Abstract
Although all three Vav family members are expressed in T lymphocytes, the role that Vav3 plays in T cell activation is poorly defined. Here we show that, like Vav1, Vav3 undergoes rapid tyrosine phosphorylation after T cell receptor (TCR) cross-linkage and interacts with the adaptor molecules SLP76 and 3BP2 in a SH2-dependent manner. However, depletion of Vav1 but not Vav3 protein by RNA interference affects TCR-mediated IL-2 promoter activity. In contrast, Vav3 function is specifically required for coupling TCR stimulation to serum response element–mediated gene transcription. These data indicate that, although both Vav proteins are biochemically coupled to the TCR, they regulate distinct molecular pathways leading to defined gene transcriptional events.
Keywords: T cell, signal transduction, Vav, GEF, SRE
Introduction
The engagement of the TCR with MHC–peptide complexes on antigen-presenting cells leads to the activation of proximal protein tyrosine kinases, resulting in the tyrosine phosphorylation of multiple membrane-bound and cytosolic proteins (1). One signaling protein that undergoes phosphorylation within seconds of TCR cross-linkage is the Vav1 proto-oncogene (2). Vav1 is a member of the Dbl superfamily of Rho/Rac guanine nucleotide exchange factors that is expressed primarily in cells of the hematopoietic lineage (3). The importance of Vav1 in the regulation of T cell activation is highlighted by defects in both thymocyte development and peripheral T cell activation in vav1−/− mice (4). Moreover, studies in a Vav1-deficient Jurkat T cell line, J.Vav1, demonstrate that Vav1 couples TCR/CD28 signals to pathways leading to transcriptional regulation of the IL-2 promoter (5). Two additional members of the Vav family of guanine nucleotide exchange factor (Vav2 and Vav3) have been identified that are also expressed in hematopoietic cell lineages and undergo TCR-induced tyrosine phosphorylation (6–9). However, whether human Vav2 or Vav3 act as functionally redundant or distinct signaling proteins during T cell activation is not known.
Although previous studies have demonstrated that murine Vav3 undergoes TCR-induced tyrosine phosphorylation in the Jurkat T cell line when ectopically expressed (8), the role of human Vav3 in the regulation of T cell activation is poorly understood. To begin to elucidate the role of Vav3 in signaling downstream of the TCR, we have studied Vav3 in the Jurkat T cell line using biochemical approaches and small-interfering RNA-mediated gene silencing. The results of these studies indicate that Vav1 and Vav3 couple TCR-induced signals to distinct molecular pathways leading to IL-2 promoter activation and serum response element (SRE)–dependent gene transcription, respectively.
Materials and Methods
Reagents and Plasmids.
All reagents are from Sigma-Aldrich unless otherwise specified. The antisera against Vav, SLP76, and 3BP2 have been described previously (5, 9–11). The anti-CD28 mAb was purchased from BD Biosciences, and the antiphosphotyrosine mAb (4G10) was purchased from Upstate Biotechnology.
The expression plasmids for Vav3, SLP76, and 3BP2 and the luciferase reporter constructs have been described previously (5, 10, 11). The SRE- and ternary complex factor (TCF)–luciferase reporter plasmids were provided by Dr. Ralf Janknecht (Mayo Clinic). The serum response factor (SRF)–luciferase reporter plasmid was obtained from Stratagene. The RNA targeting vector was generated as previously described (12). The 19-nucleotide sequence used to target Vav1 and Vav3 mRNA is TCTCTACCAGGTCTTCATC and GCTTTGTCTAACATAAGAC, respectively. Mutant targeting constructs were also generated to Vav1 (TgTCTAaCAGGTCTcCATC) and Vav3 (GCTaTGTcTAtCATAAGAC) as controls.
Cell Culture, Transfection, and Stimulation.
All cells were grown and transfected as described previously (5). For luciferase reporter assays, cells (106 cells) were distributed in triplicate in 24-well plates and stimulated as indicated. Samples were harvested and prepared for luciferase assays according to the protocol suggested by the manufacturer (Promega). All reporter assays were cotransfected with a pRL-TK reporter plasmid (Promega) to control for intersample variations in transfection efficiency. In the latter case, firefly and pRL-TK–derived Renilla luciferase activities were measured in each sample with a Dual Luciferase Assay kit (Promega).
Immunoblot Analysis.
