Abstract
In B lineage progenitors, V(D)J recombination occurs only during distinct stages of development and is restricted to immunoglobulin loci. This process is thought to be controlled by both regulated expression of the V(D)J recombinase and by limited accessibility of target loci to the recombinase complex. However, it is unknown whether these two processes occur concomitantly in developing B lineage progenitors or whether these events are temporally distinct and, therefore, potentially independently regulated. To distinguish between these possibilities, we developed a transgenic V(D)J recombination substrate that is not governed by the same chromatin remodeling constraints as endogenous immunoglobulin heavy chain (IgH) loci and examined the requirements for V(D)J recombination to initiate in early B lineage progenitors. We find that single B lineage precursors express an active V(D)J recombinase in vivo before the stage when IgH rearrangements are frequently detectable. Our results indicate that the onset of recombinase activity and the initiation of IgH recombination are developmentally distinct events in the B lineage.
Keywords: B lymphopoiesis, V(D)J recombination, chromatin, immunoglobulin, heavy chain, hematopoiesis
Introduction
V(D)J recombination, the site-specific DNA recombination process that assembles genes encoding variable regions of antigen receptor loci, produces the extraordinary diversity characteristic of the adaptive immune response. During B and T lymphopoiesis, two key events must occur in order for V(D)J recombination to initiate: (a) developmentally regulated expression of an active V(D)J recombinase (1, 2) and (b) permissive changes in chromatin accessibility of antigen receptor loci (3–7). Although great strides have been made toward understanding the role of chromatin modification in enabling V(D)J recombination, it is unknown whether induction of recombinase expression occurs concomitant with changes in locus accessibility or whether these events are temporally distinct and, therefore, potentially independently regulated.
The V(D)J recombinase is composed of recombinase-activating gene (RAG)1 and RAG2 as well as Ku70 and Ku80, DNA-PKcs, DNA ligase IV, XRCC4, and Artemis. The RAG proteins are the only components of the recombinase currently known to exhibit lineage specificity. rag genes are transcribed in both a lymphoid-restricted (8, 9) and stage-specific (2, 10–13) manner and are thought to limit V(D)J recombinase activity to developing B and T cells.
It is critical to appreciate at least two lines of evidence suggesting that expression of rag transcripts alone cannot be a definitive indicator of the presence of recombinase activity. First, B and T cell precursors that express rag transcripts but lack RAG protein have been identified (2, 14). Second, although RAG2 (but not RAG1) levels fluctuate greatly during cell division, transcription of both rag genes remains relatively steady state (14, 15). Moreover, because RAG degradation is tied to the cell cycle recombinase activity depends not only on accumulation of RAG proteins but also relocalization of RAG2 to the nucleus subsequent to each cell division (14–16). Finally, it is also unknown whether the presence of RAG protein accurately indicates activity of the entire V(D)J recombinase complex. For these reasons, the developmental expression of recombinase activity in the earliest lymphoid subsets remains unclear.
Within the B cell compartment, rag transcripts are readily detectable in the fraction B (IgM−B220+CD43+CD24+) stage of development (11) when ∼60% of cells have D-JH joins (1, 13). In contrast, only rare (<4% of cells) D-JH rearrangements are detectable in fraction A2 (17). Recent evidence suggests that rag genes may be expressed in fraction A2 or even earlier stages of development. In fraction A2 (B220+AA4.1+CD4−CD24−), rag2 locus transcription was detectable using a green fluorescence protein (GFP) reporter construct (12) but not by PCR analysis (13, 18). As another example, rag1 and rag2 expression is detectable in lin−ckitlo progenitors, bipotential cells that retain the capacity to give rise to B cells and NK cells in vitro (19), and DH-JH rearrangements are detectable among CLPs (20).
Currently, it is unknown when V(D)J recombinase expression and activity occurs relative to the changes in accessibility that enable recombination of Ig antigen receptor loci. For example, finding only rare D-JH rearrangements in fraction A2 could reflect lack of recombinase activity or inability to recombine Ig heavy chain (IgH) due to constrained chromatin. This important issue bears not only on our understanding of basic control of V(D)J recombination initiation but also on understanding extrinsic signals that may control these two processes separately. To begin to address this problem, we developed a flow cytometric assay for V(D)J recombinase activity in vivo in which V(D)J recombination is indicated by a fluorescent reporter gene. This approach enables us to examine in vivo the relationship between V(D)J recombinase activity and antigen receptor locus rearrangement in developing lymphocyte precursors.
