Abstract
The immunoglobulin κ light chain intronic enhancer (iEκ) activates κ rearrangement and is required to maintain the earlier or more efficient rearrangement of κ versus lambda (λ). To understand the mechanism of how iEκ regulates κ rearrangement, we employed homologous recombination to mutate individual functional motifs within iEκ in the endogenous κ locus, including the NF-κB binding site (κB), as well as κE1, κE2, and κE3 E boxes. Analysis of the impacts of these mutations revealed that κE2 and to a lesser extent κE1, but not κE3, were important for activating κ rearrangement. Surprisingly, mutation of the κB site had no apparent effect on κ rearrangement. Comparable to the deletion of the entire iEκ, simultaneous mutation of κE1 and κE2 reduces the efficiency of κ rearrangement much more dramatically than either κE1 or κE2 mutation alone. Because E2A family proteins are the only known factors that bind to these E boxes, these findings provide unambiguous evidence that E2A is a key regulator of κ rearrangement.
Keywords: B cell development, V(D)J recombination, accessibility, monospecificity, transcription factor
Introduction
Each B lymphocyte generates a unique set of immunoglobulin heavy (IgH) and light (IgL) chain genes through the somatic rearrangement of V, D, and J gene segments. To ensure the monospecificity of each B cell, V(D)J recombination is regulated in such a lineage- and stage-specific manner that IgH and IgL rearrangement occur at distinct stages of development (1). Because V(D)J rearrangement of all Ig loci involves the same recombination machinery, cis elements within these antigen receptor loci must play decisive roles in regulating the accessibility of each locus to V(D)J recombination machinery (2). Within the κ locus, several cis-acting elements were initially identified by their ability to activate transcription of κ reporter constructs in B cell lines, including two enhancers, one within the Jκ-Cκ intron (iEκ) and one 3′ of Cκ (3′Eκ) (3–8). More recently, a putative third enhancer 8 kb downstream of 3′Eκ (Ed) was also discovered (9). The deletion of the intronic enhancer and matrix attachment region (MiEκ) led to decreased κ rearrangement and a lower κ:λ ratio. More importantly, MiEκ is required for the earlier or more efficient rearrangement of κ versus λ loci. (10, 11). The matrix attachment region (MAR) within MiEκ does not contribute to these activities since the deletion of the κ MAR alone does not have an inhibitory effect on the overall level of κ rearrangement (12). The deletion of the 3′ enhancer (3′Eκ) results in a similar, though less dramatic decrease in the ratio of κ:λ B cells and κ rearrangement (13). In addition, 3′Eκ appears to play an important role in activating κ transcription in mature B cells (11, 13). Although the loss of either enhancer alone does not eliminate κ rearrangement, deletion of both enhancers from endogenous κ loci results in a complete block of κ rearrangement (11). Therefore, MiEκ and 3′Eκ together are the necessary elements for κ rearrangement.
Several functional motifs within the intronic enhancer were identified through a battery of biochemical and cell line transfection studies. One such functional motif is the NF-κB binding site, denoted κB (14). The potential role of the κB site in κ rearrangement was suggested by the finding that LPS could induce κ germline transcription and rearrangement through an NF-κB–dependent pathway (15–18). iEκ also contains a class of protein-binding motifs referred to as E boxes, which are also identified in enhancers of other antigen receptor genes (19, 20). iEκ contains three E boxes, labeled κE1, κE2, and κE3. κE2 was found to bind the E2A gene products E12 and E47, which are required for B cell development (21–23). E2A gene products can also bind to κE1 but not to κE3 (Murre, C., personal communication). E2A can induce germline transcription and κ rearrangement when cotransfected with the RAG genes in a nonlymphoid cell line (24). However, the role of E2A in the regulation of κ rearrangement in the physiological context remains unclear.
To delineate the mechanism through which iEκ activates κ rearrangement, we employed homologous recombination and Cre/loxP-mediated deletion to produce independently targeted deletions of four functional motifs within the endogenous κ intronic enhancer. Analysis of the effects of these mutations on κ rearrangement revealed that the E2A-binding E boxes, but not the κB site, were quantitatively important for the activation of κ rearrangement. In addition, the simultaneous mutation of both κE1 and κE2 sites impacts κ rearrangement as severely as the deletion of the entire iEκ, indicating that E2A-dependent pathways are the major mediator of iEκ's activity in activating κ rearrangement.
Materials and Methods
Generation of mκEX Embryonic Stem Cells.
