Abstract
How hematopoietic stem cells (HSCs) commit to a particular lineage is unclear. A high degree of HSC purification enabled us to address this issue at the clonal level. Single-cell transplantation studies revealed that 40% of the CD34−/low, c-Kit+, Sca-1+, and lineage marker− (CD34−KSL) cells in adult mouse bone marrow were able, as individual cells, to reconstitute myeloid and B- and T-lymphoid lineages over the long-term. Single-cell culture showed that >40% of CD34−KSL cells could form neutrophil (n)/macrophage (m)/erythroblast (E)/megakaryocyte (M) (nmEM) colonies. Assuming that a substantial portion of long-term repopulating cells can be detected as nmEM cells within this population, we compared differentiation potentials between individual pairs of daughter and granddaughter cells derived in vitro from single nmEM cells. One of the two daughter or granddaughter cells remained an nmEM cell. The other showed a variety of combinations of differentiation potential. In particular, an nmEM cell directly gave rise, after one cell division, to progenitor cells committed to nm, EM, or M lineages. The probability of asymmetric division of nmEM cells depended on the cytokines used. These data strongly suggest that lineage commitment takes place asymmetrically at the level of HSCs under the influence of external factors.
Keywords: hematopoiesis, cell differentiation, cell lineage, cell division, cytokines
Introduction
One of the central tasks of stem cell biology is to understand the mechanisms that regulate lineage commitment of stem cells (1–3). Despite the fact that hematopoietic stem cells (HSCs) are the best characterized stem population (4), how they differentiate is poorly understood. Several models have been proposed to explain HSC fate determination. Analyses of in vivo as well as in vitro colony-forming cells (CFCs), particularly CFCs obtained from blast cell colonies, have suggested a stochastic model for HSC behavior (5–13). Consistent with this model, a permissive role for cytokines has also been suggested (12, 14, 15). The classic hematopoietic inductive microenvironment (16) and stem cell competition models (17) have held that extrinsic factors play an instructive role. This idea has been supported by lineage analysis of the progeny of CFCs (18–20). Because no previous paper has verified these working models by directly examining HSCs, whether extrinsic signaling plays any role in HSC lineage commitment remains controversial.
Fate decision in HSCs (self-renewal, differentiation, or apoptosis) takes place through their cell division. Therefore, to study lineage commitment in HSCs, we sought to determine the differentiation potential of the immediate progeny of HSCs at the clonal level. That HSCs are highly enriched in a population of CD34−/low, c-Kit+, Sca-1+, and lineage marker− (CD34−KSL) cells among bone marrow cells of the adult mouse has enabled both in vitro and in vivo clonal analyses of HSCs (21, 22). Cytokines such as IL-3 and thrombopoietin (TPO) together with stem cell factor (SCF) directly acted on these cells and induced their division (22). After CD34−KSL cells underwent one cell division, stem cell activity became undetectable in their progeny except in limited cases, where this activity was maintained in one of the two daughter cells (22, 23). We assumed that lineage commitment could be responsible for loss of stem cell activity in this setting. To verify this hypothesis, we rigorously examined the differentiation potential of paired daughter cells arisen from single CD34−KSL cells in vitro.
Despite a lack of information on lymphoid differentiation potential, in vitro colony assay permits quantitative evaluation of differentiation potential along neutrophil (n), macrophage (m), erythroblast (E), and megakaryocyte (M) lineages at the single-cell level. In this work, using in vitro colony assays, we demonstrated that lineage commitment of multipotent CD34−KSL cells occurs at the initial stage of their cell divisions, and that this lineage commitment is asymmetric. Treatment with certain cytokines increases the chance that CD34−KSL cells will divide in an asymmetric rather than a symmetric manner, with one daughter cell committed to lineage-specific differentiation.
A preferred model of lineage restriction proposes that nm and EM progenitor cells separately split from a common myeloid progenitor (CMP; reference 24). A multipotent CD34−KSL cell never gave rise to an EM–nm colony pair throughout these studies; in effect, events predicted by the CMP model were not observed. We propose an alternative model for nm and EM lineage restrictions in the HSC compartment.
Materials and Methods
Purification of CD34−KSL Cells.
Bone marrow cells were obtained from 8 to 10-wk-old male C57BL/6 mice congenic for the Ly5 locus (B6-Ly5.1 mice). CD34−KSL cells were purified from bone marrow cells as described previously (25). Bone marrow cells were stained with a lineage marker cocktail consisting of biotinylated anti–Gr-1, –Mac-1, -B220, -CD4, -CD8, and -Ter119 monoclonal antibodies. Lineage marker+ cells were depleted using streptavidin (SA)-coupled magnetic beads (M-280; Dynal). The remaining cells were stained with FITC-conjugated anti-CD34, PE-conjugated anti–Sca-1, and allophycocyanin-conjugated anti–c-Kit antibodies, followed by development with SA–Texas red (Invitrogen). All antibodies were purchased from Becton Dickinson. Four-color analysis and sorting were performed on a FACSVantage™ SE System (Becton Dickinson). CD34−KSL cells were directly sorted into a 96-well plate at 1 cell/well using a FACSVantage™ SE System. To confirm the presence of 1 cell/well, the plate was centrifuged at 1,800 revolutions/min for 5 min, placed in a 37°C incubator for at least 2 h, and observed under inverted microscopy.
Single-cell Reconstitution.
