Abstract
Autoimmune regulator (AIRE) gene mutation is responsible for the development of autoimmune-polyendocrinopathy-candidiasis ectodermal dystrophy, an organ-specific autoimmune disease with monogenic autosomal recessive inheritance. AIRE is predominantly expressed in medullary epithelial cells of the thymus and is considered to play important roles in the establishment of self-tolerance. AIRE contains two plant homeodomain (PHD) domains, and the novel role of PHD as an E3 ubiquitin (Ub) ligase has just emerged. Here we show that the first PHD (PHD1) of AIRE mediates E3 ligase activity. The significance of this finding was underscored by the fact that disease-causing missense mutations in the PHD1 (C311Y and P326Q) abolished its E3 ligase activity. These results add a novel enzymatic function for AIRE and suggest an indispensable role of the Ub proteasome pathway in the establishment of self-tolerance, in which AIRE is involved.
Keywords: APECED, self-tolerance, transcriptional regulator, ubiquitylation, PHD domain
Introduction
Autoimmune disease is a pathological condition in which the immune system turns on itself and causes serious damage to the organism's tissues by as yet unknown mechanisms (1). Although a propensity to appear in families is one of the common features of autoimmune diseases, only a few genes relevant to the pathogenetic processes that underlie the development of autoimmune diseases per se are actually known (2). Because autoimmune regulator (AIRE) gene mutation is responsible for the development of autoimmune-polyendocrinopathy-candidiasis ectodermal dystrophy (APECED: OMIM 240300; references 3–6), an organ-specific autoimmune disease with monogenic autosomal recessive inheritance, understanding the relationship between AIRE gene malfunction and the breakdown of self-tolerance promises to help unravel the pathogenesis of not only APECED, but also other types of autoimmune disease. Supporting this notion, AIRE is predominantly expressed in medullary epithelial cells of the thymus (7, 8).
The ubiquitin (Ub) proteasome pathway plays an essential role in diverse cell functions such as cell cycle progression, signal transduction, cell differentiation, DNA repair, and apoptosis (9, 10). The system responsible for Ub attachment consists of several components that act in concert. A Ub-activating enzyme (E1) catalyzes the formation of a thioester bond between itself and Ub, and then transfers the activated Ub to a Ub-conjugating enzyme (E2). E3 ligase interacts with both E2 and the substrate, and facilitates polyubiquitylation of the substrate (9, 10). The plant homeodomain (PHD) is found in many proteins involved in chromatin-mediated transcriptional regulation (11). It is noteworthy that PHD resembles the RING finger, which functions as an E3 ligase (12) in both sequence and structure (13, 14). The RING finger and PHD contain a motif defined by an octet of cysteines and histidines with similar spacing that coordinates two zinc ions (15). Although the exact definition of PHD requires further study (16), recent studies have demonstrated that a subset of PHD-containing proteins such as Kaposi's sarcoma-associated herpes virus proteins (17, 18) and MAP kinase/ERK kinase (MEKK1) mediates E3 ligase activity (19).
The AIRE gene encodes a predicted 58-kD protein carrying two PHDs together with a homogeneously staining region and SAND domains (3, 4). Although the homogeneously staining region and SAND domains have been suggested to function in homodimerization and DNA binding, respectively (20, 21), the exact function of PHDs in the AIRE remains elusive. Significantly, Finnish mutation R257X and the 13-bp deletion in exon 8 are the most frequent mutations found, both producing truncated AIRE proteins lacking both PHD1 and PHD2, or lacking PHD2 with a disrupted PHD1, respectively (5, 6). Recent observations demonstrating the E3 ligase activity of PHD-containing proteins have prompted us to test whether AIRE may act as an E3 ligase. Here we show that the PHD1 of AIRE mediates E3 ligase activity. Of note, we further show that disease-causing missense mutations in the PHD1 (C311Y and P326Q) abolished its E3 ligase activity, suggesting an indispensable role of the Ub proteasome pathway in the establishment of self-tolerance, in which AIRE is involved.
Materials and Methods
Construction of the Expression Plasmids and Mutagenesis for AIRE.
