Abstract
Base substitutions, deletions, and duplications are observed at the immunoglobulin locus in DNA sequences involved in class switch recombination (CSR). These mutations are dependent upon activation-induced cytidine deaminase (AID) and present all the characteristics of the ones observed during V gene somatic hypermutation, implying that they could be generated by the same mutational complex. It has been proposed, based on the V gene mutation pattern of patients with the cancer-prone xeroderma pigmentosum variant (XP-V) syndrome who are deficient in DNA polymerase η (pol η), that this enzyme could be responsible for a large part of the mutations occurring on A/T bases. Here we show, by analyzing switched memory B cells from two XP-V patients, that pol η is also an A/T mutator during CSR, in both the switch region of tandem repeats as well as upstream of it, thus suggesting that the same error-prone translesional polymerases are involved, together with AID, in both processes.
Keywords: somatic mutation, pol η, translesional DNA polymerases, xeroderma pigmentosum variant syndrome
Introduction
Since the discovery of activation-induced cytidine deaminase (AID) and of its central role in the initiation of the three postrearrangement B cell diversification processes, somatic hypermutation (SHM), class switch recombination (CSR), and gene conversion, a major question is to unravel what are the common and the specific partners involved in these different pathways (1, 2). SHM implies the introduction mainly of base substitutions, but also of deletions and duplications into V genes to generate an antigen receptor with better affinity for the immunizing antigen. Translesional DNA polymerases, which are able to bypass specific DNA lesions in an error-free mode at the replication fork but are highly error-prone when copying undamaged DNA, have been suspected to be involved in an error-prone short patch DNA synthesis taking place during SHM (3). It has since been reported, based on a study of SHM in patients with xeroderma pigmentosum variant (XP-V), a cancer-prone disease corresponding to a deficiency in DNA polymerase η (pol η; 4), that this enzyme might be responsible for a large part of the mutations occurring at A/T basepairs (5).
In CSR, the heavy chain μ constant region is switched to downstream constant region genes that are responsible for various effector functions once the antibody has bound its cognate antigen. The heavy chain μ locus region that is targeted for CSR, encompasses a large DNA region starting downstream of the Iμ exon up to a region of repetitive pentamer motifs constituting the core μ switch region (Sμ; 6). Base substitutions, deletions, and duplications have been observed around switch junctions (7). More recently, mutations that display many characteristics of SHM, that is, base substitutions on RGYW hotspot motifs (with R for purine, Y for pyrimidines, and W for A or T; 8), bias for transitions and AID dependence have been described downstream of the Iμ exon, outside the switch core region (9, 10). Here we show, by studying hypermutation at the heavy chain locus in memory B cells of two XP-V patients, that pol η is involved in hypermutation taking place during SHM and CSR as well, implying that the same mutational complex is probably recruited during these two DNA transactions.
Materials and Methods
Characteristics of XP-V Patients and Preparation of Cell Samples.
Blood samples were obtained from four healthy donors (34, 37, 41, and 50 yr old) and two French XP-V patients (55 and 56 yr old) after informed consent. The two XP-V patients studied were diagnosed clinically and biologically. Patient 1 showed numerous skin tumors since the age of 26. Patient 2 had her first malignant melanoma at 30 and numerous melanomas, basal cell carcinomas, and squamous cell carcinomas since then. Their cultured fibroblasts exhibit normal UV-C–induced unscheduled DNA synthesis and are UV-C sensitive in the presence of caffeine, a hallmark of XP-V cells (11). Heterozygous mutations of the POL H gene were found for both of them: a stop codon at amino acid 303 and a 4-bp deletion at amino acid 408 for patient 1, and a Gly to Arg amino acid substitution at position 295 and a 2-bp deletion (1,727–1,728) at amino acid 576 for patient 2. CD19+ CD27+ IgD− memory B cells were purified by cell sorting as previously described (12) and DNA was extracted using proteinase K digestion.