A single CD34−KSL cell isolated from a B6-Ly5.1 mouse was mixed with 2 × 105 bone marrow cells from a B6-Ly5.1/Ly5.2 F1 mouse. The cell mixture was injected into a B6-Ly5.2 mouse irradiated at a dose of 950 cGy. Peripheral blood cells of the recipient mice were stained with biotinylated anti-Ly5.1, FITC-conjugated anti-Ly5.2, allophycocyanin-conjugated anti-B220 antibodies, and a mixture of PE-conjugated anti–Mac-1 and anti–Gr-1 antibodies or a mixture of PE-conjugated anti-CD4 and anti-CD8 antibodies. After biotinylated antibody was developed with SA–Texas red, cells were analyzed by FACS® as described previously (25). The extent of single-cell reconstitution was expressed as (% Ly5.1 cells × 100)/(% Ly5.1 cells + % F1 cells), or percent chimerism. When percent chimerism was >1.0 for all myeloid, B-lymphoid, and T-lymphoid lineages, a test donor cell was judged to have been a long-term multilineage repopulating cell.
Single-cell Culture.
Single-cell cultures of CD34−KSL cells were performed as described previously (22). For induction of early cell divisions, single cells were incubated in serum-free medium (Stempro-34; Invitrogen) with 2 mM l-glutamine, 5 × 10−5 M 2-β-mercaptoethanol, and the following cytokines: 100 ng/ml of mouse SCF, 10 ng/ml of mouse IL-3, and 100 ng/ml of human TPO. For in vitro colony formation, CD34−KSL cells or their progeny were individually cultured for 14 d in the same serum-free medium with SCF, IL-3, and TPO in the same concentrations as aforementioned, additionally with 10% FCS and 2 U/ml human erythropoietin (EPO). Based on colony size, the number of cells per colony was estimated to be 50–100, >100, >103, >104, or >105, followed by cell counts for representative colonies. Single-cell culture has been shown to have an advantage over conventional semisolid culture when frequencies of a variety of CFCs are to be determined. This is because the number of plated cells can be known exactly, and cells composing a colony can be efficiently collected for morphological examination using Cytospin (Shandon) without cross-contamination among colonies.
Micromanipulation of Daughter Cells.
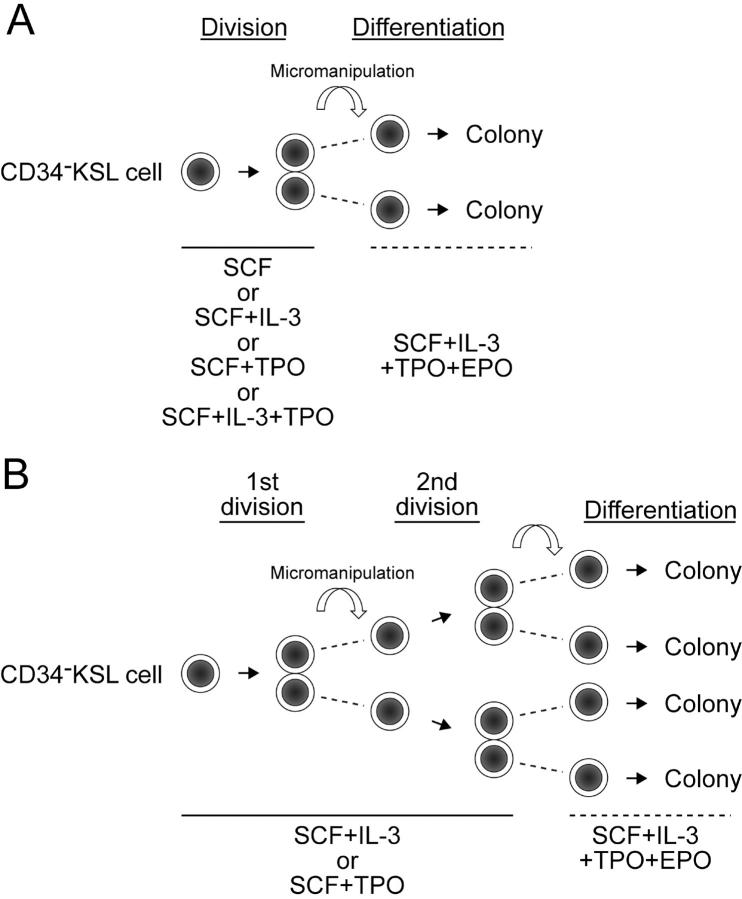
After single cells underwent cell division in the presence of SCF, SCF + IL-3, SCF + TPO, or SCF + IL-3 + TPO by day 5 of culture, one member of the pair of daughter cells was transferred into another well using a micromanipulator (Fig. 1 A). Individual members of daughter cell pairs, in different wells, were cultured in parallel in the presence of SCF + IL-3 + TPO + EPO for 10–14 d. For serial micromanipulation, after single cells gave rise to daughter cells in the presence of either SCF + IL-3 or SCF + TPO, one of the two daughter cells remained in the same well where culture had been initiated, and the other was transferred into a new well containing the same combination of cytokines. After daughter cells underwent cell division, the two cells derived from each daughter cell (granddaughter cells) were separated and continuously cultured in the presence of SCF + IL-3 + TPO + EPO for 10–14 d (Fig. 1 B).
Figure 1.
Micromanipulation of daughter cell pairs and granddaughter cell pairs derived from single CD34−KSL cells in vitro. (A) After single CD34−KSL cells underwent first divisions in the presence of SCF, SCF + IL-3, SCF + TPO, or SCF + IL-3 + TPO, members of daughter cell pairs were separated by micromanipulation and further cultured in the presence of SCF + IL-3 + TPO + EPO to permit full differentiation along myeloid lineage. (B) After single CD34−KSL cells underwent first divisions in the presence of SCF + IL-3 or SCF + TPO, members of daughter cell pairs were separated into wells containing SCF + IL-3 or SCF + TPO. After each daughter cell underwent second division, granddaughter cells were separated and individually cultured in the presence of SCF + IL-3 + TPO + EPO.
