Abstract
Epstein-Barr virus (EBV)–encoded nuclear antigen (EBNA)1 is thought to escape cytotoxic T lymphocyte (CTL) recognition through either self-inhibition of synthesis or by blockade of proteasomal degradation by the glycine-alanine repeat (GAr) domain. Here we show that EBNA1 has a remarkably varied cell type–dependent stability. However, these different degradation rates do not correspond to the level of major histocompatibility complex class I–restricted presentation of EBNA1 epitopes. In spite of the highly stable expression of EBNA1 in B cells, CTL epitopes derived from this protein are efficiently processed and presented to CD8+ T cells. Furthermore, we show that EBV-infected B cells can readily activate EBNA1-specific memory T cell responses from healthy virus carriers. Functional assays revealed that processing of these EBNA1 epitopes is proteasome and transporter associated with antigen processing dependent. We also show that the endogenous presentation of these epitopes is dependent on the newly synthesized protein rather than the long-lived stable EBNA1. Based on these observations, we propose that defective ribosomal products, not the full-length antigen, are the primary source of endogenously processed CD8+ T cell epitopes from EBNA1.
Keywords: EBNA1, CD8+ T cell, antigen processing, DRiPs, proteasome
Introduction
The interaction of CTLs with the MHC–peptide complex is a critical step toward the initiation and propagation of specific immune responses against viral infection (1). The specificity of this interaction is determined by two distinct components, namely, MHC-restricted presentation of a peptide epitope, and a heterodimeric αβ cell surface protein called the TCR (2). It is now well recognized that a successful latent infection by herpes viruses such as EBV is dependent on various strategies to evade this potent antiviral CTL response in the immune-competent host (3). These escape mechanisms include limited gene expression during latent infection, virus replication in immune-privileged tissues, down-regulation of MHC and adhesion molecules, and sequence variation affecting peptide binding to MHC molecules or recognition by the TCR of CTLs (4).
Previous studies have shown that the latent growth-transforming infection by EBV elicits a strong CD8+ CTL response directed against all the nuclear antigens except EBV-encoded nuclear antigen (EBNA)1 (5). These observations were consistent with other studies that showed that mammary carcinoma cells transfected with EBNA1 are poorly immunogenic in mice, whereas strong immunogenicity was induced by expression of other EBV proteins that are highly immunogenic in humans, suggesting that CTL evasion occurs across species (6, 7). The failure of EBNA1-specific T cells to recognize EBV-infected B cells has been attributed to an internal glycine-alanine repeat (GAr) domain of EBNA1 that acts as a cis inhibitor of MHC class I–restricted presentation (8). It was proposed that the GAr sequence within EBNA1 forms β sheets that are resistant to unfolding and affect its capacity to associate with various components of the ubiquitin(Ub)/proteasome pathway, including Ub conjugation enzymes and/or regulatory subunits of the proteasome (9, 10). However, previous studies from our laboratory demonstrated that proteasomal targeting in conjunction with the N-end rule can override the GAr-mediated inhibitory effect and allow endogenous processing of EBNA1 through the MHC class I pathway (11). More recently, Yin et al. (12) showed that the GAr domain also inhibits mRNA translation of EBNA1 in cis and thus argued that low antigen expression in virus-infected cells, rather than proteasomal blockage, is the primary mechanism preventing antigen presentation on MHC class I molecules.
However, neither of these models are able to explain the mechanisms by which occasional ex vivo EBNA1-specific T cell responses can be detected in EBV-infected individuals (13). Blake et al. (14) proposed that these EBNA1-specific responses might be generated by the cross-presentation of EBNA1 protein through the exogenous pathway, which is not affected by the GAr sequence. Alternatively, the GAr sequence is unlikely to prevent presentation of peptides from EBNA1 that are derived from defective ribosomal products (DRiPs) that are estimated to contribute up to 30% of newly synthesized mRNA (15–17). To explore this possibility and further delineate the mechanisms by which the EBNA1-specific T cell responses are generated, we conducted an extensive analysis of EBNA1 expression in different cell types and compared endogenous presentation of CTL epitopes in these cells. Here we report that EBV-infected cells can directly present EBNA1 epitopes to T cells and this presentation proceeds through a pathway in which endogenous processing of CTL epitopes is not dependent on the degradation of full-length antigen.
Materials and Methods
Cell Lines.
All cell lines were routinely maintained in RPMI 1640 supplemented with 2 mM l-glutamine, 100 IU/ml penicillin, and 100 μg/ml streptomycin plus 10% FCS (referred to as growth medium), unless otherwise stated. EBV-transformed lymphoblastoid cell lines (LCLs) were established from healthy seropositive donors by exogenous virus transformation of peripheral B cells using the B95.8 virus isolate as previously described (18). HLA class I–deficient LCLs (C1R; reference 19) transfected with HLA B*0801 (C1R.B8) and C1R.B8 cells transfected with HSV-1–encoded transporter associated with antigen processing (TAP) inhibitor, ICP47 (C1R.B8.ICP47; provided by J. McCluskey, University of Melbourne, Melbourne, Australia), were also used in this work. These C1R.B8 transfectants were maintained in growth medium supplemented with 500 μg/ml Geneticin (Invitrogen), whereas growth medium for C1R.B8.ICP47 was supplemented with 500 μg/ml Geneticin and 4 μg/ml Puromycin (Sigma-Aldrich). In addition, HLA class I–deficient 721.221 LCLs (referred to as 0.221; reference 20) and 0.221 LCLs transfected with HLA B*3501 (0.221.B35) were also used in this work. Three different human primary epithelial cell lines (SVMR6, HaCaT, and HEK 293; references 11, 21, and 22) were used for EBNA1 half-life analysis or CTL assays (see below).
Inhibitors.
Brefeldin A (BFA) and chloroquine were purchased from Sigma-Aldrich. The protease inhibitors, leupeptin and pepstatin, were from Boehringer. The proteasome inhibitors, lactacystin and Cbz-L3, were provided by E. Wiertz (Leiden University, Leiden, Netherlands).
Expression Constructs.
Full-length EBNA1 was cloned into the expression vector pcDNA3.1 (Invitrogen) to generate the expression construct EBNA1 (11). In addition, a modified EBNA1 construct in which the (GAr) domain was deleted was generated to give EBNA1ΔGA (11). The above constructs included insertion of a well-defined, HLA-B8–restricted EBNA-3 CTL epitope, FLRGRAYGL (referred to as FLR; reference 23), into the SacII site at nucleotide position 1853 of EBNA1, to allow assessment of endogenous processing of EBNA1 by FLR-specific CTL clones (see Fig. 1 A). To assess expression of EBNA1 and EBNA1ΔGA, the inserts from the above pcDNA3.1 constructs were subcloned in frame with a sequence coding for green fluorescent protein (GFP; pEGFP-N1; CLONTECH Laboratories, Inc.): EBNA1-GFP and EBNA1ΔGA-GFP.
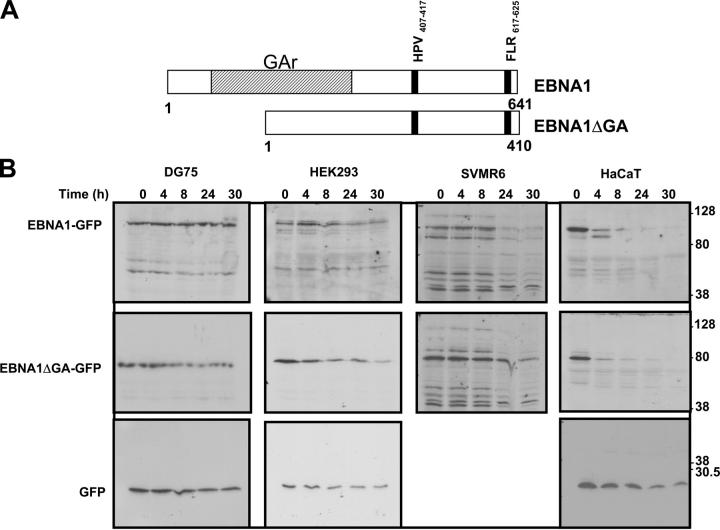
Figure 1.