Human AIRE cDNA was amplified by PCR from Marathon-Ready human thymus cDNA (CLONTECH Laboratories, Inc.). In brief, AIRE cDNA was first amplified with adaptor primers according to the manufacturer's instructions, and then nested PCR was performed with AIRE-specific primers to which EcoRI and SalI restriction sites were added at the 5′ and 3′ ends, respectively. The following primers were used. AIRE-U1: CGGAATTCATGGCGACGGACGCGGCGCTAC; AIRE-D1: ACGCGTCGACTCAGGAGGGGAAGGGGGCCG. The resulting fragment containing the human AIRE open reading frame was ligated into the cloning site downstream of the FLAG-tag in the pCR3 vector (Invitrogen) or His6-tag in pFastBac HTa (Invitrogen). Site-directed mutagenesis for the generation of C299A, C311Y, P326Q, C434A, and C299A/C434A mutants was performed using human AIRE cDNA cloned into pBluescript II SK+ (Stratagene) as a template with a Quick Change (Stratagene). The sequence of each constructed vector was confirmed by the dideoxy chain termination method with automated sequencing (Applied Biosystems).
Production of Recombinant Proteins in Bacteria.
The plasmids containing various human E2s (i.e., Ubc2A, Ubc2B, Ubc3, Ubc4, UbcH5A, UbcH5B, UbcH5C, UbcH6, UbcH7, and UbcH8) in pT7-7 or pET30a (Novagen) were constructed as previously described (22). These His6-tagged proteins were expressed in Escherichia coli strain BL21(DE3)pLysS (Novagen) cultured in the presence of 0.1 mM isopropyl-β-D-thiogalactopyranoside. The recombinant E2 proteins were purified with the use of ProBand resin (Invitrogen) as previously described (22).
Baculovirus Expression System.
The plasmid pFastBac HTa containing the relevant cDNA was subjected to recombination with the baculoviral genome in HB10BAC, and the resulting recombinant viral genome was introduced into Sf21 cells by transfection to generate recombinant baculovirus. The infected Sf21 cells were lysed and the recombinant proteins were purified by the protocol described for bacterially expressed His6-tagged proteins (22).
In Vitro Ubiquitylation Assay.
The PHD1 (amino acids 280–374) and PHD2 (amino acids 415–509) domains were PCR amplified and fused to the carboxyl terminus of the pGEX-4T-1 vector (Amersham Biosciences). The constructs were introduced into E. coli strain BL21-CodonPlus(DE3)-RP (Stratagene) and expression of glutathione S-transferase (GST) fusion proteins was induced by 0.1 mM isopropyl-β-D-thiogalactopyranoside. Bacterial pellets were sonicated in PBS containing protease inhibitors, 1% Triton X-100, and 0.1% sarcosyl. After being cleared by centrifugation, the bacterial lysates were incubated with glutathione-Sepharose 4B beads (Amersham Biosciences) for purification. The in vitro ubiquitylation assays were performed as previously described (22). In brief, reaction mixtures (20 μl) containing either 1 μg of the full-sized recombinant AIRE or 5 μg GST-PHD fusion proteins, together with 0.1 μg recombinant rabbit E1 (Boston Biomedica), 1 μl crude E. coli lysate containing various E2s, 0.5 U phosphocreatine kinase, 1 μg Ub (Sigma-Aldrich), 25 mM Tris-HCl, pH 7.5, 120 mM NaCl, 2 mM ATP, 1 mM MgCl2, 0.3 mM dithiothreitol, and 1mM creatine phosphate were incubated for 2 h at 30°C. In some assays, rabbit reticulocyte lysate (Promega) was used as a source of E1 and E2. The reaction was terminated by the addition of SDS sample buffer containing 4% 2-ME and heating at 95°C for 5 min. Samples were resolved by SDS-PAGE on a 6% gel and then subjected to immunoblot analysis with a mouse monoclonal antibody against Ub (clone 1B3; MBL International Corporation) and a horseradish peroxidase–conjugated rabbit polyclonal antibody against mouse immunoglobulin (Southern Biotechnology Associates, Inc.). Signals were detected with ECL (Amersham Biosciences).
Reporter Gene Assay.
Gal4-binding assays were performed with a CheckMate Mammalian Two-Hybrid System (Promega) according to the manufacturer's instructions. In brief, human AIRE cDNAs were subcloned into the pBIND vector containing the Renilla luciferase gene driven by the SV40 early enhancer/promoter. 0.2 μg pBIND-AIRE vectors were transfected into mouse epithelial cell line of thymic medulla origin (mTEC; 1C6; provided by M. Kasai, National Institute of Infectious Diseases, Tokyo, Japan; reference 23) with 0.2 μg of the pG5luc vector using LIPOFECTAMINE 2000 Reagent (GIBCO BRL). After 48 h, the cells were harvested and activities were determined with the dual luciferase reporter gene assay system (Promega).
RT-PCR.