Determination of Differentiation Potential.
Cytospin preparations were made for all colonies consisting of ≥50 cells. Cells were stained with May-Gruenwald-Giemsa solution and morphologically identified as n, m, E, M, or blastlike (bl) cells. Two independent persons examined a total of 1,000 cells/colony in most cases; M colonies were an exception, identified in situ when 8–50 typical Ms were detected. The differentiation potential of parent cells was extrapolated backward from the differentiation potentials of daughter cells. For example, when one or two daughter cells gave rise to nmEM colonies, their parent cell was judged to have had the potential to differentiate along nmEM lineages. When one daughter cell gave rise to an nmE colony, and the other gave rise to an nmM colony, the differentiation potential of their parent cell was also considered to have been nmEM. We defined cells with nmEM differentiation potential as uncommitted cells. By contrast, commitment was defined as the event causing cells to lose the potential to differentiate along one or more lineages from among the set of n, m, E, and M. Asymmetric division was defined as cell division, resulting in production of two daughter cells with different differentiation potentials (26). To compare probabilities that asymmetric division would occur under different conditions, Fisher's exact test was used.
Results
Long-Term Multilineage Repopulating Activity in Single CD34−KSL Cells.
Consistent with our previous observations (22), single-cell transplantation assays identified 40% of CD34−KSL cells as long-term multilineage repopulating cells (Table I). In addition, 25% of the cells appeared to be myeloid, B-lymphoid, or T-lymphoid lineage-restricted repopulating cells. When CD34+KSL cells were used as rescue cells, 20% of the CD34−KSL cells contributed to long-term repopulation (21). A certain number of recipient mice died before analysis in such a rescue experiment, perhaps resulting in this lower effective reconstitution rate.
Table I.
Long-Term Repopulation with Single CD34−KSL Cells
| Chimerism in
|
||||
|---|---|---|---|---|
| Repopulated lineage |
No. of mice |
Myeloidlineage | B-lymphoidlineage | T-lymphoidlineage |
| (%) | % | % | % | |
| My/B/T | 8/20 (40) | 53.3 ± 32.8 (n = 8) |
28.4 ± 21.8 | 32.1 ± 23.3 |
| My | 3/20 (15) | 4.1 ± 2.7 (n = 3) |
– | – |
| B | 1/20 (5) | – | 1.5 | – |
| T | 1/20 (5) | – | – | 8.6 |
A single CD34−KSL cell was transplanted into a lethally irradiated mouse together with 2 × 105 competitor cells. Lineage contribution was evaluated 4 mo after transplantation. All myeloid, B-lymphoid, and T-lymphoid lineages (My/B/T) were repopulated with a single cell in 8 out of 20 recipient mice. Repopulation only in myeloid (My), B-lymphoid (B), or T-lymphoid (T) lineage was also observed. Percent chimerism in each lineage is expressed as mean ± SD.
CD34−KSL Cells with nmEM Differentiation Potential.
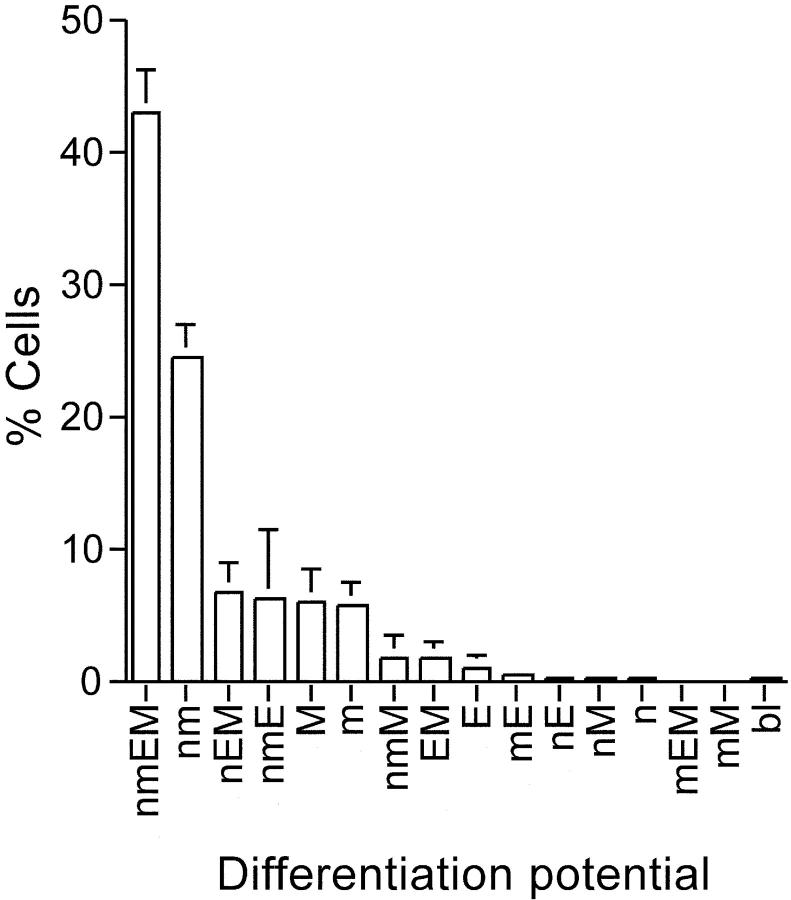
Colony formation by single CD34−KSL cells was examined in the presence of a combination of SCF + IL-3 + TPO + EPO. On average, cells were not found in 2% of the wells due to sorting failure, immediate apoptosis, or adhesion of sorted cells to the wall of the plate. The differentiation potential could not be determined in 2–3% of the cells because they gave rise either to <50 cells or to as many as 1,000 cells with bl cell morphology. The remaining cells formed a variety of colonies as shown in Fig. 2. Approximately 40% of the colonies were classified as nmEM colonies; uni-, bi-, and tripotent progenitor cells were detected less frequently. On average, 98% of the nmEM colonies consisted of >104 cells (unpublished data), suggesting that these CFCs constitute a highly proliferative subset among CD34−KSL cells.