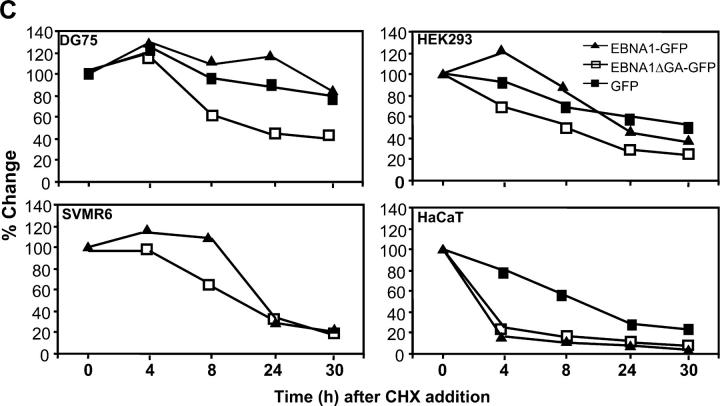
(A) Schematic description of EBNA1 and EBNA1ΔGA expression constructs showing localization of FLR and HPV epitopes. (B) Intracellular degradation of EBNA1-GFP in different cell types. DG75 B cells, HEK293 epithelial cells, SVMR6 keratinocytes, and HaCaT keratinocytes were transfected with expression constructs EBNA1-GFP, EBNA1ΔGA-GFP, or the control plasmid pEGFP-N1. At 36 h after transfection, the cells were degraded over a 30-h time course in the presence of 50 μg/ml cycloheximide as described in Materials and Methods. Molecular weight standards are indicated at the side of each panel. (C) Densitometric analysis of EBNA1-GFP, EBNA1ΔGA-GFP, and GFP expression. Band intensities were quantified by analysis of the imaging data and plotted as a relative percentage of the signal at time 0 for EBNA1-GFP, EBNA1ΔGA-GFP, and GFP.
Construction of Recombinant Ad5F35-EBNA1 Adenovirus.
The assembly and production of recombinant Ad5F35-based adenoviruses was completed in three stages using a highly efficient, ligation-based protocol of Adeno-X System (CLONTECH Laboratories, Inc.; reference 24). First, full-length EBNA1 and EBNA1ΔGA in pcDNA3.1 were digested with restriction enzymes NheI and XbaI, and subcloned into the pShuttle expression vector to create EBNA1-pShuttle and EBNA1ΔGA-pShuttle. Second, after amplification in Escherichia coli, the expression cassettes from both pShuttle vectors were excised and ligated into an Ad5F35 expression vector. Finally, the recombinant Ad5F35-EBNA1 and Ad5F35 EBNA1ΔGA vectors were transfected into HEK 293 cells, and recombinant adenoviruses were harvested from transfected cells by freeze thawing.
Transfection of EBNA1 Expression Constructs.
DG75 cells were grown to log phase. 5 × 106 cells were transfected in RPMI1640/10% FCS growth medium with 10 μg of expression constructs EBNA1-GFP, EBNA1ΔGA-GFP, or the GFP vector control using the BioRad Gene Pulser (960 μF, 250 V, 0.4 cm gap electrode, 300 μl assay volume, 25°C; reference 11). For the adherent cell lines (HaCaT, SVMR6, or HEK293), 5 × 105 cells were seeded into T25 flasks. At 60–80% confluency, the cells were transfected with 1 μg EBNA1-GFP, EBNA1ΔGA-GFP, or the GFP vector control using Effectene (QIAGEN) according to the manufacturer's instructions.
Degradation of EBNA1 Proteins.
All EBNA1 degradation studies were performed as previously described (11). In brief, at 36 h after transfection, 50 μg/ml cycloheximide (Sigma-Aldrich) was added to 8 × 106 cells. Equal aliquots of cells were removed at time points 0 min, 4, 8, 24, and 30 h, lysed in SDS-PAGE sample dye, and resolved under reducing conditions on a 7.5% SDS polyacrylamide gel. For adherent cells, transfectants were harvested by scraping and processed as described for suspension cells. The proteasome inhibitor, lactacystin, was added to the cells at a final concentration of 10 μg/ml at 24 h after transfection.
Detection of EBNA1 by Immunoblotting.
After gel electrophoresis, proteins were transferred to a nitrocellulose membrane (Hybond-C; Amersham Biosciences) and blocked in 5% milk powder/0.5% Tween/PBS. To detect GFP fusion proteins, membranes were probed with a GFP antibody (Molecular Probes) at 1:2,000, followed by a polyclonal sheep anti–rabbit horseradish peroxidase–conjugated antibody. Protein bands were detected using Chemiluminescence Reagent Plus (PerkinElmer). Protein levels were compared by densitometric analysis using Imagequant software (Molecular Dynamics). To determine the ubiquitination status of EBNA1 and EBNA1ΔGA, 5 × 105 SVMR6 and HeLa cells were cotransfected with expression vectors encoding 0.8 μg hemagglutinin (HA)-tagged 8XUb (pMT123; provided by M. Treier, University of California San Diego, San Diego, CA) and 0.6 μg EBNA1-GFP, EBNA1ΔGA-GFP, or latent membrane protein 1 (LMP1)-GFP. 36 h after transfection, the cells were lysed and any resulting Ub-EBNA1 or Ub-LMP1 complexes were immunoprecipitated with an anti-HA–specific mAb (Roche Diagnostics). These complexes were resolved by SDS-PAGE followed by immunoblotting with an anti-GFP antibody.
CTL Cultures.
CTL clones were generated by agar cloning as follows: 2 × 106 PBMCs from HLA B*3501+ EBV-exposed individuals were stimulated in 2 ml growth medium with autologous PBMCs that had been precoated with the EBNA1 epitope, HPVGEADYFEY (referred to as HPV; 1 μM for 1 h, responder/stimulator ratio of 2:1). After 3 d, cells were dispersed and seeded in 0.35% agarose (Seaplaque; BioWhittaker Molecular Applications) containing RPMI 1640, 20% FCS, 25% supernatant from MLA-144 cultures, and 50 U/ml rIL-2. Colonies were harvested after an additional 3–5 d and amplified in culture with biweekly restimulation with rIL-2, supernatant from the Gibbon lymphosarcoma T cell line, MLA-144 (86102901; European Collection of Cell Cultures) as a source of T cell growth factor, and the γ-irradiated (8,000 rads) autologous LCLs.
Short-term CTL bulk cultures were used as effectors in cytotoxicity assays. These were generated by culturing PBMCs (2 × 106 per 2-ml well) in growth medium with γ-irradiated (8,000 rads) LCLs (responder/stimulator ratio of 20:1). LCL-stimulated CTL cultures were split and restimulated with additional irradiated autologous LCLs on day 7. The CTL bulk cultures were used in chromium release assays on day 10.
In Vitro Inhibition Studies.
SVMR6 keratinocytes or LCLs were pretreated with either class I inhibitors, 10 μg/ml lactacystin, 1 μg/ml BFA, and 10 μg/ml Cbz-L3, or class II inhibitors, 80 μM chloroquine, 100 μM leupeptin, and 50 μM pepstatin for 45 min. The cells were then infected with a recombinant adenovirus expressing full-length EBNA1. At 18 h after infection, the cells were used as targets in a standard 51Cr-release assay. Inhibitors were kept at the above concentrations during the 5-h assay except for chloroquine, which was lowered to a final concentration of 20 μM. The FLRGRAYGL-specific CTL clone, LC13, was used to assess CTL activity of EBNA1 with the inserted HLA B8–restricted epitope.
Intracellular Cytokine Staining.
PBMCs from HLA B35+ individuals were incubated overnight for 14–16 h at 37°C in growth medium supplemented with 20 U/ml recombinant IL-2 with and without the following cell types at a 20:1 ratio: autologous LCLs, 721.221, 7221.221.B35, and allogeneic LCL. GolgiPlug™ (BD Biosciences) was added to the samples according to the manufacturer's instructions during the second hour of incubation. After the incubation, cells were washed and resuspended in staining buffer consisting of Dulbecco's PBS with GolgiPlug™ and 1% FCS. Cells were then stained with PE-conjugated HPV-B*3501 tetramer (ProImmune) and Tricolor-conjugated anti-CD8 (Caltag) for 30 min at 4°C in the dark. The cells were then fixed for 20 min at 4°C with Cytofix/Cytoperm™ (BD Biosciences) and resuspended in Perm/Wash™ permeabilization buffer (BD Biosciences) according to the manufacturer's protocol. The cells were then stained with FITC-conjugated anti–human IFN-γ (BD Biosciences) for 30 min at 4°C in the dark. Cells were washed in permeabilization buffer and resuspended in staining buffer before analysis by three-color flow cytometry on a Becton Dickinson FACSCalibur™ cytometer.