Jurkat T cells were used directly or were electroporated with the indicated expression constructs. Following electroporation, the cells were lysed as described previously (5). Endogenous or FLAG-tagged proteins were immunoprecipitated from the lysate, washed, eluted in 40 μl of SDS sample buffer, resolved by SDS-PAGE, and transferred to Immobilon-P membranes (Millipore). FLAG-tagged and tyrosine-phosphorylated proteins were detected using anti-pTyr and anti-FLAG mAb followed by goat anti–mouse IgG coupled to horseradish peroxidase (Santa Cruz Biotechnology, Inc.) and the SuperSignal detection system was from Pierce Chemical Co. Endogenous Vav, SLP-76, and GST-bound proteins were detected using specific polyclonal rabbit antisera followed by protein A linked to horseradish peroxidase (Amersham Biosciences) and SuperSignal. In some instances, GST fusion proteins containing the SH2 domain of either, Vav1, Vav2, or Vav3 were used to immunoprecipitate interacting proteins from TCR-stimulated Jurkat T cells as described previously (13).
Results and Discussion
Vav3 Links to the TCR and Interacts with SLP-76.
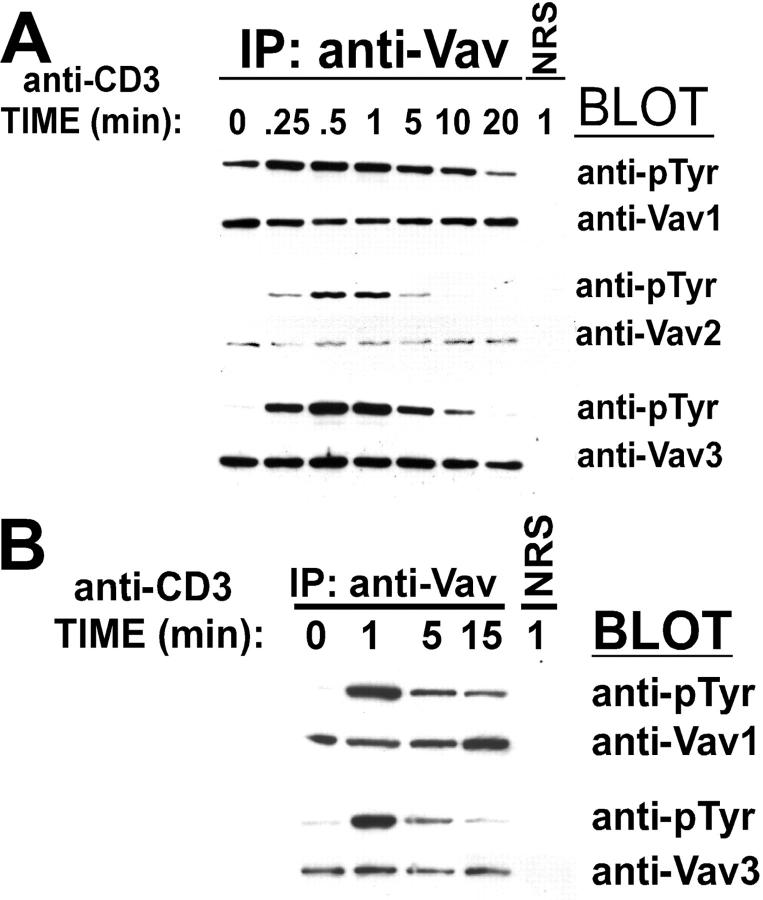
To determine the role of Vav3 in TCR signaling, we initially compared the kinetics of tyrosine phosphorylation of the three distinct Vav isoforms after TCR cross-linkage in the Jurkat T cell line. Although basal levels of Vav1 tyrosine phosphorylation are apparent, there is little or no detectable basal tyrosine phosphorylation of Vav2 or Vav3 (Fig. 1 A). However, upon TCR cross-linking all three Vav family members undergo rapid tyrosine phosphorylation with peak phosphorylation attained at 1 min poststimulation and a gradual decline in phosphotyrosine content observed at later time points. Similar results were observed when Vav1 and Vav3 tyrosine phosphorylation was measured after TCR cross-linkage in a CD4+ human T cell clone (Fig. 1 B). Thus, all three Vav proteins couple to the TCR and show similar kinetics of tyrosine phosphorylation.
