Abstract
Background
Shiga toxins 1 and 2 (Stx1 and Stx2) are bacteriophage-encoded proteins that have been associated with hemorrhagic colitis, hemolytic uremic syndrome and other severe disease conditions. Stx1 and Stx2 are genetically and immunologically distinct but share the same compound toxin structure, method of entry and enzymatic function.
Results
Phylogenetic analysis was performed using Stx1 and Stx2 amino acid and nucleotide sequences from 41 strains of Escherichia coli, along with known stx sequences available from GenBank. The analysis confirmed the Stx1 and Stx2 divergence, and showed that there is generally more sequence variation among stx2 genes than stx1. The phylograms showed generally flat topologies among our strains' stx1 and stx2 genes. In the stx2 gene, 39.5% of the amino acid sites display very low nonsynonymous to synonymous substitution ratios.
Conclusion
The stx1 and stx2 genes used in this phylogenetic study show sequence conservation with no significant divergence with respect to place or time. These data could indicate that Shiga toxins are experiencing purifying selection.
Background
Shiga toxin was discovered in Shigella dysenteriae serotype 1 by Kiyoshi Shiga in 1898 [1]. The cytotoxic effects of Escherichia coli-produced Shiga toxin on Vero cells were first described 29 years ago [2]. A few years later, the toxin was closely associated with hemorrhagic colitis, hemolytic uremic syndrome (HUS) and other severe disease conditions [3,4]. Shiga toxin producing E. coli have been implicated in food borne, waterborne and airborne outbreaks in studies all over the world [5-7]. Much of the focus of identification and characterization of Shiga toxin has been on E. coli O157:H7 strains, even though many cases of Shiga toxin associated disease were caused by other serotypes of E. coli [8]. The toxin has also been observed in other bacterial genera, including Citrobacter, Enterobacter and Acinetobacter [9-11].
Shiga toxin 1 (Stx1) and Shiga toxin 2 (Stx2) are encoded on a lambdoid bacteriophage. Stx1 is genetically and immunologically distinct from Stx2, showing 55–60% genetic and amino acid identity [12]. Stx1 is very similar to the Shiga toxin (Stx) found in Shigella dysenteriae type 1 [13]. Several variants of Stx1 (Stx1c and Stx1d) and Stx2 (Stx2c, Stx2d, Stx2e, Stx2f, and Sxt2g) have been described [14-17]. Both Stx1 and Stx2 are compound toxins made up of one 32 kDa A subunit and five identical 7.7 kDa B subunits[18,19]. The B subunits form a pentameric hollow ring that encircles the carboxyl end of the A amino acid chain. The B subunits of Stx1, and most Stx2 type toxin molecules bind to specific glycosphingolipid globotriaosylceramide (Gb3) receptors in eukaryotic cell membranes. Stx2e B subunits preferentially bind to globotetraosylceramide (Gb4) [20], which allows the toxin to target different cell types. Once bound, receptor mediated endocytosis produces toxin-containing vesicles that travel through the Golgi apparatus and endoplasmic reticulum. The A subunit is then proteolytically cleaved into A1 (27.5 kDa) and A2 (4.5 kDa), but A1 and A2 remain covalently connected through a disulfide bond between two cysteine residues. When the cysteines are reduced, the catalytically active A1 enzyme cleaves a specific adenine from the 28S rRNA of the 60S ribosomal subunit [21]. Without this adenine, the GTP/elongation factor Tu/amino acyl-tRNA complex is unable to associate properly with the ribosome. Amino acid chain elongation is stopped, usually resulting in cell death.
Shiga toxin was shown to have the same N-glycosidase depurinating enzymatic function and active site conformation found in another ribosomal inhibiting protein (RIP), ricin [22,23]. Comparison of the crystal structures of Stx from Shigella dysenteriae [19] and Stx2 from E. coli [24] showed that the active sites were similar to that of ricin. Both ricin and Shiga toxin are considered type II RIPs because of the lectin property of the B subunit that allows the enzymatic A subunit to be internalized for access to the ribosome. Abrin, modeccin, volkensin and viscumin are examples of other RIPs that use the same method of entry and mechanism of action [25-28].
Despite their similarities, Stx1 and Stx2 produce different degrees and types of tissue damage. Enterohemorrhagic E. coli that produce Stx2 are more likely to cause hemolytic uremic syndrome than are Stx1 producers [29]. This could be due to accessibility of the active site, differences in the carboxyl end of the A subunit, or differences in binding affinities of the B subunit pentamer to Gb3 [24].
Other studies have shown how different types of pathogenic E. coli and Shigella have developed using multilocus sequence typing (MLST) of several housekeeping genes [30-32], genomic hybridization by microarray [33] and comparative genomic sequencing by microarray [34]. Since Shiga toxin is encoded on a mobilizable bacteriophage, we focused on the phylogenetic diversity of the gene rather that the bacterium that happens to carry it. In this study we compared Shiga toxin gene nucleotide and amino acid sequences from twenty nine reference strains from the National Food Safety & Toxicology Center at Michigan State University and twelve clinical isolates from the Idaho Department of Health that contained one or both Stx genes. The E. coli strains used are both temporally and spatially distinct (Table 1). Included in the analysis were unique Shiga gene sequences that were available from Genbank. We confirmed that Stx2 shows more sequence variation than Stx1 among our reference and clinical isolates. We also showed that our isolates' stx1 and stx2 genes each respectively showed little divergence and may display purifying selection.
Table 1.
