Abstract
Preeclampsia is a serious complication of pregnancy in which the fetus receives an inadequate supply of blood due to failure of trophoblast invasion. There is evidence that the condition has an immunological basis. The only known polymorphic histocompatibility antigens on the fetal trophoblast are HLA-C molecules. We tested the idea that recognition of these molecules by killer immunoglobulin receptors (KIRs) on maternal decidual NK cells is a key factor in the development of preeclampsia. Striking differences were observed when these polymorphic ligand: receptor pairs were considered in combination. Mothers lacking most or all activating KIR (AA genotype) when the fetus possessed HLA-C belonging to the HLA-C2 group were at a greatly increased risk of preeclampsia. This was true even if the mother herself also had HLA-C2, indicating that neither nonself nor missing-self discrimination was operative. Thus, this interaction between maternal KIR and trophoblast appears not to have an immune function, but instead plays a physiological role related to placental development. Different human populations have a reciprocal relationship between AA frequency and HLA-C2 frequency, suggesting selection against this combination. In light of our findings, reproductive success may have been a factor in the evolution and maintenance of human HLA-C and KIR polymorphisms.
Keywords: natural killer cells, HLA, placenta, killer immunoglobulin receptors, trophoblast
Introduction
Although the fetus is allogeneic to its mother, there is little evidence that alloimmune interactions at the maternal–fetal interface in the uterine wall can compromise human pregnancy outcome. Most attention has been given to possible maternal T cell responses to fetal histocompatibility or other antigens. However, T cells are a minor component of the immune cells in the early pregnancy decidua, and placental trophoblast cells do not express the strong histocompatibility antigens, HLA-A, -B, or -D (1). Instead, there is a profusion of CD56bright NK cells that are in close contact with the allogeneic extravillous trophoblast (EVT) cells. These invade the decidua, subjacent myometrium, and the walls of the spiral arteries during the first half of human pregnancy (2). EVT expresses a unique combination of three MHC class I molecules: HLA-G, HLA-E, and HLA-C (3, 4, 5). Of these, only HLA-C is polymorphic, and the paternal allele is expressed on the trophoblast cell surface. Therefore, in terms of allorecognition in the context of placentation, trophoblast HLA-C is the relevant MHC class I molecule to consider.
NK cells are effector lymphocytes that act by production of cytokines and chemokines. They express a diverse complement of receptors mediating both activating and inhibitory signals (6). Uterine CD56bright NK cells express a variety of these receptors, including the killer immunoglobulin receptor (KIR) family that includes members known to recognize HLA-C molecules (7). The KIR multigene family exhibits an extensive degree of diversity, achieved by a combination of presence or absence of particular genes combined with allelic polymorphism of individual KIR genes. This means that almost all unrelated individuals have a different KIR genotype (7, 8, 9). On the basis of gene content, two distinct KIR haplotypes can be distinguished, termed A and B. The A haplotype has seven KIR loci with only one activating receptor, KIR2DS4. However, the most common allele of KIR2DS4 has a deletion so individuals with two A haplotypes generally have no activating receptors. The B haplotypes are mainly characterized by the presence of extra loci that are not present in the A haplotype, most of which encode activating receptors (KIR2DS1, 2, 3, 5, and KIR3DS1) as well as two inhibitory receptors, KIR2DL2 and KIR2DL5. When considering HLA-C molecules as ligands for KIR receptors on NK cells, all HLA-C allotypes can be grouped into two major KIR epitopes, C1 and C2, on the basis of a dimorphism at position 80 of the α1 domain (10, 11). Group C1 (HLA-Casn80) are ligands for inhibitory KIR2DL2/3 and activating KIR2DS2 and group C2 (HLA-Clys80) are ligands for inhibitory KIR2DL1 and activating KIR2DS1. Other activating KIR2DS receptors (KIR2DS3, 2DS4, and 2DS5) with very similar sequences may also bind HLA-C molecules (7, 8, 9).
Uterine NK (uNK) cells are phenotypically and functionally distinct from NK cells in peripheral blood (2). From gene expression studies, a case has been made that uNK cells are a distinct NK cell lineage (12), although an alternative explanation could be differentiation in the unique decidual microenvironment. Functionally, their most likely role is to modify the uterine spiral arteries to increase the blood supply to the feto-placental unit. In humans, this could occur by NK cells influencing the extent of trophoblast invasion into the arterial wall, whereas in mice their effect may be directly on the vessels. In keeping with this, mice with no uNK cells do not show the normal vascular adaptations associated with pregnancy (13, 14). The phenotype of decidual NK cells is skewed toward a higher proportion expressing KIRs with specificities for HLA-C allotypes than is found in peripheral blood from the same individual (15). Moreover, because pregnancy is a natural allograft, the mother could possess KIR genes for paternal HLA-C allotypes that she herself does not have. Conversely, the fetus could lack HLA-C KIR ligands that are present in the mother. The dominant influence of HLA-C on NK recognition and the profusion of NK cells at the implantation site indicate that maternal NK cell–trophoblast HLA-C interaction deserves particular attention.
Inadequate placentation and poor placental perfusion are features of preeclampsia, a specific disorder of human pregnancy that is associated with a significant maternal mortality and morbidity (16). Preeclampsia develops in stages, the maternal systemic illness being a late manifestation of the placental ischaemia that is secondary to the reduced utero-placental blood flow (17). Thus, the primary pathological defect in this disease is the failure of transformation of the uterine spiral arteries (2, 18). Genetic studies in preeclampsia have implicated a contribution from both maternal and paternal genotypes. A family history in maternal relatives is associated with a fourfold risk (19). Despite this, no concordance is found in monozygotic twins (20). In population-based studies, a contribution of both paternal as well as maternal factors in the development of the disease is apparent (21). That immune recognition is involved is suggested by the increased risk in first pregnancies and in multiparae, but only after changing partners (22). Importantly, there is also a greatly increased risk in women who have received donated oocytes (∼30%; reference 23). In this scenario, the fetus will share neither maternal haploid gene set with the recipient mother.
We suggest that control of the fetal vascular supply line may depend on the allogeneic interaction between maternal KIR on uNK cells and paternal HLA-C expressed by trophoblast. Therefore, uNK cell function during pregnancy is likely to be influenced by a combination of two polymorphic sets of genes; the maternal KIR genotype as well as the trophoblast (fetal) HLA-C allotypes. In this work, we have tested this hypothesis and analyzed how maternal KIR and fetal HLA-C genes combine to influence the risk of preeclampsia and reproductive success. Our results indicate that certain combinations of paternal and maternal innate immune genes are unfavorable for placentation.
Materials and Methods
Patients.