Figure 1.
Vav proteins couple to the T cell receptor. (A) Jurkat T cells or (B) a CD4+ T cell clone were treated as indicated. The different Vav isoforms were specifically immunoprecipitated (Vav1, top; Vav2, middle; and Vav3, bottom) using Vav-specific polyclonal rabbit antisera. The immunoprecipitates were resolved by SDS-PAGE, transferred to a nylon membrane, and probed with anti-pTyr (top panel in each set) or anti-Vav (bottom panel in each set).
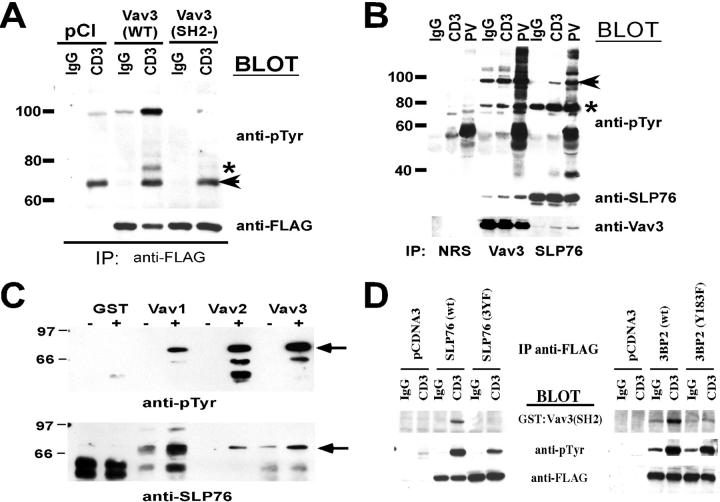
To determine if the SH2 domain of Vav3 is required for its tyrosine phosphorylation downstream of the TCR, we transfected Jurkat T cells with either a WT or SH2 domain mutant of Vav3 and analyzed TCR-induced tyrosine phosphorylation of the two proteins. As shown in Fig. 2 A, the mutant SH2 domain–containing Vav3 protein fails to undergo TCR-mediated tyrosine phosphorylation. This was not the result of different levels of protein expression since both Vav3 proteins were equivalently expressed.
Figure 2.
Vav3 interacts with SLP76 and 3BP2. (A) Jurkat T cells were transfected with the indicated expression vectors. Cells were stimulated as indicated, cell lysates were prepared, and FLAG-tagged protein was immunoprecipitated. Immunoprecipitates were analyzed as described above and probed for anti-pTyr (top) and anti-FLAG (bottom). The asterisk denotes an interacting protein of ∼76 kD. The arrow denotes a nonspecific band. (B) Jurkat T cells were treated as indicated, and cell lysates were prepared and subject to immunoprecipitation with NRS, anti-Vav3, or anti-SLP76 polyclonal rabbit antisera. Proteins were detected using anti-pTyr (top), anti-SLP76 (middle), or anti-Vav3 (bottom). In the anti-pTyr blot, the asterisk denotes SLP76, and the arrow identifies Vav3. (C) GST or GST fusion proteins containing the Vav1, Vav2, or Vav3 SH2 domains were used to immunoprecipitate interacting proteins from unstimulated (−) or CD3-stimulated (+) Jurkat T cells. Interacting proteins were analyzed using anti-pTyr (top) or anti-SLP76 (bottom). The arrow denotes the SLP76 band. (D) Jurkat T cells were transfected with the indicated expression vectors. Cells were treated as in B, and the membrane was subsequently probed with the GST–Vav3–SH2 (1 μg/ml) and detected with anti-GST (top), anti-pTyr (middle), and anti-FLAG (bottom).