Figure 2.
Colony-forming ability of single CD34−KSL cells. CD34− KSL cells were individually cultured in the presence of SCF, IL-3, TPO, and EPO for 2 wk. Percentages of CFCs with different differentiation potentials are shown based on three independent experiments. Colony cells were morphologically identified as neutrophils (n), macrophages (m), erythroblasts (E), or megakaryocytes (M). Otherwise, unidentified immature cells were designated as blastlike cells (bl). The nmEM cells constituted 43.2 ± 3.2% (mean ± SD; n = 3) of the colony-forming CD34−KSL cells.
Asymmetric Division of nmEM Cells.
Single CD34−KSL cells were incubated in the presence of different cytokines: SCF alone, SCF + IL-3, SCF + TPO, or SCF + IL-3 + TPO. After they divided once, the two resultant daughter cells were separated by micromanipulation (Fig. 1 A). Individual daughter cells were subsequently allowed to form colonies in the presence of SCF + IL-3 + TPO + EPO. A total of 340 daughter cells (170 pairs) were successfully micromanipulated; their differentiation potentials were then examined. Daughter cells showed a variety of differentiation potentials. The most frequently observed differentiation potential of daughter cells was nmEM, regardless of the cytokines used for induction of parent–cell division. In this work, we operationally defined retrospectively identifiable nmEM cells as HSCs and examined the differentiation potential of their immediate progeny.
Table II lists all the pairs whose parent cells were inferred to have had nmEM differentiation potential. In these cases, daughter cells (other than M-unipotent daughter cells) gave rise to colonies large enough for cytospin preparation. Two cases in which a nmEM daughter cell had no identifiable pair were excluded from analysis because a technical error in micromanipulation might have been responsible.
Table II.
Differentiation Potentials of Paired Daughter Cells
| Differentiation potential
|
|||
|---|---|---|---|
| Cytokine | One | The other | No. of pairs |
| SCF | nmEM | nmEM | 11 |
| nmEM | nm | 4 | |
| nmEM | nmM | 3 | |
| nmEM | nEM | 2 | |
| nmEM | M | 2 | |
| nmE | EM | 1 | |
| SCF + IL-3 | nmEM | nmEM | 8 |
| nmEM | nEM | 4 | |
| nmEM | nmE | 2 | |
| nmEM | EM | 2 | |
| nmEM | M | 2 | |
| nmEM | nmM | 1 | |
| nmEM | nm | 1 | |
| nEM | nm | 1 | |
| SCF + TPO | nmEM | nmEM | 19 |
| nmEM | nmE | 2 | |
| nmEM | nEM | 1 | |
| nmEM | m | 1 | |
| SCF + IL-3 + TPO | nmEM | nmEM | 10 |
| nmEM | nmM | 2 | |
| nmEM | nmE | 1 | |
| nmEM | nm | 1 | |
| nmEM | mE | 1 | |
| nmEM | EM | 1 | |
| nmEM | nE | 1 | |
| nmEM | M | 1 | |
| nmE | nmM | 1 | |
| nEM | mM | 1 | |
Differentiation potential along myeloid lineage was determined for each of the members of paired daughter cells. Data on three independent experiments for each culture condition are summarized. In total, 40, 37, 52, and 41 pairs were examined after the treatment with SCF, SCF + IL-3, SCF + TPO, and SCF + IL-3 + TPO, respectively. Only the pairs whose parental cells should have had neutrophil (n), macrophage (m), erythroblast (E), and megakaryocyte (M) differentiation potential are presented. The probability of asymmetric division was 0.52 (12/23), 0.62 (13/21), 0.17 (4/23), or 0.50 (10/20) in the case of SCF, SCF + IL-3, SCF + TPO, or SCF + IL-3 + TPO, respectively. The probability of asymmetric division in the presence of SCF + IL-3 was significantly greater than that in the presence of SCF + TPO (P = 0.0047).
Regardless of what kind of cytokine mix was used, one of the two daughter cells showed differentiation potential, in most cases along nmEM lineage. These nmEM cells had tri-, bi-, and unipotent daughter cells as partners. Thus, lineage commitment in nmEM cells occurred through asymmetric division. Interestingly, progenitor cells committed to M, m, EM, or nm lineages were directly derived from nmEM cells via only one division.
When only SCF was used to induce cell division, asymmetric division took place in 52% of the cases. When IL-3 was used together with SCF, the probability of asymmetric division increased only slightly (P = 0.62). After an nmEM cell underwent asymmetric division in the presence of SCF alone or SCF + IL-3, one of the two daughter cells showed a variety of differentiation potentials. In contrast, when TPO was used with SCF, the number of different combinations of differentiation potential exhibited by individual pairs of daughter cells was limited, and the probability of asymmetric division dropped to 0.17. This represented a significant difference in the probability of asymmetric division between cells treated with SCF + IL-3 and cells treated with SCF + TPO (P < 0.005). This result is consistent with our previous observation that SCF + TPO maintained repopulating activity in pairs of daughter cells derived from single CD34−KSL cells more efficiently than did SCF + IL-3 (22). In the case of SCF + IL-3 + TPO, the probability of asymmetric division was 0.5, similar to that of SCF alone. Together, these data show that one asymmetric division of multilineage CD34−KSL cells leads to lineage commitment in their progeny and that the likelihood of asymmetric division is strongly biased by cytokine administration.