The 740-bp κ intronic enhancer was cloned into pBluescript. The four sites were deleted and replaced with diagnostic EcoRI sites independently by site-directed mutagenesis. The primers used to replace each site are as follows: mκB, 5′-TGGCATCTCAACGAATTCAGAGCCATCTGG-3′; mκE1, 5′-GACTTTCCGAGAGAATTCAGTTGCTTAAGA-3′; mκE2, 5′-CAGTTCCTCCGAATTCGATTACAG-3′; and mκE3, 5′-TGGCTAAAAATTGA-ATTCAAACCATTAGAC-3′.
The mκE1/2 double mutations were generated by PCR-mediated mutagenesis. The κE1 and κE2 sequences are as follows, with the underlined C nucleotides mutated to A: κE1, CATCTGGC; κE2, CAGGTGGC. The MiEκ deletion (mκD) was generated as described (10). All mκEX clones were inserted into the mκD targeting construct. 20 μg of each targeting construct was linearized with PvuI and transfected into embryonic stem (ES) cells. Transfectants were selected with G418 (300 μg/ml) and gancyclovir (1 μM). Homologous recombination was screened by Southern blot, and positive clones were then transiently transfected with 10–20 μg of a plasmid containing the Cre gene to remove the PGK-neo r gene as described (10). PGK-neo r–deleted ES clones were subcloned, confirmed by Southern blot, and analyzed by RAG-2–deficient blastocyst complementation as described (10). mκE1/2 ES cells were microinjected into blastocysts before Cre transfection and then bred to Cre-expressing mice to generate mκE1/2 knock-in mice.
Quantitative Analysis of Vκ Jκ Rearrangements.
To detect Vκ Jκ rearrangements, genomic DNA from mature B cells (CD19+/IgM+) was purified and amplified by PCR using the degenerate Vκ primer (VκD) (15) and a primer within the kappa intronic enhancer (K1). PCR reactions were conducted under the following conditions: 95°C for 5 min, followed by 26 cycles of 94°C for 1 min, 60°C for 1 min, and 72°C for 3.5 min, followed by an additional 10 min at 72°C. To control for the total amount of B cell DNA used in each PCR reaction, VDJ H rearrangement was also analyzed using a primer for the J558 family (J558) and a primer that can bind to all four J H gene segments (J H (1+4)). The intensity of each PCR product was quantified with a Storm phosphorimager. Primers used are as follows: VκD, 5′-GGCTGCAGSTTCAGTGGCAGTGGRTCWGGRAC-3′; K1, 5′-CCAT-GACTTTTGCTGGCTGTAGATTTTACCTC-3′; J558, 5′-CTTCAGTGAAGCTGTCCTGCAAGGCTT-3′; and J H (1+4), 5′-CAGCTTACCTGAGGAGACGGTGA-3′. Hybridomas were generated and analyzed as described (10). The University of California, San Diego Animal Subject Committee approved all experiments that involved mice.
Quantitative Analysis of κ Transcription by Real-Time PCR.
B cells from the spleens of WT and homozygous mκE1/2 mutant mice were purified by magnetic-activated cell sorting using αCD43 microbeads according to the manufacturer's protocol (Miltenyi Biotec). In mκE1/2 mutants, λ+ B cells were removed by costaining with 0.5 μg/million cells biotin-conjugated α-mouse Igλ antibodies (clone R26-46; BD Biosciences) and then, after washing, staining with 0.5 μg/million cells α-biotin microbeads (Miltenyi Biotec). RNA from 1 million sorted cells was purified using the RNAeasy kit (QIAGEN) combined with on-column DNase digestion (QIAGEN) and converted into cDNA using the Superscript First Strand System (Invitrogen) according to the manufacturers' protocols. Primers for amplifying the constant regions of μ (C μ 1 and C μ 2) and κ (Cκ1 and Cκ2) were designed using Primer Express software (Applied Biosystems) and used at concentrations of 200 and 400 nM, respectively. Real-time PCR reactions were performed using the SYBR Green PCR Master Mix (Applied Biosystems) in an ABI Prism 7000 Sequence Detection System according to the manufacturer's protocols. Relative transcription levels were calculated using ABI Prism 7000 SDS software using the standard curve method. Primer sequences are as follows: Cμ1, 5′-ACACCTGCCGTGTGGATCA-3′; Cμ2, 5′-GAGGAAGATGTCGGCAAAGG-3′; Cκ1, 5′-CAACTGTATCCATCTTCCCACCA-3′; and Cκ2, 5′-GGCACCTCCAGATGTTAACTGCT-3′.
Results and Discussion
Generation of mκEX ES Cells.