RNAs were extracted from mouse organs with TRIzol (Invitrogen) and treated with DNase to eliminate any contaminating DNA. After phenol/chloroform extraction and ethanol precipitation, 5 μg total RNA was subjected to oligo(dT)-primed reverse transcription with a cDNA cycle kit (Invitrogen). The primer pairs used for PCR were 5′-CAAGGAATTGAATGACCTGG-3′ and 5′-GAGTCCATTCCCGAGCTATT-3′ for Ubc4 and 5′-TGGAATCCTGTGGCATCCATGAAAC-3′ and 5′-TAAAACGCAGCTCAGTAACAGTCCG-3′ for β-actin. PCR was performed in a final volume of 30 μl with 1.5 U ExTaq DNA polymerase (Takara Biomedicals) and 250 nM of each primer. Cycling conditions were a single denaturing step at 94°C for 10 min followed by 30 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 1.5 min, followed by a final extension step of 72°C for 10 min.
Online Supplemental Material.
Fig. S1 shows Ubc4 expression in the thymus, and Fig. S2 shows that AIRE mediates ubiquitylation with UbcH5B. These figures are available at http://www.jem.org/cgi/content/full/jem.20031291/DC1.
Results and Discussion
PHD1 of AIRE Mediates E3 Ligase Activity.
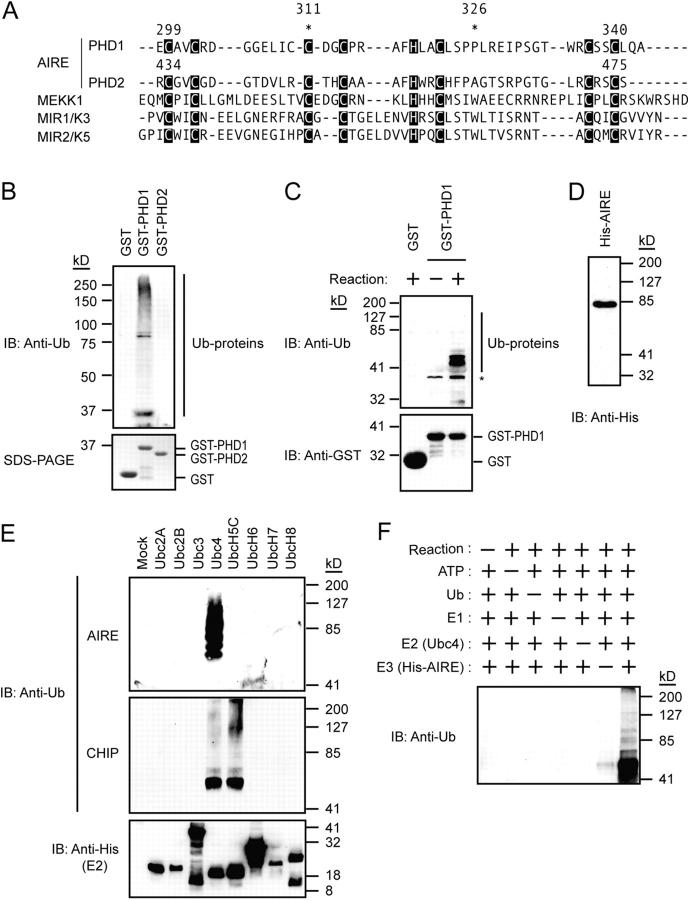
When aligned with other PHDs showing E3 ligase activity, both PHD1 and PHD2 of AIRE apparently demonstrated homology with the PHDs from those proteins (Fig. 1 A). To determine whether the PHD1 and/or PHD2 of AIRE have E3 ligase activity, we performed in vitro ubiquitylation assays. PHD1 and PHD2 were fused to the carboxyl terminus of GST and the purified fusion proteins were subjected to the assay using rabbit reticulocyte lysate as a source of E1 and E2. Polyubiquitylated proteins were observed from the GST-PHD1 fusion protein, whereas GST alone and the GST-PHD2 fusion protein did not have this form, suggesting that the PHD1 of AIRE mediates E3 ligase activity (Fig. 1 B). E3 ligase activity mediated through PHD1 was also demonstrated by the assay using recombinant E1 and E2 (Ubc4, see below), and this activity was incubation dependent (Fig. 1 C).
Figure 1.