PCR and Sequence Analysis.
The JH4 intronic sequence was amplified using an FR3 consensus primer CACGGCYGTGTATTACTGTGC and a primer upstream from JH5, AGGACCCCAGGCAAGAAC (at 94°C for 45 s, 55°C for 1 min, and 72°C for 2 min for 40 cycles with Pfu polymerase; Stratagene). The sequence upstream from the Sμ core was amplified using the following primers: 5′-Sμ, GAATGATTCCATGCCAAAGC and 3′-Sμ, AGCTGGATGGAGTTGTCATGGC (at 94°C for 45 s, 56°C for 1 min, and 72°C for 3 min for 35 cycles with Pfu polymerase), and the Sμ–α switch region (Sα) switch junctions with Sμ-ext, GGGGACCTGCTCATTTTTATC and Sα-ext, CCCTCAGAACCCCTAAGAAC (at 94°C for 45 s, 60°C for 1 min, and 72°C for 3 min for 40 cycles using Pfu Turbo polymerase). In this last case, multiple PCRs were performed on aliquots of 2,000–5,000 cells, and PCR products below 1 kb were selected for cloning. PCR products were cloned using the TOPO TA cloning kit (Invitrogen) and sequences were determined on an ABI prism 3100 Analyzer (Applera). All amplifications were performed using Pfu polymerase with a number of cycles to keep background mutations at a minimal level. Mutations induced during amplification by polymerases devoid of proofreading show an A/T over G/C bias that may confuse analysis, notably in the case of switch junction mutations. Error rate of Pfu polymerase is in our assay around 5 × 10−5 for 40 cycles of amplification.
Results and Discussion
DNA pol η has been proposed as an A/T mutator of Ig V genes (5). This was based on the analysis of the mutation pattern of functional VH6 genes from three XP-V patients, with characterized mutations of pol η for two of them. However, the question of the exact contribution of pol η to hypermutation has been challenged by Dorner and Lipsky (13). By studying one of these three patients by a single cell PCR approach, designed at amplifying both functional and nonfunctional VH genes, they failed to observed such a deficiency at A/T targeting and concluded that it might be a bias introduced by the study of selected V gene sequences (14).
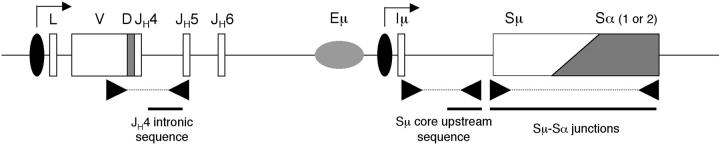
To readdress this question, CD27+ IgD− memory B cells from two XP-V patients and from four normal individuals were purified by cell sorting and mutations were analyzed in three noncoding regions of the heavy chain locus: the JH4 intron, the Sμ core upstream region, and Sμ–Sα switch junctions (Fig. 1) .
Figure 1.
Schematic representation of the regions selected for mutation analysis at the Ig heavy chain locus. The configuration selected represents a VH gene rearranged to JH4 and switched to the α-constant region (α1 or α2). Horizontal arrows above the locus represent transcription initiation sites. Dotted lines between arrows mark the DNA regions amplified and the bold line represents the DNA regions sequenced. Eμ, heavy chain enhancer; L, leader; V, variable; D, diversity; and J, joining coding elements; Iμ, intronic leader exon; Sμ and Sα, core switch regions.