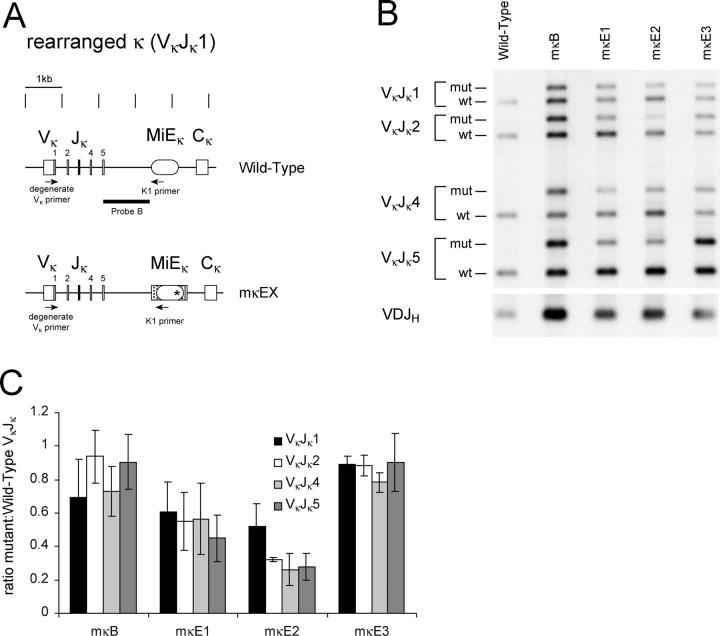
The 740-bp HindIII–AflII region spanning the κ intronic enhancer and associated matrix attachment region (MiEκ) was cloned into pBluescript. The NF-κB, κE1, κE2, or κE3 sites were each individually replaced with a diagnostic EcoRI restriction site by site–directed mutagenesis (Fig. 1 F). These iEκ mutants, collectively referred to as mκEX and individually as mκB, mκE1, mκE2, and mκE3, were independently inserted into the targeting construct used previously to delete MiEκ, so that homologous recombination between the targeting vector and the endogenous locus would replace the endogenous MiEκ with mκEX (10) (Fig. 1 B). Homologous recombination events were screened by Southern blotting with EcoRI digestion and hybridization to probe A (Fig. 1, A and C; not depicted). The PGK-neo r gene was excised from the targeted allele by transient expression of the Cre gene in positive ES clones (Fig. 1 D). ES clones with the PGK-neo r deleted were subcloned and confirmed by Southern blot with EcoRI digestion and hybridization to probe A (Fig. 1 E, lanes 2–5).
Figure 1.
Generation of mκEX mutant ES cells. (A) The endogenous WT κ locus. The lengths of the diagnostic restriction fragments and probes are shown. Markings above the diagram are spaced 1 kb apart. (B) The mκEX targeting construct. The mutated intronic enhancer (*) was inserted into the targeting construct just upstream of the loxP-flanked PGK-neo r gene. Additional polylinker sequence upstream of the mutated intronic enhancer is represented by a box with diagonal lines. (C) Targeted locus with the PGK-neo r gene inserted. Size of the diagnostic EcoRI fragment is shown. (D) Final configuration with the PGK-neo r gene removed by Cre/loxP-mediated deletion. The size of the EcoRI restriction fragment is shown. (E) Southern blot analysis of the genomic DNA from WT ES cells and ES clones from the final configuration of each of the four heterozygous mκEX mutants. WT (13.5 kb) and neo r-deleted (9.4 kb) bands are indicated. (F) Diagram of the intronic enhancer and its functional motifs. The MAR, κB site, and the three E boxes are shown. The sequences spanning each of the four sites that were replaced by EcoRI sites are shown in boxes. The consensus sequences of the each of the four sites are underlined.
Analysis of Vκ Jκ Recombination in mκEX B Cells.
The effects of mκEX mutations on κ rearrangement in B cells were assayed by recombination activating gene-2–deficient (RAG-2−/−) blastocyst complementation as described (10). Since heterozygous mutant ES cells were used, this enabled us to compare the rearrangement frequency between the WT and mutant κ alleles. B cells were sorted from mκEX–RAG-2−/− mice and analyzed for Vκ Jκ recombination using a quantitative PCR assay as previously described (11, 12). This assay could detect rearrangement to all four functional Jκ gene segments and distinguish PCR products derived from the WT allele from those from the mutant allele due to an additional 150 bp within the mutant allele (Fig. 2 A). Therefore, the efficiency of Vκ Jκ1-5 rearrangement of the mutant κ allele could be compared with that of the WT allele in the same PCR reaction (Fig. 2 C). Our analysis indicated that neither the NF-κB (mκB) nor κE3 (mκE3) site mutations had any apparent effect on κ rearrangement (Fig. 2 B). However, mutation of the κE1 (mκE1) or κE2 (mκE2) site reduced the rearrangement efficiency of the mutant κ allele. The mutation of the κE2 site had a more inhibitory effect on κ rearrangement (Fig. 2 B, lane 4). In this context, the ratio of the rearrangement frequency of V κ to J κ 1, J κ 2, J κ 4, and J κ 5 gene segments of the mutant allele versus that of the WT allele was 52.1, 31.9, 26.2, and 27.9%, respectively (Fig. 2 C). In mκE1 B cells, the rearrangement efficiency of the mutant allele was approximately twofold reduced for each of the four possible Vκ Jκ rearrangements compared with that of the WT allele (Fig. 2 B, lane 3, and C).