It is of interest that this mutant protein fails to interact with a tyrosine-phosphorylated protein with apparent molecular weight of 76 kD (Fig. 2 B, asterisk). Since Vav1 and Vav2 have been observed to interact with SLP76 (14, 15), we questioned whether Vav3 also interacts with SLP76 downstream of the TCR. Anti-Vav3 or anti-SLP76 antibodies were incubated with detergent extracts from Jurkat T cells that were left unstimulated or stimulated by anti-CD3 or treatment with pervanadate. As shown in Fig. 2 B, immunoprecipitation of Vav3 after TCR cross-linkage results in an inducible association with SLP76. In addition, Vav3 was also observed in SLP76 immunoprecipitates (Fig. 2 B, bottom), indicating that these two proteins become physically associated after TCR cross-linking. Given that the Vav1 SH2 domain is important for linking Vav1 to the TCR and SLP76 (14), we determined if the interaction of Vav3 with SLP76 was mediated through the Vav3 SH2 domain using GST fusion proteins. Consistent with previous results, GST fusion proteins containing the Vav1 and Vav2 SH2 domains captured SLP76 in extracts from anti-CD3–stimulated T cells (Fig. 2 C). Similarly, the Vav3 SH2 domain fusion protein interacted with SLP76 after TCR cross-linking (Fig. 2 C), indicating that SLP76 is a common target for the SH2 domain of all three Vav isoforms. Note that some SLP76 was pulled down in the absence of stimulation but was not detected on the anti-pTyr blot, most likely due to the level of tyrosine phosphorylation. Therefore, like Vav1 and Vav2, Vav3 has the potential to interact with SLP76. It is likely, that the SH2 domain of all three Vav proteins interact with an overlapping set of tyrosine molecules that conform to the (p)YXEP motif (where X is either M, L, or E) (3).
Lastly, in order to demonstrate that this interaction is direct we performed a “Far-Western” blot using the GST–Vav3–SH2 fusion protein as a probe on SLP76 immunoprecipitates from TCR-stimulated cells that had been transfected with either WT SLP76 or a mutant SLP76 in which all three tyrosines within the acidic region were mutated to phenylalanine (3YF). As shown in Fig. 2 D, the Vav3 SH2 domain GST fusion protein specifically interacted with the WT SLP76 protein. However, consistent with the mechanism of the Vav1-SLP76 interaction, Vav3 did not interact with the 3YF mutant protein. Thus, similar to what has been described for Vav1, the interaction of Vav3 with SLP76 is mediated through tyrosine-phosphorylated residues within the SLP76 acidic region (Y112, Y128, and Y145) (16, 17). In addition to SLP76, we also found that the SH2 domain of Vav3 directly interacts with the adaptor protein 3BP2, a protein involved in TCR-mediated nuclear factor of activated T cells (NFAT)/AP-1 activity and NK cell-mediated natural cytotoxicity (11, 18). Consistent with our previous observations regarding the interaction of Vav1 and Vav2 with 3BP2 (11), the Vav3–3BP2 interaction primarily occurs through Y183, since we observed a significant reduction in binding of the Vav3-SH2 domain when this residue was mutated to phenylalanine (Fig. 2 D). The significance of Y183 in 3BP2 is highlighted by the inability of the Y183F mutant protein to augment cell-mediated killing when expressed in NK cells (11). Whether Vav3 couples to 3BP2 in NK cells and regulates NK cell killing remains to be established. Together, these data show that the SH2 domain of Vav3, like that of Vav1 and Vav2, can couple this family of signaling mediators with an overlapping set of adaptor proteins downstream of the TCR.
Suppression of Vav3 Has No Effect on TCR-induced IL-2 Promoter Activity.
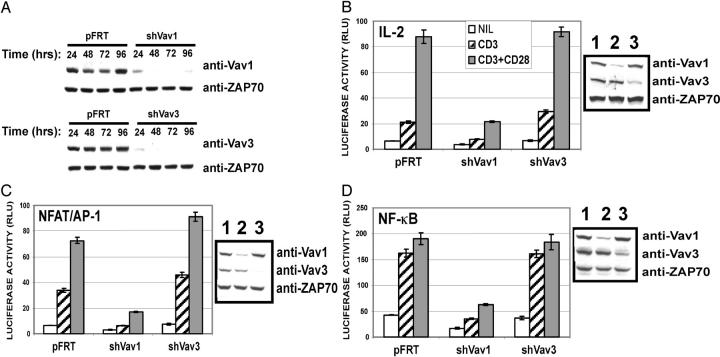
The above results suggest that Vav1 and Vav3 couple to overlapping pathways in T cells after TCR cross-linkage. However, it is clear that Vav3 is not functionally redundant with Vav1, since T cell activation pathways are severely compromised in vav1−/− murine T cells and IL-2 promoter activity is severely impaired in J.Vav1 cells after activation (5). Thus, it is possible that Vav3 does not normally regulate the molecular pathways that activate the IL-2 promoter after TCR ligation. To test this, we generated short-hairpin RNA interference vectors that specifically target either Vav1 (shVav1) or Vav3 (shVav3). As shown in Fig. 3 A, when Vav1 or Vav3-targeting vectors are transiently transfected into Jurkat T cells we observe a significant loss of Vav1 and Vav3 protein up to 96 h posttransfection.