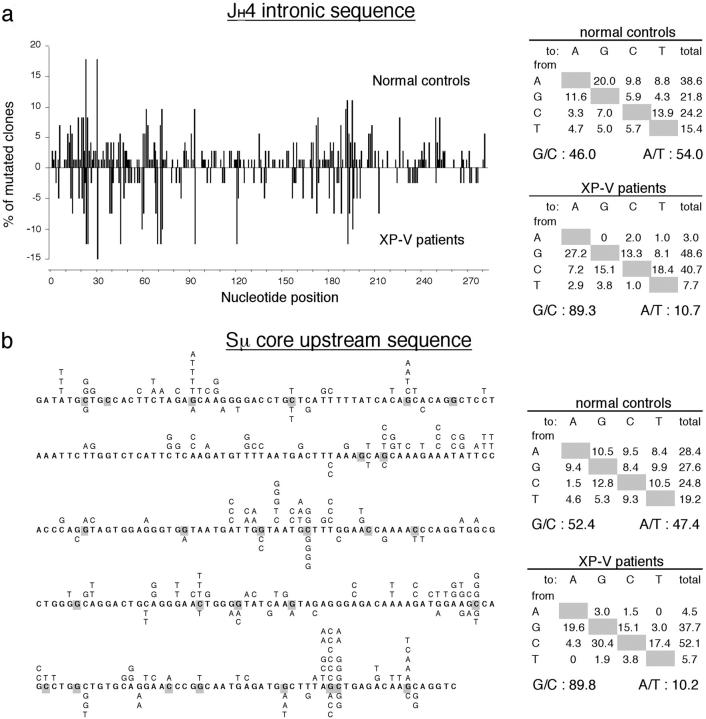
The JH4 intron downstream from rearranged VH genes was amplified using a 5′ FR3 primer consensus for all human VH genes, and 283 bp starting at the border of JH4 intron were sequenced. Only sequences corresponding to different V-D-J junctions were considered. The JH4 segment was used in 50% of rearranged VH genes; such an approach allows a large sampling of unselected sequences (Fig. 2 a; reference 15). The mutation frequency was 2.0/100 bp for the controls, and 1.5 for the XP-V patients (Table I). In both groups, mutations bore the hallmark of somatic mutations: prevalence of base substitutions (with 6–9% of total mutations representing deletions or duplications), bias for transitions (47–51% of all mutations), and targeting of RGYW motifs (Table II). This targeting was calculated taking into account mutations occurring at the G position of the hotspot motif only (considered for both strands of DNA), as compared with mutations at other G/C positions. Moreover, normal controls showed a marked preference for mutations at A over T positions. In accordance with Zeng et al. (5), we observed a drastic reduction of mutations on A/T bases in XP-V patients as compared with the controls (10.7 vs. 54.0%; Fig. 2 a), thus confirming the implication of pol η. The lower number of events on A/T bases also correlates with a slight reduction in the total amount of mutations, obviously only noticeable on such unselected targets.
Figure 2.
Mutation pattern of JH4 intronic sequences and Sμ core upstream regions in normal controls and XP-V patients. (a) Distribution and nucleotide substitution preference of mutations in JH4 intronic sequences. The distribution of mutations is represented along the 283 bp of intronic JH4 sequence, with the proportion of mutated clones in ordinates. Mutations from normal controls are represented above the nucleotide position, and those from XP-V patients are below. Nucleotide substitution preferences are corrected for base composition. A, 18.0%; G, 31.1%; C, 32.2%; T, 18.7%. (b) Distribution and nucleotide substitution preference of mutations in Sμ core upstream sequences. Mutations obtained for normal controls are represented above the 295 nucleotides of the Sμ core upstream sequence, and those from XP-V patients are below. The G position of the RGYW motif (and the C position of the complementary WRCY motif on the other strand) is highlighted in gray. Nucleotide substitution preferences are corrected for base composition. A, 28.5%; G, 28.8%; C: 20.0%; T, 22.7%.
Table I.