Cell Surface MHC–Peptide Stripping and Antigen Presentation Assays.
For MHC–peptide stripping, HLA B35+ LCLs were washed in PBS and pellets were resuspended in a citrate buffer phosphate, pH 3, (0.131 M citric acid, 0.066 M Na2HPO4) for 2 min on ice. The suspension was then neutralized by a 100-fold dilution with RPMI/10% FCS, and cells washed twice. Aliquots of cells were resuspended in 2 ml complete medium and incubated in the presence or absence of 50 μM cycloheximide for 5 h. To assess the surface MHC class I expression, these LCLs were preincubated with anti–MHC class I mAb (W6/32; American Type Culture Collection) followed by incubation with FITC-labeled anti–mouse Ig. The fluorescence intensity was measured by FACSCalibur™ and data were analyzed by CELLQuest™ software (Becton Dickinson). PBMCs from HLA B35+ individuals were incubated overnight for 14–16 h at 37°C in growth medium supplemented with 20 U/ml recombinant IL-2 with and without the stimulator cells at a responder/stimulator ratio of 20:1. These stimulator cells included autologous LCLs, the citrate buffer–treated HLA B35+ LCLs after incubation in the absence or presence of cycloheximide and the untreated HLA B35+ control LCL after cycloheximide treatment. The responding cells were assessed for intracellular IFN-γ expression as described above.
In Vitro Cytotoxicity Assays.
EBV-specific CTL clones or polyclonal T cell lines were used as effectors in the CTL assays. In some experiments, target cells were transfected with EBNA1 expression vectors EBNA1-GFP or EBNA1ΔGA-GFP for 36 h or infected with recombinant adenovirus encoding EBNA1 or EBNA1ΔGA and incubated for 14–16 h at 37°C. After incubation, cells were washed in growth medium, labeled with 51Cr for 60 min. After incubation, these cells were washed with growth medium and used as targets in standard 5-h 51Cr-release assays (25).
Results
EBNA1 Displays Differential Intracellular Stability in Different Cell Types.
To determine the intracellular kinetics of EBNA1 in different cell types, an EBV− B cell line (DG75) and a number of epithelial cell lines (HEK293, HaCaT, and SVMR6) were transiently transfected with expression vectors pEGFP-N1, EBNA1-GFP, or EBNA1ΔGA-GFP, and protein expression was analyzed by SDS-PAGE after incubation with 50 μg/ml cycloheximide. The intensity of the EBNA1-GFP band was measured by densitometric analysis. Representative data from this analysis is shown in Fig. 1 B. Full-length EBNA1-GFP and GFP were highly stable in DG75 cells and even after 30 h >80% of the protein was detectable. The stability of EBNA1 was also determined in another B cell line (AS LCL) and also observed to have a half-life of >30 h (not depicted). In contrast, epithelial cells transfected with EBNA1-GFP showed a rapid reduction in EBNA1 levels with >60% of the protein being degraded within 24 h (Fig. 1 B). Densitometric analysis indicated that the half-life for full-length EBNA1 in B cells was >30 h, whereas the half-life was as low as <4 h in HaCaT cells and ∼20–24 h in SVMR6 and HEK293 cells (Fig. 1 C). Interestingly, the half-life of full-length EBNA1 versus a truncated version of the antigen with GAr sequence deleted (EBNA1ΔGA) was quite comparable in the three epithelial cell lines tested, whereas there was a significant difference in the half-life of these proteins in B cells. Another interesting feature of the EBNA1 expression observed in epithelial cells was the presence of multiple low molecular weight expression products, which may represent either the degraded forms of the full-length protein or DRiPs. To differentiate between DRiPs and postlysis artifacts, the lysates from each time point were added to hot (95°C) sample buffer (2% SDS/1% β mercaptoethanol/1% glycerol/65 mM Tris HCL, pH 6.5) and boiled for 10 min (26). These samples were separated on 7.5% SDS-PAGE and displayed a similar pattern of multiple low molecular weight expression products (not depicted). These results further support our contention that these low molecular weight expression products might be EBNA1 DRiPs. To demonstrate that the expression of these recombinant proteins does not effect cell viability, the immunoblots were also probed with a specific antibody to demonstrate the stability of actin over the time course of the experiment. A similar pattern of stable actin expression was observed for all the transfectants and demonstrated equal loading of lanes (not depicted).
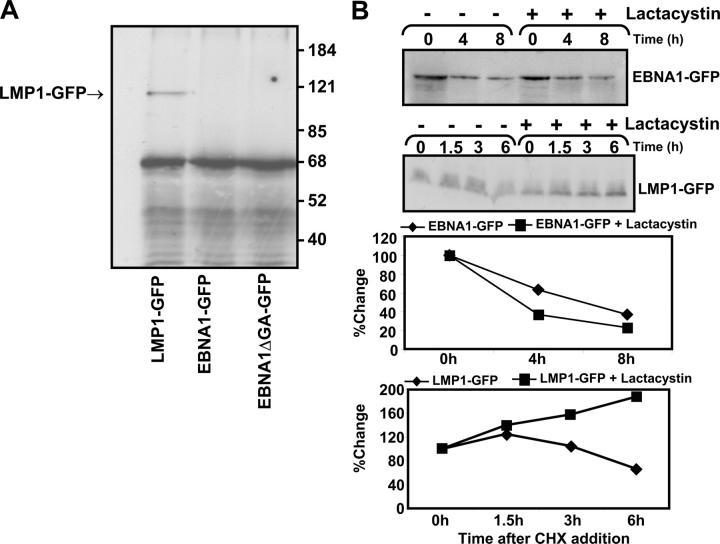
To delineate the pathway for the rapid degradation of EBNA1 in epithelial cells, we first tested the possibility that this protein is targeted through the Ub-dependent pathway. SVMR6 cells were transiently cotransfected with expression vectors encoding HA-tagged 8xUb and EBNA1-GFP, EBNA1ΔGA-GFP, or LMP1-GFP. Previous studies have shown that the EBV-LMP1 is degraded through the Ub/proteasome-dependent pathway (27) and thus LMP1-GFP was included as a positive control. 36 h after transfection, these cells were lysed and Ub–EBNA1 or Ub–LMP1 complexes were immunoprecipitated with an anti-HA–specific mAb. These complexes were then resolved by SDS-PAGE, followed by immunoblotting with a GFP-specific antibody. Data presented in Fig. 2 A clearly shows that although LMP1-GFP was ubiquitinated, no ubiquitination was observed for EBNA1-GFP or EBNA1ΔGA-GFP. These results are supported by recent studies by Holowaty et al. (28) who showed that EBNA1 interacts with a Ub-specific protease, HAUSP/USP7, which is demonstrated to deubiquitinate EBNA1. Furthermore, the pattern of intracellular degradation of EBNA1 in epithelial cells remained unaffected even after the addition of the proteasome inhibitor, lactacystin. This was not the case when the same concentration of lactacystin (10 μM) was used to inhibit proteasome-dependent degradation of another EBV protein, LMP1 (Fig. 2 B). Surprisingly, these inhibitory effects do not prevent EBNA1 degradation in epithelial cells, suggesting that the rapid degradation of EBNA1 in epithelial cells does not proceed through the classical Ub/proteasome-dependent pathway.
Figure 2.
Ubiquitination analysis of EBNA1-GFP and EBNA1ΔGA-GFP in vitro. (A) SVMR6 keratinocytes were transiently cotransfected with expression vectors encoding HA-tagged 8xUb and LMP1-GFP, EBNA1-GFP, or EBNA1ΔGA-GFP. Ubiquitinated complexes were immunoprecipitated with an anti-HA–specific mAb and immunoblotted with anti-GFP. The ubiquitinated LMP1+ control is indicated. (B) Effect of the proteasomal inhibitor lactacystin on the stability of EBNA1-GFP and LMP1-GFP in epithelial cells. Duplicate aliquots of HaCaT cells were transfected with the expression construct EBNA1-GFP or LMP1-GFP. At 36 h after transfection, the proteasome inhibitor lactacystin was added at a final concentration of 10 μg/ml for 12 h to one of the duplicates. Both duplicates were then subjected to treatment with 50 μg/ml cycloheximide over a 6–8-h time course. Cell lysates at the indicated time points were separated by SDS-PAGE for immunoblotting with a GFP-specific antibody. The absence (−) or presence (+) of lactacystin is indicated. Densitometric analysis of the EBNA1-GFP, LMP1-GFP, EBNA1-GFP plus lactacystin, and LMP1-GFP plus lactacystin expression products are shown.