E. coli reference strains used in this study. Information for the strains obtained from the National Food Safety & Toxicology Center at Michigan State University was taken directly from their web site.
| Michigan State Name | Name Used in This Study1 | Shiga Toxin Gene | GenBank Accession Number | O antigen | H antigen | Host | Location | Date |
| EDL933 | stx1 | 157 | 7 | Food | Michigan | 1982 | ||
| stx2 | ||||||||
| 93–111 | E12 | stx1 | EF441572 | 157 | 7 | Human | Washington | 1993 |
| stx2 | EF441599 | |||||||
| OK-1 | E13 | stx1 | EF441573 | 157 | 7 | Human | Japan | 1996 |
| stx2 | EF441600 | |||||||
| 86-24 | E14 | stx2 | EF441601 | 157 | 7 | Human | Washington | 1986 |
| 2886-75 | E15 | stx1 | EF441574 | 157 | 7 | Human | California | 1975 |
| 493/89 | E16 | stx2 | EF441602 | 157 | - | Human | Germany | 1989 |
| E32511 | E17 | stx2 | EF441603 | 157 | - | Human | No Data | 1985 |
| G5101 | E18 | stx1 | EF441575 | 157 | 7 | Human | Washington | 1995 |
| stx2 | EF441604 | |||||||
| 5905 | E19 | stx2 | EF441605 | 55 | 7 | Food | No data | 1994 |
| DEC8B | E22 | stx1 | EF441576 | 111 | 8 | Human | Idaho | 1986 |
| stx2 | EF441606 | |||||||
| 3007-85 | E23 | stx1 | EF441577 | 111 | - | Human | Nebraska | 1985 |
| stx2 | EF441607 | |||||||
| TB226A | E24 | stx2 | EF441608 | 111 | - | Human | Washington | 1991 |
| 928/91 | E25 | stx1 | EF441578 | 111 | - | Human | Germany | 1991 |
| stx2 | EF441609 | |||||||
| 412/55 | E26 | stx1 | EF441579 | 111 | No data | Human | Germany | 1955 |
| DEC8C | E27 | stx1 | EF441580 | 111 | - | Calf | Scotland | No data |
| C412 | E28 | stx1 | EF441581 | 111 | No data | Calf | S. Dakota | 1986 |
| H19 | E29 | stx1 | EF441582 | 26 | 11 | Human | Canada | 1977 |
| DEC10B | E210 | stx1 | EF441583 | 26 | 11 | Human | Australia | 1986 |
| TB285C | E213 | stx1 | EF441584 | 26 | - | Human | Washington | 1991 |
| BCL19 | E216 | stx1 | EF441585 | No data | - | Cow | California | No data |
| DEC10J | E217 | stx1 | EF441586 | 70 | 11 | Human | Canada | 1988 |
| ED-31 | E218 | stx1 | EF441587 | 111 | - | Human | Italy | No data |
| stx2 | EF441610 | |||||||
| CL-3 | S1 | stx2 | EF441618 | 113 | 21 | Human | Canada | 1980 |
| G5506 | S3 | stx2 | EF441619 | 104 | 21 | Human | Montana | 1994 |
| B2F1 | S4 | stx2 | EF441620 | 91 | 21 | Human | Canada | 1985 |
| TB154A | S7 | stx1 | EF441595 | 103 | 6 | Human | Washington | 1991 |
| 88–1509 | S8 | stx1 | EF441596 | 15 | 27 | Human | Canada | 1988 |
| stx2 | EF441621 | |||||||
| M2113 | S10 | stx1 | EF441597 | 156 | 21 | Cow | Germany | No data |
| BCL17 | S11 | stx1 | EF441598 | 5 | No data | Cow | California | No data |
| 90–1787 | S12 | stx2 | EF441622 | X03 | - | Cow | Canada | No data |
1These names used only in the alignments.
Results and Discussion
Similarity of Shiga Toxin 1 A and B subunit genes
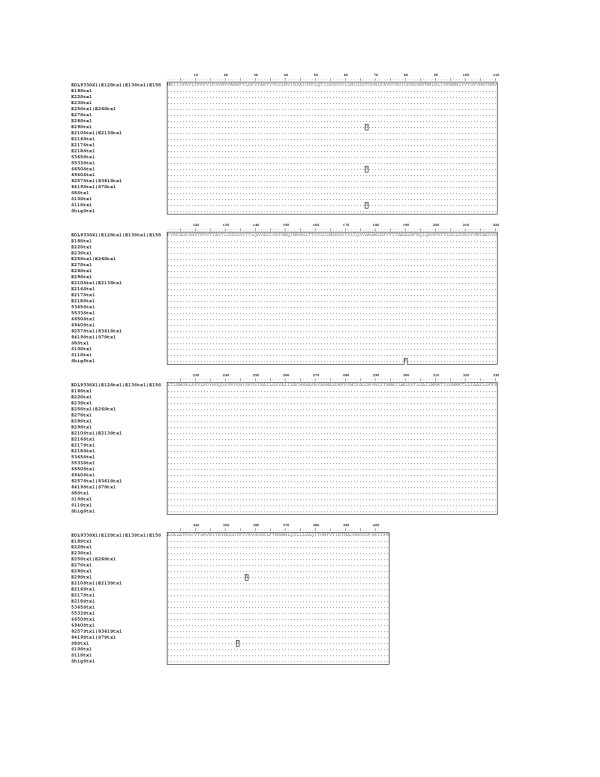
The Shiga Toxin 1 A and B subunit genes showed very little difference in amino acid sequence (Figure 1). There were only four amino acid differences among the stx1 genes in the entire alignment. The change from threonine to serine at A67 in strains H19, BCL17 and I6650 would predictably have little effect on the enzymatic activity of the A subunit. The B354 change from threonine to serine in 88–1509 and the B357 change from valine to alanine in H19 are also unlikely to change the binding function of the B subunit.
Figure 1.
Stx1 A and B Subunit Amino Acid Alignment. Sequences that are identical are on the same line and are pipe-delimited.
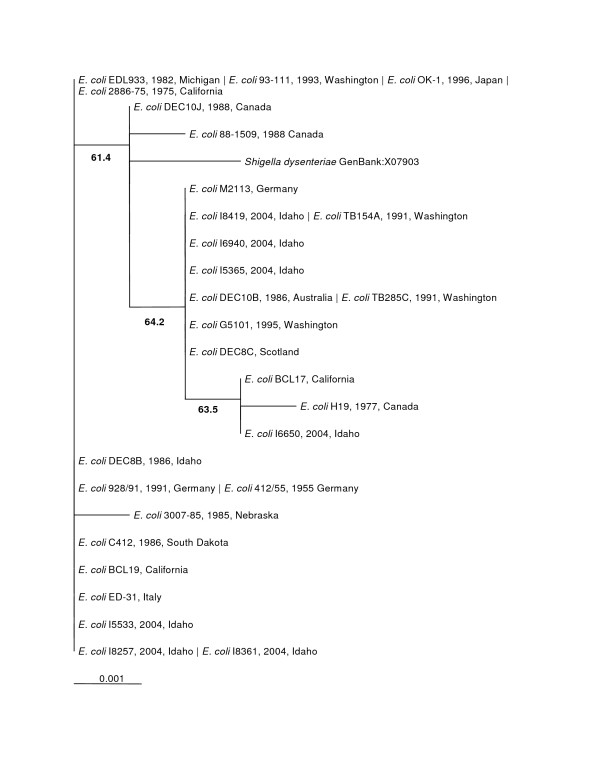
The phylogenetic trees made from the nucleotide sequences (Figure 2) and the amino acid sequences (Figure 3) were topologically the same with very short branch lengths. Bootstrap replicates in both trees supported the H19/BCL17/I6650 clade and the Shigella dysenteriae/88–1509 clade. The short branch lengths and relatively low bootstrap values can be directly attributed to the low number of informative characters in the analysis.