Materials and Methods
Mice.
C57BL/6 and RAG1−/− mice were obtained from the Jackson Laboratories. RAG2 GFP reporter NG BAC mice (FVBN) (12) were backcrossed to C57BL/6 in our laboratory for 12 generations. All mice were treated humanely in accordance with federal and state government guidelines and UMMS institutional animal committees.
Construction of H2-SVEX Transgenic Mice.
The H2-SVEX transgene was constructed by placing the RSS-VEX-RSS fragment (Fig. 1 A) into the H2K (HIL) transgenic vector using a unique NotI restriction site located between the H2 promoter and the H2 exon fragment as described previously (21). The H2K cassette vector expresses genes under the control of the H2K promoter/enhancer and Moloney MuLV enhancer/poly(A), typically at high levels in HSC and all hematolymphoid cells (21). Inbred C57BL/6 transgenic mice were generated using standard procedures (21).
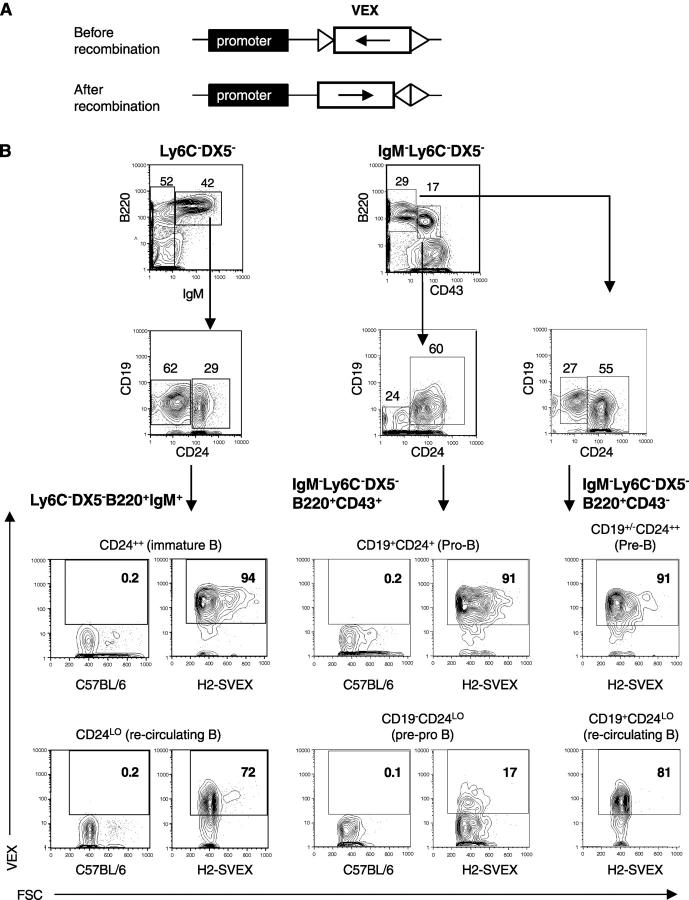
Figure 1.
V(D)J recombinase activity in early B lineage precursors. (A) The transgenic H2-SVEX substrate contains VEX (white rectangle) driven by the murine H2K promoter (black rectangle). VEX within the substrate is initially in the antisense orientation and is flanked by V(D)J recombination signal sequences (triangles) which direct inversional recombination. (B) Bone marrow from SB110 H2-SVEX transgenic mice was stained with antibodies to Ly6C, DX5, B220, CD43, CD19, CD24, and IgM in order to examine VEX expression throughout B cell development. The percentage of cells in each region is given. The data are representative of three independent experiments.
Single Cell PCR.