Figure 2.
PCR analysis of Vκ Jκ rearrangement in mκEX heterozygous mice. (A) PCR strategy. Examples of rearranged Vκ Jκ1 WT and mκEX alleles are shown. Annealing locations of the degenerate Vκ primer and K1 primer are shown with arrowheads, and probe B is shown with a horizontal bar. Markers are placed at 1 kb intervals for scale. (B) Representative PCR products of the rearrangement assay from B cells of WT and each of the four mκEX heterozygous mutants are shown. Bands corresponding to mutant (mut) and WT Vκ Jκ1, Vκ Jκ2, Vκ Jκ4, and Vκ Jκ5 rearrangements are indicated. Heavy chain rearrangement was also assayed by PCR as a control for the number of B cells, using primers J558 and J H (1+4) as described (11). Band shown corresponds to rearrangement from J558 V H family members to either J H 1 or J H 4 (VDJ H). Only the smallest band produced by this primer set is shown. (C) Statistical analysis of the ratio of the rearrangement efficiency of mκEX allele versus that of the WT allele. Rearrangement efficiency was calculated by dividing the intensity of bands derived from the mutant allele by that of the WT allele. Means values from at least three independent experiments are presented with error bars. Total number of mice (n) analyzed for each mutation were; mκB (n = 3), mκE1 (n = 3), mκE2 (n = 5), and mκE3 (n = 2).
Analysis of Vκ Jκ Recombination in mκEX κ+ Hybridomas.
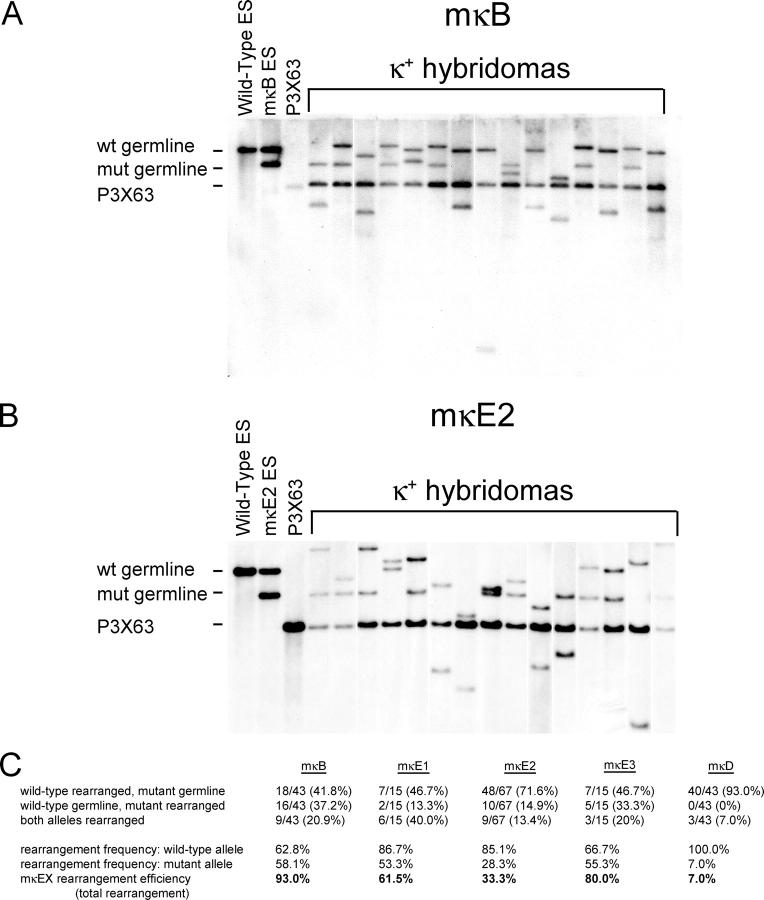
To further confirm the impact of mκEX mutations on κ rearrangement efficiency and analyze the rearrangement frequency of mutant and WT alleles in individual B cells, we generated hybridomas from the spleen cells of all four mκEX–RAG-2−/− mice as described (10). Genomic DNA derived from κ+ hybridomas was analyzed by Southern blotting to detect Vκ Jκ rearrangements at the WT and mutant alleles. Consistent with the data from the PCR analysis, we observed the most dramatic decrease in the rearrangement frequency of the mutant allele in mκE2 hybridomas (Fig. 3 B). In this context, of the 67 κ+ hybridomas analyzed 48 had rearrangements on only the WT allele, 10 had rearrangements on only the mutant allele, and 9 had rearrangements on both alleles (Fig. 3 C). The ratio of the rearrangement frequency of the WT allele versus that of the mutant allele was ∼3:1. A less dramatic decrease was observed in mκE1 hybridomas (Fig. 3 C). Consistent with the data derived from the PCR analysis, little difference was detected in the rearrangement frequency of the WT and mutant alleles in hybridomas derived from the mκB and mκE3 B cells (Fig. 3, A and C).