Figure 2.
stx1 Maximum Likelihood Nucleotide Phylogenetic Tree (unrooted). Bootstrap values over 50% are displayed. Pipe-delimited strain names indicate identical sequences. Dates and locations of the isolates are shown if available. The horizontal bar shows 0.001 nucleotide substitutions per site.
Figure 3.
Stx1 Distance Amino Acid Phylogenetic Tree (unrooted). Bootstrap values over 50% are displayed. Pipe-delimited strain names indicate identical sequences. Dates and locations of the isolates are shown if available. The horizontal bar shows 0.001 amino acid substitutions per site.
Similarity of Shiga Toxin 2 A and B subunit genes
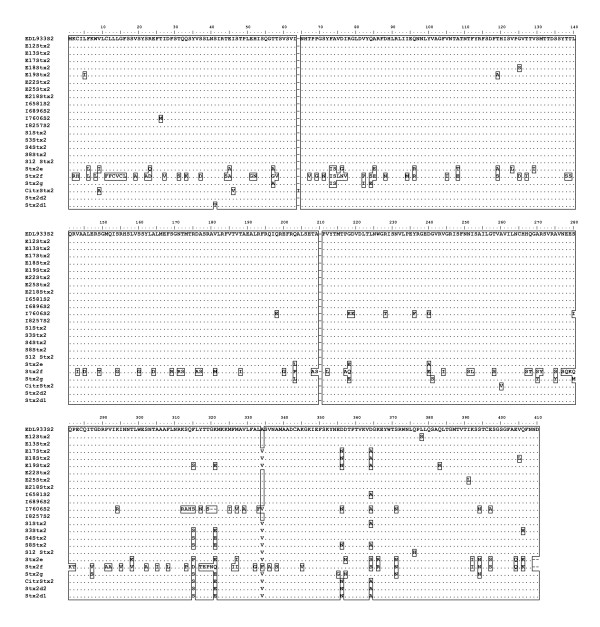
The Shiga toxin 2 A and B subunit sequences showed much more sequence diversity than did the Stx1 group. None of the putative substitutions replaced key residues used in the active site [24] or either of the cysteines that form the disulfide bridge between A1 and A2. There were 56 total amino acid differences from EDL933 Stx2 in this study's sequences, 28 in the A subunit and 28 in the B subunit. Twenty-five of these changes occurred in I7606 Stx2. The number of positions in each subunit that were affected was 17 for the A subunit and 15 for the B subunit. The Stx2 alignment (Figure 4) shows all of the amino acid differences among this study's sequences, along with Stx2d1, Stx2d2, Stx2e, Stx2f, Stx2g and Citrobacter freundii Stx2 sequences from GenBank.
Figure 4.
Stx2 A and B Subunit Amino Acid Alignment. Sequences that are identical are on the same line and are pipe-delimited.
Strains E32511 and B2F1 have each been shown to contain two copies of the stx2 gene [35,36]. E32511 carries one stx2 gene that is almost identical to the EDL933 sequence and a second that differs by 15 nucleotides. Three of these nucleotide differences result in three amino acid changes in the B subunit only. The stx2 amplified from E32511 matches the Genbank accession number M59432 with no equivocal peaks in the chromatogram. B2F1 carries the stx2d1 and stx2d2 genes represented by the accessions AF479828 and AF479829, respectively. These sequences differ from EDL933 stx2 by 10 nucleotides with one amino acid change. The sequencing reaction chromatogram from our stx2 amplicon of the B2F1 strain showed nucleotide differences between both stx2d1 and stx2d2 with no ambiguities. These differences resulted in three amino acid changes along the sequence. In both E32511 and B2F1, the few sequence differences did not affect our analysis. However, until further analysis of our E32511 and B2F1 strains is complete to determine whether they contain one or two stx2 genes, our sequences EF441603 and EF441620 should be held in question.
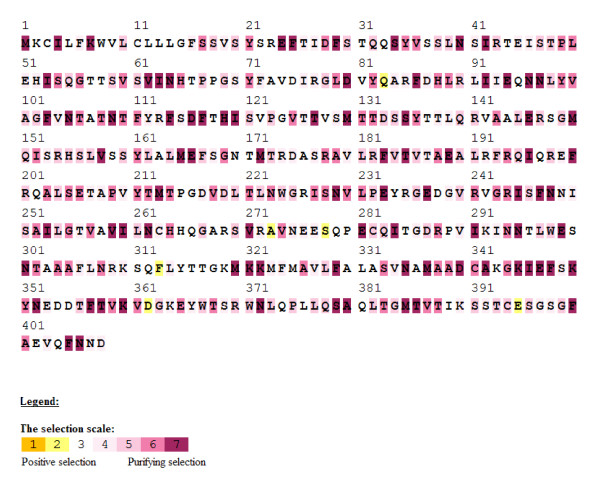
The ratio non-synonymous (dN) to synonymous (dS) substitutions was used to estimate whether positive or purifying selection had occurred at each amino acid site. A nonsynonymous:synonymous (dN:dS) ratio of greater than one indicates positive selection at that amino acid site whereas a dN:dS ratio of less than one shows purifying selection. Forty percent of the amino acid sites in the full alignment showed purifying selection using the Selecton program [37]. The Stx2 A and B subunits showed 37.3% and 44.9%, respectively, of their amino acid sites were well conserved. (Figure 5)
Figure 5.
Color Coded Selecton Results for stx2 Sequences. Nonsynonymous:synonymous (dN:dS) ratios using the output from the Selecton program. Shades of yellow (1 and 2) indicate a dN:dS ratio of > 1 or positive selection. Any shade of bordeaux (4–7) indicates a dN:dS ratio of < 1 or purifying selection.