AIRE mediates E3 ligase activity through PHD1. (A) The PHD1 and PHD2 domains of AIRE showed homology with the PHDs from other proteins showing E3 ligase activity. *, the position of disease-causing mutations (C311Y and P326Q) used in the study. (B) The GST-PHD1, but not GST-PHD2, fusion protein exhibited polyubiquitylated proteins when rabbit reticulocyte lysate was used for in vitro ubiquitylation assays (top). SDS-PAGE of GST-fusion proteins used in the assay (10% input) is shown on the bottom. (C) Recombinant E1 and E2 (Ubc4) were used for in vitro ubiquitylation assays (top). The ubiquitylation reactions were subjected to immunoblot analysis with antibodies against GST (bottom). E3 ligase activity mediated through PHD1 is incubation dependent. *, nonspecific bands. (D) The full-sized recombinant AIRE protein was successfully and homogeneously expressed in Sf21 cells using a baculovirus expression system. The purified recombinant AIRE protein was subjected to immunoblot analysis with an antibody against His6-tag. (E) Ubc4 E2 preference by the full-sized recombinant AIRE in in vitro ubiquitylation assays (top). CHIP, a U-box–type E3 ligase, is included to verify the specificity and biological activities of E2s (middle; reference 22). Equal amount of E2s were used in the reaction (bottom). (F) Specificity of E3 ligase activity mediated by the full-sized recombinant AIRE in in vitro ubiquitylation assays.
To further confirm the E3 ligase activity of AIRE, the recombinant full-sized AIRE protein was expressed in Sf21 cells using a baculovirus expression system (Fig. 1 D). Because most E3s exhibit specificity for a relatively narrow range of E2 enzymes, we simultaneously examined the preference of the AIRE proteins for eight different human E2 enzymes in the presence of E1. The wild-type full-sized recombinant AIRE exhibited polyubiquitylated proteins when Ubc4 was present as E2 in the reaction (Fig. 1 E). Consistent with this functional association, Ubc4 was abundantly expressed in the thymus, as is the case with AIRE (see Fig. S1, available at http://www.jem.org/cgi/content/full/jem.20031291/DC1). This reaction was dependent on the incubation and the presence of AIRE (Fig. 1 F), confirming the specificity of the E3 ligase activity. Further examination has revealed that AIRE also mediates ubiquitylation with UbcH5B, which is structurally very close to Ubc4 (see Fig. S2, available at http://www.jem.org/cgi/content/full/jem.20031291/DC1).
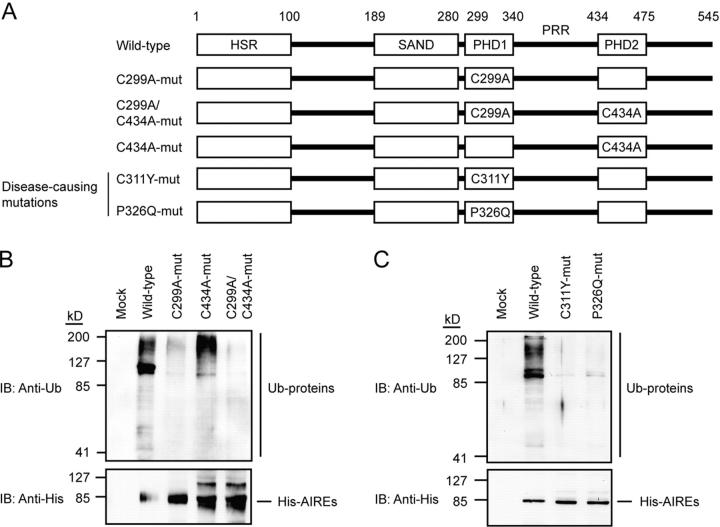
The indispensability of PHD1 for E3 ligase activity was demonstrated with PHD mutants in which the first cysteines were mutated to alanines in either PHD1 (C299A), PHD2 (C434A), or both PHDs (C299A/C434A) of the full-sized recombinant AIRE (Fig. 2 A). Corresponding amino acid substitution in PHD of MEKK1 has demonstrated the significance of this cysteine for its E3 ligase activity (19). Although the wild-type and the PHD2 mutant (C434A) mediated polyubiquitylation, the PHD1 mutant (C299A) and the PHD1/PHD2 double mutant (C299A/C434A) showed no obvious polyubiquitylation (Fig. 2 B). Together with the data obtained from GST-PHD fusion proteins, these results indicate that AIRE mediates E3 ligase activity primarily through PHD1. Although we cannot completely exclude the possibility that PHD2 is involved in the full enzymatic activity, the absence of the tryptophan residue usually located between cysteines 6 and 7 of ligase-type PHDs might be consistent with the lack of E3 ligase activity from PHD2 (Fig. 1 A; reference 18).
Figure 2.
Mutations in PHD1 affect E3 ligase activity. (A) Schematic representation of the PHD mutants used in the study. (B) Substitutions of the first cysteines into alanines in PHD1, or PHD1 and PHD2, but not in PHD2 alone, abolished E3 ligase activity (top). The ubiquitylation reactions were subjected to immunoblot analysis with antibodies against His6-tag (bottom). (C) Disease-causing mutations of PHD1 (C311Y and P326Q) resulted in loss of E3 ligase activity as assessed with in vitro ubiquitylation assays.