Figure 3.
Southern blot analysis of κ rearrangement in mκEX hybridomas. Representative data from mκB (A) and mκE2 (B) hybridomas. Genomic DNA was digested with BamHI and hybridized to probe B. Lanes 1, 2, and 3 on each blot are genomic DNA from WT ES, mutant ES, and P3X63 fusion partner, respectively. All other lanes represent individual κ+ hybridoma lines. Bands representing germline WT (12.5 kb) and mutant (9.7 kb) alleles and the contribution of the P3X63 fusion partner are indicated. Unspecified bands of various sizes represent rearranged alleles. White lines indicate that intervening lanes have been spliced out. (C) Analysis of κ rearrangement frequency in κ-expressing hybridomas. Hybridomas were scored for rearrangement of the WT allele, mutant allele, or both alleles. Rearrangement frequencies are the number of WT or mutant rearranged alleles divided by the total number of hybridomas. Rearrangement efficiency is calculated by dividing the mutant rearrangement frequency by the WT rearrangement frequency.
To determine the contribution of the κE1 and κE2 sites to the full activity of iEκ, hybridomas were also generated from B cells in which the entire intronic enhancer was deleted from one allele (mκD). Only 3 of 43 κ+ mκD hybridomas analyzed harbored a V κ J κ rearrangement on the mutant allele, whereas the WT allele was rearranged in all 43 hybridomas (Fig. 3 C). Therefore, the reduction of the rearrangement efficiency caused by the deletion of the entire iEκ was much more dramatic than that caused by κE1 or κE2 mutations alone. In addition, these data indicate that the rearrangement of the mutant allele occurs only after the rearrangement of the WT allele in mκD B cells.
Critical Roles of κE1 and κE2 in Mediating iEκ's Function in Activating κ Rearrangement.
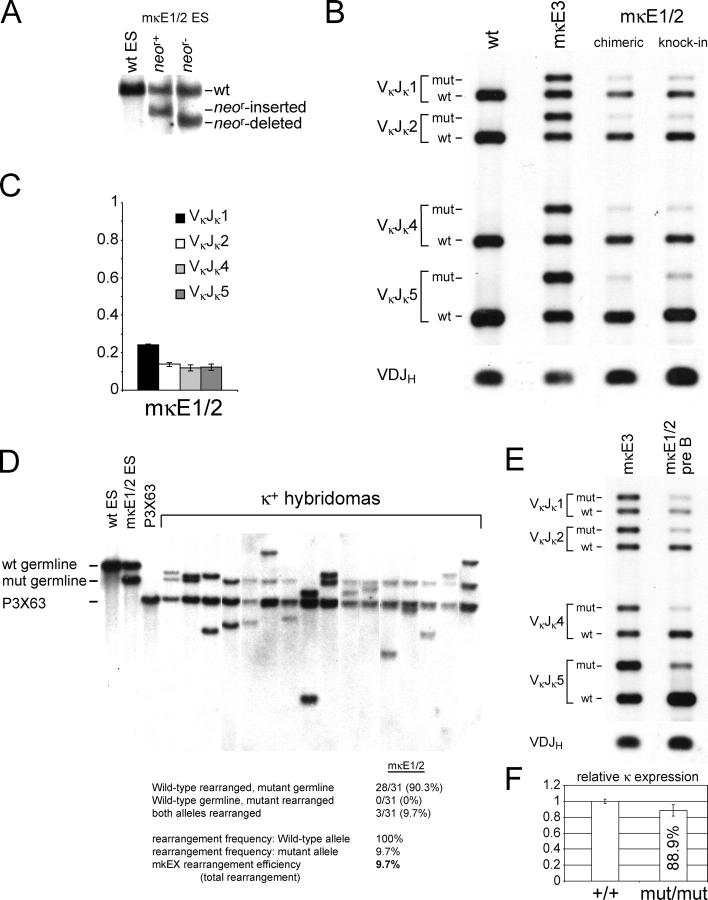
Although the reduction of κ rearrangement caused by the κE1 or κE2 mutations alone was significantly less than that caused by the deletion of the entire enhancer, it is possible that these two sites have redundant functions since both sites can be bound by E2A family proteins. To test this hypothesis, we mutated both κE1 and κE2 sites simultaneously through homologous recombination and assayed for effects on κ rearrangement. The mutation introduced at both sites was a single nucleotide (C to A) mutation that destroyed the canonical basic helix-loop-helix binding site (CANNTG) (Fig. 4 A). To determine the effects of the κE1/2 mutation on κ rearrangement, we used the same PCR assay described in Fig. 2 to analyze κ rearrangement of WT and mutant alleles in κ+ mκE1/2 B cells purified from the spleens of RAG−/− chimeric mice or heterozygous mutant mice. The reduction in κ rearrangement caused by the mκE1/2 mutation was much more severe than that caused by either κE1 or κE2 mutations alone (Fig. 4, B and C). Analysis of κ+ hybridomas derived from mκE1/2 B cells revealed a more than 10-fold reduction in the rearrangement efficiency of the mutant allele, comparable to that caused by the deletion of the entire enhancer (Fig. 4 D). In addition, similar to findings in mκD hybridomas, the mκE1/2 allele only rearranged after the WT allele had already rearranged.