DNA was obtained from the blood of 200 pregnant women with preeclampsia and from 201 matched women who had normal pregnancies. Cord samples or mouth swabs were obtained from the babies for genomic DNA isolation. The women were recruited from three hospitals in the United Kingdom: St. James University Hospital (Leeds), John Radcliffe Hospital (Oxford), and Addenbrooke's Hospital (Cambridge). Informed written consent and local Ethics Committees' approval were obtained. Preeclampsia was defined as new hypertension after week 20 of pregnancy (140/90 mm Hg) together with new proteinuria >300 mg/24 h.
DNA Isolation.
Maternal genomic DNA was isolated from 5 ml of blood using the QIAamp DNA Maxi Blood Kit (QIAGEN), and the babies' DNA was isolated from cord samples after overnight incubation with proteinase K (Roche Diagnostics), phenol/chloroform purification and ethanol precipitation. Occasionally, mouth-swab samples were taken and processed using whole genome amplification performed with N15 random primers (24).
KIR Genotyping.
KIR genotyping was based on the primers and methods described previously (reference 25 and the online supplemental Materials and Methods, available at http://www.jem.org/cgi/content/full/jem.20041214/DC1). The KIR genes were typed for presence or absence by PCR-SSP using two pairs of primers per gene or allele and choosing primers that gave relatively short amplicons of 100–800 bp (Table S1, available at http://www.jem.org/cgi/content/full/jem.20041214/DC1). Where there were problems with specificity, primers with mismatched bases next to the specific 3′ terminal base were used. Initially, PCR product identity was checked by sequencing. KIR genes typed were 2DL1, 2DL2, 2DL3, 2DL5, 2DS1, 2DS2, 2DS3, 2DS4, KIR1D (2DS4 allele with a 22-bp deletion), and 3DL3.
HLA-C Genotyping.
HLA-C typing was performed by the Tissue Typing laboratories at Addenbrooke's Hospital or by the Anthony Nolan Trust. Medium resolution PCR-SSP or PCR-SSOP was used for typing, and all HLA-C group C1 alleles could be distinguished from group C2.
Statistical Analysis.
Frequency differences between the preeclampsia and control groups were tested for significance using two-by-two tables and Fisher's Exact statistic test, and the magnitude of effect was estimated by odds ratios and their 95% confidence intervals (Windows 11.0.0.2001; SPSS Inc.). Where multiple tests were involved, the p-values are given as an indication of possible associations, not as definitive analyses. This involves the testing of the separate KIR genes and the separate subsets of mother/child pairs with the mothers' AA genotype. A linear model with a logistic link was used to test the association between increasing numbers of activating KIR and reduction in prevalence of preeclampsia (LogXact software; Cytel Software Corp.). For all analyses, p-values were exact and two sided.
Online Supplemental Material.
The detailed methods of KIR genotyping are presented in a table of the primers used (Table S1). HLA-C allele frequencies in preeclampsia and control patients are shown in Table S2. KIR genotypes found in control and preeclampsia mothers are shown in descending order of frequency (Table S3). The number of activating and inhibitory KIR is given in Table S4. References relating to the online supplemental material are included. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20041214/DC1.
Results
The Frequencies of HLA-C Groups and Alleles Are Similar in Preeclampsia Patients and Controls.
First, we analyzed the frequency of HLA-C groups, C1 and C2, in preeclampsia and control pregnancies. The HLA-C1 and C2 frequencies in preeclampsia and control mothers and their babies were not significantly different and were similar to the results from an Irish population (Table I and reference 26). We also evaluated whether individual HLA-C alleles were associated with preeclampsia. All mothers and babies were typed by medium resolution PCR-SSP and PCR-SSOP down to allele level. There were no significant differences in allele frequencies in preeclampsia patients (n = 200) and their babies compared with the control mothers (n = 201) and their babies (Table S2, available at http://www.jem.org/cgi/content/full/jem.20041214/DC1).
Table I.
HLA-C Genotype Groups for Control and Preeclampsia Cases
| Controls
|
Preeclampsia
|
Controla(N. Ireland)
|
|||
|---|---|---|---|---|---|
| HLA-C groups | Mother | Fetus | Mother | Fetus | Individuals |
| % (n = 201) | % (n = 201 + 4)b | % (n = 200) | % (n = 201 + 10)b | % (n = 1,000) | |
| 1 + 1 | 50.6 (102) | 45.3 (91) | 44.0 (88) | 45.5 (91) | 43 (430) |
| 1 + 2 | 36.9 (77) | 44.3 (89) | 44.0 (88) | 43.0 (86) | 46.4 (464) |
| 2 + 2 | 12.5 (22) | 10.4 (21) | 12.0 (24) | 11.5 (23) | 10.6 (106) |
| C1 | 69.9 | 67.7 | 66.0 | 67.0 | 66.2 |
| C2 | 30.1 | 32.3 | 33.0 | 33.0 | 33.8 |
This independent, large dataset confirms the frequencies in our control samples (reference 26).
Multiple births, only fetus no. 1 was counted.
The Frequency of the Maternal AA Genotype Is Increased in Preeclampsia Pregnancies.
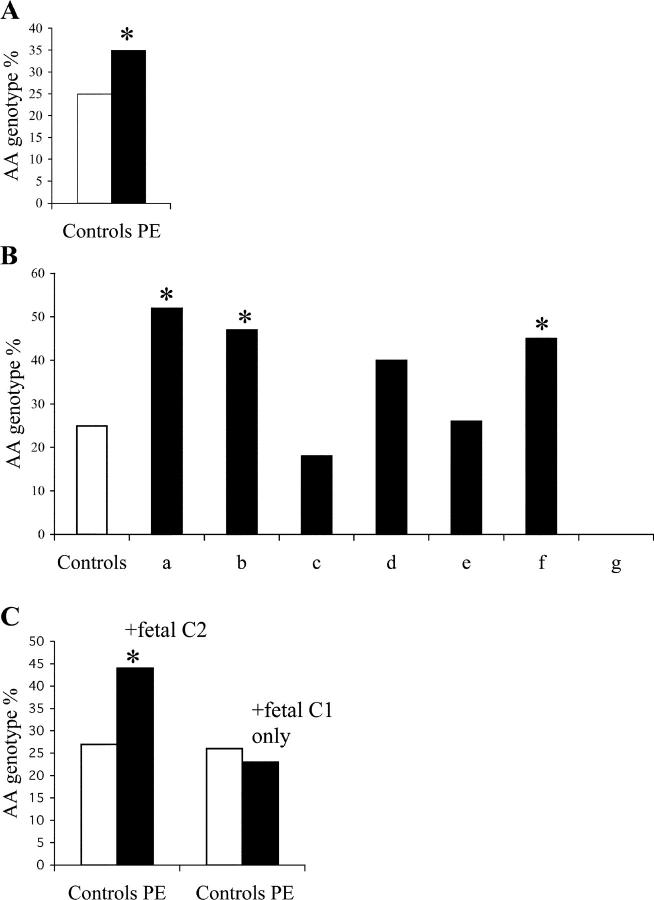
All mothers were also typed for the KIR genes: inhibitory, 2DL1, 2DL2, 2DL3, 2DL5, and 3DL3; and activating, 2DS1, 2DS2, 2DS3, 2DS4, and 2DS5. On the basis of the presence of activating receptors characteristic of the B haplotypes, the mothers were designated as having either an AA genotype or an AB/BB genotype. 25% of our control population had two A haplotypes (AA genotype) in agreement with previous reports in caucasoid populations (8, 27). Considering all preeclampsia patients, there was a statistically significant increase in the frequency of the AA genotype to 35% (P = 0.038, OR = 1.59, CI = 1.03–2.45; Fig. 1 A). When individual KIR genes were analyzed, several were less frequent in preeclampsia mothers but only the KIR2DL5 gene frequency reached statistical significance (Table II). Consistent with previous reports we identified considerable variation in frequency of the KIR leading to a great diversity in genotypes (reference 8 and Table S3, available at http://www.jem.org/cgi/content/full/jem.20041214/DC1). A total of 48 genotypes could be identified, a few of which were common but most were present in only one or two individuals.