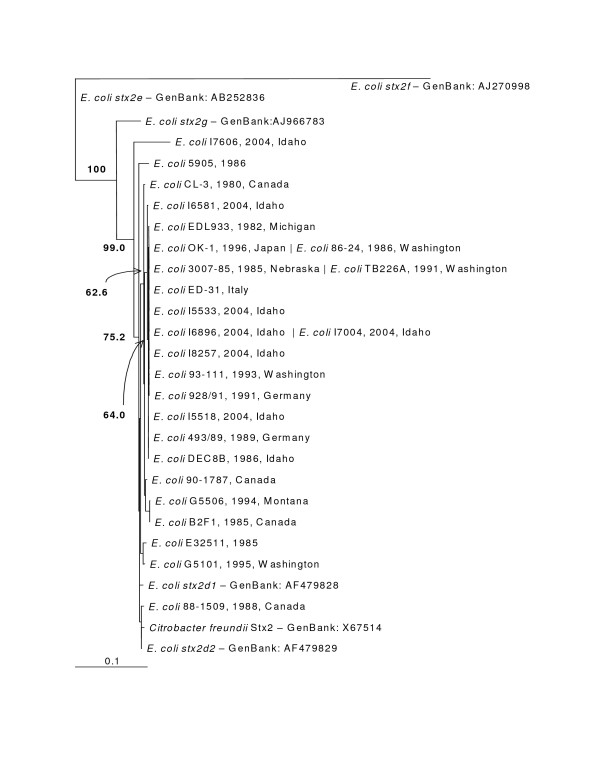
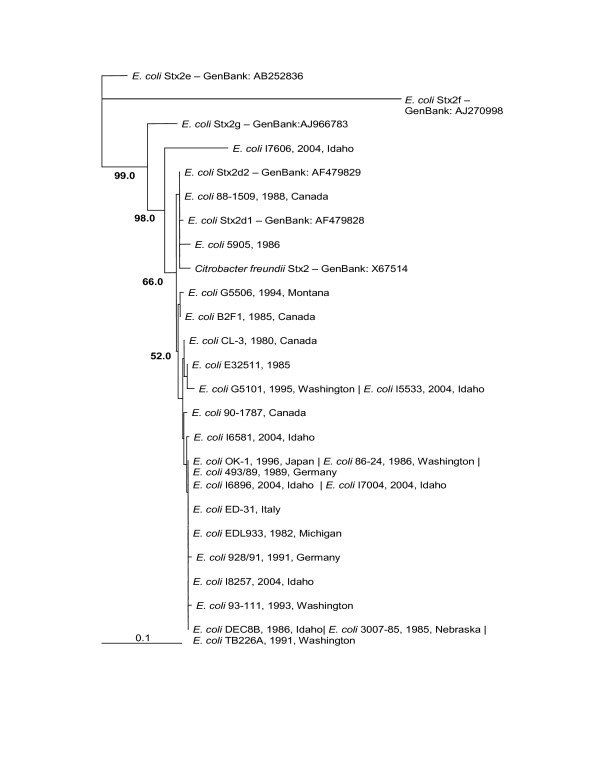
The nucleotide phylogenetic tree (Figure 6) and the amino acid phylogenetic tree (Figure 7) showed almost the same topology with short branch lengths for our strains. The obvious dissimilarity of Stx2e, Stx2f and Stx2g is striking in both the alignment and phylogenetic trees and is well supported by the high bootstrap values. Idaho strain I7606 had the most divergent sequence among the strains obtained from the Idaho State Health Department and is most closely related to Stx2d A subunit [38]. The lower bootstrap values in the figures are characteristic of highly similar sequences in the branches. Interestingly, the C. freundii Stx2 grouped with the Stx2d group of elastase activated Shiga toxins [39].
Figure 6.
stx2 Maximum Likelihood Nucleotide Phylogenetic Tree (unrooted). Bootstrap values over 50% displayed. Pipe-delimited strain names indicate identical sequences. Dates and locations of the isolates are shown if available. The horizontal bar shows 0.1 nucleotide substitutions per site.
Figure 7.
Stx2 Distance Amino Acid Phylogenetic Tree (unrooted). Bootstrap values over 50% are displayed. Pipe-delimited strain names indicate identical sequences. Dates and locations of the isolates are shown if available. The horizontal bar shows 0.1 amino acid substitutions per site.
Conclusion
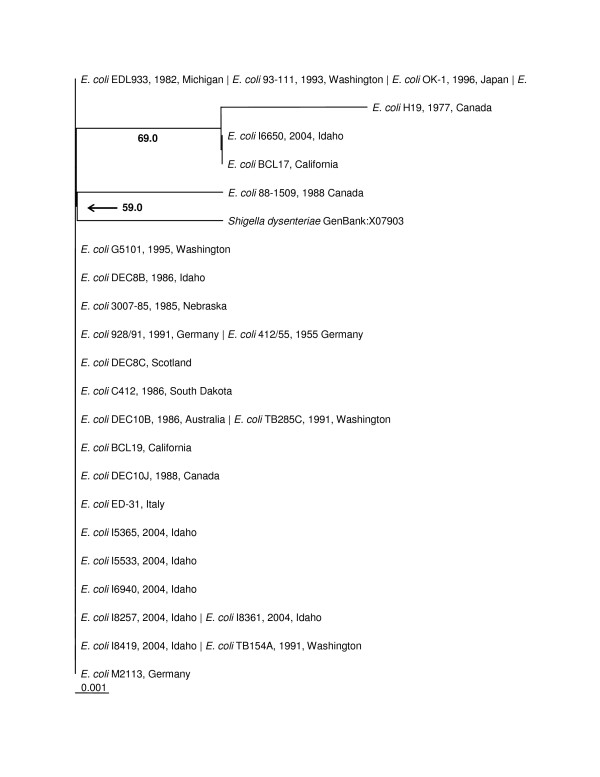
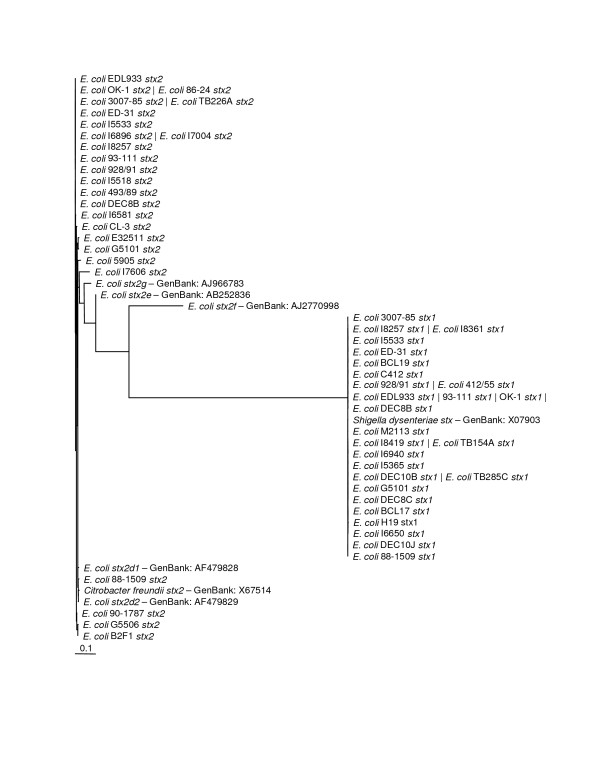
This investigation into the phylogenetic relationships among Shiga toxins confirmed that the two major groups, Stx1 and Stx2, are easily differentiated by both their amino acid and nucleotide sequences. The phylogram in Figure 8 shows stx1 and stx2 forming two distinct clades with stx2f branching off significantly from both. The stx1 group displayed an almost flat phylogeny whereas the stx2 group has much more sequence diversity. Stx2d, Stx2e and Stx2g all show at least 91% similarity to the EDL933 stx2 gene. The Stx2f amino acid and nucleotide sequences are only 72.5 and 70.4 percent similar to EDL933 Stx2 sequences, respectively, making it the most divergent Stx2 found to date. The greater diversity of Stx2 sequences could be the result of sampling bias. Since the majority of the gene sequences came from human outbreak pathogens and because Stx2 is associated with more severe disease, this could have an effect on what strains are recovered. Despite the fact that diarrheal illness is a leading causes of death worldwide [40], most sources providing isolates used in this study are located in areas with well established public health systems. This source location could also introduce bias into the isolates investigated.
Figure 8.
Unrooted Maximum Likelihood Nucleotide Phylogenetic Tree of all Shiga Toxin Sequences. The horizontal bar shows 0.1 nucleotide substitutions per site.