Somatic Mutations in JH4 Intronic Sequences and Sμ Core Upstream Sequences from Normal Individuals and XP-V Patients
| JH4 intronic sequence (283 bp)
|
Sμ core upstream sequence (295 bp)
|
|||
|---|---|---|---|---|
| Control | XP-V | Control | XP-V | |
| Number of sequences | 73 | 41 | 199 | 101 |
| Total length sequenced (bp) | 20,659 | 11,603 | 58,705 | 29,795 |
| Unmutated sequences (percent) | 2.7 | 2.4 | 54 | 59 |
| Total number of mutations | 414 | 168 | 180 | 63 |
| (Number of deletions and duplications) | (23) | (15) | (11) | (8) |
| Mutations range per mutated sequence | 1–25 | 1–19 | 1–11 | 1–5 |
| Mutation frequency (per 100 bp) | 2.0 | 1.5 | 0.31 | 0.21 |
Table II.
Somatic Mutations at G/C Positions of RGYW (or WRCY) Motifsa
| JH4 intronic sequences(283 bp) | Sμ core upstream sequences(295 bp) | Sμ core junctional sequencesb(250 bp) | |
|---|---|---|---|
| G/C within RGYW motifs | 14% | 19% | 35% |
| Per total G/C bases | (22/157) | (27/144) | (50/142) |
| G/C mutated within RGYW motifs | 36% | 49% | 79% |
| Per total G/C bases in sequences from normal controls |
(89/246) | (45/86) | (57/72) |
| G/C mutated within RGYW motifs | 31% | 57% | 89% |
| Per total mutated G/C bases in sequences from XP-V patients |
(45/147) | (28/49) | (40/45) |
Only the underlined G or C base within the motif is considered.
Sequences listed in Fig. 3 are tabulated.
To find out whether the mutations observed during CSR and SHM were generated by the same mutational complex, we analyzed two regions of the heavy chain locus that undergo hypermutation during CSR in memory B cells of both groups: the region upstream of the Sμ core region and Sμ–Sα junctions. AID-dependent mutagenesis of the Sμ core upstream region has been shown to be induced after in vitro induction of switch in mouse splenic B cells (9, 10). Mutations in this region have also been detected in human chronic lymphocytic leukemia (16).
A region of 900 bp upstream from the core repetitive sequences of the Sμ region was amplified from controls and XP-V patients, and the sequence of the 3′ 295 bp was determined (16). A mutation frequency of 0.31/100 and 0.21/100 bp was found for the controls and the XP-V individuals, respectively, i.e., a value six- to sevenfold lower than for the JH4 intron (Table I). It should be noted that both alleles are scored in this assay and that the precise fraction of nonfunctional alleles that undergo isotype switch in memory B cells in vivo is not known. The pattern of mutation was very similar to the one observed at the VDJ locus considering the ratio of base substitutions over deletions and duplications, the bias for transitions and for A over T mutations (although somewhat reduced), and the targeting to G/C bases of the RGYW (WRCY) motifs (Table II and Fig. 2 b). This similarity extends to the selective deficiency of mutations at A and T positions observed for the two XP-V patients in this genomic sequence as well (10.2% in XP-V patients compared with 47.4% in controls).