Lineage Commitment of nmEM Cells through Serial Division.
To discover whether lineage commitment of nmEM cells occurs in the same manner through serial division, we examined the differentiation potential of individual granddaughter cells generated from single CD34−KSL cells in the presence of SCF + IL-3 or SCF + TPO (Fig. 1 B). 118 and 94 paired granddaughter cells in SCF + IL-3 and SCF + TPO regimens, respectively, were successfully micromanipulated. Like daughter cells, granddaughter cells showed wide varieties of differentiation potential, but the capacity for nmEM differentiation remained most frequent. Table III shows the differentiation potentials of granddaughter cells that were considered to be derived from nmEM cells. Lineage commitment of nmEM cells also took place via asymmetric division at their second division, independent of whether IL-3 or TPO was the cytokine administered. When SCF + IL-3 were used, the second division of nmEM cells had a probability of 0.58 of generating cells asymmetric for differentiation potential. This probability was similar to that on the first division in the presence of SCF + IL-3 (Tables II and III). When SCF + TPO were used, the probability that a second division would generate cells asymmetric for differentiation potential was 0.32, higher than that on first division. Despite this increase, the difference in probability of asymmetric division between cells treated with SCF + IL-3 and cells treated with SCF + TPO remained significant. Once nmEM cells are regenerated on first division, these daughter cells behave like their parent nmEM cells.
Table III.
Differentiation Potential of Paired Granddaughter Cells
| Differentiation potential
|
|||
|---|---|---|---|
| Cytokine | One | The other | No. of pairs |
| SCF + IL-3 | nmEM | nmEM | 15 |
| nmEM | nm | 6 | |
| nmEM | mEM | 5 | |
| nmEM | nmM | 2 | |
| nmEM | nmE | 1 | |
| nmEM | mM | 1 | |
| nmEM | M | 1 | |
| nmM | nmE | 2 | |
| nmM | nEM | 1 | |
| nm | nEM | 2 | |
| SCF + TPO | nmEM | nmEM | 21 |
| nmEM | nm | 5 | |
| nmEM | nmM | 2 | |
| nmEM | nmE | 1 | |
| nmEM | M | 1 | |
| nEM | m | 1 | |
After single CD34−KSL cells divided in the presence of the cytokines shown, the two daughter cells were separated. After individual daughter cells underwent division under the same condition, the granddaughter cells were again separated and were allowed to form colonies in the presence of SCF + IL-3 + TPO + EPO (Fig. 1 B). In total, 59 and 47 cell pairs generated in the presence of SCF + IL-3 and SCF + TPO were serially manipulated. Only the pairs derived from nmEM cells are presented. The probability of asymmetric division induced by SCF + IL-3 (0.58, 21/36) was significantly greater than that induced by SCF + TPO (0.32, 10/31; P = 0.0492).
Discussion
Precise analysis of the mode of lineage commitment in HSCs has been hampered because HSCs are very rare in the bone marrow and probably are functionally heterogeneous. Moreover, their differentiation process is mostly associated with cell division. We addressed this issue by analyzing daughter cells derived from single nmEM cells among highly purified HSCs.
The success rate of long-term reconstitution with single CD34−KSL cells ranged from 20 to 50% in our series of studies, presumably depending on the degree of purification in each experiment (unpublished data). As shown in Table I, 40 and 25% of the CD34−KSL cells were detected as multilineage and unilineage repopulating cells in this work. The seeding efficiency of repopulating cells in total appeared to be from 65 to 100%, supporting the proposition that HSCs can engraft much more efficiently than previously thought (27). By calculation, the frequency of long-term multilineage repopulating cells in this population can be corrected as 40–60%. Because >90% of the cells showed colony-forming activity (Fig. 2), most HSCs, if not all, should be able to form colonies in vitro.
To study the commitment process of HSCs while excluding from our analyses HSCs' committed progeny, we further selected for study, by retrospective inference, cells retaining the full range of capacities for differentiation (nmEM differentiation potential). The proportion of long-term repopulating cells that can be identified as nmEM progenitor cells remains uncertain. However, nmEM cells appeared to be a major subpopulation, consisting of highly proliferative cells, among CD34−KSL cells. Retrospectively identified nmEM cells indeed constituted a major subpopulation among CD34−KSL cells, but not among CD34+KSL cells (unpublished data).
In most daughter cell and granddaughter cell pairs, one of the two cells inherited nmEM differentiation potential from its parent cell. The other daughter or granddaughter cell either also remained capable of giving rise to an nmEM colony or became committed to some lineage (Tables II and III). These analyses relied on identification of lineage components in well-formed colonies. One of the components originally present could have been lost during the numerous cell divisions required for colony formation. In such a case, the differentiation potential of daughter cells (as assessed by identification of their descendants' phenotypes) would be underestimated. However, daughter cell pairs and granddaughter cell pairs were always assayed in parallel under exactly the same conditions. Bias owing to underestimation of differentiation potential should be equal for each of the two cells. Perhaps variation in differentiation potential between the two cells in each pair was inherent from the first division onward.