Endogenous Processing of CTL Epitopes from EBNA1 in EBV-infected B Cells.
Previous studies have demonstrated high frequencies of T cells in the peripheral circulation of HLA B35+ EBV-exposed individuals that recognize the EBNA1-derived CTL epitope HPV (13). These T cells can be readily expanded in vitro by stimulation with autologous LCLs. Blake et al. (14) proposed that this in vivo and in vitro stimulation of T cells proceeds through the cross-presentation of exogenous EBNA1 antigen, rather than direct stimulation by the EBV-infected B cells.
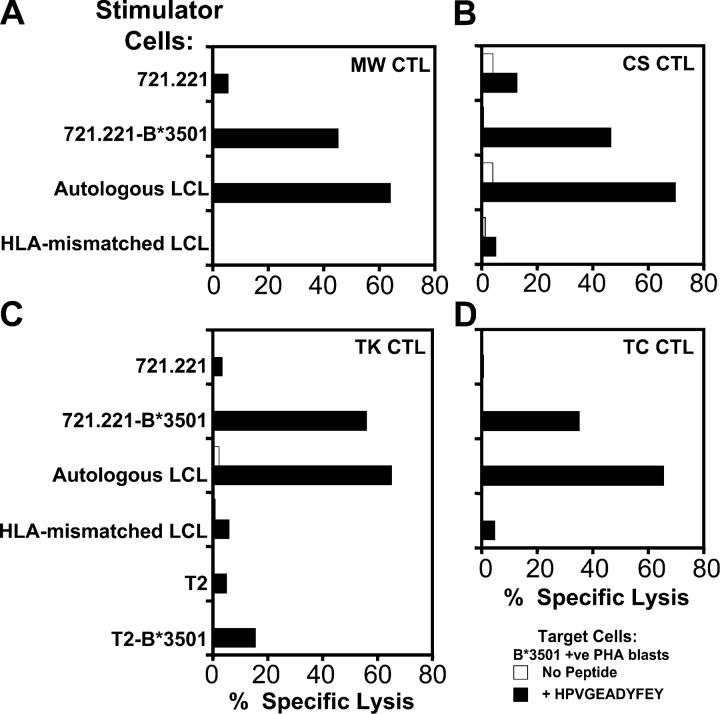
To delineate the possible mechanisms involved in the induction of EBNA1-specific T cell responses, PBMCs from four HLA B*3501+ EBV-exposed individuals (MW, CS, TK, and TC) were stimulated with either B*3501+ or B*3501− LCLs to generate CTL cultures that were then tested for reactivity with the HPV epitope. These stimulator cells included the autologous LCLs, an HLA-mismatched LCL, the class I− LCL 721.221, or the 721.221 cell line that had been transfected with the HLA B*3501 gene. In all cases, the HLA B*3501+ stimulator cells were shown to be highly efficient at stimulating EBNA1-specific CTLs, whereas the CTL cultures stimulated with B*3501− cell lines showed negligible peptide-specific cytotoxicity (Fig. 3). This stimulation of an HPV-specific memory response was clearly dependent on the expression of TAP in the stimulator cells as TAP 1– and TAP 2–deficient T2 cells transfected with B*3501 were unable to stimulate a significant HPV-specific response (Fig. 3 C). These data suggest that the HPV-specific T cell response can be directly activated by EBV-infected B cells and this EBNA1 epitope is efficiently processed through the TAP-dependent pathway.
Figure 3.
Direct stimulation of EBNA1-specific CTL responses in vitro using LCL stimulators. CTL bulk cultures were generated from the HLA B*3501+ EBV-seropositive donors MW, CS, TK, and TC by incubating PBMCs with irradiated LCLs (responder/stimulator ratio of 20:1). CTL cultures were split and restimulated with additional irradiated LCLs on day 7. Stimulator cells were the class I− LCL 721.221, HLA B*3501-transfected 721.221 cells, the autologous LCL for each donor, or an LCL from the B*3501− donor DM. CTL cultures were also generated from donor TK after stimulation with the T2 cell line or B*3501-transfected T2 cells. On day 10, each CTL bulk culture was screened in chromium release assays for lysis of HLA B*3501+ PHA blasts that had been pretreated with 1 μM of the HPV peptide for 1 h or left untreated. An E/T ratio of 20:1 was used in each of these assays. These data are a representation of two separate experiments.
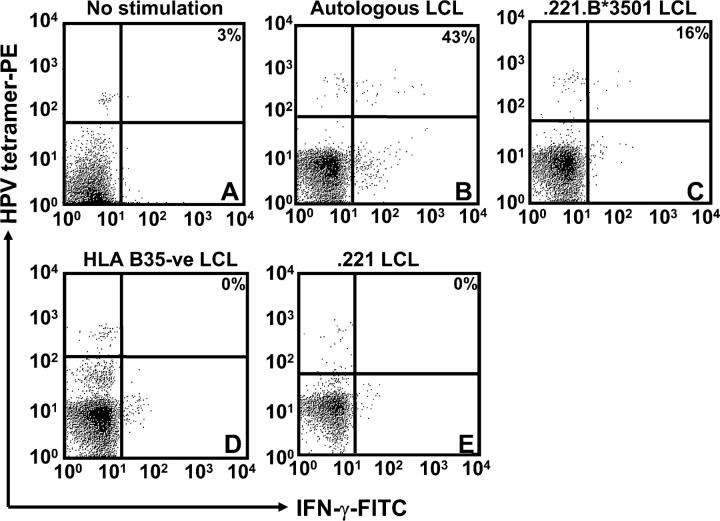
To further confirm these observations, we established an ex vivo stimulation assay, in which PBMCs from three unrelated HLA B35–seropositive individuals were stimulated with autologous LCLs, 721.221 LCL, 721.221 LCL transfected with HLA B*3501 (referred to as 0.221.B*3501 LCL), or HLA B35− LCLs. After overnight incubation, these cells were assessed for binding to an HPV-HLA B*3501 tetramer and intracellular IFN-γ expression. Data presented in Fig. 4 clearly demonstrate that a large proportion of HPV-HLA B*3501 tetramer+ T cells showed strong intracellular IFN-γ expression after stimulation with autologous LCL or the 0.221.B*3501 LCL. On the other hand, no ex vivo IFN-γ response was observed within the HPV tetramer+ cells when the PBMCs were stimulated with the HLA B35− LCLs or left unstimulated. A similar pattern of IFN-γ response was also observed in other donors when stimulated with HLA B35+ LCLs (not depicted) These observations further confirm our conclusions drawn from Fig. 3, which indicated that EBV-infected B cells can endogenously process and present CTL epitopes directly to the virus-specific T cells.
Figure 4.
Ex vivo intracellular IFN-γ production by EBNA1-specific T cells after stimulation with LCLs. PBMCs from an HLA B35+ donor were incubated alone (A) or with autologous LCL (B), 0.221.B35 LCL (C), HLA B35− LCL (D), or 0.221 LCL (E). Samples shown were gated on the CD8+ population, and then the percentage of CD8+ and HPV tetramer+ cells that were producing IFN-γ was assessed. The percentage of HPV-specific T cells producing IFN-γ after LCL stimulation is shown on the top right hand corner of each of the panels. These data are a representation of two separate experiments.