Figure 3.
Vav1 but not Vav3 links TCR-induced signals to IL-2 promoter activity. (A) Jurkat T cells were transfected with either a control suppression vector (pFRT) or a Vav1 (shVav1) or Vav3 (shVav3) RNA targeting vector. Cell lysates were prepared at the indicated time intervals posttransfection, and 100 μg of total protein was immunoblotted with anti-Vav1 (top) and anti-Vav3 (bottom) or anti-ZAP70 as a loading control. (B–D) Jurkat T cells were transfected with luciferase reporter containing the IL-2p (B), NFAT/AP-1 (C), or NF-κB (D), and the indicated RNA suppression constructs. 36 h posttransfection, cells were treated, and luciferase activity was measured as indicated in Materials and Methods. Bars represent the mean ± SD from triplicate samples. Western blotting of 100 μg of protein prepared from the indicated transfected cell population was performed to demonstrate the depletion of the indicated Vav isoform.
To investigate if depletion of Vav3 has a similar effect on IL-2 promoter activity to that of Vav1, we cotransfected Jurkat T cells with the full-length IL-2 promoter reporter construct and the indicated RNA targeting vectors and measured luciferase activity after CD3 or CD3/CD28 cross-linking. Consistent with data from the J.Vav1 cell line, depletion of Vav1 severely compromises CD3 and CD3/CD28-mediated IL-2 promoter activity when compared with the control transfected population (Fig. 3 A) (5). In stark contrast, Jurkat T cells in which Vav3 protein had been depleted by RNA interference showed no demonstrable defect in either CD3 or CD3/CD28-mediated IL-2 promoter activity (Fig. 3 A). Moreover, whereas depletion of Vav1 impacted on NFAT/AP-1–, NF-κB–, and AP-1–mediated gene transcription, Vav3 depletion had no effect (Fig. 3, C and D; not depicted). Previous data suggest that when overexpressed Vav3 is capable of augmenting NF-κB activity downstream of the TCR in Jurkat T cells (8) and functionally substituting for Vav1 in the J.Vav1 cell line (5). Yet, the results presented here suggest that Vav3 does not share with Vav1 the ability to transduce TCR-initiated signals leading to the activation of transcription factors that are required for IL-2 gene transcription. Moreover, these results suggest that caution be taken when analyzing data using overexpression, since such experimental systems may not always fairly recapitulate the normal role of a signaling protein. However, using the J.Vav1 cell line we have been able to further diminish TCR-induced IL-2p activity after the suppression of Vav3 by RNA interference (unpublished data), indicating that Vav1 is the dominant Vav family member in this pathway but that Vav3 can couple, although weakly, some TCR-induced signals leading to IL-2 promoter activity. Consistent with this notion, a recent study of Vav-deficient animals has demonstrated that the vav1/3−/− T cell phenotype is worse than vav1−/− T cells (19).
Vav3 Couples TCR-induced Signaling Pathways to SRE-mediated Gene Transcription.