The terms “symmetric” and “asymmetric” were also defined based on components of well-formed colonies. Cell numbers and differential counts within colonies were not taken into account when evaluating lineage commitment. The pioneer work on paired daughter cells done by Suda et al. has suggested the progressive and stochastic lineage restriction of HSCs (8, 9). Suda et al. also have observed that multipotent cells asymmetrically give rise to unipotent cells. It has been shown that these blast colonies contain very few long-term repopulating cells (28). Their observations likely reflect the behavior of progenitor cells at an early stage of differentiation. Together, our data suggest that HSCs and their early progeny similarly undergo lineage commitment in an asymmetric manner.
The Drosophila melanogaster neuroblast asymmetrically divides to produce both neuroblast and ganglion mother cell (26). This similarity to mouse HSC behavior suggests that asymmetric division is a mechanism for generating cellular diversity common to stem and progenitor cells in the nervous and hematopoietic systems. Asymmetric division is considered to involve both unequal segregation of determinants and cell–cell interaction (26). Numb protein has been implicated as one such determinant in neural development. The primary function of Numb seems not the direct specification of fate, but rather modulation of environmental cues like Notch signaling (29). Although such a determinant has not been recognized in hematopoietic systems, a distinct expression pattern of particular genes in one of the two daughter cells may lead to lineage commitment as proposed by Cross et al. (30): different regions of the cell sap at the moment of division may contain unequally distributed gene products, and this inequality in distribution may tip the balance from one commitment pattern to another.
Expression of transcriptional factors such as SCL, GATA-2, Gfi-1B, and PU.1 has been detected in CD34−KSL cells (31, 32). If all these genes are expressed in a daughter cell, that cell may retain nmEM differentiation potential. If PU.1 is predominantly expressed, a daughter cell may commit to nm lineage. In contrast, a daughter cell may commit to EM lineage if expression levels of SCL, GATA-2, and Gfi-1B in concert dominate PU.1. Cross-antagonism can be an alternative mechanism for dominant action of a certain transcriptional factor over an opposing one (33).
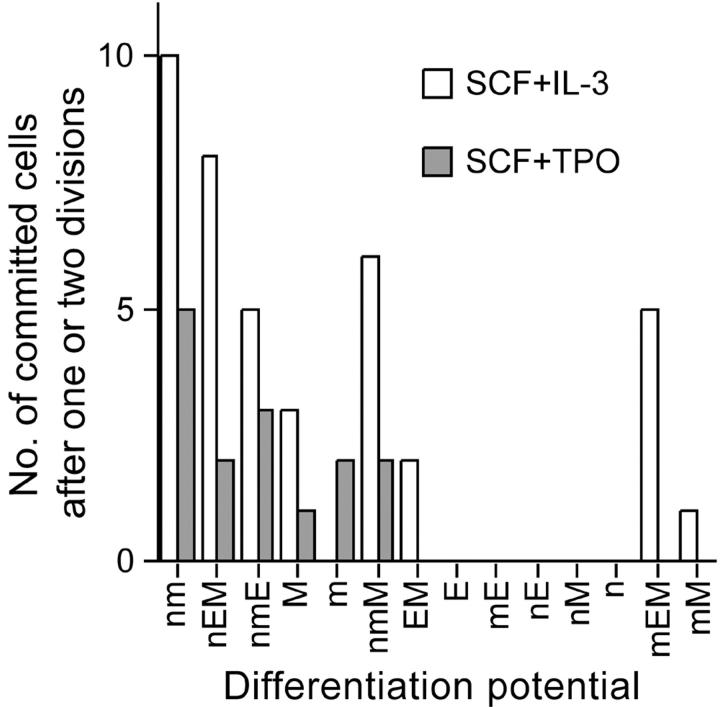
This work clearly demonstrates an instructive role of cytokines in lineage commitment at the level of HSCs (Tables II and III). The probability of lineage commitment by nmEM cells in the presence of SCF + TPO was significantly lower than that in the presence of SCF + IL-3. The probability remained similar through the initial two divisions with SCF + IL-3 (0.62 and 0.58), but not with SCF + TPO (0.17 and 0.32). Lineage commitment by HSCs can be controlled to some extent by external factors. Whether cytokines also influence which lineage potential is chosen by HSCs upon their lineage commitment remains unclear. Fig. 3 presents the numbers of committed progenitor cells that appeared after the first and second divisions of nmEM cells. In the presence of either IL-3 or TPO, nmEM cells seemed to have a preference in lineage choice, suggesting that the fate decision of HSCs is not a random event. Alternatively, this may simply indicate postcommitment selection in given culture conditions.
Figure 3.
Distribution of lineage-committed daughter and granddaughter cells. The numbers of daughter and granddaughter cells that lost the capacity to differentiate along one or more lineages are graphed, using data in Tables II and III. All possible combinations of differentiation potential are shown in the same order as that presented in Fig. 2.
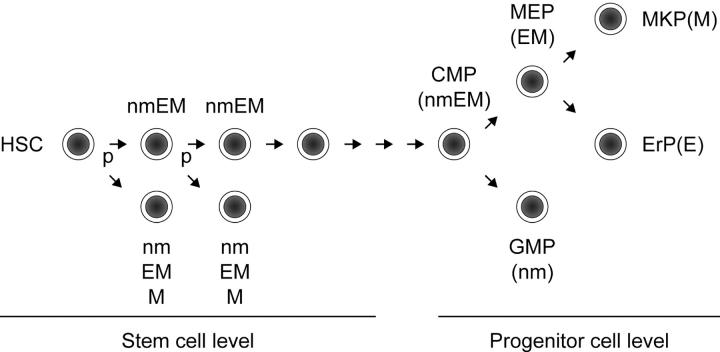
Interestingly, we did not observe the combination of nm lineage and EM lineage in a total of 154 pairs of daughter and granddaughter cells (Tables II and III). A CMP has been described as giving rise to separate colonies of nm lineage and EM lineage in culture, with EM lineage–committed progenitors, in turn, giving rise to E lineage–committed progenitors and M lineage–committed progenitors, supporting a progressive loss of differentiation potential (34). Our data suggest an alternative pathway of myeloerythroid differentiation in which nm or EM progenitor cells can asymmetrically develop from an HSC population with no need for a CMP intermediate (Fig. 4). The modes of lineage commitment possibly differ between stem and progenitor cell compartments.