The significance of compiling these sequences became clear when both the Stx1 tree and Stx2 tree showed generally flat topologies. Our further analysis of the nucleotide sequences for Stx2 A and B subunits showed that a large proportion of the amino acid sites had undergone purifying selection. Our hypothesis when undertaking this project was that Shiga toxin sequences from different places and collected at different times would show divergence. Our data analysis indicates that there is no significant divergence among either the Stx1 or Stx2 gene sequences based on collection time or location. The Stx1 phylogeny (Figure 2 and 3) shows that these genes do not vary greatly and are perhaps undergoing purifying selective pressure. The Stx2 phylogeny (Figure 4 and 5) is also generally flat among the strains we tested. This would indicate that among our strains, both the Stx1 and Stx2 gene sequences are closely conserved. One reason for the conservation of this sequence could be that intestinal Shiga toxin producing E. coli in the bovine host are mitigating Bovine Leukemia Virus induced disease [41]. Another recent finding suggests that the Shiga toxin might play a role in helping E. coli survive predation from bacteriovores outside of the cow [42]. Since many of our strains can be related to illness produced from bovine feces, it is possible that a Shiga toxin gene that is beneficial to the bacterium that carries it has undergone purifying selection to the benefit of both their bacterial and mammalian host.
Methods
Culture and Colony Isolation
Forty-one strains of Escherichia coli were used, including 29 outbreak reference strains provided by the National Food Safety & Toxicology Center at Michigan State University, and twelve clinical isolates from patients reported to the Idaho Department of Health. The characteristics and sources of E. coli strains used are described in Table 1. The specimens were received as 25% glycerol stocks on wet ice. E. coli strains were grown at 37°C in Luria-Bertani (LB) broth and isolated on LB agar.
DNA Extraction
After isolation on LB agar, each strain was grown in 11 ml of LB broth for 18–24 hours at 37°C with shaking. All DNA extractions were performed using the PureGene DNA Isolation Kit (Gentra Systems, Minneapolis, MN). To maximize the yield of DNA, the following modifications to the PureGene protocol were made: 1) The time for cell lysis was increased to 10 minutes; 2) The DNA was precipitated using 300 μl 100% Isopropanol (tubes were inverted 50 times, incubated at -20°C for 1 hour and then centrifuged for 5 minutes at 13–16,000 × g); 3) DNA was dissolved in 100 μl sterile filtered 10 mM Tris, pH 8.3.
Amplification of Stx genes
Primer sequences targeting specific stx1 and stx2 genes are listed in Table 2, along with PCR conditions used. The stx1 primers amplified 1178 bp of the 1227 bp sequence. The stx2 primers amplified the entire 1241 bp sequence. PCR master mix was made to final concentration of 1 × ThermoPol Reaction Buffer (New England BioLabs, Beverly, MA), 500 μM dNTP's (New England BioLabs), 0.02 units/μl Vent exo(-) polymerase (New England BioLabs), and 1 μM of each of the stx1 primers or stx2 primers. Five microliters of each amplification reaction was analyzed by 1% agarose gel electrophoresis. All gels were run at 80–90 V for 90 minutes in 1 × TAE buffer. Both gel and buffer contained 0.5 mg/ml ethidium bromide.
Table 2.