For single cell PCR analysis, cells were sorted directly into Eppendorf 96-well plates containing 10 μl/well of lysis buffer (1× Promega Mg2+-free PCR buffer, 0.5 mg/ml proteinase K, 9.2 μg/ml tRNA [Sigma-Aldrich]). Plates were incubated at 55°C for 1 h, 95°C for 10 min, and stored at −80°C. The PCR protocol involves two rounds of amplification with heminested primers (1, 22). Primers used were DFLe1 (5′), ACAAGCTTCAAAGCACAATGCCTGGCT; DFLe(G+C) (5′), ACGTCGACTTTT(G or C)TCAAGGGATCTACTACTGT; JH4e1 (3′), AGGCTCTGAGATCCCTAGACAG; JH4e2 (3′), GGGTCTAGACTCTCAGCCGGCTCCCTCAGGG; and J1 (3′), AATGTGCAGAAAGAAAAAAGCCAG. In the first round, PCR amplification was performed with 10 μl single cell lysate (or 1 μl of control DNA + 9 μl ddH2O) in 25 μl total volume containing 0.25 μM dNTP, 0.5 U KlenTaq LA, 1× KlenTaq LA buffer (40 mM Tricine-KOH [pH 9.2 at 25], 15 mM KOAc, 3.5 mM Mg(OAc)2, 7.5 μg/μl BSA), 0.5 mg/ml BSA, and 2 μM of each first round primer (DFLe1, 5′JH1, JH4e1). In the second round, 1 μl of first round product was added to a 25-μl reaction volume using the same conditions described above except that second round primers (DFLe[G+C] and 5′JH4 to detect D-JH; 5′JH1 and JH4e2 to detect germline) were used. Both rounds of PCR used the following conditions: 95°C for 1 min, 41 cycles of 94°C for 1 min, 60°C for 1 min, 72°C for 90 s, and 72°C for 10 min. Only samples producing either a germline product or a specific rearrangement product are included in the analysis (∼68% of sorted single cells yielded a PCR product).
Cell Sorting and Flow Cytometry.
Freshly isolated cells were resuspended to 3 × 107/ml in staining media containing biotin-, flavin-, and phenol red–deficient RPMI 1640 (Irvine Scientific), 10 mM Hepes, pH 7.2, 0.02% sodium azide, 1 mM EDTA, and 3% newborn calf serum, and treated with 2.4G2 Fc block for 10 min on ice. Cells were incubated with primary antibodies for 20 min, then washed three times, incubated with streptavidin reagents for 15 min, and then washed three more times. After the final wash, samples were resuspended in 1 μg/ml propidium iodide to exclude dead cells. VEX was detected using 407-nm excitation (23), and GFP from NG transgenics was detected using 488-nm excitation. Primary antibodies included AA4.1 biotin, B220 APC or Cy5PE or FITC, BP-1 biotin (6C3), CD4 Cy5PE or PE, CD19 Cy5PE or FITC, CD24 (clone 30-F1) Alexa 594, Cascade blue or FITC, CD43 PE, Ly6C biotin or FITC, DX5 biotin or FITC, and IgM (clone 331) biotin or FITC. Secondary reagents were SA-Cy5PE, SA-Alexa 594, SA-Cy7PE, or SA-APC. Antibodies were purchased from BD Biosciences, eBioscience, Southern Biotechnology Associates, Inc., or CALTAG, or purified and conjugated as described (23). Flow cytometry was performed on a 3 laser, 7 detector DIVA FACSVantage or a 3 laser 9 detector LSRII (Becton Dickinson). Data were analyzed with FlowJo software (Tree Star).
Results and Discussion
The H2-SVEX Transgenic Recombination Substrate.
The V(D)J recombination substrate H2-SVEX expresses VEX-GFP (23) as a consequence of V(D)J recombination. As shown in Fig. 1 A, the VEX gene is initially in the antisense orientation and is flanked by V(D)J recombination signal sequences (RSSs). RSSs in this orientation mediate inversion such that after recombination VEX is in the correct orientation for expression. Cells that undergo V(D)J recombination are VEX+; these cells are easily detected and quantified by FACS®. VEX expression and H2-SVEX rearrangement completely depends on the presence of rag1, and coding joint and signal joint recombination products have the expected sequence (21).
H2-SVEX recombination is not governed by the same chromatin remodeling constraints as endogenous IgH or TCR antigen receptor loci for two reasons. First, the H2-SVEX construct is driven by an active H2K promoter/MoMLV enhancer that enables robust transgene expression, and presumably, accessibility in primary pro-B and pro-T cells (21). Second, the H2-SVEX substrate does not contain Ig (or TCR) locus transcription regulatory regions and, therefore, cannot be regulated in the same manner as endogenous IgH (or TCR) loci. Thus, the absence of common regulatory motifs between the H2-SVEX substrate and endogenous antigen receptor loci enables us to distinguish cells with an active V(D)J recombinase but inaccessible IgH loci from cells that have an active recombinase and accessible loci.
V(D)J Recombinase Activity in the Earliest B Lineage Precursors.