Figure 4.
Myeloid lineage restriction model. The mode of lineage commitment at the level of HSCs may differ from that at the level of progenitor cells. Our model for HSCs is presented in combination with the model proposed by Weissman's group (34). An HSC can directly give rise to lineage-committed progenitor cells such as nm, EM, or M progenitor cells through initial HSC division in asymmetric manner. It may give rise to a common myeloid progenitor (CMP) after a certain number of divisions. The CMP gives rise to a megakaryocyte/erythrocyte lineage–restricted progenitor (MEP) and to a granulocyte/macrophage lineage-restricted progenitor (GMP). The MEP progressively gives rise to a megakaryocyte-committed progenitor (MKP) and to an erythrocyte lineage-committed progenitor (ErP). P, probability of asymmetric division.
HSCs, progenitor cells of CMPs and common lymphoid progenitors, and CMPs could have all been detected as nmEM cells in this work because self-renewal activity and B- and T-lymphoid differentiation potentials were not examined. In the presence of SCF + TPO, 42 out of 46 daughter cells (91%) and 51 out of 62 granddaughter cells (82%) retained the capacity to differentiate along nmEM lineage after, respectively, their first and second divisions (Tables II and III). The nmEM cells apparently regenerated themselves in their early divisions. On the other hand, a substantial loss in the number of long-term repopulating cells under the same conditions has been observed (22). It is assumed that a majority of nmEM cells generated in culture in this work might have lost self-renewal activity, but still maintained the capacity to differentiate along multiple lineages. Common progenitor cells giving rise to both CMPs and common lymphoid progenitors have not been identified. After a certain number of divisions, HSCs should give rise to such progenitor cells at a certain stage of differentiation. To verify our model of lineage commitment, both myeloid and lymphoid differentiation potentials should be examined for intermediate cells between HSCs and CMPs at the clonal level. To do this, an assay sensitive and efficient enough for detection of all potentials in individual cells needs to be developed. Our working model, nonetheless, may help in studying the molecular mechanism of lineage commitment in HSCs.
Acknowledgments
The authors thank K. Fujii and T. Suda for helpful discussion and A.S. Knisely for critical reading of the manuscript.
This work was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology.
The present address of H. Takano is the Dept. of Hematology, Musashino Red Cross Hospital, 1-26-1 Kyonan, Musashino, 180-8610 Tokyo, Japan.
Abbreviations used in this paper: bl, blastlike; CFC, colony-forming cell; CMP, common myeloid progenitor; E, erythroblast; EPO, erythropoietin; HSC, hematopoietic stem cell; m, macrophage; M, megakaryocyte; n, neutrophil; SA, streptavidin; SCF, stem cell factor; TPO, thrombopoietin.
References
- 1.Lansdorp, P.M. 1997. Self-renewal of stem cells. Biol. Blood Marrow Transplant. 3:171–178. [PubMed] [Google Scholar]
- 2.Metcalf, D. 1998. Lineage commitment and maturation in hematopoietic cells: The case for extrinsic regulation. Blood. 92:354–352. [PubMed] [Google Scholar]
- 3.Ogawa, M. 1999. Stochastic model revisited. Int. J. Hematol. 69:2–5. [PubMed] [Google Scholar]
- 4.Weissman, I.L. 2000. Stem cells: Units of development, units of regeneration, and units in evolution. Cell. 100:157–168. [DOI] [PubMed] [Google Scholar]
- 5.Till, J.E., E.A. McCulloch, and L. Siminovitch. 1964. A stochastic model of stem cell proliferation, based on the growth of spleen colony-forming cells. Proc. Natl. Acad. Sci. USA. 51:29–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Humphries, R.K., A.C. Eaves, and C.J. Eaves. 1981. Self-renewal of hemopoietic stem cells during mixed colony formation in vitro. Proc. Natl. Acad. Sci. USA. 78:3629–3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakahata, T., A.J. Gross, and M. Ogawa. 1982. A stochastic model of self-renewal and commitment to differentiation of the primitive hemopoietic stem cells in culture. J. Cell. Physiol. 113:455–458. [DOI] [PubMed] [Google Scholar]
- 8.Suda, T., J. Suda, and M. Ogawa. 1984. Disparate differentiation in mouse hemopoietic colonies derived from paired progenitors. Proc. Natl. Acad. Sci. USA. 81:2520–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suda, J., T. Suda, and M. Ogawa. 1984. Analysis of differentiation of mouse hemopoietic stem cells in culture by sequential replating of paired progenitors. Blood. 64:393–399. [PubMed] [Google Scholar]
- 10.Leary, A.G., L.C. Strauss, C.I. Civin, and M. Ogawa. 1985. Disparate differentiation in hemopoietic colonies derived from human paired progenitors. Blood. 66:327–332. [PubMed] [Google Scholar]
- 11.Tsuji, K., and T. Nakahata. 1989. Stochastic model for multipotent hemopoietic progenitor differentiation. J. Cell. Physiol. 139:647–653. [DOI] [PubMed] [Google Scholar]