Idaho E. coli strains used in this study. Information on the Idaho strains was obtained from the Idaho Department of Health.
| Strain Name | Shiga Toxin Gene | GenBank Accession Number | O antigen | H antigen | Host | Location | Date |
| I5365 | stx1 | EF441588 | 26 | 11 | Human | Idaho | 2004 |
| I5518 | stx2 | EF441611 | 121 | 19 | Human | Idaho | 2004 |
| I5533 | stx1 | EF441589 | 157 | - | Human | Idaho | 2004 |
| stx2 | EF441612 | ||||||
| I6581 | stx2 | EF441613 | 157 | 7 | Human | Idaho | 2004 |
| I6650 | stx1 | EF441590 | No data | No data | Human | Idaho | 2004 |
| I6896 | stx2 | EF441614 | 145 | - | Human | Idaho | 2004 |
| I6940 | stx1 | EF441591 | 145 | - | Human | Idaho | 2004 |
| I7004 | stx2 | EF441615 | 157 | 7 | Human | Idaho | 2004 |
| I7606 | stx2 | EF441616 | 146 | 21 | Human | Idaho | 2004 |
| I8257 | stx1 | EF441592 | 157 | 7 | Human | Idaho | 2004 |
| stx2 | EF441617 | ||||||
| I8361 | stx1 | EF441593 | 111 | - | Human | Idaho | 2004 |
| I8419 | stx1 | EF441594 | 103 | 25 | Human | Idaho | 2004 |
Purification and Sequencing of Amplification Products
Amplicons were extracted from agarose gels using the QIAquick Gel Extraction Kit (Qiagen, Valencia, CA) according to the manufacturer's specifications. The amplicons were further purified for sequencing using Millipore Montage PCR filters (Billerica, MA). All sequencing was performed by the Idaho State University Molecular Research Core Facility using primers listed in Table 2. Accession numbers for nucleotide sequences submitted to GenBank are shown in Table 3.
Table 3.
Primer sequences
| Primer Name | Nucleotide Sequence 5'-3' | Target | PCR Conditionsa | ||
| PCR Primers | Denature | Anneal | Extension | ||
| BGR1U | TCAACGAAAAATAACTTCGCTGAATCCC | Stx1 Base 1–28 | 95°C, | 61°C, | 72°C, |
| BGR1D | CAGTTAATGTGGTTGCGAAGGAATTTACC | Stx1 Base (1150–1178) | 60 sec | 60 sec | 240 sec |
| BGR2U | ATGAAGTGTATATTATTTAAATGGGTACTGTG | Stx2 Base 1–32 | 95°C, | 61°C, | 72°C, |
| BGR2D | TCAGTCATTATTAAACTGCACTTCAG | Stx2 Base (1201–1226) | 60 sec | 60 sec | 240 sec |
| Sequencing Primers | Target | ||||
| Stx1SeqF | TGTAACCGCTGTTGTACCTGG | Stx1 Base (367–387) | |||
| Stx1SeqR | TTAATACTGAATTGTCATCATCATGC | Stx1 Base 778–803 | |||
| Stx2SeqF | TTGCATTAGCTTCTGTTAATGCA | Stx2 Base 1000–1022 | |||
| Stx2SeqR | CATTCCGGAACGTTCCAG | Stx2 Base (433–450) | |||
a PCR was performed for 30 cycles followed by a final extension step of 10 min at 72°C.
Sequence Alignment and Phylogenetic Analysis
All amplicons were sequenced on both strands except the first and last 30 bases that were sequenced in only one direction in duplicate from internal primers. Raw sequence alignment was performed using Vector NTI 9 (InforMax, Invitrogen, Carlsbad, CA). Reference sequences for Shiga toxins used in the analysis were obtained from GenBank and are indicated in the phylogenetic tree by their accession numbers. Consensus sequences were entered into BioEdit 7.0.5.2 [43], aligned using ClustalW and then manually trimmed so that the nucleotide sequence accurately reflected the amino acid sequence. Amino acid sequence phylogenetic trees were created by distance analysis of the alignment using PAUP 4.0 Beta 10 [44]. Nucleotide phylogenetic trees were generated by maximum likelihood analysis of the alignment using PAUP 4.0 Beta 10 [44]. The Hasegawa-Kishino-Yano or HKY (stx1) and HKY with gamma likelihood models (stx2) were chosen after the data were analyzed using ModelTest [45] in PAUP. Construction of the nucleotide trees was done using the neighbor joining method with 1000 bootstrap replicates. The Selecton program [37] was used to determine the ratio of non-synonymous to synonymous substitutions. The top two purifying selection categories of 6 and 7 on the selection scale were used to calculate percentage of amino acids that were undergoing purifying selection. The maximum likelihood nucleotide phylogenetic tree produced above was inputted and the M8 evolutionary model [46] was used with EDL933 stx2 as the query sequence. Other evolutionary models available (M8a, M7, M5, MEC) produced results that were not significantly different. (data not shown) The full table of non-synonymous to synonymous ratios for stx2 sequences is available in additional file 1.
Authors' contributions
JEL completed many of the amplifications, performed the alignments and phylogenetic analyses and drafted the manuscript. JR performed the majority of the amplifications, prepared the amplicons for sequencing, edited sequences and wrote portions of the methods. KMS and LDF participated in conceptual design of the study and helped to draft portions of the manuscript. MSS participated in design and coordination of the project and helped to revise portions of the manuscript. PPS participated in the study design, provided critical assistance with alignments and phylogenetic analyses, and helped draft portions of the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Selecton Output. Selecton output file showing non-synonymous to synonymous ratio and confidence intervals of the stx2 sequences.
Acknowledgments
Acknowledgements
This work was partially supported by grant number F04-107 from the Graduate Student Research and Scholarship Committee of Idaho State University, Pocatello, ID. We wish to thank the Idaho State University Molecular Core Research Facility for their fast service and high quality sequence. We would also like to acknowledge Vivian Lockary of the Idaho Department of Health for providing strains. Acknowledgement is also made to the National Food Safety & Toxicology Center at Michigan State University for providing strains.
Contributor Information
James E Lee, Email: james.e.lee@amedd.army.mil.
Junelina Reed, Email: jreed96@hotmail.com.
Malcolm S Shields, Email: shiemalc@isu.edu.
Kathleen M Spiegel, Email: spiekath@isu.edu.
Larry D Farrell, Email: farrlarr@isu.edu.
Peter P Sheridan, Email: sherpete@isu.edu.
References
- Shiga K. Zentralblatt fur Bakteriologie. 1898. pp. 913–918.