Figure 1.
AA genotype and activating KIR frequencies in control and preeclampsia mothers. (A) There was a significant difference in the AA genotype frequencies between controls (white bar, n = 201) and preeclampsia patients (black bar, n = 200). *, P = 0.038. (B) The AA genotype frequencies in subsets of patients with preeclampsia categorized depending on the HLA-C groups 1 and 2 of mother (M) and fetus (F) (see Table III for details and patient numbers). (a) M 1 + 1/F 1 + 2. *, P 5 0.005. OR = 3.22; CI = 95%; 1.49–6.98. (b) M 2 + 2/F 1 + 2. *, P = 0.034. (c) M 1 + 2/F 1 + 1. (d) M 1 + 2/F 2 + 2. (e) M 1 + 1/F 1 + 1. (f) M 1 + 2/F 1 + 2. *, P = 0.011. (g) M 2 + 2/F 2 + 2. The subsets in which the fetus had a C2 allotype were a, b, d, f, and g. The subsets in which the fetus only had a C1 allotype were c and e. The subset in which C2 was absent from the mother, but present in the fetus, was a. The subsets with both mother and fetus having C2 were b, d, f, and g. Subset g is an exception to the increase in frequency of maternal AA genotype when the fetus had a C2 allotype but the numbers were small (n = 6). (C) The patients from Fig. 1 A have been divided into two subsets as follows: those in which the fetus presents a C2 allotype (n = 109; *, P = 0.001; OR = 2.38; 95% CI = 1.45 − 3.90) and those in which the fetus only had a C1 allotype (n = 91; NS).
Table II.
KIR Gene/Genotype Frequencies in Control and Preeclampsia Mothers and Those Preeclampsia Mothers Presented with Fetal C2 in Homozygous (2 + 2) or Heterozygous (1 + 2) Form
| Gene/genotype | Control mothers | Preeclampsia mothers | Preeclampsia subsets with fetal C2 (a, b, d, f, g)a |
|---|---|---|---|
| % (n = 201) | % (n = 200) | % (n = 109) | |
| 2DL2 | 53 (107) | 52 (103) | 40b (44) P = 0.033 |
| 2DS1 | 47 (94) | 38 (76) | 34b (37) P = 0.031 |
| 2DS2 | 53 (108) | 52 (104) | 42 (46) P = 0.058 |
| 2DS3 | 29 (58) | 24 (47) | 15b (16) P = 0.005 |
| 2DS5 | 39 (83) | 32 (83) | 33 (36) P = 0.179 |
| 2DL5 | 58 (116) | 47b (94) P = 0.036 |
41b (45) P = 0.006 |
| Genotype AA | 25 (50) | 35b (69) P = 0.038 |
44b(48) P = 0.001 |
The B haplotypes KIR, 2DL2, 2DL5, 2DS1, 2DS3, and 2DS5 all occur at lower frequency in the preeclampsia mothers presented with HLA-C2 as compared with the controls.
Significantly different frequencies between preeclampsia and control mothers.
The KIR AA Genotype Is Significantly Increased in Preeclampsia When the Fetus Has an HLA-C2 Gene.
Continuing to work from our original hypothesis, we analyzed combinations of maternal KIR genotypes with fetal HLA-C subgroups C1 and C2 that are relevant to KIR2D recognition. We found that the increased frequency of the aforementioned AA genotype was only found in mothers who were presented by a C2 allotype in the fetus (Fig. 1 B). This increase in frequency of AA genotype was seen even when the fetus was heterozygous for C2 (Fig. 1 B), indicating that the expression of C2 has a dominant effect in the presence of C1. In contrast, if the fetus was homozygous for HLA-C1, the frequency of the AA genotype in preeclampsia was similar to controls (Fig. 1 B). Merging the data from Fig. 1 B, to distinguish between pregnancies where a fetal C2 is presented compared with those in which C1 only is presented, there was a highly significant difference in the frequency of the maternal AA genotype when combined with fetal C2 (P = 0.001; OR = 2.38; CI = 1.45–3.90; Fig. 1 C).
