor. WormBook. 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kao G, Tuck S, Baillie D, Sundaram MV. C. elegans SUR-6/PR55 cooperates with LET-92/protein phosphatase 2A and promotes Raf activity independently of inhibitory Akt phosphorylation sites. Development. 2004;131:755–765. doi: 10.1242/dev.00987. [DOI] [PubMed] [Google Scholar]
- 34.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 35.Zhang H, Christoforou A, Aravind L, Emmons SW, van den Heuvel S, Haber DA. The C. elegans Polycomb gene SOP-2 encodes an RNA binding protein. Mol Cell. 2004;14:841–847. doi: 10.1016/j.molcel.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 36.Grishok A, Sinskey JL, Sharp PA. Transcriptional silencing of a transgene by RNAi in the soma of C. elegans. Genes Dev. 2005;19:683–696. doi: 10.1101/gad.1247705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang T, Sun Y, Tian E, Deng H, Zhang Y, Luo X, Cai Q, Wang H, Chai J, Zhang H. RNA-binding proteins SOP-2 and SOR-1 form a novel PcG-like complex in C. elegans. Development. 2006;133:1023–1033. doi: 10.1242/dev.02275. [DOI] [PubMed] [Google Scholar]
- 38.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 39.Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, Hammond SM. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev. 2006;20:2202–2207. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ungar D, Oka T, Krieger M, Hughson FM. Retrograde transport on the COG railway. Trends Cell Biol. 2006;16:113–120. doi: 10.1016/j.tcb.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 41.Reeves JE, Fried M. The surf-4 gene encodes a novel 30 kDa integral membrane protein. Mol Membr Biol. 1995;12:201–208. doi: 10.3109/09687689509027508. [DOI] [PubMed] [Google Scholar]
- 42.Hamelin M, Scott IM, Way JC, Culotti JG. The mec-7 beta-tubulin gene of Caenorhabditis elegans is expressed primarily in the touch receptor neurons. Embo J. 1992;11:2885–2893. doi: 10.1002/j.1460-2075.1992.tb05357.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hayes GD, Frand AR, Ruvkun G. The mir-84 and let-7 paralogous microRNA genes of Caenorhabditis elegans direct the cessation of molting via the conserved nuclear hormone receptors NHR-23 and NHR-25. Development. 2006;133:4631–4641. doi: 10.1242/dev.02655. [DOI] [PubMed] [Google Scholar]
- 44.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 45.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 46.Timmons L, Fire A. Specific interference by ingested dsRNA. Nature. 1998;395:854–854. doi: 10.1038/27579. [DOI] [PubMed] [Google Scholar]
- 47.Reinhart BJ, Ruvkun G. Isoform-specific mutations in the Caenorhabditis elegans heterochronic gene lin-14 affect stage-specific patterning. Genetics. 2001;157:199–209. doi: 10.1093/genetics/157.1.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. let-7 enhancement screen details
A. let-7(mg279) is sensitized for defects in miRNA processing. Feeding of dsRNA targeting dcr-1 induces significant bursting in the let-7(mg279) background as compared to wild-type N2. Dcr-1 RNAi also induces bursting in eri-1(mg366) and this bursting is significantly increased in the eri-1(mg366); let-7(mg279) background (by chi square test, p value = 9.15 × 10-9). Each of these worm strains fed empty vector control showed negligible bursting.
B. Positive and negative dsRNA feeding controls. Empty vector negative controls were consistently scored 0 in the let-7 enhancement screen. dcr-1 positive controls were mostly scored as strong inducers of bursting (3 or 4), but a minority of these control clones were scored as inducing weak or no bursting. All controls were scored blindly.
C. let-7 enhancement screen summary. The whole-genome library consists of the Ahringer library supplemented with clones from the Vidal Orfeome library for a total of ~17,900 clones and was screened in the second generation. The ~2,700 sterile/lethal sub-library clones have been previously described [15] and were screened in the first generation. The primary screen was done in a single pass, the re-testing was done in triplicate. A total of 8 tests (for most clones) were used to calculate an average burst score (see Table S1) and those clones with an average of 2 or greater or that were scored as 3 or 4 on at least 2 separate occasions make up the strict positive class.
Figure S2. Bursting enhancement test with the general vulval development mutant lin-1(e1777).
RNAi clones that induced let-7(mg279)-dependent bursting were fed to the lin-1(e1777) mutant, which has a multiple vulva phenotype and bursts at a low level on its own. The percent burst after dsRNA treatment was normalized to that seen for lin-1(e1117) fed empty vector control. None of the dsRNA treatments significantly enhanced the bursting of this vulval development mutant.
Figure S3. let-7 Northern blots.
Total RNA was isolated from young adults fed RNAi. Each blot contains an empty vector-fed negative control and a dcr-1 (RNAi) positive control. Blots were re-probed with a U6 snRNA probe as a loading control. Only dcr-1 (RNAi) shows a significant increase in precursor accumulation indicating a defect in let-7 miRNA processing.
Table S1. Re-tested positives from the eri-1(mg366); let-7(mg279) whole-genome RNAi screen for bursting.
Listed are all genes identified from the primary screen whose inactivation resulted in an average burst score >0 when re-tested in triplicate. The burst score shown is a more complete average score encompassing 8 separate burst tests (for most genes). Genes classified as strict positives are indicated with a “+”. These include all those with an average score of 2 or greater and those that scored a 3 or 4 in at least 2 separate tests. Gene descriptions are from WormBase version WS170 and were acquired using WormMart.