We have shown previously that VEX expression is both efficient and restricted to lymphocytes. For example, 84–86% of CD19+IgM+ splenic cells and 91–93% of CD3+ splenic T cells are VEX+, whereas <0.5% of myeloid and granulocytes express VEX (21). As bone marrow pro-B cells are actively rearranging IgH genes and preB cells are actively rearranging IgL genes (1, 11, 22), pro- and preB cells should express an active V(D)J recombinase capable of rearranging the H2-SVEX substrate, and we examined VEX expression at these stages of development.
As shown in Fig. 1 B, within the DX5−Ly6C−IgM− subset of bone marrow, VEX is expressed in 91% of both B220+CD43+CD19+CD24+ pro-B cells (third row, middle panels) and B220+CD43−CD19+CD242+ preB cells (third row, right panel). Similar results were obtained with two independent H2-SVEX founder lines (SB110 and SB88), indicating position-independent recombination of the transgenic substrate (not depicted). Importantly, the B220+CD43+CD19−CD24LO pre-pro-B cell population (24) has 17% VEX+ cells (Fig. 1 B, fourth row, middle panels). This pre-pro-B cell population has been shown to produce B lineage lymphocytes with extraordinarily high cloning efficiency and is one of the earliest defined B lineage–restricted subsets (24).
Because VEX is driven by a constitutively active promoter, VEX acts a permanent marker of cells that have, or had, V(D)J recombinase activity. Therefore, typically >90% of immature bone marrow B cells (Fig. 1 B, third row, left panels) are VEX+, whereas rag1 and rag2 expression are down-regulated in this B cell population (2). Interestingly, a small decrease in the percentage of VEX+ cells is consistently detected within more mature B cell subsets, since 84% of IgM+CD19+ splenic B cells (21) and 72% of recirculating IgM+ bone marrow B cells (Fig. 1 B, fourth row, left panels) are VEX+. This decrease is unlikely to be due to reduced expression of the recombination reporter transgene because expression of a second transgene, H2-BEX (which is analogous to H2-SVEX except that V(D)J recombination is not required for GFP expression), is expressed at uniformly high levels (>95%) throughout all stages of B cell development (not depicted). Alternatively, it is possible that somatic hypermutation or other late stage events may influence H2-SVEX recombination or expression. Thus, H2-SVEX may be a useful tool for examining molecular processes characteristic of mature B cells. Together, these experiments indicate that H2-SVEX is efficiently recombined in the earliest B lineage progenitors in vivo and that VEX expression is readily detectable in these developmental populations.
Simultaneous Analysis of rag Expression and Recombinase Activity in B Lineage Progenitors.
Progenitor B cells can be divided into specific developmental subsets in which D-JH rearrangements are not readily detectable. Fraction A cells largely retain germline configuration of endogenous IgH loci (11, 12, 17), and there are two possible explanations for this observation: (a) the V(D)J recombinase is not yet expressed or (b) the V(D)J recombinase is expressed but IgH loci are not accessible to the recombinase complex due to chromatin restrictions. We predicted that if the V(D)J recombinase is active as early as the fraction A stage of development, it will recombine the H2-SVEX target regardless of endogenous IgH loci susceptibility.
For these experiments, we analyzed VEX expression in progenitor B subsets derived from the progeny of crosses between H2-SVEX and NG transgenic RAG2 reporter mice (12). This approach enables us to examine both RAG2 gene transcription (GFP) and V(D)J recombinase activity (VEX) in single lymphocyte progenitors. VEX and GFP have distinct excitation requirements, and thus can be readily resolved (23). Fig. 2 A depicts GFP versus VEX (within B220+CD43+DX5−Ly6C−IgM− cells) for pre-pro-B (CD19−CD24−/LO), early pro-B (CD19−CD24+), and late pro-B (CD19+CD24+) cells.