Having identified that PHD1 of AIRE functions as an E3 ligase, we addressed whether the loss of this function would have any relevance to the development of APECED. Our approach was to introduce missense mutations (Figs. 1 A and 2 A, C311Y and P326Q) into the PHD1 of the full-sized recombinant AIRE, both of which are found in patients with APECED (6, 24). Both of the disease-causing mutants showed dramatically reduced E3 ligase activity in in vitro ubiquitylation assays (Fig. 2 C), strongly suggesting that loss of E3 ligase activity has relevance to the disease process of APECED.
Essential Role for PHD2 in Transcriptional Activation.
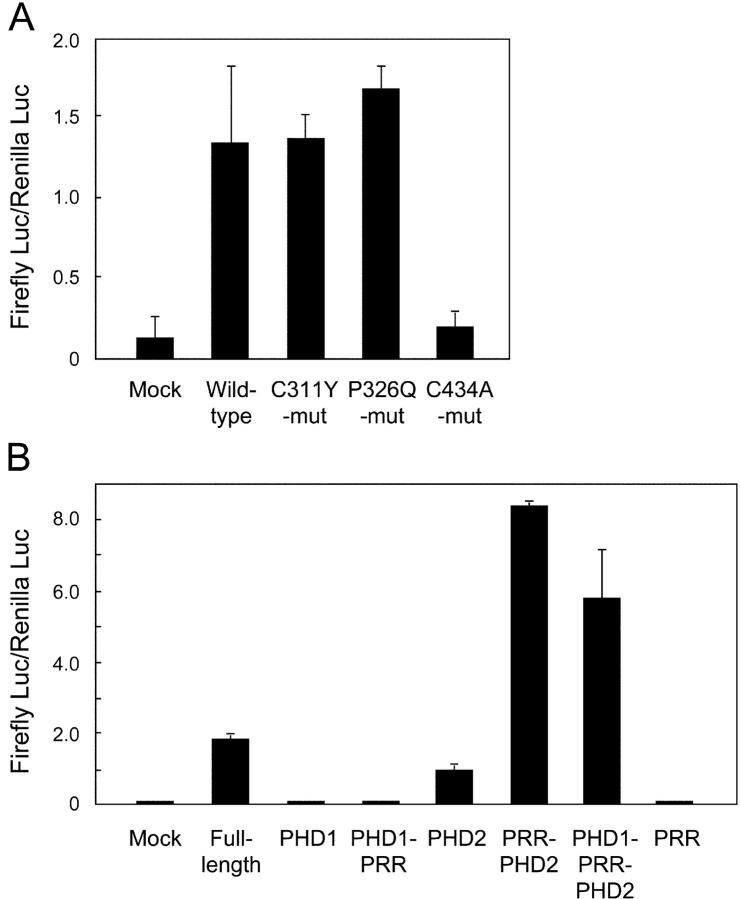
It has been previously demonstrated that AIRE has transcription-activating properties when fused to a heterologous DNA-binding domain (20, 24), and many PHD-disrupting mutations result in decreased transcriptional activity. We investigated whether the loss of E3 ligase activity resulting from the disease-causing mutations C311Y and P326Q is associated with alterations in the transcription-activating properties assessed with a series of Gal4-binding assays. In light of the fact that AIRE expression is most prominent in the thymic medulla (7, 8), we used mouse mTEC (1C6; reference 23), which does not express endogenous AIRE as assessed by standard RT-PCR (unpublished data), for the transfection. As expected, the full-length wild-type AIRE induced activation of the reporter genes in mTEC (Fig. 3 A). The disease-causing mutant forms of PHD1 (C311Y and P326Q), which lack E3 ligase activity, exhibited normal transcriptional activation. In contrast, a PHD2 mutant (C434A) with normal E3 ligase activity showed dramatically reduced transcriptional activation.
Figure 3.
Effect of PHD mutations on the transcriptional transactivating properties of AIRE. (A) Various forms of full-length AIRE including disease-causing mutations were fused with Gal4-binding domain and transfected into mTEC cells together with a reporter plasmid. Firefly luciferase activities were normalized with respect to the Renilla luciferase activities used for assessment of the transfection efficiency. No significant reduction of Gal4 activation was observed from the disease-causing mutations of PHD1 (C311Y and P326Q), whereas the PHD2 mutant (C434A) showed loss of transcriptional activation. The results are expressed as the mean ± SEM for triplicate wells during one representative experiment from a total of five repeat experiments. (B) Isolated PHDs were tested for the transcriptional activation with the same system described in A. The fragments used are as follows (see Figs. 1 A and 2 A): PHD1, amino acids 292–341; PHD1 plus PRR, 292–432; PHD2, 433–545; PRR plus PHD2, 342–545; PHD1 plus PRR plus PHD2, 292–545; and PRR, 342–432. PHD2 suffices for the transcriptional activation in this assay, whereas PHD1 has no such activities. The results are expressed as the mean ± SEM for triplicate wells during one representative experiment from a total of three repeat experiments.