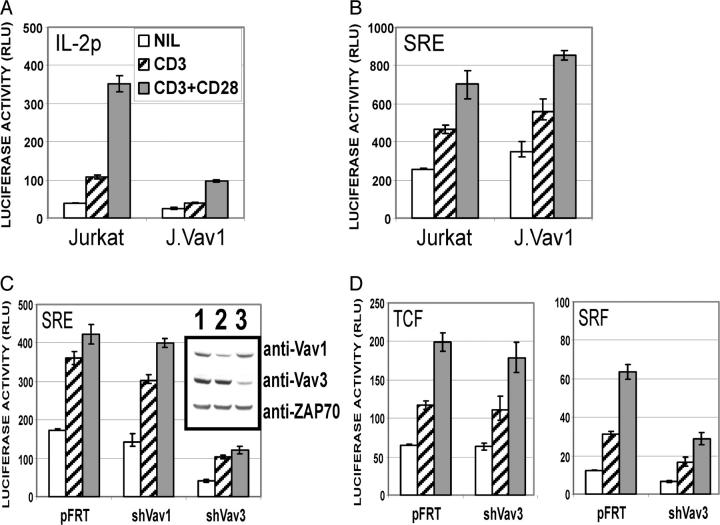
Vav proteins have been suggested to activate an overlapping set of Rho family GTP-binding proteins (20). This family of GTP-binding proteins regulate downstream effector pathways that modulate SRE-dependent gene transcription (21, 22). Efficient transactivation of the SRE requires the binding of SRF and the ternary TCF complex (23). It is of interest that the activation of both transcription factors requires phosphorylation events mediated in part by members of the MAPK family (24). Indeed, T cells from vav1−/− mice fail to activate ERK in response to low levels of TCR stimulation (25). Moreover, previous studies in Jurkat have shown that SRE-dependent transcription after TCR cross-linking is enhanced by the overexpression of WT but not dominant-negative forms of murine Vav1 and murine Vav2 (15, 26). To more directly assess the role of Vav1 in controlling SRE-dependent gene transcription downstream of the TCR, we initially assessed SRE-dependent gene transcription in the J.Vav1 cell line. As shown in Fig. 4 A, although IL-2 promoter activity induced by CD3 or CD3/CD28 ligation is severely compromised in the J.Vav1 cell line (Fig. 4 A, compare Jurkat to J.Vav1), we observed no defect in either basal level or TCR-induced SRE-mediated gene transcription (Fig. 4 B, compare Jurkat to J.Vav1). However, it remained possible that the J.Vav1 clone retained TCR-induced SRE-mediated gene transcription as a consequence of selection during the generation of this cell line. To determine if this was the case, we analyzed SRE-mediated gene transcription in Jurkat T cells in which Vav1 was depleted by RNA interference. Consistent with the J.Vav1 cells, depletion of Vav1 by RNA interference in the Jurkat T cell line had minimal to no effect on TCR-induced SRE-mediated gene transcription (Fig. 4 C). These data indicate that caution must be taken when overexpressing dominant-negative forms of Vav proteins as they have the potential to “poison” normal signaling complexes and nonspecifically block signaling pathways in which they are not a normal component.
Figure 4.
Vav3 couples TCR-initiated signaling to the regulation of SRE-mediated gene transcription. (A) Jurkat and J.Vav1 T cells were transfected with luciferase reporter containing the IL-2 promoter, stimulated, and analyzed as indicated in Fig. 3. (B) Jurkat and J.Vav1 T cells were transfected with luciferase reporter containing the c-fos SRE stimulated and analyzed as indicated in Fig. 3. (C and D) Jurkat were transfected with luciferase reporter containing the SRE (C), TCF, or SRF (D) and the indicated RNA suppression constructs, stimulated, and luciferase activity was measured as described in Fig. 3. Bars represent the mean ± SD from triplicate samples. Western blotting of protein lysates is as described in Fig. 3.
To investigate the potential role of Vav3 in SRE-mediated gene transcription, we depleted Vav3 by RNA interference and measured SRE-luciferase activity. As shown in Fig. 4 C, RNA interference of Vav3 severely compromises basal and TCR-induced SRE-dependent gene transcription (Fig. 4 C). To further delineate the mechanism by which Vav3 suppression was affecting SRE-mediated gene transcription, we assessed the ability of control-transfected or Vav3-suppressed Jurkat T cells to regulate either TCF-dependent or SRF-dependent gene transcription. As shown in Fig. 4 D, depletion of Vav3 protein had no effect on the TCF reporter but did diminish the basal and TCR-induced activation of the SRF-specific promoter. The data presented in Fig. 4, B and C, would suggest that Vav1 is not necessary to couple TCR-initiated signaling pathways to the regulation of SRE-mediated gene transcription, whereas Vav3 is centrally involved in regulating the SRF transcription factor after TCR cross-linkage. In fact, it has been shown recently that SRF-dependent gene transcription in B cells is JNK independent but ERK dependent (27). Although our data clearly link Vav3-mediated signaling events to SRF activity, we have not observed any defect in ERK activation after TCR cross-linking of Vav3-depleted Jurkat T cells (unpublished data). However, it remains possible that Vav3 is controlling a signaling pathway downstream of the TCR that regulates SRF activity in a MAPK-independent manner. Thus, it will be important to determine which pathway leading to SRF activation is impaired in Vav3-depleted Jurkat T cells.