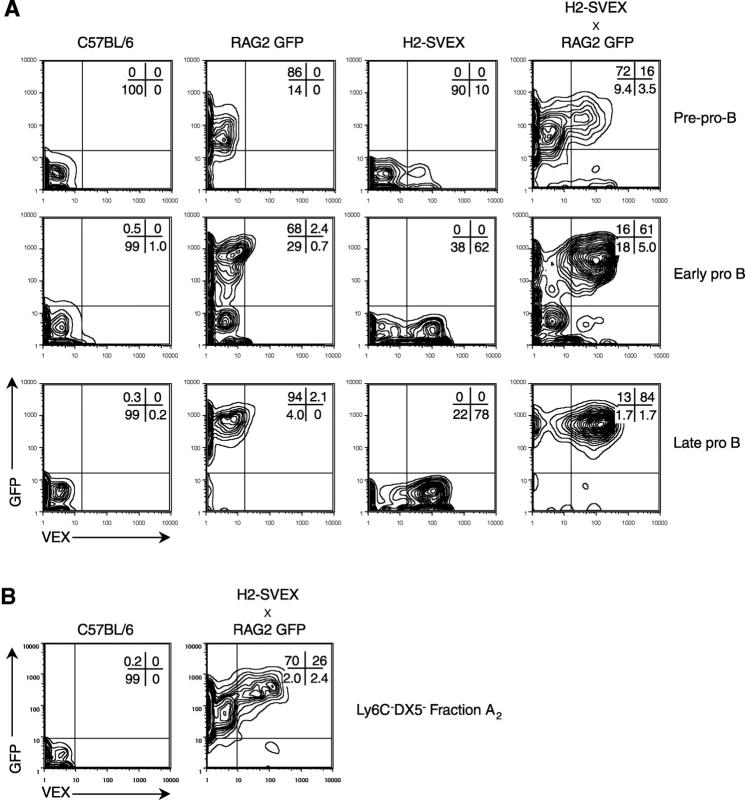
Figure 2.
Simultaneous analysis of rag2 transcription and recombinase activity in B lineage progenitors. (A) Bone marrow obtained from the F1 progeny of RAG2 GFP × H2-SVEX (SB110) animals (fourth column) was stained with antibodies to identify B lineage progenitors. The B220+CD43+DX5−Ly6C−IgM− subset of bone marrow was then gated to examine CD19−CD24− pre-pro-B cells (first row), CD19−CD24+ early pro-B (second row), and CD19+CD24+ late pro-B (third row) cells. Gated samples were simultaneously examined for GFP expression, an indicator of rag2 transcription, and VEX expression, an indicator of V(D)J recombinase activity. Because the GFP expression in RAG2 GFP NG animals is so bright (12), a small percentage (<3%) of the GFPBRIGHTVEX− cells fall into the GFP+VEX+ quadrant due to limitations of compensation. Therefore, bone marrow from single transgenic RAG2 GFP NG mice (second column), single transgenic H2-SVEX transgenic mice (third column), and nontransgenic B6 control mice is provided for comparison. The percentage of cells in each region is given. (B) VEX and GFP expression was similarly examined in Ly6C−DX5− fraction A2(AA4.1+B220+CD24−CD4−) bone marrow cells. The data are representative of two to five independent experiments.
Within pre-pro-B cells from SB110 H2-SVEX animals, 10–20% of the cells are VEX+ (Fig. 2 A, top row) indicating the presence of an active V(D)J recombinase complex at this stage in development. Similar results are observed with SB88 H2-SVEX mice (not depicted). These data are supported by the fact that nearly 90% of pre-pro-B cells express the GFP reporter of rag2 (Fig. 2 A, top row, second and fourth columns). Moreover, 16–20% of this subset and 26% of Ly6C−DX5− fraction A2 cells simultaneously express GFP and VEX (compare the top row, fourth column of Fig. 2 A with Fig. 2 B; and not depicted), providing evidence using independent transgenic strains that the V(D)J recombination machinery is both present and active in this developmental subset. It remains unclear whether the modest percentage (9.4–18%) of GFP−VEX− pre-pro-B and pro-B cells reflects a population of precursors that simply has not yet received the appropriate developmental signals or a population of non–B lineage contaminants. The ability to sort these subsets based on GFP and VEX expression will provide a useful tool for evaluating their contribution to the B lineage.
Although the percentage of GFP+ cells remains uniformly high (>70%) across the pro-B subsets, the percentage of cells that express both GFP and VEX progressively increases from 16% in pre-pro-B to 61% in early pro-B and 84% in late pro-B. This is consistent with previous detection of rag2 transcripts at very low levels in fraction A cells and markedly higher levels of transcripts in later pro-B cells. These observations provide direct evidence that the V(D)J recombinase is expressed and active in pre-pro-B/fraction A2 cells, despite the fact that the majority of these cells are not rearranging endogenous IgH loci (13, 17, 18).