The distinct role of PHD1 and PHD2 in the function of AIRE was further examined with the isolated PHD fragments in the same Gal4-binding assays (Fig. 3 B). Neither PHD1 alone nor PHD1 with a proline-rich region (PRR) showed transcriptional activation. In contrast, PHD2 alone was almost sufficient for the transcriptional activation, and this activity was further pronounced when PRR was added. These results further suggest a distinct role for each PHD domain in the function of AIRE. PHD1 mediates E3 ligase activity, whereas PHD2 is essential for the transcriptional activation.
Apparently normal transactivating properties in the absence of E3 ligase activity as a result of the disease-causing mutations (Fig. 3 A) might be due to enforced recruitment of AIRE to the target DNA in the Gal4-binding assay system. It is possible to speculate that under physiological conditions, E3 ligase activity is necessary for the organization and/or modification of AIRE-interacting proteins in order for AIRE to be located to the transcriptional complex. In contrast, PHD2 might be an important platform required for the recruitment of transcriptional coactivators and the structural integrity of the PHD2 for this action might be disrupted by the C434A mutation. However, it also remains possible that AIRE is a multifunctional protein with E3 ligase activity and transcription-activating properties mediated primarily through PHD1 and PHD2, respectively, and that loss of E3 ligase activity may cause the disease independently from the transcription-activating function of AIRE.
One characteristic features of AIRE is its subcellular localization. Immunocytochemical staining has revealed its presence in the nucleus, appearing as a speckled pattern known as a nuclear body (7, 8, 25). However, to a lesser extent, AIRE also exists in the cytoplasm. To test whether PHD domains are involved in the targeting of AIRE subcellular localization, we have established mTEC lines stably transfected with various PHD mutants using a retrovirus expression system. Wild-type AIRE was observed in both the cytoplasm and nucleus mainly as scattered fine dots, and a similar distribution pattern of AIRE was observed in the cells transfected with either the PHD1 mutant (C299A) or PHD2 mutant (C434A; unpublished data). These results indicate that targeting of AIRE to its defined subcellular localization does not depend on either E3 ligase activity or transcription-activating properties, and further suggest that loss of E3 ligase activity caused by the PHD1 mutations is relevant to the development of the disease.
Despite the growing numbers of known PHD proteins with E3 ligase activity, a defect of PHD E3 ligase had not been suggested for the development of a human disease until very recently. During the processing of this work, Meetei et al. (26) reported a novel PHD-type E3 ligase with defective function in a patient with Fanconi anemia. Here we have demonstrated that disease-causing mutations in the PHD1 of AIRE result in the loss of its E3 ligase activity. Because there are several naturally occurring mutations or deletions in the PHDs implicated in human diseases, we suspect that loss of E3 ligase activity might account for the pathogenesis of some other diseases as well. These include ATRX protein in α-thalassemia and mental retardation (27), AF10 in myeloid leukemias (28), and ING1 in squamous cell carcinomas of the head and neck (29). The prevalence of disease-causing mutations in PHDs suggests that E3 ligase activity mediated through the PHDs could play an essential role in the normal function of human cells (19), although other aspects of PHD function need to be tested (30). Identification of the specific target(s) of ubiquitylation by AIRE is one of the most important tasks remaining in order to clarify the molecular mechanisms by which AIRE establishes and maintains self-tolerance.
Acknowledgments
We thank Drs. P. Peterson, J. Pitkänen, S. Ishii, N. Yamakawa, T. Obata, and K. Hofmann for valuable suggestions. We also thank S. Matsushita and K. Shinohara for technical assistance.
This work was supported in part by Special Coordination Funds for Promoting Science and Technology from the Ministry of Education, Culture, Sports, Science and Technology, the Japanese Government (MEXT; to M. Matsumoto), Grant-in-Aid for Scientific Research from the MEXT (to S. Hatakeyama and M. Matsumoto), the Yasuda Research Foundation (to S. Hatakeyama), the Japan Rheumatism Foundation and the Uehara Memorial Foundation (to M. Matsumoto), Grant-in-Aid for Scientific Research (A) and Fund for “Research for the Future” Program from the Japan Society for the Promotion of Science and MEXT (to N. Shimizu), and the CNRS (to V. Doucas).