Figure 4.
Impacts of the κE1/κE2 double mutation (mκE1/2) on κ rearrangement. (A) Southern blotting analyses of mκE1/2 heterozygous ES cells. The enzymes and probe used are the same as in Fig. 1. Genomic DNA derived from WT, PGK-neo r–inserted and PGK-neo r–deleted mκE1/2 ES cells are shown in lanes 1, 2, and 3, respectively. (B) PCR analysis of Vκ Jκ rearrangement at the WT and mutant alleles. The strategy for the rearrangement PCR assay is identical to that described in Fig. 2. Representative data derived from κ+ splenic B cells from mκE1/2–Rag2−/− chimeric mice and heterozygous germline mice are shown in lane 3 and 4, respectively. As PCR controls, PCR products of genomic DNA derived from WT splenic B cells and mκE3 splenic B cells are shown in lanes 1 and 2, respectively. (C) Statistical analysis of the ratio of the rearrangement frequency of the mutant allele versus that of the WT allele. Mean values derived from four independent experiments are shown with error bars. (D) Representative Southern blotting analysis of hybridomas derived from spleen mκE1/2 B cells. Total numbers of the rearranged WT and mutant allele in 31 hybridomas are shown. White lines indicate that intervening lanes have been spliced out. (E) PCR analysis of Vκ Jκ rearrangement of the WT and mutant allele in mκE1/2 pre–B cells (lane 2). As a control, the rearrangement of WT and mutant alleles in mκE3 splenic B cells is also shown (lane 1). (F) Relative κ expression in κ1 B cells derived from WT and mκE1/2 homozygous mutant B cells. κ transcripts were analyzed by quantitative real-time PCR. mRNA levels were normalized to μ expression. Average of three RNA purifications are shown with error bars.
To rule out the possibility that the greatly reduced rearrangement efficiency of mκE1/2 alleles observed in splenic B cells is due to the impaired expression of rearranged κ at mκE1/2 alleles during the transition from pre–B cells to immature B cells, we examined the rearrangement frequency of WT and mutant alleles in pre–B cells (B220+/CD43−/IgM−) sorted from the BM of mκE1/2 mice (Fig. 4 E). Similar to that observed in splenic B cells, the rearrangement frequency of the mκE1/2 allele was significantly reduced in pre–B cells compared with the WT allele. To further determine whether the mκE1/2 mutation affects κ expression in B cells, we analyzed κ expression in κ1 B cells purified from the spleens of WT mice and homozygous mκE1/2 mutant mice. Similar levels of κ mRNA were detected in WT and mκE1/2 κ1 B cells, indicating that the mκE1/2 mutation had no apparent effect on κ transcription (Fig. 4 F). Therefore, κE1 and κE2 play synergistic roles in iEκ's function in activating κ rearrangement.
Sequential rearrangement of the IgH and IgL chain genes is critical to ensure B cell monospecificity. The κ intronic enhancer plays an important role in this process (10, 11). By analyzing the functional motifs within iEκ in vivo, we demonstrated that the κB site mutation had no apparent impact on κ rearrangement. The κB site is bound by NF-κB family members and was found to play an important role in activating κ rearrangement of recombination substrates in cell line transfection and transgenic studies (16, 25). However, the findings that the impacts of κE1 and κE2 double mutation on κ rearrangement are very similar to the deletion of the entire iEκ rule out any important roles of other protein-binding motifs within iEκ, such as the NF-κB binding site, in activating κ rearrangement. Based on the findings that iEκ and 3′Eκ are the essential elements activating κ rearrangement and that there is no NF-κB binding site within 3′Eκ, it remains unclear how NF-κB family transcription factors might activate κ rearrangement. Based on the findings that NF-κB is activated during B cell activation induced by cross-linking of the antigen receptor (for review see reference 26), the potential roles of the κB site in receptor editing and somatic hypermutation of κ should be examined.