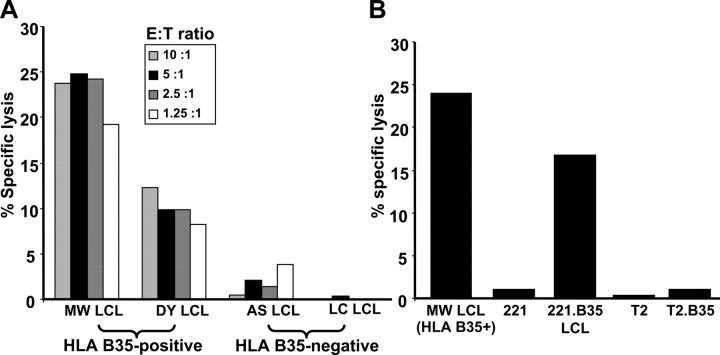
In the next set of experiments, we further assessed the endogenous processing of EBNA1 CTL epitopes by different cell types using in vitro cytotoxicity assays. In the first instance, we used a CTL clone(s) specific for the HLA B35–binding HPV epitope to assess CTL lysis of B cells expressing EBNA1. Blake et al. (13) have previously shown that T cells specific for this epitope are unable to recognize HLA-matched EBV-transformed LCLs. Contrary to these observations, we found that HPV-specific CTL clones do recognize HLA B35+ LCLs (Fig. 5 A), although there was some degree of variation in the level of CTL lysis amongst different HLA B35+ LCLs. It is important to mention here that the level of lysis observed for EBNA1-specific CTLs is quite comparable to that seen for CTL clones specific for other latent antigens (EBNA2-6; references 5 and 23). The fine specificity of this CTL recognition was further confirmed by the recognition of 721.221 LCLs transfected with HLA B35 (referred to as 0.221.B35 LCL), whereas no CTL lysis was observed for 721.221 LCLs or other LCLs that were negative for HLA B35 (Fig. 5, A and B). We have also demonstrated that the endogenous presentation of the HPV epitope was clearly dependent on the expression of TAP as T2 cells expressing HLA B35 were not recognized by these CTL clones (Fig. 5 B). In addition, the killing of HLA B35 LCLs was completely blocked by the pretreatment of these cells with BFA (not depicted). Taken together, these results suggest that in spite of the highly stable expression of EBNA1 in B cells, CTL epitopes within this protein can be directly presented by EBV-transformed LCLs, and this presentation proceeds through a TAP-dependent pathway.
Figure 5.
CTL recognition of endogenously processed EBNA1 epitopes. (A) HLA B35+ and HLA B35− LCLs were used as targets in a standard 51Cr-release assay to assess endogenous processing of EBNA1. (A) A CTL clone specific for the HLA B35-binding HPVGEADYFEY epitope was added to target cells at the E/T ratios indicated. (B) An HLA B35+ LCL, 721.221 LCLs, 721.221 LCLs transfected with HLA B*3501, T2 LCLs, and T2 LCLs transfected with HLA B*3501 were used as targets in a standard 51Cr-release assay to assess CTL activity to an HPVGEADYFEY-specific CTL clone at an E/T ratio of 5:1. These data are a representation of three separate experiments.
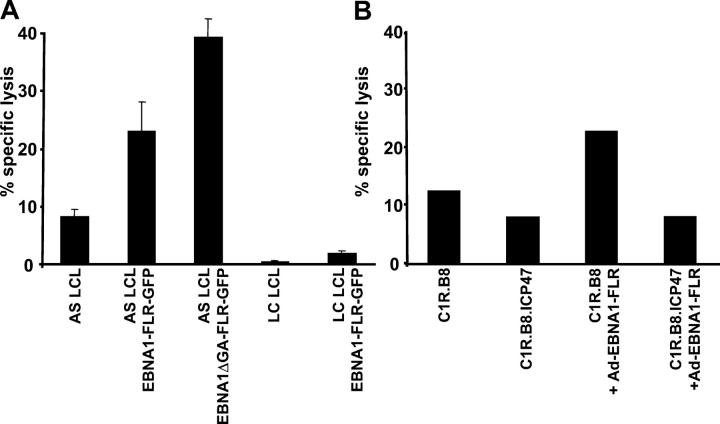
Although data presented in Fig. 5 clearly showed that EBNA1 epitopes can be endogenously processed through the class I pathway, it was important to further confirm these observations by using another human class I–restricted epitope. To address this issue, we tested the endogenous presentation of an inserted HLA B8–restricted CTL epitope, FLRGRAYGL (referred to as FLR), from an EBV-encoded EBNA3 protein within the EBNA1 COOH-terminal sequence (referred to as EBNA1-FLR), which allowed the study of endogenous processing of EBNA1 using an FLR-specific human CTL clone (11). The target cells were either transfected with expression vectors (EBNA1-FLR-GFP or EBNA1ΔGA-FLR-GFP) or infected with adenovirus encoding full-length EBNA1-FLR protein. It should be noted that target LCLs used in these assays were transformed with the B95.8 strain of EBV, which carries a mutation within the FLR epitope (29). As in the case of the HPV epitope, data presented in Fig. 6 shows that this epitope was endogenously processed by some HLA B8+ cells (AS LCL and C1R.B8) expressing EBNA1-FLR, whereas other HLA B8+ cells (LC LCLs) failed to present this epitope. The specificity of this recognition was confirmed by the inhibition of CTL killing of target cells coexpressing on HSV-1–encoded TAP inhibitor ICP47 (Fig. 6 B). These observations further confirm that CTL epitopes within EBNA1 can be endogenously processed and directly presented to CD8+ T cells through a TAP-dependent pathway.
Figure 6.
Endogenous processing of an inserted HLA B8–restricted CTL epitope within EBNA1. (A) Two HLA B8+ LCLs and the same LCLs transfected with either the EBNA1-FLR-GFP or EBNA1ΔGA-FLR-GFP expression constructs were used as targets in a standard 51Cr-release assay to assess CTL activity to a B8-specific CTL clone, LC13. An E/T ratio of 5:1 was used in this assay. (B) C1R.B8 LCLs, C1R.B8.ICP47 LCLs, and the same LCLs infected with a recombinant adenovirus encoding full-length EBNA1 (Ad-EBNA1-FLR) were used as targets in a standard 51Cr-release assay to assess CTL activity. An HLA B8–restricted FLR-specific CTL clone, LC13, was used as an effector in this assay. An E/T ratio of 5:1 was used in this assay. These data are a representation of two separate experiments.
Endogenous Processing of EBNA1 CTL Epitopes in Epithelial Cells.
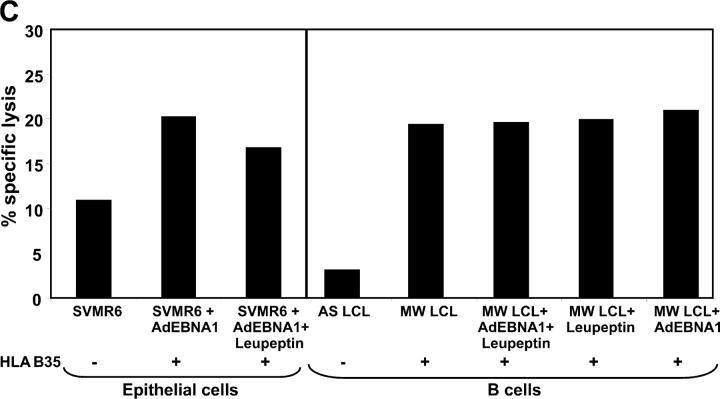
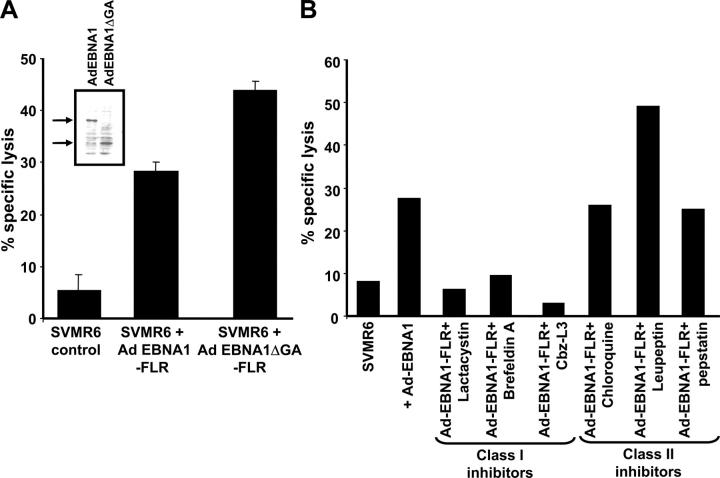
Our intracellular kinetic studies had shown that the EBNA1 protein was highly unstable in epithelial cells. To test the hypothesis that the rapid degradation of EBNA1 in these cells may enhance CTL recognition in vitro, SVMR6 cells were infected with a recombinant adenovirus expressing either full-length EBNA1-FLR or EBNA1ΔGA-FLR, and then exposed to the CTL clone (LC13) specific for the FLR epitope. Cells infected with these recombinant adenoviruses showed similar levels of protein expression (Fig. 7 A, inset). The data presented in Fig. 7 A shows that SVMR6 cells expressing either full-length EBNA1-FLR or EBNA1ΔGA-FLR were efficiently recognized by an LC13 CTL clone and the level of lysis was comparable to that seen in LCLs expressing EBNA1 constructs (Fig. 6). As with the endogenous processing of the EBNA1 epitopes in B cells, the CTL lysis of SVMR6 cells was also blocked in the presence of BFA. However, in contrast to the degradation of full-length EBNA1, the presentation of the FLR CTL epitope in SVMR6 cells was blocked by the proteasome inhibitors, lactacystin and Cbz-L3 (Fig. 7 B). These observations strongly suggest that the full-length EBNA1 protein is unlikely to be the primary source of endogenously processed epitopes. We hypothesize that EBNA1 CTL epitopes are primarily derived from DRiPs and this presentation is dependent on the proteasomal pathway. The endogenous presentation of the FLR epitope within EBNA1 was not affected by the addition of chloroquine or pepstatin. Unexpectedly, pretreatment of target cells with leupeptin significantly enhanced the CTL recognition of the FLR epitope within the COOH terminus of EBNA1 (Fig. 7 B). This increase in CTL lysis was not observed when leupeptin-treated target cells were presensitized with synthetic FLR peptide (not depicted). To determine whether this enhanced killing of target cells after leupeptin treatment occurred for another EBNA1 epitope, HLA B*3501-transfected SVMR6 cells and LCLs were infected with AdEBNA1 and then exposed to the HPV-specific CTL clone, DY1. Data presented in Fig. 7 C shows that leupeptin treatment of these cells had no effect on the CTL recognition by an HPV-specific CTL clone.