D. Uchida and S. Hatakeyama contributed equally to this work.
The online version of this article contains supplemental material.
Abbreviations used in this paper: AIRE, autoimmune regulator; APECED, autoimmune-polyendocrinopathy-candidiasis ectodermal dystrophy; GST, glutathione S-transferase; mTEC, medullary thymic epithelial cell; PHD, plant homeodomain; PRR, proline-rich region; Ub, ubiquitin.
References
- 1.Kamradt, T., and N.A. Mitchison. 2001. Tolerance and autoimmunity. N. Engl. J. Med. 344:655–664. [DOI] [PubMed] [Google Scholar]
- 2.Wanstrat, A., and E. Wakeland. 2001. The genetics of complex autoimmune diseases: non-MHC susceptibility genes. Nat. Immunol. 2:802–809. [DOI] [PubMed] [Google Scholar]
- 3.Nagamine, K., P. Peterson, H.S. Scott, J. Kudoh, S. Minoshima, M. Heino, K.J. Krohn, M.D. Lalioti, P.E. Mullis, S.E. Antonarakis, et al. 1997. Positional cloning of the APECED gene. Nat. Genet. 17:393–398. [DOI] [PubMed] [Google Scholar]
- 4.The Finnish-German APECED Consortium. 1997. An autoimmune disease, APECED, caused by mutations in a novel gene featuring two PHD-type zinc-finger domains. Nat. Genet. 17:399–403. [DOI] [PubMed] [Google Scholar]
- 5.Björses, P., J. Aaltonen, N. Horelli-Kuitunen, M.L. Yaspo, and L. Peltonen. 1998. Gene defect behind APECED: a new clue to autoimmunity. Hum. Mol. Genet. 7:1547–1553. [DOI] [PubMed] [Google Scholar]
- 6.Pitkänen, J., and P. Peterson. 2003. Autoimmune regulator: from loss of function to autoimmunity. Genes Immun. 4:12–21. [DOI] [PubMed] [Google Scholar]
- 7.Heino, M., P. Peterson, J. Kudoh, K. Nagamine, A. Lagerstedt, V. Ovod, A. Ranki, I. Rantala, M. Nieminen, J. Tuukkanen, et al. 1999. Autoimmune regulator is expressed in the cells regulating immune tolerance in thymus medulla. Biochem. Biophys. Res. Commun. 257:821–825. [DOI] [PubMed] [Google Scholar]
- 8.Björses, P., M. Pelto-Huikko, J. Kaukonen, J. Aaltonen, L. Peltonen, and I. Ulmanen. 1999. Localization of the APECED protein in distinct nuclear structures. Hum. Mol. Genet. 8:259–266. [DOI] [PubMed] [Google Scholar]
- 9.Hochstrasser, M. 1996. Ubiquitin-dependent protein degradation. Annu. Rev. Genet. 30:405–439. [DOI] [PubMed] [Google Scholar]
- 10.Pickart, C.M. 2001. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70:503–533. [DOI] [PubMed] [Google Scholar]
- 11.Aasland, R., T.J. Gibson, and A.F. Stewart. 1995. The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem. Sci. 20:56–59. [DOI] [PubMed] [Google Scholar]
- 12.Joazeiro, C.A., and T. Hunter. 2000. Biochemistry. Ubiquitination–more than two to tango. Science. 289:2061–2062. [DOI] [PubMed] [Google Scholar]
- 13.Pascual, J., M. Martinez-Yamout, H.J. Dyson, and P.E. Wright. 2000. Structure of the PHD zinc finger from human Williams-Beuren syndrome transcription factor. J. Mol. Biol. 304:723–729. [DOI] [PubMed] [Google Scholar]
- 14.Capili, A.D., D.C. Schultz, I.F. Rauscher, and K.L. Borden. 2001. Solution structure of the PHD domain from the KAP-1 corepressor: structural determinants for PHD, RING and LIM zinc-binding domains. EMBO J. 20:165–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Borden, K.L., J.M. Lally, S.R. Martin, N.J. O'Reilly, E. Solomon, and P.S. Freemont. 1996. In vivo and in vitro characterization of the B1 and B2 zinc-binding domains from the acute promyelocytic leukemia protooncoprotein PML. Proc. Natl. Acad. Sci. USA. 93:1601–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scheel, H., and K. Hofmann. 2003. No evidence for PHD fingers as ubiquitin ligases. Trends Cell Biol. 13:285–288. [DOI] [PubMed] [Google Scholar]