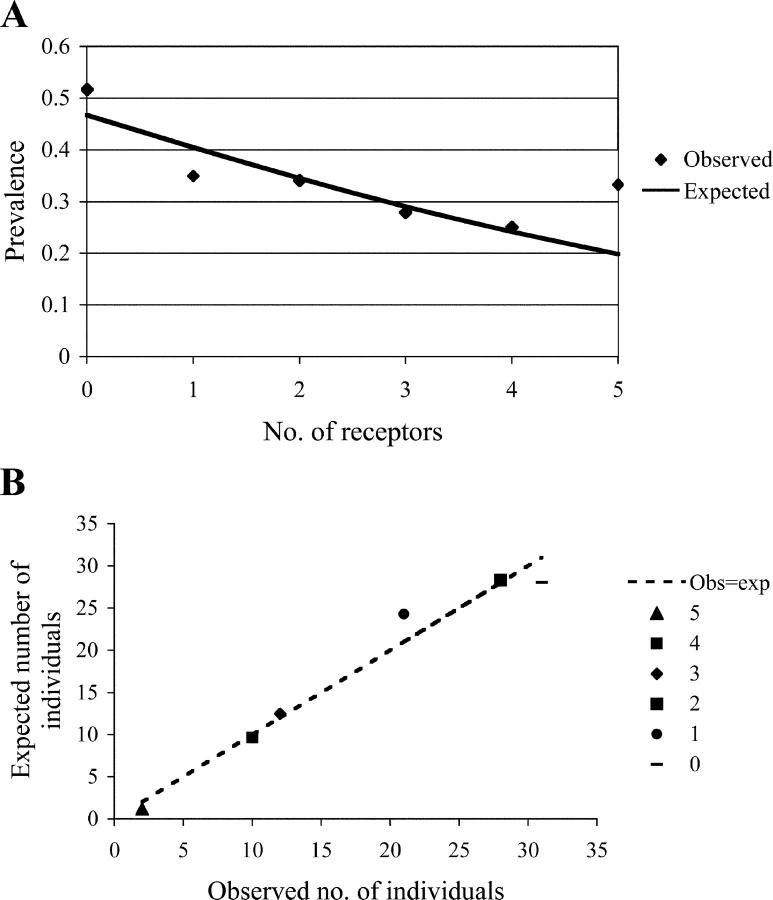
We looked in detail at individual maternal KIR genes that differed significantly between normal and preeclampsia pregnancies. As we saw no effect in preeclampsia pregnancies when only fetal C1 was present, we have omitted these patients (Table II). Most of the B haplotype KIRs were present at significantly lower frequencies in these mothers as would be expected from the AA genotype frequencies. To test whether the number of activating receptors (characteristic of the B haplotypes) possessed by the mother was associated with preeclampsia, we used a linear logistic model (Fig. 2, A and B). A 1.2-fold reduction in prevalence with each extra activating receptor was seen and this was highly significant (P = 0.0063). Data on the frequency of KIR activating and inhibitory receptors in control and preeclampsia mothers can be found in Table S4 (available at http://www.jem.org/cgi/content/full/jem.20041214/DC1). To summarize, we conclude that a combination of maternal AA genotype with a fetal HLA-C2 group is associated with preeclampsia and poor placentation.
Figure 2.
(A) The addition of each activating receptor is associated with a 1.2-fold reduction in prevalence. A linear logistic model was used to relate the prevalence of preeclampsia to the numbers of activating receptors possessed by the mother (KIR2DS1, 2DS2, 2DS3, 2DS4wt, and 2DS5). Control pregnancies were compared with affected mothers presented with fetal C2 as before. The model suggests an approximate 1.2-fold reduction in prevalence with each additional activating receptor. The sample size for the mothers with five activating receptors was very small (n = 6 for control and preeclampsia mothers combined). (B) A multiplicative model is a good fit to the data. A plot of the expected versus observed numbers of affected mothers with increasing numbers of activating receptors shows that the logistic model provides a good fit.
Maternal KIR Recognition of Fetal HLA-C Is Not in Terms of Nonself or Missing Self.
Next, we looked at whether the C2 group was perceived as a nonself or missing self molecule by the mother. The fetus may have an HLA-C allele that is in a different group than the mother's (Table III, subsets a and b, “nonself”), or, in contrast, the mother could have an HLA-C allele that is in a group that the fetus lacks (Table III, subsets c and d, “missing self”). In the remaining pregnancies, the fetus and mother will have HLA-C from the same groups (Table III, subsets e, f, and g, “self”). We assigned both preeclampsia and control pregnancies to these three possible scenarios to see if any occurred more frequently in preeclampsia compared with control pregnancies. There was no significant increase in risk with these three groups. We infer that, in classical immunological terms, the fetal C1 and C2 groupings of nonself, missing self, and self, in relation to the mother, probably have no effect on reproductive success.
Table III.
HLA-C Groups C1 and C2 in Control and Preeclampsia Mother/Child Pairs Categorized as Nonself, Missing Self, and Self
| HLA-C group
|
||||
|---|---|---|---|---|
| Subsets | Mother | Fetus | Control | Preeclampsia |
| % (n = 201) | % (n = 200) | |||
| Nonself | ||||
| a | 1 + 1 | 1 + 2 | 18.4 (37) | 15.5 (31) |
| b | 2 + 2 | 1 + 2 | 7.0 (14) | 9.0 (18) |
| Missing self | ||||
| c | 1 + 2 | 1 + 1 | 12.4 (25) | 16.5 (33) |
| d | 1 + 2 | 2 + 2 | 5.5 (11) | 7.7 (15) |
| Self | ||||
| e | 1 + 1 | 1 + 1 | 32.3 (65) | 29.0 (58) |
| f | 1 + 2 | 1 + 2 | 19.9 (40) | 19.5 (39) |
| g | 2 + 2 | 2 + 2 | 4.5 (9) | 3.0 (6) |
There were no significant differences between numbers of control and preeclampsia pairs within these categories.
When considering the effect of C2 in the fetus in combination with the maternal AA genotype, we attempted to dissect whether the presence of C2 in the mother as well as the fetus had any influence. The effect of a maternal AA genotype was particularly striking when HLA-C2 was absent from the mother and was only present in the fetus (Fig. 1 B, subset a). The frequency of the AA genotype in the mother was 52% compared with 25% in controls (P = 0.005). Importantly, an increase in the frequency of the AA genotype was seen even when the mother herself also possessed a C2 allele (Fig. 1 B). This indicates that the fetal HLA-C molecules are probably not being perceived by the maternal KIR in terms of nonself or missing self. Rather, it suggests that it is the presence of the HLA-C2 molecules on trophoblast (as opposed to fetal or maternal somatic cells) that is important in KIR recognition.
Group 2 HLA-C Frequencies and AA Genotypes in Different Populations.
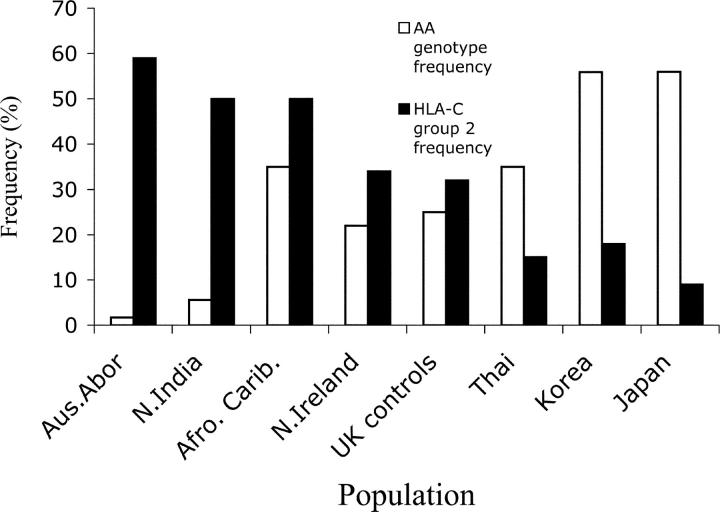
Given the dramatic influence of the KIR AA/HLA-C2 combinations on reproductive success, we investigated if their effects might be detectable at the population level. According to the model we have presented, and assuming that preeclampsia is disadvantageous in any population, we would predict an inverse relationship between the population prevalence of HLA-C2 and AA genotypes. To test this, we tabulated the relative frequencies of HLA-C2 and the KIR AA genotype in populations for which data were available (Fig. 3 and references 26–35). There was an apparent inverse relationship between HLA-C2 and AA frequencies (correlation coefficient (r) = −0.82). The more frequent C2, the less frequent was the AA genotype. The most obvious association of maternal KIR AA genotype with HLA-C2 was seen in the two historically isolated populations from Japan and the Australian Aborigines who represent the extreme ends of the spectrum.