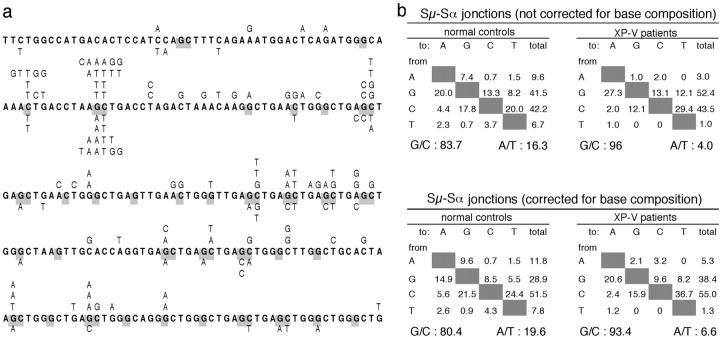
Sμ-Sα switch junctions from either Sα1 or Sα2 loci were amplified and PCR products below 1 kb were isolated and sequenced. An average of 170 bp on the Sμ side and 211 bp on the Sα1 or Sα2 side were determined for control sequences, and 200 bp on the Sμ side and 317 bp on the Sα side were determined for the XP-V individuals (Table III). A similar mutation frequency was observed for both control and XP-V patients (0.43/100 and 0.50/100 bp, respectively; Table III). Mutations were mainly base substitutions (deletions over 25 bp were not included and considered as an internal switch event), with a bias for transitions (51.8 and 59.4 for controls and XP-V patients, respectively; Fig. 3) . The switch core region, which is made of pentamer repeats, differs from other regions of the heavy chain locus that are targeted for mutations under two aspects: first an extreme incidence of G/C bases within RGYW hotspots, as the switch repeat motif at both Sμ and Sα loci GAGCTGGGCT includes three G/C bases out of seven in such a context. Accordingly, 79–89% of all G/C mutations are targeted to such hotspot positions. The second specific feature is a high G/C content with a marked G/C imbalance (40% G and 20% C in the reference sequence of the nontranscribed strand). Therefore, mutations have been tabulated in two different ways: as raw proportions, thus reflecting their real occurrence in the sequence studied, and also after correction for base composition, reflecting then their mechanistic occurrence (Fig. 3). The G/C targeting was higher than in the other regions of the heavy chain locus for the controls, even after correction for the base composition. Nevertheless, there was still a clear diminution of mutations on A/T bp in the XP-V patients with a threefold reduction compared with control values (6.6 vs. 19.6%), a figure similar to the one found for either the JH4 intron or the Sμ core upstream region. The moderate intervention of pol η can probably also explain why the frequency of mutations is approximately similar for the two groups of individuals in this region. We also observed that cytosines were twice as often targeted than guanines after correction for base composition, but that the absolute number of Cs and Gs mutated was the same. The Iμ sterile transcript has been shown to form an R loop with its template strand (17), which would favor the attack of the single stranded nontranscribed strand of DNA by AID according to its in vitro substrate preference (18–22). Assuming that most of the G mutations scored are due to an AID-mediated attack of cytosines on the template strand, it is tempting to propose that the lower accessibility to AID of this strand that forms a DNA–RNA hybrid is compensated by its C richness, whereas the C poorness of the accessible strand is compensated by its preferential targeting. Both factors, AID substrate preference and evolution of target sequences, would ensure approximately the same amount of AID-mediated deamination on each strand, thus allowing for efficient generation of double stranded breaks, necessary for the switch recombination process (23).
Table III.
Somatic Mutations in Sequences Flanking Sμ–Sα Junctions from Normal Individuals and XP-V Patients
| Controls
|
XP-V
|
|||||
|---|---|---|---|---|---|---|
| Sμ | Sα | Sμ | Sα | |||
| Total | Total | |||||
| Number of sequences | 88 | 38 | ||||
| Total length sequenced (bp) | 14,992 | 18,591 | 7,588 | 12,054 | ||
| Unmutated sequences (percent) | 47 | 60 | 34 | 29 | ||
| Total number of mutations | 84 | 61 | 57 | 43 | ||
| (Number of deletions and duplications) | (9) | (4) | (6) | (1) | ||
| Mutation range per mutated sequence | 1–8 | 1–4 | 1–13 | 1–4 | ||
| Mutation frequency (per 100 bp) | 0.56 | 0.33 | 0.75 | 0.36 | ||
| 0.43 | 0.50 | |||||
Figure 3.
Mutation pattern of Sμ–Sα junctions in normal controls and XP-V patients. (a) Mutation distribution along the first 250 bp of the Sμ region in Sμ–Sα junctions. Mutations collected from normal controls are listed above the reference sequence, and mutations from XP-V patients are below. The G position of the RGYW motif (and the C position of the complementary WRCY motif on the other strand) is highlighted in gray. (b) Nucleotide substitution preference of mutations in Sμ–Sα junctions. Mutations collected on both Sμ and Sα regions are tabulated, first as raw proportions and then after correction for base composition, taking into account the base composition of the average region sequenced in each case. Sμ controls: 170 bp, 25.9% A, 30.0% G, 22.3% C, and 21.8% T; Sα controls: 211 bp, 12.6% A, 47.6% G, 18.6% C, and 21.2% T; Sμ-XP-V: 200 bp, 25.5% A, 31% G, 22% C, and 21.5% T; Sα-XP-V: 350 bp, 12.9% A, 46.1% G, 19.6% C, and 21.4% T.