By examining the H2-SVEX and RAG2 reporter transgenes, we also find that rag expression is proportional to recombinase activity in pro-B cells but only to a degree. As the mean fluorescence intensity of GFP within the GFP+ subset increases from 484 to 666 as cells progress from pro-pre-B to early pro-B, indicating a higher per cell level of GFP, the percentage of cells that express VEX also increases (Fig. 2 A, pre-pro-B compared with early pro-B). However, even within the GFP-expressing subset not all GFP+ pro-B cells recombine the substrate (Fig. 2 A). That VEX expression within pro-B cells is proportional to rag transcript levels is also clearly shown by our analysis of Erag mice in which VEX is modestly decreased in pro-B cells lacking one copy of the Erag enhancer of rag expression and severely decreased in mice lacking both copies of the enhancer (21). Together, these results suggest that (a) V(D)J recombination occurs most efficiently in cells that express high levels of rag1 and rag2 and (b) rag gene transcription may not be the only factor that dictates or limits efficient V(D)J recombination.
V(D)J Recombinase Activity in B Lymphoid Progenitors with Germline IgH Loci.
To directly compare the developmental stages at which recombination activity and IgH rearrangement is induced, we characterized IgH rearrangement status in VEX+ and VEX− early B lineage progenitors. According to one model, the earliest B cells arise from the CD4−AA4.1+ subset (fraction A2) of total fraction A, and the majority of these cells have germline IgH (13, 17). Typically, 20–30% of Ly6C−DX5− fraction A2 cells isolated from H2-SVEX animals express VEX (Fig. 2 B, and not depicted) versus 0% from control B6. Moreover, 26% of Ly6C−DX5− fraction A2 cells simultaneously express VEX and GFP (Fig. 2 B). Then, we sorted single fraction A2 cells on the basis of VEX expression and directly examined IgH rearrangement status by single cell PCR.
All IgH recombination events within fraction A2 are contained within the VEX+ subset. We found that 22/43 VEX+ and 0/34 VEX− fraction A2 progenitors have detectable D-JH joins (Table I). According to another model (24), B lineage development proceeds through a B220+CD43+CD19−DX5−Ly6C− stage. When single cells from this subset were purified, as with fraction A2, we found that all D-JH recombination events are contained within the VEX+ subset (11 out of 25 in VEX+ and 0 out of 33 in VEX− cells; Table I). Both models predict developmental progression through the fraction B(B220+CD43+CD24+BP-1−) stage, and we found nearly all VEX+ fraction B cells (28 out of 29 cells analyzed; Table I) had D-JH joins.
Table I.
Single Cell Detection of D-J Joins at IgH Loci
| Progenitor subset | VEX | No. of D-JH +/N cells |
|---|---|---|
| Fraction A2 | + − |
22/43 0/34 |
| Pre-pro-B cells | + − |
11/25 0/33 |
| Fraction B | + − |
28/29 15/34 |
Single cells sorted from H2-SVEX transgenics (SB110 or SB88) were analyzed by PCR to detect D-JH joins (see Materials and Methods). Fraction A2 is defined as AA4.1+B220+CD24−CD4−, pre-pro-B cells are B220+CD43+CD19−DX5−Ly6C−, and fraction B is B220+CD43+CD24+BP-1−.
These data highlight two important findings. First, our data showing that only half of VEX+ early B cell progenitors (defined according to either of two developmental models) have D-JH joins provides evidence that V(D)J recombinase expression and V(D)J recombination initiation of antigen receptor loci are separable events. This observation suggests not only that recombinase expression may precede chromatin remodeling in B cell development but also that recombinase activity and chromatin accessibility may be separately regulated. IL-7 influences VH locus accessibility in pro-B cells (4); however, factors that control D-JH accessibility in the earliest stages of B lineage development remain uncharacterized.
Second, one of the most striking characteristics of the H2-SVEX system is the accuracy with which VEX expression predicts V(D)J recombination of IgH. All of the fraction A2 progenitors or B220+CD43+CD19−DX5−Ly6C− pre-pro-B cells that had detectable D-JH recombination events were contained within the VEX+ subset (Table I). This is an important observation as it is clear that the earliest B lineage precursor subsets including fraction A are not homogenous populations (25, 26); yet few tools are available for dividing these subsets on the basis of relevant molecular characteristics. One prediction drawn from our findings is that VEX+ fraction A2 or pre-pro-B cells contain a subset of cells that are in the process of chromatin remodeling of the D-JH region of IgH.