Figure 3.
KIR genotype AA and HLA-C group 2 frequencies in different populations. Most of the HLA-C and KIR data have been taken from different cohorts within each population (references 26–35). The correlation coefficient (r) was −0.82.
Discussion
We have found that the combination of maternal KIR AA genotype with a fetal HLA-C2 is associated with an increased risk of preeclampsia. In this scenario, the C2 on trophoblast will only engage the inhibitory KIR2DL1 receptor. Therefore, the simplest interpretation is that the primary defect in preeclampsia is caused by too much inhibition of uNK cells leading to poor trophoblast invasion into the uterine arteries. Because there is no association with preeclampsia in pregnancies where the fetus is homozygous for C1 with a maternal KIR AA genotype, it is presumed that in this situation, the inhibitory signal is less strong to uNK cells and trophoblast invasion is unaffected. This hypothesis is supported by previous data on C1 and C2 interactions with their relevant inhibitory KIR that show a stronger interaction of KIR2DL1 with C2 than KIR2DL2/3 with C1. Thus, analysis of the crystal structures of C1 and C2 with their cognate KIR ligands revealed the KIR2DL1–C2 complex to be energetically more favorable than the KIR2DL2–C1 complex (36). Binding studies using KIR–Fc fusion proteins or surface plasmon resonance also indicate stronger binding of KIR2DL1 to C2 allotypes (37, 38, 39). Clinically, HLA-C1 and C2 have been shown to have diverse effects in both HLA-identical stem cell transplantation, and Hepatitis C infection (40, 41). In both these settings, a stronger inhibition of C2 via KIR2DL1 than C1 via KIR2DL3 could explain the different outcomes.
In the situation in which the fetal C2 allotype is combined with maternal activating KIR (AB/BB genotypes), the strong inhibition generated by engagement of KIR2DL1 would be balanced by activating signals to uNK cells, which results in better trophoblast invasion. In keeping with this, we found that every additional activating KIR incrementally decreases the incidence of preeclampsia when the fetus has C2. Given the extreme diversity of the KIR B haplotypes (7, 8), more patients will be needed to discern which particular activating KIRs are important.
From our results, we would predict that the KIR AA genotype–C2 combination would be selected against in populations given the deleterious effect these genetic variables have on reproductive success. Indeed, this seems to be the case, so that we have noted populations with a high AA genotype frequency have a low frequency of C2 alleles and vice versa. Therefore, the selective pressures for optimum reproductive performance could be a major factor in driving the evolution and maintenance of KIR and HLA-C polymorphisms. In infectious disease, such as HIV, acquisition of activating KIR–HLA combinations has also been shown to be advantageous (42). This must be offset by the potential NK cell overexuberance of activating KIR that could result in autoimmune disease as seen in psoratic arthropathy (25) and rheumatoid arthritis (43), thus providing balancing selection for AA genotypes.
This association of maternal KIR AA–fetal C2 combination in preeclampsia is seen whether or not the mother herself has a C2 gene. Therefore, this interaction does not fit the well-established models of nonself and missing self recognition. Because our results show that the essential interaction is between maternal NK cell KIR and fetal trophoblast HLA-C2 independent of the presence of maternal C2, this implies that the C2 molecules displayed by trophoblast must be special and distinct from those found on somatic cells. We have preliminary evidence that this is indeed the case (4, 44). For example, trophoblast HLA-C molecules are found to occur both as β2m-associated forms and as free heavy chains. Like HLA-G, trophoblast HLA-C could also form dimers or oligomers and be unusually glycosylated (45–48). Specificity of KIR interactions does depend on the peptide bound to HLA-C (49, 50). These peptides are likely to be different when derived from EVT because trophoblast are extraembryonic cells with many unusual properties, such as endogenous retroviral protein expression (51).
Previous studies have shown that both maternal and paternal genotypes contribute to the risk of developing preeclampsia (21, 52). Our data now extend these epidemiological findings by identifying for the first time some of the genes involved. Despite the strong family history found in maternal relatives (19), the lack of concordance in identical twins indicates that maternal genes cannot be solely responsible (20). Preeclampsia is essentially a disease of primigravidae (53–55), and it is difficult to explain why a first pregnancy protects against the development of the disease in subsequent pregnancies with the same partner. NK cells are not known to exhibit memory, but, if our model is correct, in subsequent pregnancies with the same AA-C2 combination, we would predict that weaker inhibition would occur via KIR2DL1. A possible way to achieve this, given that the maternal KIR genotype is fixed, is for fewer KIR2DL1+ cells to be present at the implantation site or a reduced level of expression of KIR2DL1 in the second pregnancy. This could result from diminished influx of blood KIR2DL1+ NK cells to the uterine mucosa. Alternatively, if KIR acquisition takes place in utero, a previous pregnancy might affect this so that fewer KIR2DL1+ cells or cells with lower expression levels are generated. This is obviously speculative, but we have found phenotypic differences in KIR expression between decidual and blood NK cells in the same woman (15), although it is not yet clear how this relates to the KIR genotype or obstetric history. How and why the decidual KIR phenotype is different, given the genetically and temporally fixed KIR phenotype of blood NK cells (56, 57), is unknown. Understanding this might explain why a first pregnancy protects against preeclampsia in subsequent pregnancies with the same father.
The KIRs specific for MHC-C (lineage III) are confined to humans and the great apes (7). Interestingly, MHC-C is seen in only half of orangutans and all are C1 group, indicating that C2 is a later addition (58). Comparisons of these KIR and MHC-C genes indicate that these two gene families may have coevolved (59). This coordinated evolution in different species suggests that they perform specialized functions such as defense against species-specific pathogens. Our paper raises the question whether species differences in placentation is another stimulus driving KIR and MHC coevolution. The size of the human brain relative to body weight far exceeds any other primate (60). During the early stage of simian primate evolution, a discoid invasive hemochorial type of placenta emerged. With this placental model, the only way to further increase the blood supply necessary for development of a larger brain was for EVT to invade the arteries in the placental bed more vigorously (61). This is a dangerous strategy and balancing mechanisms must have evolved concomitantly to prevent uterine penetration and maternal death. It is significant that preeclampsia occurs only in humans and represents pregnancies where the balance is tilted toward maternal protective mechanisms resulting in placental ischaemia and frequently low birth weight.