In conclusion, we would like to propose that the mutations observed during CSR most probably involve the SHM mutasome. The pattern of mutation is clearly more G/C targeted in the Sμ core region. This G/C targeting might be explained by the G/C richness within pentameric hotspots in this region and the unique accessibility provided by the R loop structure, so that less nuclease digestion, and/or less error-prone short patch DNA synthesis would be required to generate adjacent cleavage sites, i.e., double stranded breaks that can be engaged into a successful switch recombination event. A similar conclusion was drawn from a study on the role of MSH2 during CSR (24) showing that this molecule is more specifically required for switching occurring in the Sμ upstream region. In this region, the lower density of hotspots, as compared with the Sμ core, may require more processing of the DNA ends to render them suitable for recombination, a processing that would rely on the strand displacement activity of MSH2.
This work also brings the confirmation that the translesion DNA synthesis (TLS) polymerase pol η is an A/T mutator. Nevertheless, there are still mutations on A/T bases in both SHM and CSR. Therefore, it is probable that other TLS polymerases, such as pol ι that interacts with pol η at replication foci, could be involved in both processes (25). Accordingly, in a human Burkitt's B cell line inducible for SHM and whose pattern of mutations is strongly G/C biased, induction of SHM was shown to be dependent on pol ι (26, 27). This result remains controversial because it was recently reported that 129/SvJ mice that lack a functional pol ι did not show any significant change in the frequency and pattern of Ig mutations (28). To what extent the human in vitro and mice in vivo systems differ and to what extent TLS polymerases may compensate for each other remains to be explored.
The rationale for using error-prone polymerases during CSR is not clearly perceptible. Here we propose that these enzymes participate in an AID-triggered, short patch DNA synthesis ensuring the efficient fill-in of staggered double strand breaks. However, we cannot exclude that the mutations observed may represent the bystander recruitment of AID partners involved in the mutagenesis of Ig V genes, and that the involvement of TLS polymerases as cofactors of the nonhomologous end joining process might be dispensable. An in vitro system that would enable to assess the impact of these polymerases on the strict efficiency of switching may help resolve this issue.
Acknowledgments
We thank Dr. M.F. Avril for providing blood samples from XP-V patients, C. Debacker for cell cultures, A. Van der Linden, and the laboratory of Alan Lehmann for the determination of pol η mutations in XP-V patients 1 and 2, respectively. We thank Jérome Mégret for performing cell sorting, Claire Soudais and Claire Fieschi for their contribution, and Guillaume Dighiero for stimulating discussions.
This work was supported by the Ligue contre le Cancer (Equipe Labelisée) and the Fondation Princesse Grace de Monaco.
C.-A. Reynaud and J.-C. Weill are senior authors on this paper.
Abbreviations used in this paper: AID, activation-induced cytidine deaminase; CSR, class switch recombination; pol η, polymerase η; TLS, translesion DNA synthesis; Sα, α switch region; SHM, somatic hypermutation; Sμ, μ switch region; XP-V, xeroderma pigmentosum variant.