- Konowalchuk J, Speirs JI, Stavric S. Vero response to a cytotoxin of Escherichia coli. Infect Immun. 1977;18:775–779. doi: 10.1128/iai.18.3.775-779.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade WG, Thom BT, Evans N. Cytotoxic enteropathogenic Escherichia coli. Lancet. 1979;2:1235–1236. doi: 10.1016/S0140-6736(79)92349-3. [DOI] [PubMed] [Google Scholar]
- Wilson MW, Bettelheim KA. Cytotoxic Escherichia coli serotypes. Lancet. 1980;1:201. doi: 10.1016/S0140-6736(80)90682-0. [DOI] [PubMed] [Google Scholar]
- Bruce MG, Curtis MB, Payne MM, Gautom RK, Thompson EC, Bennett AL, Kobayashi JM. Lake-associated outbreak of Escherichia coli O157:H7 in Clark County, Washington, August 1999. Arch Pediatr Adolesc Med. 2003;157:1016–1021. doi: 10.1001/archpedi.157.10.1016. [DOI] [PubMed] [Google Scholar]
- Howie H, Mukerjee A, Cowden J, Leith J, Reid T. Investigation of an outbreak of Escherichia coli O157 infection caused by environmental exposure at a scout camp. Epidemiol Infect. 2003;131:1063–1069. doi: 10.1017/S0950268803001250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahali S, Sarkar B, Chakraborty S, Macaden R, Deokule JS, Ballal M, Nandy RK, Bhattacharya SK, Takeda Y, Ramamurthy T. Molecular epidemiology of diarrhoeagenic Escherichia coli associated with sporadic cases and outbreaks of diarrhoea between 2000 and 2001 in India. Eur J Epidemiol. 2004;19:473–479. doi: 10.1023/B:EJEP.0000027353.70796.25. [DOI] [PubMed] [Google Scholar]
- Bettelheim KA. Role of non-O157 VTEC. Symp Ser Soc Appl Microbiol. 2000:38S–50S. doi: 10.1111/j.1365-2672.2000.tb05331.x. [DOI] [PubMed] [Google Scholar]
- Schmidt H, Montag M, Bockemuhl J, Heesemann J, Karch H. Shiga-like toxin II-related cytotoxins in Citrobacter freundii strains from humans and beef samples. Infect Immun. 1993;61:534–543. doi: 10.1128/iai.61.2.534-543.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paton AW, Paton JC. Enterobacter cloacae producing a Shiga-like toxin II-related cytotoxin associated with a case of hemolytic-uremic syndrome. J Clin Microbiol. 1996;34:463–465. doi: 10.1128/jcm.34.2.463-465.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotiuz G, Sirok A, Gadea P, Varela G, Schelotto F. Shiga Toxin 2-Producing Acinetobacter haemolyticus Associated with a Case of Bloody Diarrhea 10.1128/JCM.00407-06. J Clin Microbiol. 2006;44:3838–3841. doi: 10.1128/JCM.00407-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein DL, Jackson MP, Samuel JE, Holmes RK, O'Brien AD. Cloning and sequencing of a Shiga-like toxin type II variant from Escherichia coli strain responsible for edema disease of swine. J Bacteriol. 1988;170:4223–4230. doi: 10.1128/jb.170.9.4223-4230.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson MP, Newland JW, Holmes RK, O'Brien AD. Nucleotide sequence analysis of the structural genes for Shiga-like toxin I encoded by bacteriophage 933J from Escherichia coli. Microb Pathog. 1987;2:147–153. doi: 10.1016/0882-4010(87)90106-9. [DOI] [PubMed] [Google Scholar]
- Burk C, Dietrich R, Acar G, Moravek M, Bulte M, Martlbauer E. Identification and characterization of a new variant of Shiga toxin 1 in Escherichia coli ONT:H19 of bovine origin. J Clin Microbiol. 2003;41:2106–2112. doi: 10.1128/JCM.41.5.2106-2112.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich AW, Bielaszewska M, Zhang WL, Pulz M, Kuczius T, Ammon A, Karch H. Escherichia coli harboring Shiga toxin 2 gene variants: frequency and association with clinical symptoms. J Infect Dis. 2002;185:74–84. doi: 10.1086/338115. [DOI] [PubMed] [Google Scholar]
- Leung PHM, Peiris JSM, Ng WWS, Robins-Browne RM, Bettelheim KA, Yam WC. A Newly Discovered Verotoxin Variant, VT2g, Produced by Bovine Verocytotoxigenic Escherichia coli. Appl Environ Microbiol. 2003;69:7549–7553. doi: 10.1128/AEM.69.12.7549-7553.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt H. Shiga-toxin-converting bacteriophages. Res Microbiol. 2001;152:687–695. doi: 10.1016/S0923-2508(01)01249-9. [DOI] [PubMed] [Google Scholar]
- Stein PE, Boodhoo A, Tyrrell GJ, Brunton JL, Read RJ. Crystal structure of the cell-binding B oligomer of verotoxin-1 from E. coli. Nature. 1992;355:748–750. doi: 10.1038/355748a0. [DOI] [PubMed] [Google Scholar]
- Fraser ME, Chernaia MM, Kozlov YV, James MN. Crystal structure of the holotoxin from Shigella dysenteriae at 2.5 A resolution. Nat Struct Biol. 1994;1:59–64. doi: 10.1038/nsb0194-59. [DOI] [PubMed] [Google Scholar]
- DeGrandis S, Law H, Brunton J, Gyles C, Lingwood CA. Globotetraosylceramide is recognized by the pig edema disease toxin. J Biol Chem. 1989;264:12520–12525. [PubMed] [Google Scholar]
- Endo Y, Tsurugi K, Yutsudo T, Takeda Y, Ogasawara T, Igarashi K. Site of action of a Vero toxin (VT2) from Escherichia coli O157:H7 and of Shiga toxin on eukaryotic ribosomes. RNA N-glycosidase activity of the toxins. Eur J Biochem. 1988;171:45–50. doi: 10.1111/j.1432-1033.1988.tb13756.x. [DOI] [PubMed] [Google Scholar]
- Saxena SK, O'Brien AD, Ackerman EJ. Shiga toxin, Shiga-like toxin II variant, and ricin are all single-site RNA N-glycosidases of 28 S RNA when microinjected into Xenopus oocytes. J Biol Chem. 1989;264:596–601. [PubMed] [Google Scholar]
- Endo Y, Mitsui K, Motizuki M, Tsurugi K. The mechanism of action of ricin and related toxic lectins on eukaryotic ribosomes. The site and the characteristics of the modification in 28 S ribosomal RNA caused by the toxins. J Biol Chem. 1987;262:5908–5912. [PubMed] [Google Scholar]