Figure 7.
Endogenous processing of EBNA1 CTL epitopes in epithelial cells and the effect of MHC class I and II inhibitors on endogenous processing of EBNA1 epitopes. (A) SVMR6 cells were infected with a recombinant adenovirus expressing either full-length EBNA1-FLR or EBNA1ΔGA-FLR. These cells were used as targets in a standard 51Cr-release assay to assess endogenous processing of an HLA B8–restricted FLR epitope encoded within EBNA1. The FLR-specific CTL clone LC13 was used as effector cells in the assay. An E/T ratio of 5:1 was used in the assay. The inset gel photo shows relative expression levels of full-length AdEBNA1 and AdEBNA1ΔGA after infection of SVMR6 cells. (B) SVMR6 keratinocytes were pretreated with either class I inhibitors, 10 μg/ml lactacystin, 1 μg/ml BFA, and 10 μg/ml Cbz-L3, or class II inhibitors, 80 μM chloroquine, 100 μM leupeptin, and 50 μM pepstatin, for 45 min. The cells were then infected with a recombinant adenovirus expressing full-length EBNA1 (Ad EBNA1-FLR). At 18 h after infection, the cells were used as targets in a standard 51Cr-release assay to assess endogenous presentation of the FLR epitope. FLR-specific CTL clone LC13 was used as effector cells in the assay. An E/T ratio of 5:1 was used in the assay. This data is a representation of three separate experiments. (C) CTL recognition of EBNA1-expressing SVMR6 cells and LCLs (HLA B35+, MW LCL; or HLA B35−, AS LCL) by HLA B35–restricted HPV-specific CTL clone (DY1). SVMR6 cells were transfected with an expression vector encoding the HLA B*3501 allele. Target cells were either pretreated with 100 μM leupeptin or left untreated. An E/T ratio of 5:1 was used in the assay.
Newly Synthesized Protein Provides the Primary Source of Endogenously Processed CD8+ Epitopes from EBNA1.
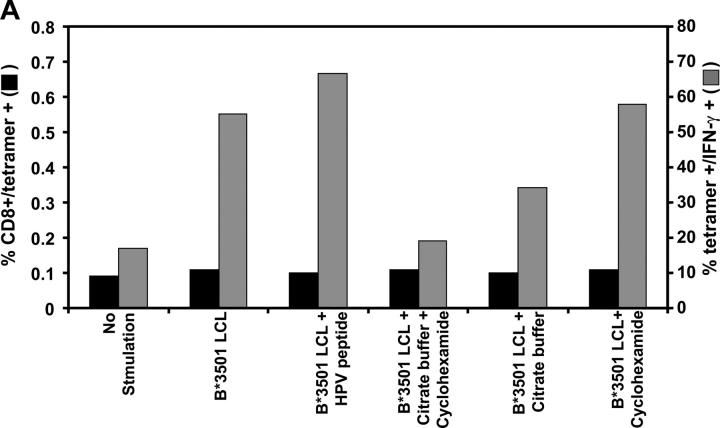
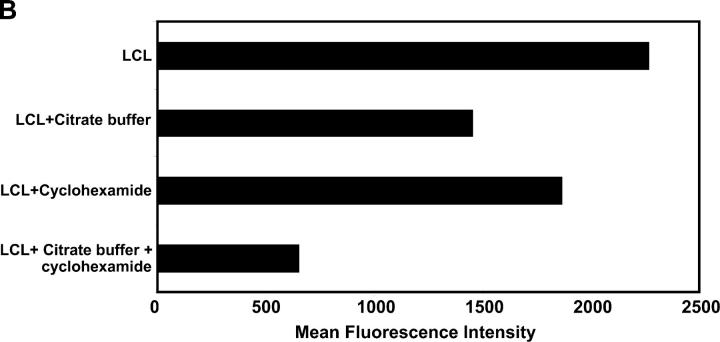
To determine whether the endogenously processed epitopes are derived from newly synthesized protein or from the long-lived stable EBNA1, we used the ex vivo stimulation assay in which PBMCs from an HLA B*3501+ EBV-seropositive individual (MW) were stimulated with MHC–peptide-stripped B*3501+ LCLs that had been incubated in the presence or absence of cycloheximide to block fresh protein synthesis. After overnight incubation, these cells were assessed for binding to an HPV HLA B*3501 tetramer and for intracellular IFN-γ expression. Data presented in Fig. 8 A demonstrate that >50% of HPV HLA B*3501 tetramer+ T cells showed strong intracellular IFN-γ expression after stimulation with untreated HLA B*3501+ LCLs. However, pretreatment of the LCLs with citrate buffer followed by incubation with cycloheximide significantly reduced the proportion of intracellular IFN-γ–expressing T cells within the HPV HLA B*3501 tetramer+ population (18%). On the other hand, LCLs treated with either citrate buffer (34.3%) or cycloheximide (53.1%) alone had a less significant effect on the stimulating capability of these cells. This data is consistent with the levels of surface MHC class I expression on LCLs after each of these treatments (Fig. 8 B). Furthermore, stimulation of PBMCs with PHA in the presence or absence of cycloheximide and/or citrate buffer showed comparable levels of IFN-γ expression (not depicted). These observations indicate that endogenously processed EBNA1 epitopes, which stimulate antigen-specific T cells, are primarily derived from newly synthesized protein rather than the long-lived stable EBNA1.
Figure 8.
(A) Effect of MHC–peptide stripping and cycloheximide treatment on ex vivo intracellular IFN-γ production by EBNA1-specific T cells. PBMCs from an HLA B35+ donor were incubated alone or with either an HLA B*3501+ LCL, an HLA B*3501+ LCL plus 0.01 μM HPV peptide, an HLA B*3501+ LCL treated with citrate buffer and 50 μM cycloheximide, an HLA B*3501+ LCL treated with citrate buffer, or an HLA LHLA B*3501+ LCL treated with cycloheximide. Data shown represents the CD8+ and tetramer+ population (solid bars) and the tetramer+ population producing IFN-γ (shaded bars). This data is a representation of two separate experiments. (B) Surface MHC class I expression on untreated LCLs or LCLs treated with either cycloheximide, citrate buffer alone, or cycloheximide and citrate buffer. LCLs were initially incubated with MHC class I–specific mAb (W6/32) followed by incubation with FITC-labeled anti–mouse Ig. The fluorescence intensity was measured by FACSCalibur™ and data were analyzed by CELLQuest™ software. The results are expressed as mean fluorescence intensity.
Discussion
This work provides a new perspective on the T cell recognition of EBNA1 and raises important questions on the current models to explain EBV persistence and the outgrowth of EBV-associated malignancies. A number of previous studies have proposed that EBV maintains latent infection of host cells by restricting its gene expression to a single protein, EBNA1 (30). This protein includes a unique GAr sequence that is thought to protect these virus-infected cells from immune recognition. The GAr sequence has a cis-inhibitory effect on its own degradation and blocks synthesis of EBNA1 protein in vitro and in vivo (8, 12). In spite of these inhibitory mechanisms, ex vivo analysis of EBV-specific T cell responses revealed that not only could EBNA1-specific T cells be readily detected, but in some individuals these responses constituted a major component of the EBV-specific T cell memory response (13, 14). The precursor frequency for these T cells was in the same range as that seen for epitopes within the immunodominant EBNA3 proteins (14). Because these EBNA1-specific CTLs showed no direct recognition of EBV-infected B cells, Blake et al. (13, 14) proposed that much of this antigen-specific memory response is generated through the cross-presentation of this antigen through a TAP-independent pathway. However, data presented in this work suggest that EBNA1 epitopes can be directly presented by EBV-infected B cells through a proteasome/TAP-dependent pathway.