- 17.Boname, J.M., and P.G. Stevenson. 2001. MHC class I ubiquitination by a viral PHD/LAP finger protein. Immunity. 15:627–636. [DOI] [PubMed] [Google Scholar]
- 18.Coscoy, L., and D. Ganem. 2003. PHD domains and E3 ubiquitin ligases: viruses make the connection. Trends Cell Biol. 13:7–12. [DOI] [PubMed] [Google Scholar]
- 19.Lu, Z., S. Xu, C. Joazeiro, M.H. Cobb, and T. Hunter. 2002. The PHD domain of MEKK1 acts as an E3 ubiquitin ligase and mediates ubiquitination and degradation of ERK1/2. Mol. Cell. 9:945–956. [DOI] [PubMed] [Google Scholar]
- 20.Pitkänen, J., V. Doucas, T. Sternsdorf, T. Nakajima, S. Aratani, K. Jensen, H. Will, P. Vahamurto, J. Ollila, M. Vihinen, et al. 2000. The autoimmune regulator protein has transcriptional transactivating properties and interacts with the common coactivator CREB-binding protein. J. Biol. Chem. 275:16802–16809. [DOI] [PubMed] [Google Scholar]
- 21.Kumar, P.G., M. Laloraya, C.Y. Wang, Q.G. Ruan, A. Davoodi-Semiromi, K.J. Kao, and J.X. She. 2001. The autoimmune regulator (AIRE) is a DNA-binding protein. J. Biol. Chem. 276:41357–41364. [DOI] [PubMed] [Google Scholar]
- 22.Hatakeyama, S., M. Yada, M. Matsumoto, N. Ishida, and K.I. Nakayama. 2001. U box proteins as a new family of ubiquitin-protein ligases. J. Biol. Chem. 276:33111–33120. [DOI] [PubMed] [Google Scholar]
- 23.Kasai, M., K. Hirokawa, K. Kajino, K. Ogasawara, M. Tatsumi, E. Hermel, J.J. Monaco, and T. Mizuochi. 1996. Difference in antigen presentation pathways between cortical and medullary thymic epithelial cells. Eur. J. Immunol. 26:2101–2107. [DOI] [PubMed] [Google Scholar]
- 24.Björses, P., M. Halonen, J.J. Palvimo, M. Kolmer, J. Aaltonen, P. Ellonen, J. Perheentupa, I. Ulmanen, and L. Peltonen. 2000. Mutations in the AIRE gene: effects on subcellular location and transactivation function of the autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy protein. Am. J. Hum. Genet. 66:378–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pitkänen, J., P. Vahamurto, K. Krohn, and P. Peterson. 2001. Subcellular localization of the autoimmune regulator protein. Characterization of nuclear targeting and transcriptional activation domain. J. Biol. Chem. 276:19597–19602. [DOI] [PubMed] [Google Scholar]
- 26.Meetei, A.R., J.P. de Winter, A.L. Medhurst, M. Wallisch, Q. Waisfisz, H.J. van de Vrugt, A.B. Oostra, Z. Yan, C. Ling, C.E. Bishop, et al. 2003. A novel ubiquitin ligase is deficient in Fanconi anemia. Nat. Genet. 35:165–170. [DOI] [PubMed] [Google Scholar]
- 27.Gibbons, R.J., S. Bachoo, D.J. Picketts, S. Aftimos, B. Asenbauer, J. Bergoffen, S.A. Berry, N. Dahl, A. Fryer, K. Keppler, et al. 1997. Mutations in transcriptional regulator ATRX establish the functional significance of a PHD-like domain. Nat. Genet. 17:146–148. [DOI] [PubMed] [Google Scholar]
- 28.Linder, B., R. Newman, L.K. Jones, S. Debernardi, B.D. Young, P. Freemont, C.P. Verrijzer, and V. Saha. 2000. Biochemical analyses of the AF10 protein: the extended LAP/PHD-finger mediates oligomerisation. J. Mol. Biol. 299:369–378. [DOI] [PubMed] [Google Scholar]
- 29.Gunduz, M., M. Ouchida, K. Fukushima, H. Hanafusa, T. Etani, S. Nishioka, K. Nishizaki, and K. Shimizu. 2000. Genomic structure of the human ING1 gene and tumor-specific mutations detected in head and neck squamous cell carcinomas. Cancer Res. 60:3143–3146. [PubMed] [Google Scholar]
- 30.Gozani, O., P. Karuman, D.R. Jones, D. Ivanov, J. Cha, A.A. Lugovskoy, C.L. Baird, H. Zhu, S.J. Field, S.L. Lessnick, et al. 2003. The PHD finger of the chromatin-associated protein ING2 functions as a nuclear phosphoinositide receptor. Cell. 114:99–111. [DOI] [PubMed] [Google Scholar]