In conclusion, by using a transgenic V(D)J recombination reporter substrate that lacks Ig or TCR locus transcriptional control elements, we assayed recombinase activity at a transgenic locus subject to different accessibility constraints than IgH. Our findings demonstrate that the V(D)J recombinase is active in early B lineage progenitors with germline IgH status and provide direct evidence that developmentally regulated expression of the V(D)J recombinase is not the limiting step to initiation of recombination at antigen receptor loci. The major implications of these data are that recombinase expression and IgH recombination are developmentally distinct events. We have already begun to dissect control of recombinase activity in lymphoid progenitors (21), and we expect the H2-SVEX model system to be similarly valuable for investigators studying the signaling pathways and specific mechanisms of chromatin modifications that contribute to “active” versus “inactive” recombination targets (7). For example, it would be useful to compare the relative frequency of histone modifications at distinct recombination targets (e.g., IgH and H2-SVEX) that we predict would have varying levels of accessibility during the pre-pro-B stage of development.
Acknowledgments
We gratefully acknowledge Ranjan Sen, Dipanjan Chowdhury, Michel Nussenzweig, David Allman, Mark Schlissel, Richard Hardy, and Joe Labrie for thoughtful discussions, and Kim Martin, Julie O'Connor, Angela Ariza, Neal Gehani, and Danielle Davignon for invaluable assistance with experiments. Naomi Rosenberg (Tufts University School of Medicine, Boston, MA) and Chris Roman (State University of New York-Downstate Medical Center, Brooklyn, NY) kindly provided the 300-35 and A12 cell lines, respectively. We are grateful to Michel Nussenzweig (The Rockefeller University, New York, NY) for the NG RAG2 GFP transgenic. Finally, we thank the UMMS Flow Cytometry Facility and UMMS Transgenic Facility for their excellent help.
This work is supported by National Institutes of Health grant RO1 AI043534-03 (to R.M. Gerstein), training grant AI007349-14 (L. Borghesi), and grant NIDDK 5 P30 DK32520 to the UMMS Diabetes and Endocrinology Center.
Abbreviations used in this paper: GFP, green fluorescence protein; IgH, Ig heavy chain; RAG, recombinase-activating gene; RSS, recombination signal sequence.
References
- 1.Ehlich, A., V. Martin, W. Muller, and K. Rajewsky. 1994. Analysis of the B-cell progenitor compartment at the level of single cells. Curr. Biol. 4:573–583. [DOI] [PubMed] [Google Scholar]
- 2.Grawunder, U., T.M. Leu, D.G. Schatz, A. Werner, A.G. Rolink, F. Melchers, and T.H. Winkler. 1995. Down-regulation of RAG1 and RAG2 gene expression in preB cells after functional immunoglobulin heavy chain rearrangement. Immunity. 3:601–608. [DOI] [PubMed] [Google Scholar]
- 3.Johnson, K., C. Angelin-Duclos, S. Park, and K.L. Calame. 2003. Changes in histone acetylation are associated with differences in accessibility of V(H) gene segments to V-DJ recombination during B-cell ontogeny and development. Mol. Cell. Biol. 23:2438–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chowdhury, D., and R. Sen. 2001. Stepwise activation of the immunoglobulin {micro} heavy chain gene locus. EMBO J. 20:6394–6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Durum, S.K., S. Candeias, H. Nakajima, W.J. Leonard, A.M. Baird, L.J. Berg, and K. Muegge. 1998. Interleukin 7 receptor control of T cell receptor gamma gene rearrangement: role of receptor-associated chains and locus accessibility. J. Exp. Med. 188:2233–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McMurry, M.T., and M.S. Krangel. 2000. A role for histone acetylation in the developmental regulation of VDJ recombination. Science. 287:495–498. [DOI] [PubMed] [Google Scholar]
- 7.Morshead, K.B., D.N. Ciccone, S.D. Taverna, C.D. Allis, and M.A. Oettinger. 2003. Antigen receptor loci poised for V(D)J rearrangement are broadly associated with BRG1 and flanked by peaks of histone H3 dimethylated at lysine 4. Proc. Natl. Acad. Sci. USA. 100:11577–11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schatz, D.G., M.A. Oettinger, and D. Baltimore. 1989. The V(D)J recombination activating gene, RAG-1. Cell. 59:1035–1048. [DOI] [PubMed] [Google Scholar]