- Fraser ME, Fujinaga M, Cherney MM, Melton-Celsa AR, Twiddy EM, O'Brien AD, James MN. Structure of shiga toxin type 2 (Stx2) from Escherichia coli O157:H7. J Biol Chem. 2004;279:27511–27517. doi: 10.1074/jbc.M401939200. [DOI] [PubMed] [Google Scholar]
- Barbieri L, Ciani M, Girbes T, Liu W, Van Damme EJM, Peumans WJ, Stirpe F. Enzymatic activity of toxic and non-toxic type 2 ribosome-inactivating proteins. FEBS Letters. 2004;563:219–222. doi: 10.1016/S0014-5793(04)00286-8. [DOI] [PubMed] [Google Scholar]
- Olsnes S, Stirpe F, Sandvig K, Pihl A. Isolation and characterization of viscumin, a toxic lectin from Viscum album L. (mistletoe) J Biol Chem. 1982;257:13263–13270. [PubMed] [Google Scholar]
- Chambery A, Di Maro A, Monti MM, Stirpe F, Parente A. Volkensin from Adenia volkensii Harms (kilyambiti plant), a type 2 ribosome-inactivating protein: Gene cloning, expression and characterization of its A-chain. Eur J Biochem. 2004;271:108–117. doi: 10.1046/j.1432-1033.2003.03909.x. [DOI] [PubMed] [Google Scholar]
- Endo Y, Tsurugi K. Mechanism of action of ricin and related toxic lectins on eukaryotic ribosomes. Nucleic Acids Symp Ser. 1986:187–190. [PubMed] [Google Scholar]
- Siegler RL, Obrig TG, Pysher TJ, Tesh VL, Denkers ND, Taylor FB. Response to Shiga toxin 1 and 2 in a baboon model of hemolytic uremic syndrome. Pediatr Nephrol. 2003;18:92–96. doi: 10.1007/s00467-002-1035-7. [DOI] [PubMed] [Google Scholar]
- Whittam TS. Escherichia coli and Salmonella: Cellular and Molecular Biology. Vol. 1. Washington D.C., ASM Press; 1996. Genetic variation and evolutionary processes in natural populations of Escherichia coli; pp. 2708–2720. [Google Scholar]
- Lacher DW, Steinsland H, Blank TE, Donnenberg MS, Whittam TS. Molecular evolution of typical enteropathogenic Escherichia coli: clonal analysis by multilocus sequence typing and virulence gene allelic profiling. J Bacteriol. 2007;189:342–350. doi: 10.1128/JB.01472-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pupo GM, Lan R, Reeves PR. Multiple independent origins of Shigella clones of Escherichia coli and convergent evolution of many of their characteristics. Proc Natl Acad Sci U S A. 2000;97:10567–10572. doi: 10.1073/pnas.180094797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick LM, Qi W, Lacher DW, Whittam TS. Evolution of genomic content in the stepwise emergence of Escherichia coli O157:H7. J Bacteriol. 2005;187:1783–1791. doi: 10.1128/JB.187.5.1783-1791.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Qi W, Albert TJ, Motiwala AS, Alland D, Hyytia-Trees EK, Ribot EM, Fields PI, Whittam TS, Swaminathan B. Probing genomic diversity and evolution of Escherichia coli O157 by single nucleotide polymorphisms. Genome Res. 2006;16:757–767. doi: 10.1101/gr.4759706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt CK, McKee ML, O'Brien AD. Two copies of Shiga-like toxin II-related genes common in enterohemorrhagic Escherichia coli strains are responsible for the antigenic heterogeneity of the O157:H- strain E32511. Infect Immun. 1991;59:1065–1073. doi: 10.1128/iai.59.3.1065-1073.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teel LD, Melton-Celsa AR, Schmitt CK, O'Brien AD. One of two copies of the gene for the activatable shiga toxin type 2d in Escherichia coli O91:H21 strain B2F1 is associated with an inducible bacteriophage. Infect Immun. 2002;70:4282–4291. doi: 10.1128/IAI.70.8.4282-4291.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doron-Faigenboim A, Stern A, Mayrose I, Bacharach E, Pupko T. Selecton: a server for detecting evolutionary forces at a single amino-acid site 10.1093/bioinformatics/bti259. Bioinformatics. 2005;21:2101–2103. doi: 10.1093/bioinformatics/bti259. [DOI] [PubMed] [Google Scholar]
- Zhang W, Bielaszewska M, Friedrich AW, Kuczius T, Karch H. Transcriptional Analysis of Genes Encoding Shiga Toxin 2 and Its Variants in Escherichia coli. Appl Environ Microbiol. 2005;71:558–561. doi: 10.1128/AEM.71.1.558-561.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton-Celsa AR, Darnell SC, O'Brien AD. Activation of Shiga-like toxins by mouse and human intestinal mucus correlates with virulence of enterohemorrhagic Escherichia coli O91:H21 isolates in orally infected, streptomycin-treated mice. Infect Immun. 1996;64:1569–1576. doi: 10.1128/iai.64.5.1569-1576.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray CJL, Lopez AD. Mortality by cause for eight regions of the world: Global Burden of Disease Study. The Lancet. 1997;349:1269–1276. doi: 10.1016/S0140-6736(96)07493-4. [DOI] [PubMed] [Google Scholar]
- Ferens WA, Cobbold R, Hovde CJ. Intestinal Shiga toxin-producing Escherichia coli bacteria mitigate bovine leukemia virus infection in experimentally infected sheep. Infect Immun. 2006;74:2906–2916. doi: 10.1128/IAI.74.5.2906-2916.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg KM, Levin BR. Grazing protozoa and the evolution of the Escherichia coli O157:H7 Shiga toxin-encoding prophage. Proc Biol Sci. 2007;274:1921–1929. doi: 10.1098/rspb.2007.0245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Swofford DL. Phylogenetic Analysis Using Parsimony (*and Other Methods). 4.0b10. Sunderland, Massachusetts, Sinauer Associates; 2000. [Google Scholar]
- Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution 10.1093/bioinformatics/14.9.817. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R, Goldman N, Pedersen AM. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics. 2000;155:431–449. doi: 10.1093/genetics/155.1.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Selecton Output. Selecton output file showing non-synonymous to synonymous ratio and confidence intervals of the stx2 sequences.