Our findings provide unambiguous evidence that the κE1 and κE2 sites play critical roles in activating κ rearrangement. E2A family transcription factors are the primary transcription factors binding to the κE1/2 sites, and when coexpressed with the RAG proteins, are able to activate κ rearrangement in a non–B cell line (24). Therefore, our findings provide a mechanism for how E2A family transcription factors might function in activating κ rearrangement. Based on the findings that E2A can interact with histone acetyltransferases such as p300/CBP and the SAGA complex (27–29), κE1/2 might activate κ rearrangement by increasing histone acetylation of the κ locus.
Acknowledgments
The authors thank David Baltimore, Cornelis Murre, and Ann Feeney for helpful discussion and critical reading of the manuscript.
This work was supported by a National Institutes of Health grant (AI44838) to Y. Xu.
The authors have no conflicting financial interests.
References
- 1.Bassing, C.H., W. Swat, and F.W. Alt. 2002. The mechanism and regulation of chromosomal V(D)J recombination. Cell. 109:S45–S55. [DOI] [PubMed] [Google Scholar]
- 2.Yancopoulos, G.D., T.K. Blackwell, H. Suh, L. Hood, and F.W. Alt. 1986. Introduced T cell receptor variable region gene segments recombine in pre-B cells: evidence that B and T cells use a common recombinase. Cell. 44:251–259. [DOI] [PubMed] [Google Scholar]
- 3.Parslow, T.G., and D.K. Granner. 1983. Structure of a nuclease-sensitive region inside the immunoglobin kappa gene: evidence for a role in gene regulation. Nucleic Acids Res. 11:4775–4792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Emorine, L., M. Kuehl, L. Weir, P. Leder, and E.E. Max. 1983. A conserved sequence in the immunoglobulin J kappa-C kappa intron: possible enhancer element. Nature. 304:447–449. [DOI] [PubMed] [Google Scholar]
- 5.Queen, C., and D. Baltimore. 1983. Immunoglobulin gene transcription is activated by downstream sequence elements. Cell. 33:741–748. [DOI] [PubMed] [Google Scholar]
- 6.Picard, D., and W. Schaffner. 1984. A lymphocyte-specific enhancer in the mouse immunoglobulin kappa gene. Nature. 307:80–82. [DOI] [PubMed] [Google Scholar]
- 7.Queen, C., and J. Stafford. 1984. Fine mapping of an immunoglobulin gene activator. Mol. Cell. Biol. 4:1042–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meyer, K.B., and M.S. Neuberger. 1989. The immunoglobulin kappa locus contains a second, stronger B-cell-specific enhancer which is located downstream of the constant region. EMBO J. 8:1959–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu, Z.-M., J.B. George-Raizen, S. Li, K.C. Meyers, M.Y. Chang, and W.T. Garrard. 2002. Chromatin structural analyses of the mouse Igk gene locus reveal new hypersensitive sites specifying a transcriptional silencer and enhancer. J. Biol. Chem. 277:32640–32649. [DOI] [PubMed] [Google Scholar]
- 10.Xu, Y., L. Davidson, F.W. Alt, and D. Baltimore. 1996. Deletion of the Ig kappa light chain intronic enhancer/matrix attachment region impairs but does not abolish V kappa J kappa rearrangement. Immunity. 4:377–385. [DOI] [PubMed] [Google Scholar]
- 11.Inlay, M., F.W. Alt, D. Baltimore, and Y. Xu. 2002. Essential roles of the kappa light chain intronic enhancer and 3′ enhancer in kappa rearrangement and demethylation. Nat. Immunol. 3:463–468. [DOI] [PubMed] [Google Scholar]
- 12.Yi, M., P. Wu, K.W. Trevorrow, L. Claflin, and W.T. Garrard. 1999. Evidence that the Igk gene MAR regulates the probability of premature V-J joining and somatic hypermutation. J. Immunol. 162:6029–6039. [PubMed] [Google Scholar]
- 13.Gorman, J.R., N. vanderStoep, R. Monroe, M. Cogne, L. Davidson, and F.W. Alt. 1996. The Ig kappa 3′ enhancer influences the ratio of Ig kappa versus Ig lambda B lymphocytes. Immunity 5:241–252. [DOI] [PubMed] [Google Scholar]