In conclusion, we have identified Vav1 and Vav3 as positive regulators of distinct TCR-induced transcriptional events leading to IL-2 promoter activity and SRE-dependent gene transcription, respectively. It will be important to determine the pathways controlled by Vav3 leading to SRE-mediated gene transcription and to determine if the pathways activated by Vav3 are in fact distinct from those activated by Vav1. Moreover, our data clearly indicate that, although overexpression studies can give initial insights into protein function, acute gene silencing with small-interfering RNA represents a more definitive epigenetic strategy for the functional characterization of TCR-linked signaling proteins in human T cell lines.
Acknowledgments
This work was supported by funding from the Medical Research Council and the Biotechnology and Biological Sciences Research Council (to M. Turner), and the Mayo Foundation and a Cancer Research Institute Investigator award (to D.D. Billadeau).
References
- 1.Koretzky, G.A., F. Abtahian, G.S. Derimanov, S.A. Dmowski, A. Guerriero, M.S. Jordan, J.S. Maltzman, B.A. Olenchock, and A.L. Singer. 2003. Regulation of hematopoietic cell development and activation by adapter proteins. Immunol. Res. 27:357–366. [DOI] [PubMed] [Google Scholar]
- 2.Bustelo, X.R., J.A. Ledbetter, and M. Barbacid. 1992. Product of vav proto-oncogene defines a new class of tyrosine protein kinase substrates. Nature. 356:68–71. [DOI] [PubMed] [Google Scholar]
- 3.Turner, M., and D.D. Billadeau. 2002. VAV proteins as signal integrators for multi-subunit immune-recognition receptors. Nat. Rev. Immunol. 2:476–486. [DOI] [PubMed] [Google Scholar]
- 4.Turner, M., P.J. Mee, A.E. Walters, M.E. Quinn, A.L. Mellor, R. Zamoyska, and V.L. Tybulewicz. 1997. A requirement for the Rho-family GTP exchange factor Vav in positive and negative selection of thymocytes. Immunity. 7:451–460. [DOI] [PubMed] [Google Scholar]
- 5.Cao, Y., E.M. Janssen, A.W. Duncan, A. Altman, D.D. Billadeau, and R.T. Abraham. 2002. Pleiotropic defects in TCR signaling in a Vav-1-null Jurkat T-cell line. EMBO J. 21:4809–4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schuebel, K.E., X.R. Bustelo, D.A. Nielsen, B.J. Song, M. Barbacid, D. Goldman, and I.J. Lee. 1996. Isolation and characterization of murine Vav-2, a member of the Vav family of proto-oncogenes. Oncogene. 13:363–371. [PubMed] [Google Scholar]
- 7.Movilla, N., and X.R. Bustelo. 1999. Biological and regulatory properties of Vav-3, a new member of the Vav family of oncoproteins. Mol. Cell. Biol. 19:7870–7885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moores, S.L., L.M. Selfors, J. Fredericks, T. Breit, K. Fujikawa, F.W. Alt, J.S. Brugge, and W. Swat. 2000. Vav family proteins couple to diverse cell surface receptors. Mol. Cell. Biol. 20:6364–6373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Billadeau, D.D., S.M. Mackie, R.A. Schoon, and P.J. Leibson. 2000. The Rho family guanine nucleotide exchange factor Vav-2 regulates the development of cell-mediated cytotoxicity. J. Exp. Med. 192:381–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Binstadt, B.A., D.D. Billadeau, D. Jevremovic, B.L. Williams, N. Fang, T. Yi, G.A. Koretzky, R.T. Abraham, and P.J. Leibson. 1998. SLP-76 is a direct substrate of SHP-1 recruited to killer cell inhibitory receptors. J. Biol. Chem. 273:27518–27523. [DOI] [PubMed] [Google Scholar]
- 11.Jevremovic, D., D.D. Billadeau, R.A. Schoon, C.J. Dick, and P.J. Leibson. 2001. Regulation of NK cell-mediated cytotoxicity by the adaptor protein 3BP2. J. Immunol. 166:7219–7228. [DOI] [PubMed] [Google Scholar]