An interesting feature of this observation was that the endogenous presentation of the EBNA1 CTL epitope appears to be independent of the intracellular stability of this antigen. In spite of the highly stable expression of EBNA1 in B cells, the level of CTL lysis of these cells was quite comparable to that of epithelial cells, which showed less stable expression of EBNA1. Surprisingly, the rapid degradation of EBNA1 in epithelial cells appears to proceed through a proteasome-independent pathway, whereas the endogenous presentation of CTL epitopes within this protein was blocked in the presence of proteasome-specific inhibitors. This unexpected finding raised an important question on the source of endogenously processed EBNA1 epitopes. Recent studies have proposed a novel source of peptides associated with MHC class I molecules. These peptides are mainly derived from DRiPs, which were referred to as “unavoidable imperfections in the process of translating genetic information into functional proteins” (17). There is increasing evidence to suggest that DRiPs can account for a large fraction (∼30%) of newly synthesized proteins that are rapidly degraded by proteasomes (16, 31). The data presented in this work is consistent with the hypothesis that the primary source of endogenously processed EBNA1 epitopes is derived from newly synthesized protein rather than the long-lived stable EBNA1. Indeed, the treatment of EBV-transformed LCLs with citrate buffer and cycloheximide, which blocked the endogenous presentation of epitopes from newly synthesized EBNA1, significantly impaired the stimulation of antigen-specific T cells.
Further support for this hypothesis comes from an unexpected observation that the endogenous presentation of at least one of the epitopes (FLR) within EBNA1 was enhanced after the addition of a cysteine protease inhibitor (leupeptin). Yewdell et al. (17) have recently proposed that after the degradation of DRiPs by proteasomes, peptides of various lengths are produced and many of these peptides are substrates for various cytosolic peptidases that further degrade these peptides into single amino acids that are recycled for the synthesis of new proteins. Thus, it is possible that pretreatment of target cells with leupeptin inhibits cysteine proteases that either specifically cleave the FLR epitope sequence alone or the DRiPs that include this epitope from the extreme COOH-terminal region of EBNA1. This argument is also supported by previous studies showing that endogenous processing of some MHC class I epitopes can be influenced by both proteasomes and cysteine proteases (32).
It is important to mention here that in contrast to other proteins, most of the endogenously processed CTL epitopes from EBNA1 might be derived from DRiPs because the GAr sequence within EBNA1 blocks proteasomal degradation. Because many of these DRiPs may not include GAr sequences, the presentation of CTL epitopes from DRiPs proceeds through a classic TAP/proteasome-dependent pathway. Indeed, data presented in this work and previous studies published by Blake et al. have shown that expression of full-length EBNA1 either by vaccinia (13) or adenovirus (Fig. 7, inset) results in the expression of GAr-deleted truncated products, which is consistent with the presence of DRiPs. Furthermore, this view is compatible with recent findings that other EBNA1 epitopes are processed independent of their location relative to the GAr domain (33, 34).
The demonstration of endogenous processing of EBNA1 and the lysis of EBV-transformed LCLs by EBNA1-specific CTLs needs to be addressed in the context of previous observations that suggested that this protein escapes T cell recognition by inhibiting its proteasomal degradation. One possible reason for the discrepancy relates to the difficulty of isolating EBNA1-specific CTLs using EBV-transformed LCLs as stimulators that are known to selectively expand T cells specific for immunodominant EBNA3 family proteins. However, the use of overlapping peptides from EBNA1 to stimulate T cells in vitro, and newer and more sensitive technologies to assess immune responses, have facilitated opportunities to not only map EBNA1 epitopes, but also to investigate the issue of the endogenous processing of EBNA1. Although the data presented in this work is consistent with the previous biochemical observations that the GAr domain inhibits degradation of the full-length EBNA1 protein in B cells (9), this inhibitory effect is not absolute in blocking CD8+ CTL recognition of EBV-transformed LCLs.
Overall, this work has important implications for the immune control of EBV-associated diseases. First, these observations provide a new opportunity for the development of novel therapeutic strategies against EBV-associated malignancies. Although much of the emphasis on the development of therapeutic vaccines for EBV-associated nasopharyngeal carcinoma and Hodgkin's disease have primarily concentrated on latent membrane antigens 1 and 2, inclusion of EBNA1 epitopes may significantly enhance their therapeutic potential. These studies also open the possibility of targeting Burkitt lymphoma cells through EBNA1, although the loss of TAP expression in these malignant cells remains a major hindering factor. The second important implication relates to the latent infection of EBV in healthy virus carriers. Previous studies have proposed that because EBNA1 is required to maintain the viral genome, latently infected B cells restrict viral gene expression to EBNA1 only, thus limiting immune recognition by T cells. This assumption was based on the observations that T cell epitopes from EBNA1 are not endogenously processed and presented by EBV-infected B cells. However, the data presented in this work has raised questions on the relevance of this hypothesis. In a recent study, Hochberg et al. (35) have shown that most of the EBV-infected B cells in the blood do not express any detectable latent antigen mRNA or proteins. However, when these infected cells divide, they express EBNA1 only. These EBNA1-expressing dividing B cells might be controlled by EBNA1-specific T cells, whereas a long-term latent infection is maintained by completely switching off the expression of all EBV-latent genes.
Acknowledgments
We wish to thank Wendy Van Zuylen for technical assistance.
This work was supported by funding from the National Health and Medical Research Council (NH & MRC), Canberra, Australia.
S.R. Burrows and R. Khanna contributed equally to this work.
Abbreviations used in this paper: BFA, brefeldin A; DRiPs, defective ribosomal products; EBNA, EBV-encoded nuclear antigen; GAr, glycine-alanine repeat; GFP, green fluorescent protein; HA, hemagglutinin; LCL, lymphoblastoid cell line; LMP1, latent membrane protein 1; TAP, transporter associated with antigen processing; Ub, ubiquitin.