- 14.Sen, R., and D. Baltimore. 1986. Multiple nuclear factors interact with the immunoglobulin enhancer sequences. Cell. 46:705–716. [DOI] [PubMed] [Google Scholar]
- 15.Schlissel, M.S., and D. Baltimore. 1989. Activation of immunoglobulin kappa gene rearrangement correlates with induction of germline kappa gene-transcription. Cell. 58:1001–1007. [DOI] [PubMed] [Google Scholar]
- 16.Demengeot, J., E.M. Oltz, and F.W. Alt. 1995. Promotion of V(D)J recombinational accessibility by the intronic E kappa element: role of the kappa B motif. Int. Immunol. 7:1995–2003. [DOI] [PubMed] [Google Scholar]
- 17.Scherer, D.C., J.A. Brockman, H.H. Bendall, G.M. Zhang, D.W. Ballard, and E.M. Oltz. 1996. Corepression of RelA and c-rel inhibits immunoglobulin kappa gene transcription and rearrangement in precursor B lymphocytes. Immunity. 5:563–574. [DOI] [PubMed] [Google Scholar]
- 18.O'Brien, D.P., E.M. Oltz, and B.G. Van Ness. 1997. Coordinate transcription and V(D)J recombination of the kappa immunoglobulin light-chain locus: NF-kappaB-dependent and -independent pathways of activation. Mol. Cell. Biol. 17:3477–3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ephrussi, A., G.M. Church, S. Tonegawa, and W. Gilbert. 1985. B lineage–specific interactions of an immunoglobulin enhancer with cellular factors in vivo. Science. 227:134–140. [DOI] [PubMed] [Google Scholar]
- 20.Church, G.M., A. Ephrussi, W. Gilbert, and S. Tonegawa. 1985. Cell-type-specific contacts to immunoglobulin enhancers in nuclei. Nature. 313:798–801. [DOI] [PubMed] [Google Scholar]
- 21.Murre, C., P.S. McCaw, and D. Baltimore. 1989. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell. 56:777–783. [DOI] [PubMed] [Google Scholar]
- 22.Bain, G., E.C. Maandag, D.J. Izon, D. Amsen, A.M. Kruisbeek, B.C. Weintraub, I. Krop, M.S. Schlissel, A.J. Feeney, M. van Roon, et al. 1994. E2A proteins are required for proper B cell development and initiation of immunoglobulin gene rearrangements. Cell. 79:885–892. [DOI] [PubMed] [Google Scholar]
- 23.Zhuang, Y., P. Soriano, and H. Weintraub. 1994. The helix-loop-helix gene E2A is required for B cell formation. Cell. 79:875–884. [DOI] [PubMed] [Google Scholar]
- 24.Romanow, W.J., A.W. Langerak, P. Goebel, I.L. Wolvers-Tettero, J.J. van Dongen, A.J. Feeney, and C. Murre. 2000. E2A and EBF act in synergy with the V(D)J recombinase to generate a diverse immunoglobulin repertoire in nonlymphoid cells. Mol. Cell. 5:343–353. [DOI] [PubMed] [Google Scholar]
- 25.Coquilleau, I., P. Cavelier, F. Rougeon, and M. Goodhardt. 2000. Comparison of mouse and rabbit Ei kappa enhancers indicates that different elements within the enhancer may mediate activation of transcription and recombination. J. Immunol. 164:795–804. [DOI] [PubMed] [Google Scholar]
- 26.Gugasyan, R., R. Grumont, M. Grossmann, Y. Nakamura, T. Pohl, D. Nesic, and S. Gerondakis. 2000. Rel/NF-kappaB transcription factors: key mediators of B-cell activation. Immunol. Rev. 176:134–140. [DOI] [PubMed] [Google Scholar]
- 27.Eckner, R., T.P. Yao, E. Oldread, and D.M. Livingston. 1996. Interaction and functional collaboration of p300/CBP and bHLH proteins in muscle and B-cell differentiation. Genes Dev. 10:2478–2490. [DOI] [PubMed] [Google Scholar]
- 28.Qiu, Y., A. Sharma, and R. Stein. 1998. p300 mediates transcriptional stimulation by the basic helix-loop-helix activators of the insulin gene. Mol. Cell. Biol. 18:2957–2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Massari, M.E., P.A. Grant, M.G. Pray-Grant, S.L. Berger, J.L. Workman, and C. Murre. 1999. A conserved motif present in a class of helix-loop-helix proteins activates transcription by direct recruitment of the SAGA complex. Mol. Cell. 4:63–73. [DOI] [PubMed] [Google Scholar]