- 12.Mayani, H., W. Dragowska, and P.M. Landsdorp. 1993. Lineage commitment in human hemopoiesis involves asymmetric cell division of multipotent progenitors and does not appear to be influenced by cytokines. J. Cell. Physiol. 157:576–586. [DOI] [PubMed] [Google Scholar]
- 13.Brummendorf, T.H., W. Dragowska, J.M.J.M. Zijlmans, G. Thornbury, and P.M. Lansdorp. 1998. Asymmetric cell divisions sustain long-term hematopoiesis from single-sorted human fetal liver cells. J. Exp. Med. 188:1117–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suda, T., J. Suda, M. Ogawa, and J.N. Ihle. 1985. Permissive role of interleukin 3 (IL-3) in proliferation and differentiation of multipotential hemopoietic progenitors in culture. J. Cell. Physiol. 124:182–190. [DOI] [PubMed] [Google Scholar]
- 15.Stoffel, R., S. Ziegler, N. Ghilardi, B. Ledermann, F.J. de-Sauvage, and R.C. Skoda. 1999. Permissive role of thrombopoietin and granulocyte colony-stimulating factor receptors in hematopoietic cell fate decisions in vivo. Proc. Natl. Acad. Sci. USA. 96:698–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wolf, N.S., and J.J. Trentin. 1975. The restorative effect of erythropoietic stimulation upon the sublethally irradiated (SIL) hematopoietic stem and/or its progeny. Exp. Hematol. 3:57–64. [PubMed] [Google Scholar]
- 17.Van Zant, G., and E. Goldwasser. 1979. Competition between erythropoietin and colony-stimulating factor for target cells in mouse marrow. Blood. 53:946–965. [PubMed] [Google Scholar]
- 18.Metcalf, D., and A.W. Burgess. 1982. Clonal analysis of progenitor cell commitment to granulocyte or macrophage production. J. Cell. Physiol. 111:275–283. [DOI] [PubMed] [Google Scholar]
- 19.Metcalf, D. 1991. Lineage commitment of hemopoietic progenitor cells in developing blast cell colonies: Influence of colony-stimulating factors. Proc. Natl. Acad. Sci. USA. 88:11310–11314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Metcalf, D. 1998. Lineage commitment in the progeny of murine hematopoietic preprogenitor cells: Influence of thrombopoietin and interleukin 5. Proc. Natl. Acad. Sci. USA. 95:6408–6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Osawa, M., K.-i. Hanada, H. Hamada, and H. Nakauchi. 1996. Long-term lymphohematopoietic reconstitution by a single CD34-low/negative hematopoietic stem cell. Science. 273:242–245. [DOI] [PubMed] [Google Scholar]
- 22.Ema, H., H. Takano, K. Sudo, and H. Nakauchi. 2000. In vitro self-renewal division of hematopoietic stem cells. J. Exp. Med. 192:1281–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakauchi, H., K. Sudo, and H. Ema. 2000. Quantitative assessment of the stem cell self-renewal capacity. Ann. NY Acad. Sci. 938:18–24. [DOI] [PubMed] [Google Scholar]
- 24.Akashi, K., D. Traver, T. Miyamoto, and I.L. Weissman. 2000. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 404:193–197. [DOI] [PubMed] [Google Scholar]
- 25.Sudo, K., H. Ema, Y. Morita, and H. Nakauchi. 2000. Age-associated characteristics of murine hematopoietic stem cells. J. Exp. Med. 192:1273–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jan, Y.N., and L.Y. Jan. 2001. Asymmetric cell division in the Drosophila nervous system. Nat. Rev. Neurosci. 2:772–779. [DOI] [PubMed] [Google Scholar]
- 27.Benveniste, P., C. Cantin, D. Hyam, and N.N. Iscove. 2003. Hematopoietic stem cells engraft in mice with absolute efficiency. Nat. Immunol. 4:708–713. [DOI] [PubMed] [Google Scholar]
- 28.Tsunoda, J., S. Okada, J. Suda, K. Nagayoshi, H. Nakauchi, K. Hatake, Y. Miura, and T. Suda. 1991. In vivo stem cell function of interleukin-3-induced blast cells. Blood. 78:318–322. [PubMed] [Google Scholar]
- 29.Cayouette, M., and M. Raff. 2002. Asymmetric segregation of Numb: a mechanism for neural specification from Drosophila to mammals. Nat. Neurosci. 5:1265–1269. [DOI] [PubMed] [Google Scholar]
- 30.Cross, M.A., and T. Enver. 1997. The lineage commitment of haemopoietic progenitor cells. Curr. Opin. Genet. Dev. 7:609–613. [DOI] [PubMed] [Google Scholar]
- 31.Nakauchi, H., H. Takano, H. Ema, and M. Osawa. 1999. Further characterization of CD34-low/negative mouse hematopoietic stem cells. Ann. NY Acad. Sci. 872:57–66. [DOI] [PubMed] [Google Scholar]
- 32.Osawa, M., T. Yamaguchi, Y. Nakamura, S. Kaneko, M. Onodera, K. Sawada, A. Jegalian, H. Wu, H. Nakauchi, and A. Iwama. 2002. Erythroid expansion mediated by the Gfi-1B zinc finger protein: role in normal hematopoiesis. Blood. 100:2769–2777. [DOI] [PubMed] [Google Scholar]
- 33.Cantor, A.B., and S.H. Orkin. 2001. Hematopoietic development: a balancing act. Curr. Opin. Genet. Dev. 11:513–519. [DOI] [PubMed] [Google Scholar]
- 34.Nakorn, T.N., T. Miyamoto, and I.L. Weissman. 2003. Characterization of mouse clonogenic megakaryocyte progenitors. Proc. Natl. Acad. Sci. USA. 100:205–210. [DOI] [PMC free article] [PubMed] [Google Scholar]