References
- 1.Doherty, P.C., J.P. Christensen, G.T. Belz, P.G. Stevenson, and M.Y. Sangster. 2001. Dissecting the host response to a gamma-herpesvirus. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356:581–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benoist, C., and D. Mathis. 1992. Generation of the alpha beta T-cell repertoire. Curr. Opin. Immunol. 4:156–161. [DOI] [PubMed] [Google Scholar]
- 3.Khanna, R., and S.R. Burrows. 2000. Role of cytotoxic T lymphocytes in Epstein-Barr virus-associated diseases. Annu. Rev. Microbiol. 54:19–48. [DOI] [PubMed] [Google Scholar]
- 4.Tortorella, D., B.E. Gewurz, M.H. Furman, D.J. Schust, and H.L. Ploegh. 2000. Viral subversion of the immune system. Annu. Rev. Immunol. 18:861–926. [DOI] [PubMed] [Google Scholar]
- 5.Murray, R.J., M.G. Kurilla, J.M. Brooks, W.A. Thomas, M. Rowe, E. Kieff, and A.B. Rickinson. 1992. Identification of target antigens for the human cytotoxic T-cell response to Epstein-Barr virus (EBV): implications for the immune control of EBV-positive malignancies. J. Exp. Med. 176:157–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trivedi, P., G. Winberg, and G. Klein. 1997. Differential immunogenicity of Epstein-Barr virus (EBV) encoded growth transformation-associated antigens in a murine model system. Eur. J. Cancer. 33:912–917. [DOI] [PubMed] [Google Scholar]
- 7.Mukherjee, S., P. Trivedi, D.M. Dorfman, G. Klein, and A. Townsend. 1998. Murine cytotoxic T lymphocytes recognize an epitope in an EBNA-1 fragment, but fail to lyse EBNA-1–expressing mouse cells. J. Exp. Med. 187:445–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levitskaya, J., A. Sharipo, A. Leonchiks, A. Ciechanover, and M.G. Masucci. 1997. Inhibition of ubiquitin/proteasome-dependent protein degradation by the Gly-Ala repeat domain of the Epstein-Barr virus nuclear antigen 1. Proc. Natl. Acad. Sci. USA. 94:12616–12621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levitskaya, J., M. Coram, V. Levitsky, S. Imreh, P.M. Steigerwald-Mullen, G. Klein, M.G. Kurilla, and M.G. Masucci. 1995. Inhibition of antigen processing by the internal repeat region of the Epstein-Barr virus nuclear antigen 1. Nature. 375:685–688. [DOI] [PubMed] [Google Scholar]
- 10.Dantuma, N.P., A. Sharipo, and M.G. Masucci. 2002. Avoiding proteasomal processing: the case of EBNA1. Curr. Top. Microbiol. Immunol. 269:23–36. [DOI] [PubMed] [Google Scholar]
- 11.Tellam, J., M. Sherritt, S. Thomson, R. Tellam, D.J. Moss, S.R. Burrows, E. Wiertz, and R. Khanna. 2001. Targeting of EBNA1 for rapid intracellular degradation overrides the inhibitory effects of the Gly-Ala repeat domain and restores CD8+ T cell recognition. J. Biol. Chem. 276:33353–33360. [DOI] [PubMed] [Google Scholar]
- 12.Yin, Y., B. Manoury, and R. Fahraeus. 2003. Self-inhibition of synthesis and antigen presentation by Epstein-Barr virus-encoded EBNA1. Science. 301:1371–1374. [DOI] [PubMed] [Google Scholar]
- 13.Blake, N., S. Lee, I. Redchenko, W. Thomas, N. Steven, A. Leese, P. Steigerwald-Mullen, M.G. Kurilla, L. Frappier, and A. Rickinson. 1997. Human CD8+ T cell responses to EBV EBNA1: HLA class I presentation of the (Gly-Ala)-containing protein requires exogenous processing. Immunity. 7:791–802. [DOI] [PubMed] [Google Scholar]
- 14.Blake, N., T. Haigh, G. Shaka'a, D. Croom-Carter, and A. Rickinson. 2000. The importance of exogenous antigen in priming the human CD8+ T cell response: lessons from the EBV nuclear antigen EBNA1. J.Immunol. 165:7078–7087. [DOI] [PubMed] [Google Scholar]
- 15.Yewdell, J.W., L.C. Anton, and J.R. Bennink. 1996. Defective ribosomal products (DRiPs): a major source of antigenic peptides for MHC class I molecules? J. Immunol. 157:1823–1826. [PubMed] [Google Scholar]
- 16.Schubert, U., L.C. Anton, J. Gibbs, C.C. Norbury, J.W. Yewdell, and J.R. Bennink. 2000. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature. 404:770–774. [DOI] [PubMed] [Google Scholar]
- 17.Yewdell, J.W., E. Reits, and J. Neefjes. 2003. Making sense of mass destruction: quantitating MHC class I antigen presentation. Nat. Rev. Immunol. 3:952–961. [DOI] [PubMed] [Google Scholar]
- 18.Rickinson, A.B., and D.J. Moss. 1997. Human cytotoxic T lymphocyte responses to Epstein-Barr virus infection. Annu. Rev. Immunol. 15:405–431. [DOI] [PubMed] [Google Scholar]
- 19.Alexander, J., J.A. Payne, R. Murray, J.A. Frelinger, and P. Cresswell. 1989. Differential transport requirements of HLA and H-2 class I glycoproteins. Immunogenetics. 29:380–388. [DOI] [PubMed] [Google Scholar]
- 20.Shimizu, Y., D.E. Geraghty, B.H. Koller, H.T. Orr, and R. DeMars. 1988. Transfer and expression of three cloned human non-HLA-A,B,C class I major histocompatibility complex genes in mutant lymphoblastoid cells. Proc. Natl. Acad. Sci. USA. 85:227–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boukamp, P., R.T. Petrussevska, D. Breitkreutz, J. Hornung, A. Markham, and N.E. Fusenig. 1988. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J. Cell Biol. 106:761–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simmons, N.L. 1990. A cultured human renal epithelioid cell line responsive to vasoactive intestinal peptide. Exp. Physiol. 75:309–319. [DOI] [PubMed] [Google Scholar]
- 23.Burrows, S.R., S.J. Rodda, A. Suhrbier, H.M. Geysen, and D.J. Moss. 1992. The specificity of recognition of a cytotoxic T lymphocyte epitope. Eur. J. Immunol. 22:191–195. [DOI] [PubMed] [Google Scholar]
- 24.Yotnda, P., H. Onishi, H.E. Heslop, D. Shayakhmetov, A. Lieber, M. Brenner, and A. Davis. 2001. Efficient infection of primitive hematopoietic stem cells by modified adenovirus. Gene Ther. 8:930–937. [DOI] [PubMed] [Google Scholar]
- 25.Moss, D.J., I.S. Misko, S.R. Burrows, K. Burman, R. McCarthy, and T.B. Sculley. 1988. Cytotoxic T-cell clones discriminate between A- and B-type Epstein-Barr virus transformants. Nature. 331:719–721. [DOI] [PubMed] [Google Scholar]
- 26.Princiotta, M.F., U. Schubert, W. Chen, J.R. Bennink, J. Myung, C.M. Crews, and J.W. Yewdell. 2001. Cells adapted to the proteasome inhibitor 4-hydroxy- 5-iodo-3-nitrophenylacetyl-Leu-Leu-leucinal-vinyl sulfone require enzymatically active proteasomes for continued survival. Proc. Natl. Acad. Sci. USA. 98:513–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aviel, S., G. Winberg, M. Massucci, and A. Ciechanover. 2000. Degradation of the Epstein-Barr virus latent membrane protein 1 (LMP1) by the ubiquitin-proteasome pathway. Targeting via ubiquitination of the N-terminal residue. J. Biol. Chem. 275:23491–23499. [DOI] [PubMed] [Google Scholar]
- 28.Holowaty, M.N., M. Zeghouf, H. Wu, J. Tellam, V. Athanasopoulos, J. Greenblatt, and L. Frappier. 2003. Protein profiling with Epstein-Barr nuclear antigen-1 reveals an interaction with the herpesvirus-associated ubiquitin-specific protease HAUSP/USP7. J. Biol. Chem. 278:29987–29994. [DOI] [PubMed] [Google Scholar]
- 29.Apolloni, A., D. Moss, R. Stumm, S. Burrows, A. Suhrbier, I. Misko, C. Schmidt, and T. Sculley. 1992. Sequence variation of cytotoxic T-cell epitopes in different isolates of Epstein-Barr virus. Eur. J. Immunol. 22:183–189. [DOI] [PubMed] [Google Scholar]
- 30.Chen, F., J.Z. Zou, L. di Renzo, G. Winberg, L.F. Hu, E. Klein, G. Klein, and I. Ernberg. 1995. A subpopulation of normal B cells latently infected with Epstein-Barr virus resembles Burkitt lymphoma cells in expressing EBNA-1 but not EBNA-2 or LMP1. J. Virol. 69:3752–3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Princiotta, M.F., D. Finzi, S.B. Qian, J. Gibbs, S. Schuchmann, F. Buttgereit, J.R. Bennink, and J.W. Yewdell. 2003. Quantitating protein synthesis, degradation, and endogenous antigen processing. Immunity. 18:343–354. [DOI] [PubMed] [Google Scholar]
- 32.Lopez, D., and M. Del Val. 1997. Selective involvement of proteasomes and cysteine proteases in MHC class I antigen presentation. J. Immunol. 159:5769–5772. [PubMed] [Google Scholar]
- 33.Voo, K.S., T. Fu, H.Y. Wang, J. Tellam, H.E. Heslop, M.K. Brenner, C.M. Rooney, and R.F. Wang. 2004. Evidence for the presentation of major histocompatibility complex class I–restricted Epstein-Barr virus nuclear antigen 1 peptides to CD8+ T lymphocytes. J. Exp. Med. 199:459–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee, S.P., J.M. Brooks, H. Al-Jarrah, W.A. Thomas, T.A. Haigh, G.S. Gaylor, S. Humme, A. Schepers, W. Hammerschmidt, J. Yates, et al. 2004. CD8 T cell recognition of endogenously expressed Epstein-Barr virus nuclear antigen 1. J. Exp. Med. 199:1409–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hochberg, D., J.M. Middeldorp, M. Catalina, J.L. Sullivan, K. Luzuriaga, and D.A. Thorley-Lawson. 2004. Demonstration of the Burkitt's lymphoma Epstein-Barr virus phenotype in dividing latently infected memory cells in vivo. Proc. Natl. Acad. Sci. USA. 101:239–244. [DOI] [PMC free article] [PubMed] [Google Scholar]