- 9.Oettinger, M.A., D.G. Schatz, C. Gorka, and D. Baltimore. 1990. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science. 248:1517–1523. [DOI] [PubMed] [Google Scholar]
- 10.Monroe, R.J., K.J. Seidl, F. Gaertner, S. Han, F. Chen, J. Sekiguchi, J. Wang, R. Ferrini, L. Davidson, G. Kelsoe, and F.W. Alt. 1999. RAG2:GFP knockin mice reveal novel aspects of RAG2 expression in primary and peripheral lymphoid tissues. Immunity. 11:201–212. [DOI] [PubMed] [Google Scholar]
- 11.Hardy, R.R., C.E. Carmack, S.A. Shinton, J.D. Kemp, and K. Hayakawa. 1991. Resolution and characterization of pro-B and pre-pro-B cell stages in normal mouse bone marrow. J. Exp. Med. 173:1213–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu, W., Z. Misulovin, H. Suh, R.R. Hardy, M. Jankovic, N. Yannoutsos, and M.C. Nussenzweig. 1999. Coordinate regulation of RAG1 and RAG2 by cell type-specific DNA elements 5′ of RAG2. Science. 285:1080–1084. [DOI] [PubMed] [Google Scholar]
- 13.Li, Y.S., R. Wasserman, K. Hayakawa, and R.R. Hardy. 1996. Identification of the earliest B lineage stage in mouse bone marrow. Immunity. 5:527–535. [DOI] [PubMed] [Google Scholar]
- 14.Lin, W.C., and S. Desiderio. 1994. Cell cycle regulation of V(D)J recombination-activating protein RAG-2. Proc. Natl. Acad. Sci. USA. 91:2733–2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ross, A.E., M. Vuica, and S. Desiderio. 2003. Overlapping signals for protein degradation and nuclear localization define a role for intrinsic RAG-2 nuclear uptake in dividing cells. Mol. Cell. Biol. 23:5308–5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee, J., and S. Desiderio. 1999. Cyclin A/CDK2 regulates V(D)J recombination by coordinating RAG-2 accumulation and DNA repair. Immunity. 11:771–781. [DOI] [PubMed] [Google Scholar]
- 17.Allman, D., J. Li, and R.R. Hardy. 1999. Commitment to the B lymphoid lineage occurs before DH-JH recombination. J. Exp. Med. 189:735–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu, L.S., J. Tung, N. Baumgarth, O. Herman, M. Gleimer, and L.A. Herzenberg. 2002. Identification of a germ-line pro-B cell subset that distinguishes the fetal/neonatal from the adult B cell development pathway. Proc. Natl. Acad. Sci. USA. 99:3007–3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Igarashi, H., S.C. Gregory, T. Yokota, N. Sakaguchi, and P.W. Kincade. 2002. Transcription from the RAG1 locus marks the earliest lymphocyte progenitors in bone marrow. Immunity. 17:117–130. [DOI] [PubMed] [Google Scholar]
- 20.Allman, D., A. Sambandam, S. Kim, J.P. Miller, A. Pagan, D. Well, A. Meraz, and A. Bhandoola. 2003. Thymopoiesis independent of common lymphoid progenitors. Nat. Immunol. 4:168–174. [DOI] [PubMed] [Google Scholar]
- 21.Borghesi, L., L.-Y. Hsu, J.P. Miller, M. Anderson, L. Herzenberg, L. Herzenberg, M.S. Schlissel, D. Allman, and R.M. Gerstein. 2004. B Lineage–specific regulation of V(D)J recombinase activity is established in common lymphoid progenitors. J. Exp. Med. 199:491–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.ten Boekel, E., F. Melchers, and A. Rolink. 1995. The status of Ig loci rearrangements in single cells from different stages of B cell development. Int. Immunol. 7:1013–1019. [DOI] [PubMed] [Google Scholar]
- 23.Anderson, M.T., N. Baumgarth, R.P. Haugland, R.M. Gerstein, T. Tjioe, and L.A. Herzenberg. 1998. Pairs of violet-light-excited fluorochromes for flow cytometric analysis. Cytometry. 33:435–444. [DOI] [PubMed] [Google Scholar]
- 24.Tudor, K.S., K.J. Payne, Y. Yamashita, and P.W. Kincade. 2000. Functional assessment of precursors from murine bone marrow suggests a sequence of early B lineage differentiation events. Immunity. 12:335–345. [DOI] [PubMed] [Google Scholar]
- 25.Kouro, T., V. Kumar, and P.W. Kincade. 2002. Relationships between early B- and NK-lineage lymphocyte precursors in bone marrow. Blood. 100:3672–3680. [DOI] [PubMed] [Google Scholar]
- 26.Izon, D., K. Rudd, W. DeMuth, W.S. Pear, C. Clendenin, R.C. Lindsley, and D. Allman. 2001. A common pathway for dendritic cell and early B cell development. J. Immunol. 167:1387–1392. [DOI] [PubMed] [Google Scholar]