References
- 1.Honjo, T., K. Kinoshita, and M. Muramatsu. 2002. Molecular mechanism of class switch recombination: linkage with somatic hypermutation. Annu. Rev. Immunol. 20:165–196. [DOI] [PubMed] [Google Scholar]
- 2.Martin, A., and M.D. Scharff. 2002. AID and mismatch repair in antibody diversification. Nat. Rev. Immunol. 2:605–614. [DOI] [PubMed] [Google Scholar]
- 3.Goodman, M.F., and B. Tippin. 2000. Sloppier copier DNA polymerases involved in genome repair. Curr. Opin. Genet. Dev. 10:162–168. [DOI] [PubMed] [Google Scholar]
- 4.Masutani, C., M. Araki, A. Yamada, N. Dohmae, M. Yokoi, M. Yuasa, M. Araki, S. Iwai, K. Takio, and F. Hanaoka. 1999. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase eta. Nature. 399:700–704. [DOI] [PubMed] [Google Scholar]
- 5.Zeng, X., D.B. Winter, C. Kasmer, K.H. Kraemer, A.R. Lehmann, and P.J. Gearhart. 2001. DNA polymerase eta is an A-T mutator in somatic hypermutation of immunoglobulin variable genes. Nat. Immunol. 2:537–541. [DOI] [PubMed] [Google Scholar]
- 6.Manis, J.P., M. Tian, and F.W. Alt. 2002. Mechanism and control of class-switch recombination. Trends Immunol. 23:31–39. [DOI] [PubMed] [Google Scholar]
- 7.Dunnick, W., M. Wilson, and J. Stavnezer. 1989. Mutations, duplication, and deletion of recombined switch regions suggest a role for DNA replication in the immunoglobulin heavy-chain switch. Mol. Cell. Biol. 9:1850–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rogozin, I.B., and N.A. Kolchanov, N.A. 1992. Somatic hypermutagenesis in immunoglobulin genes. II. Influence of neighbouring base sequences on mutagenesis. Biochim. Biophys. Acta 1171:11–18. [DOI] [PubMed] [Google Scholar]
- 9.Nagaoka, H., M. Muramatsu, N. Yamamura, K. Kinoshita, and T. Honjo. 2002. Activation-induced deaminase (AID)-directed hypermutation in the immunoglobulin Smu region: implication of AID involvement in a common step of class switch recombination and somatic hypermutation. J. Exp. Med. 195:529–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reina-San-Martin, B., S. Difilippantonio, L. Hanitsch, R.F. Masilamani, A. Nussenzweig, and M.C. Nussenzweig. 2003. H2AX is required for recombination between immunoglobulin switch regions but not for intra-switch region recombination or somatic hypermutation. J. Exp. Med. 197:1767–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lehmann, A.R., S. Kirk-Bell, C.F. Arlett, M.C. Paterson, P.H. Lohman, E.A. de Weerd-Kastelein, and D. Bootsma. 1975. Xeroderma pigmentosum cells with normal levels of excision repair have a defect in DNA synthesis after UV-irradiation. Proc. Natl. Acad. Sci. USA. 172:219–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weller, S., A. Faili, C. Garcia, M.C. Braun, F. Le Deist, G. de Saint Basile, O. Hermine, A. Fischer, C.-A. Reynaud, and J.-C. Weill. 2001. CD40-CD40L independent Ig gene hypermutation suggests a second B cell diversification pathway in humans. Proc. Natl. Acad. Sci. USA. 98:1166–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dorner, T., and P.E. Lipsky. 2001. Smaller role for pol eta? Nat. Immunol. 2:982–984. [DOI] [PubMed] [Google Scholar]
- 14.Yavuz, S., A.S. Yavuz, K.H. Kraemer, and P.E. Lipsky. 2002. The role of polymerase eta in somatic hypermutation determined by analysis of mutations in a patient with xeroderma pigmentosum variant. J. Immunol. 169:3825–3830. [DOI] [PubMed] [Google Scholar]