In conclusion, we have found that a combination of a maternal AA genotype with an HLA-C2 group in the fetus is significantly associated with preeclampsia. This means that if the fetus expresses a C2 allotype, it is detrimental for placentation for the mother to have no activating KIR. The effect of HLA-C2 group in the fetus was seen even if the mother also had an HLA-C2 allele, and, therefore, neither nonself nor missing self is operative. A logical extension of this conclusion is that HLA-C2 allotypes are displayed by trophoblast in some different form than on somatic cells. That the KIR AA genotype and HLA-C2 frequencies are inversely related in populations supports the notion that reproduction provides a selective pressure for KIR and HLA-C diversity. These results open up new ways of thinking about both the maternal–fetal interaction and the functions of KIR and HLA-C genes in populations. Overall, the conclusion is that the interaction between maternal KIR on uNK cells and fetal HLA class I molecules on trophoblast has a physiological function in regulating the development of the placenta, rather than the much-vaunted idea that it is a maternal immunological defense reaction mounted against her allogeneic fetus. This would be a new view of an old problem (62).
Acknowledgments
The enthusiasm and support of Prof. C. Loke is gratefully acknowledged. We wish to thank Drs. S. Frost, R. Single, and L. Hiby for expert assistance with statistical analyses. We are indebted to all the midwives, L. Samwiil, J. Hayes, C. Simms, H. Coburn, P. de la Salle, and B. Newcombe, for dedicated collection of clinical information and samples. Thanks to J. Connor for typing the paper.
Grant support from British Heart Foundation, National Institutes of Health (5R01HD39670-02), Isaac Newton Trust, and the Wellcome Trust. This publication has been funded in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract no. NO1-CO-12400. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. government.
The authors have no conflicting financial interests.
Abbreviations used in this paper: EVT, extravillous trophoblast; KIR, killer immunoglobulin receptors; uNK, uterine NK.
References
- 1.Loke, Y.W., and A. King. 1995. Human Implantation: Cell Biology and Immunology. Cambridge University Press, Cambridge, UK. 299 pp.
- 2.Moffett-King, A. 2002. Natural killer cells and pregnancy. Nat. Rev. Immunol. 2:656–663. [DOI] [PubMed] [Google Scholar]
- 3.King, A., D.S.J. Allan, J.M. Bowen, S.J. Powis, S. Joseph, S. Verma, S.E. Hiby, A.J. McMichael, Y.W. Loke, and V.M. Braud. 2000. HLA-E is expressed on trophoblast and interacts with CD94/NKG2 receptors on decidual NK cells. Eur. J. Immunol. 30:1623–1631. [DOI] [PubMed] [Google Scholar]
- 4.King, A., T.D. Burrows, S.E. Hiby, S. Joseph, S. Verma, P.B. Lim, L. Gardner, J.M. Bowen, A. Ziegler, B. Uchanska-Ziegler, et al. 2000. Surface expression of HLA-C antigen by human extravillous trophoblast. Placenta. 21:376–387. [DOI] [PubMed] [Google Scholar]
- 5.Hiby, S.E., A. King, A. Sharkey, and Y.W. Loke. 1999. Molecular studies of trophoblast HLA-G polymorphism, isoforms, imprinting and expression in pre-implantation embryo. Tissue Ant. 53:1–13. [DOI] [PubMed] [Google Scholar]
- 6.Lanier, L.L. 1998. NK cell receptors. Annu. Rev. Immunol. 16:619–648. [DOI] [PubMed] [Google Scholar]
- 7.Vilches, C., and P. Parham. 2002. KIR: diverse, rapidly evolving receptors of innate and adaptive immunity. Annu. Rev. Immunol. 20:217–251. [DOI] [PubMed] [Google Scholar]
- 8.Carrington, M., and P. Norman. 2003. The KIR Gene Cluster. Bethesda, MD: National Library of Medicine (US), National Center for Biotechnology Information. 48 pp.
- 9.Trowsdale, J. 2001. Genetic and functional relationships between MHC and NK receptor genes. Immunity. 15:363–374. [DOI] [PubMed] [Google Scholar]
- 10.Colonna, M., G. Borsellino, M. Falco, G.B. Ferrara, and J.L. Strominger. 1993. HLA-C is the inhibitory ligand that determines dominant resistance to lysis by NK1- and NK2-specific natural killer cells. Proc. Natl. Acad. Sci. USA. 90:12000–12004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mandelboim, O., H.T. Reyburn, M. Vales-Gomez, L. Pazmany, M. Colonna, G. Borsellino, and J.L. Strominger. 1996. Protection from lysis by natural killer cells of group 1 and 2 specificity is mediated by residue 80 in human histocompatibility leukocyte antigen C alleles and also occurs with empty major histocompatibility complex molecules. J. Exp. Med. 184:913–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koopman, L.A., H.D. Kopcow, B. Rybalov, J.E. Boyson, J.S. Orange, F. Schatz, R. Masch, C.J. Lockwood, A.D. Schachter, P.J. Park, and J.L. Strominger. 2003. Human decidual natural killer cells are a unique NK cell subset with immunomodulatory potential. J. Exp. Med. 198:1201–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guimond, M.J., J.A. Luross, B. Wang, C. Terhorst, S. Danial, and B.A. Croy. 1997. Absence of natural killer cells during murine pregnancy is associated with reproductive compromise in TgE26 mice. Biol. Reprod. 56:169–179. [DOI] [PubMed] [Google Scholar]
- 14.Croy, B.A., A.A. Ashkar, K. Minhas, and J.D. Greenwood. 2000. Can murine uterine natural killer cells give insights into the pathogenesis of preeclampsia? J. Soc. Gynecol. Investig. 7:12–20. [DOI] [PubMed] [Google Scholar]
- 15.Verma, S., A. King, and Y.W. Loke. 1997. Expression of killer-cell inhibitory receptors (KIR) on human uterine NK cells. Eur. J. Immunol. 27:979–983. [DOI] [PubMed] [Google Scholar]
- 16.Roberts, J.M. 2003. Pre-eclampsia: a two-stage disorder. Pre-eclampsia. H. Critchley, A. MacLean, L. Poston, and J. Walker, editors. RCOG Press, London, UK. 66–78.
- 17.Redman, C.W. 1991. Current topic: pre-eclampsia and the placenta. Placenta. 12:301–308. [DOI] [PubMed] [Google Scholar]
- 18.Pijnenborg, R., L. Vercruysse, M. Hanssens, and A. van Assche. 2003. Incomplete trophoblast invasion: the evidence. Pre-eclampsia. H. Critchley, A. MacLean, L. Poston, and J. Walker, editors. RCOG Press, London, UK. 15–26.