- 12.Brummelkamp, T.R., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science. 296:550–553. [DOI] [PubMed] [Google Scholar]
- 13.Doody, G.M., D.D. Billadeau, E. Clayton, A. Hutchings, R. Berland, S. McAdam, P.J. Leibson, and M. Turner. 2000. Vav-2 controls NFAT-dependent transcription in B- but not T-lymphocytes. EMBO J. 19:6173–6184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu, J., D.G. Motto, G.A. Koretzky, and A. Weiss. 1996. Vav and SLP-76 interact and functionally cooperate in IL-2 gene activation. Immunity. 4:593–602. [DOI] [PubMed] [Google Scholar]
- 15.Tartare-Deckert, S., M.-N. Monthouel, C. Charvet, I. Foucault, E. Van Obberghen, A. Bernard, A. Altman, and M. Deckert. 2001. Vav2 Activates c-fos serum response element and CD69 expression but negatively regulates nuclear factor of activated T cells and interleukin-2 gene activation in T lymphocytes. J. Biol. Chem. 276:20849–20857. [DOI] [PubMed] [Google Scholar]
- 16.Fang, N., and G.A. Koretzky. 1999. SLP-76 and Vav function in separate, but overlapping pathways to augment interleukin-2 promoter activity. J. Biol. Chem. 274:16206–16212. [DOI] [PubMed] [Google Scholar]
- 17.Raab, M., A.J. da Silva, P.R. Findell, and C.E. Rudd. 1997. Regulation of Vav-SLP-76 binding by ZAP-70 and its relevance to TCR zeta/CD3 induction of interleukin-2. Immunity. 6:155–164. [DOI] [PubMed] [Google Scholar]
- 18.Deckert, M., S. Tartare-Deckert, J. Hernandez, R. Rottapel, and A. Altman. 1998. Adaptor function for the Syk kinases-interacting protein 3BP2 in IL-2 gene activation. Immunity. 9:595–605. [DOI] [PubMed] [Google Scholar]
- 19.Fujikawa, K., A.V. Miletic, F.W. Alt, R. Faccio, T. Brown, J. Hoog, J. Fredericks, S. Nishi, S. Mildiner, S.L. Moores, et al. 2003. Vav1/2/3-null mice define an essential role for Vav family proteins in lymphocyte development and activation but a differential requirement in MAPK signaling in T and B cells. J. Exp. Med. 198:1595–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bustelo, X.R. 2001. Vav proteins, adaptors and cell signaling. Oncogene. 20:6372–6381. [DOI] [PubMed] [Google Scholar]
- 21.Treisman, R., A.S. Alberts, and E. Sahai. 1998. Regulation of SRF activity by Rho family GTPases. Cold Spring Harb. Symp. Quant. Biol. 63:643–651. [DOI] [PubMed] [Google Scholar]
- 22.Hill, C.S., J. Wynne, and R. Treisman. 1995. The Rho family GTPases RhoA, Rac1, and CDC42Hs regulate transcriptional activation by SRF. Cell. 81:1159–1170. [DOI] [PubMed] [Google Scholar]
- 23.Treisman, R. 1995. Journey to the surface of the cell: Fos regulation and the SRE. EMBO J. 14:4905–4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Price, M.A., F.H. Cruzalegui, and R. Treisman. 1996. The p38 and ERK MAP kinase pathways cooperate to activate ternary complex factors and c-fos transcription in response to UV light. EMBO J. 15:6552–6563. [PMC free article] [PubMed] [Google Scholar]
- 25.Costello, P.S., A.E. Walters, P.J. Mee, M. Turner, L.F. Reynolds, A. Prisco, N. Sarner, R. Zamoyska, and V.L.J. Tybulewicz. 1999. The Rho-family GTP exchange factor Vav is a critical transducer of T cell receptor signals to the calcium, ERK, and NF-kappaB pathways. Proc. Natl. Acad. Sci. USA. 96:3035–3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Charvet, C., P. Auberger, S. Tartare-Deckert, A. Bernard, and M. Deckert. 2002. Vav1 couples T cell receptor to serum response factor-dependent transcription via a MEK-dependent pathway. J. Biol. Chem. 277:15376–15384. [DOI] [PubMed] [Google Scholar]
- 27.Hao, S., T. Kurosaki, and A. August. 2003. Differential regulation of NFAT and SRF by the B cell receptor via a PLCγ-Ca2+-dependent pathway. EMBO J. 22:4166–4177. [DOI] [PMC free article] [PubMed] [Google Scholar]