- 15.Levy, Y., N. Gupta, F. Le Deist, C. Garcia, A. Fischer, J.-C. Weill, and C.-A. Reynaud. 1998. Defect in IgV gene somatic hypermutation in common variable immuno-deficiency syndrome. Proc. Natl. Acad. Sci. USA. 95:13135–13140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oppezzo, P., F. Vuillier, Y. Vasconcelos, G. Dumas, C. Magnac, B. Payelle-Brogard, O. Pritsch, and G. Dighiero. 2003. Chronic lymphocytic leukemia B cells expressing AID display dissociation between class switch recombination and somatic hypermutation. Blood. 101:4029–4032. [DOI] [PubMed] [Google Scholar]
- 17.Yu, K., F. Chedin, C.L. Hsieh, T.E. Wilson, and M.R. Lieber. 2003. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat. Immunol. 4:442–451. [DOI] [PubMed] [Google Scholar]
- 18.Pham, P., R. Bransteitter, J. Petruska, and M.F. Goodman. 2003. Processive AID-catalysed cytosine deamination on single-stranded DNA simulates somatic hypermutation. Nature. 424:103–107. [DOI] [PubMed] [Google Scholar]
- 19.Chaudhuri, J., M. Tian, C. Khuong, K. Chua, E. Pinaud, and F.W. Alt. 2003. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature. 422:726–730. [DOI] [PubMed] [Google Scholar]
- 20.Ramiro, A.R., P. Stavropoulos, M. Jankovic, and M.C. Nussenzweig. 2003. Transcription enhances AID-mediated cytidine deamination by exposing single-stranded DNA on the nontemplate strand. Nat. Immunol. 4:452–456. [DOI] [PubMed] [Google Scholar]
- 21.Dickerson, S.K., E. Market, E. Besmer, and F.N. Papavasiliou. 2003. AID mediates hypermutation by deaminating single stranded DNA. J. Exp. Med. 197:1291–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sohail, A., J. Klapacz, M. Samaranayake, A. Ullah, and A.S. Bhagwat. 2003. Human activation-induced cytidine deaminase causes transcription-dependent, strand-biased C to U deaminations. Nucleic Acids Res. 31:2990–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reynaud, C.-A., S. Aoufouchi, A. Faili, and J.-C. Weill. 2003. What role for AID: mutator, or assembler of the immunoglobulin mutasome? Nat. Immunol. 4:631–638. [DOI] [PubMed] [Google Scholar]
- 24.Min, I.M., C.E. Schrader, J. Vardo, T.M. Luby, N. D'Avirro, J. Stavnezer, and E. Selsing. 2003. The Sμ tandem repeat region is critical for Ig isotype switching in the absence of Msh2. Immunity. 19:515–529. [DOI] [PubMed] [Google Scholar]
- 25.Kannouche, P., A.R. Fernandez de Henestrosa, B. Coull, A.E. Vidal, C. Gray, D. Zicha, R. Woodgate, and A.R. Lehmann. 2002. Localization of DNA polymerases eta and iota to the replication machinery is tightly co-ordinated in human cells. EMBO J. 21:6246–6256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Faili, A., S. Aoufouchi, Q. Gueranger, C. Zober, A. Leon, B. Bertocci, J.-C. Weill, and C.-A. Reynaud. 2002. AID-dependent somatic hypermutation occurs as a DNA single-strand event in the BL2 cell line. Nat. Immunol. 3:815–821. [DOI] [PubMed] [Google Scholar]
- 27.Faili, A., S. Aoufouchi, E. Flatter, Q. Guéranger, C.-A. Reynaud, and J.-C. Weill. 2002. Induction of somatic hypermutation in immunoglobulin genes is dependent on DNA polymerase iota. Nature. 419:944–947. [DOI] [PubMed] [Google Scholar]
- 28.McDonald, J.P., E.G. Frank, B.S. Plosky, I.B. Rogozin, C. Masutani, F. Hanaoka, R. Woodgate, and P.J. Gearhart. 2003. 129-derived strains of mice are deficient in DNA polymerase iota and have normal immunoglobulin hypermutation. J. Exp. Med. 198:635–643. [DOI] [PMC free article] [PubMed] [Google Scholar]