- 19.Cincotta, R.B., and S.P. Brennecke. 1998. Family history of pre-eclampsia as a predictor for pre-eclampsia in primigravidas. Int. J. Gynaecol. Obstet. 60:23–27. [DOI] [PubMed] [Google Scholar]
- 20.Treloar, S.A., D.W. Cooper, S.P. Brennecke, M.M. Grehan, and N.G. Martin. 2001. An Australian twin study of the genetic basis of preeclampsia and eclampsia. Am. J. Obstet. Gynecol. 184:374–381. [DOI] [PubMed] [Google Scholar]
- 21.Esplin, M.S., M.B. Fausett, A. Fraser, R. Kerber, G. Mineau, J. Carrillo, and M.W. Varner. 2001. Paternal and maternal components of the predisposition to preeclampsia. N. Engl. J. Med. 344:867–872. [DOI] [PubMed] [Google Scholar]
- 22.Trupin, L.S., L.P. Simon, and B. Eskenazi. 1996. Change in paternity: a risk factor for preeclampsia in multiparas. Epidemiology. 7:240–244. [DOI] [PubMed] [Google Scholar]
- 23.Soderstrom-Anttila, V., A. Tiitinen, T. Foudila, and O. Hovatta. 1998. Obstetric and perinatal outcome after oocyte donation: comparing with in vitro fertilisation pregnancies. Hum. Reprod. 13:483–490. [DOI] [PubMed] [Google Scholar]
- 24.Gillespie, K.M., S.J. Valovin, J. Saunby, K.M. Hunter, D.A. Savage, D. Middleton, J.A. Todd, P.J. Bingley, and E.A.M. Gale. 2000. HLA class II typing of whole genome amplified mouth swab DNA. Tissue Ant. 56:530–538. [DOI] [PubMed] [Google Scholar]
- 25.Martin, M.P., G. Nelson, J.H. Lee, F. Pellett, X. Gao, J. Wade, M.J. Wilson, J. Trowsdale, D. Gladman, and M. Carrington. 2002. Susceptibility to psoriatic arthritis: influence of activating killer Ig-like receptor genes in the absence of specific HLA-C alleles. J. Immunol. 169:2818–2822. [DOI] [PubMed] [Google Scholar]
- 26.Williams, F., A. Meenagh, C. Patterson, and D. Middleton. 2002. Molecular diversity of the HLA-C gene identified in a Caucasian population. Hum. Immunol. 63:602–613. [DOI] [PubMed] [Google Scholar]
- 27.Yawata, M., N. Yawata, K.L. McQueen, N.W. Cheng, L.A. Guethlein, R. Rajalingam, H.G. Shilling, and P. Parham. 2002. Predominance of group A KIR haplotypes in Japanese associated with diverse NK cell repertoires of KIR expression. Immunogenetics. 54:543–550. [DOI] [PubMed] [Google Scholar]
- 28.Norman, P.J., H.A. Stephens, D.H. Verity, D. Chandanayingyong, and R.W. Vaughan. 2001. Distribution of natural killer cell immunoglobulin-like receptor sequences in three ethnic groups. Immunogenetics. 52:195–205. [DOI] [PubMed] [Google Scholar]
- 29.Crum, K.A., S.E. Logue, M.D. Curran, and D. Middleton. 2000. Development of a PCR-SSOP approach capable of defining the natural killer cell inhibitory receptor (KIR) gene sequence repertoires. Tissue Ant. 56:313–326. [DOI] [PubMed] [Google Scholar]
- 30.Cook, M.A., P.A. Moss, and D.C. Briggs. 2003. The distribution of 13 killer-cell immunoglobulin-like receptor loci in UK blood donors from three ethnic groups. Eur. J. Immunogenet. 30:213–221. [DOI] [PubMed] [Google Scholar]
- 31.Toneva, M., V. Lepage, G. Lafay, N. Dulphy, M. Bussom, S. Lester, A. Vu-Trien, A. Michaylova, E. Naumova, J. McCluskey, and D. Charron. 2001. Genomic diversity of natural killer cell receptor genes in three populations. Tissue Ant. 57:358–362. [DOI] [PubMed] [Google Scholar]
- 32.Rajalingam, R., P. Krausa, H.G. Shilling, J.B. Stein, A. Balamurugan, M.D. McGinnis, N.W. Cheng, N.K. Mehra, and P. Parham. 2002. Distinctive KIR and HLA diversity in a panel of north Indian Hindus. Immunogenetics. 53:1009–1019. [DOI] [PubMed] [Google Scholar]
- 33.Whang, D.E., H. Park, and M.H. Park. 2003. Determination of KIR haplotypes in 34 Korean families. Hum. Immunol. 64:S168. [Google Scholar]
- 34.Wang, H., K. Kokunaga, Y. Ishikawa, H. Tanataka, K. Kashiwase, Y. Shibata, and T. Juji. 1997. A high-resolution genotyping method for HLA-C alleles and possible shared HLA-C-B haplotypes between Japanese and Caucasians. Tissue Ant. 50:620–626. [DOI] [PubMed] [Google Scholar]
- 35.Kitts, A., M. Feola, and W. Helmberg. 2002. The Major Histocompatibility Complex Database, dbMHC. The NCBI Handbook (Internet). J. McEntyre, editor. National Library of Medicine (US), National Centre for Biotechnology Information. 29 pp.
- 36.Fan, Q.R., E.O. Long, and D.C. Wiley. 2001. Crystal structure of the human natural killer cell inhibitory receptor KIR2DL1-HLA-Cw4 complex. Nat. Immunol. 2:452–460. [DOI] [PubMed] [Google Scholar]
- 37.Winter, C.C., J.E. Gumperz, P. Parham, E.O. Long, and N. Wagtmann. 1998. Direct binding and functional transfer of NK cell inhibitory receptors reveal novel patterns of HLA-C allotype recognition. J. Immunol. 161:571–577. [PubMed] [Google Scholar]
- 38.Vales-Gomez, M., H. Reyburn, and J. Strominger. 2000. Molecular analyses of the interactions between human NK receptors and their HLA ligands. Hum. Immunol. 61:28–38. [DOI] [PubMed] [Google Scholar]
- 39.Maenaka, K., T. Juji, T. Nakayama, J.R. Wyer, G.F. Gao, T. Maenaka, N.R. Zaccai, A. Kikuchi, T. Yabe, K. Tokunaga, et al. 1999. Killer cell immunoglobulin receptors and T cell receptors bind peptide-major-histocompatibility complex class I with distinct thermodynamic and kinetic properties. J. Biol. Chem. 274:28329–28334. [DOI] [PubMed] [Google Scholar]
- 40.Cook, M.A., D.W. Milligan, C.D. Fegan, P.J. Darbyshire, P. Mahendra, C.F. Craddock, P.A. Moss, and D.C. Briggs. 2004. The impact of donor KIR and patient HLA-C genotypes and outcome following HLA-identical sibling haematopoietic stem cell transplantation for myeloid leukaemia. Blood. 103:1521–1526. [DOI] [PubMed] [Google Scholar]
- 41.Khakoo, S.I., C.L. Thio, M.P. Martin, C.R. Brooks, X. Gao, J. Astemborski, J. Cheng, J.J. Goedert, D. Vlahov, M. Hilgartner, et al. 2004. HLA and NK cell inhibitory receptor genes in resolving Hepatitis C virus infection. Science. 305:872–874. [DOI] [PubMed] [Google Scholar]
- 42.Martin, M.P., X. Gao, J.H. Lee, G.W. Nelson, R. Detels, J.J. Goedert, S. Buchbinder, K. Hoots, D. Vlahov, J. Trowsdale, et al. 2002. Epistatic interaction between KIR3DS1 and HLA-B delays the progression of AIDS. Nat. Genet. 31:429–434. [DOI] [PubMed] [Google Scholar]
- 43.Yen, J.-H., B.E. Moore, T. Nakajima, D. Scholl, D.J. Schaid, C.M. Weyand, and J.J. Goronzy. 2001. Major histocompatibility complex class I–recognizing receptors are disease risk genes in rheumatoid arthritis. J. Exp. Med. 193:1159–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.King, A., C. Boocock, A. Sharkey, L. Gardner, and Y.W. Loke. 1996. Evidence for the expression of HLA-C class I mRNA and protein by first trimester trophoblast. J. Immunol. 156:2068–2076. [PubMed] [Google Scholar]
- 45.Barber, L.D., T.P. Patel, L. Percival, J.E. Gumperz, L.L. Lanier, J.H. Phillips, J.C. Bigge, M.R. Wormwald, R.B. Parekh, and P. Parham. 1996. Unusual uniformity of the N-linked oligosaccarides of HLA-A, -B, and -C glycoproteins. J. Immunol. 156:3275–3284. [PubMed] [Google Scholar]
- 46.McMaster, M., Y. Zhou, S. Shorter, K. Kapasi, D. Geraghty, K.H. Lim, and S. Fisher. 1998. HLA-G isoforms produced by placental cytotrophoblasts and found in amniotic fluid are due to unusual glycosylation. J. Immunol. 160:5922–5928. [PubMed] [Google Scholar]
- 47.Boyson, J.E., R. Erskine, M.C. Whitman, M. Chiu, J.M. Lau, L.A. Koopman, M.M. Valter, P. Angelisova, V. Horejsi, and J.L. Strominger. 2002. Disulfide bond-mediated dimerization of HLA-G on the cell surface. Proc. Natl. Acad. Sci. USA. 99:16180–16185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gonen-Goss, T., H. Achdout, R. Gazit, J. Hanna, S. Mizrahi, G. Markel, D. Goldman-Wohl, S. Yagel, V. Horejsi, O. Levy, et al. 2003. Complexes of HLA-G protein on the cell surface are important for leukocyte Ig-like receptor-1 function. J. Immunol. 171:1343–1351. [DOI] [PubMed] [Google Scholar]
- 49.Rajagopalan, S., and E.O. Long. 1997. The direct binding of a p58 killer cell inhibitory receptor to human histocompatibility leukocyte antigen (HLA)-Cw4 exhibits peptide selectivity. J. Exp. Med. 185:1523–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brooks, A.G., J.C. Boyington, and P.D. Sun. 2000. Natural killer cell recognition of HLA class I molecules. Rev. Immunogenet. 2:433–448. [PubMed] [Google Scholar]
- 51.Muir, A., A. Lever, and A. Moffett. 2004. Expression and functions of human endogenous retroviruses in the placenta: an update. Placenta. 25:S16–S25. [DOI] [PubMed] [Google Scholar]
- 52.Lie, R.T., S. Rasmussen, H. Brunborg, H.K. Gjessing, E. Lie-Nielsen, and L.M. Irgens. 1998. Fetal and maternal contributions to risk of preeclampsia: population based study. BMJ. 316:1343–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eskenazi, B., L. Fenster, and S. Sidney. 1991. A multivariate analysis of risk factors for preeclampsia. JAMA. 266:237–241. [PubMed] [Google Scholar]
- 54.Trogstad, L. 2003. The paternal role in preeclampsia. Pre-eclampsia. Critchley H., A. Maclean, L. Poston, and J. Walker, editors. RCOG Press, London, UK. 208–224.
- 55.Roberts, J.M., and C.W. Redman. 1993. Pre-eclampsia: more than pregnancy-induced hypertension. Lancet. 341:1447–1451. [DOI] [PubMed] [Google Scholar]
- 56.Valiante, N.M., M. Uhrberg, H.G. Shilling, K. Lienert-Weidenbach, K.L. Arnett, A. D'Andrea, J.H. Phillips, L.L. Lanier, and P. Parham. 1997. Functionally and structurally distinct NK cell receptor repertoires in the peripheral blood of two human donors. Immunity. 7:739–751. [DOI] [PubMed] [Google Scholar]
- 57.Shilling, H.G., N. Young, L.A. Guethlein, N.W. Cheng, C.M. Gardiner, D. Tyan, and P. Parham. 2002. Genetic control of human NK cell repertoire. J. Immunol. 169:239–247. [DOI] [PubMed] [Google Scholar]
- 58.Trowsdale, J., and P. Parham. 2004. Defense strategies and immunity-related genes. Eur. J. Immunol. 34:7–17. [DOI] [PubMed] [Google Scholar]
- 59.Guethlein, L.A., L.R. Flodin, E.J. Adams, and P. Parham. 2002. NK cell receptors of the orangutan (Pongo pygmaeus): a pivotal species for tracking the coevolution of killer cell Ig-like receptors with MHC-C. J. Immunol. 169:220–229. [DOI] [PubMed] [Google Scholar]
- 60.Chaline, J. 2003. Increased cranial capacity in hominid evolution and preeclampsia. J. Reprod. Immunol. 59:137–152. [DOI] [PubMed] [Google Scholar]
- 61.Martin, R.D. 2003. Human reproduction: a comparative background for medical hypotheses. J. Reprod. Immunol. 59:111–135. [DOI] [PubMed] [Google Scholar]
- 62.Moffett, A., and Y.W. Loke. 2004. The immunological paradox of pregnancy: a reappraisal. Placenta. 25:1–8. [DOI] [PubMed] [Google Scholar]