Abstract
Mutations within cytotoxic T lymphocyte (CTL) epitopes impair T cell recognition, but escape mutations arising in flanking regions that alter antigen processing have not been defined in natural human infections. In human histocompatibility leukocyte antigen (HLA)-B57+ HIV-infected persons, immune selection pressure leads to a mutation from alanine to proline at Gag residue 146 immediately preceding the NH2 terminus of a dominant HLA-B57–restricted epitope, ISPRTLNAW. Although N-extended wild-type or mutant peptides remained well-recognized, mutant virus–infected CD4 T cells failed to be recognized by the same CTL clones. The A146P mutation prevented NH2-terminal trimming of the optimal epitope by the endoplasmic reticulum aminopeptidase I. These results demonstrate that allele-associated sequence variation within the flanking region of CTL epitopes can alter antigen processing. Identifying such mutations is of major relevance in the construction of vaccine sequences.
Keywords: CD8 T cell responses, viral evolution, immune evasion, antigen presentation
Introduction
CTL activity plays an important role in the immune control of chronic viral infections such as EBV, CMV, and HIV (1–5). A variety of strategies have evolved by which these viruses successfully evade elimination by the CTL response, indicating the importance of these cells in viral containment (6). Viral escape via mutation within CTL epitopes has been well documented in acute and chronic immunodeficiency virus infection (7–15). In all of these cases, the mutation occurred within the epitope and it led either to a reduction in binding to the MHC class I molecule and/or to altered T cell receptor recognition.
Interference with intracellular antigen processing may also lead to loss of recognition. This could arise at any stage in the processing pathway, including the degradation of viral proteins by cytosolic proteases, mainly the 26S proteasomes (16, 17); the destruction of the proteasome products by peptidases in the cytosol or the ER (18–20); the binding of oligopeptides to the transporter associated with antigen processing (TAP) and their translocation into the ER (21, 22); and the trimming of precursors into antigenic peptides of the correct length to bind to the MHC class I molecule (19, 23, 24). The involvement of an ER-resident aminopeptidase termed ER aminopeptidase I (ERAPI) in the NH2-terminal trimming of N-extended antigenic peptides for loading onto class I molecules was recently demonstrated in the murine system (25–27).
Interference with antigen processing has been induced by specific viral proteins, for example in CMV and HSV infection (28), but theoretically could be achieved by viral sequence mutations. Several groups have shown that the artificial mutation of the flanking sequence of CTL epitopes can lead to loss of recognition (29–35). However, thus far there is no in vivo evidence that immune selection pressure leads to CTL escape by altered antigen processing, even though such evidence has been sought (36). Here we present evidence for allele-specific positive selection of escape mutation within the flanking region of a defined CTL epitope within the course of a natural human infection. This mutation leads to the inability of ERAPI to cleave the variant N-extended peptide to the optimal size.
Materials and Methods
Study Patients.
HIV-1 clade B– or clade C–infected individuals of different disease status were recruited at the Cato Manor Clinic, Durban, South Africa, Queen Elizabeth II Hospital, Bridgetown, Barbados, Wycombe General Hospital, UK, the Harrison Clinic, Radcliffe Infirmary, Oxford, UK, and the Massachusetts General Hospital, Boston, MA. HLA class I typing was performed at Dynal Biotech, Oxford, UK. The study was approved by the institutional review boards at the University of Natal, at Queen Elizabeth II Hospital, by the Oxford Research Ethics Committee, and the Massachusetts General Hospital Institutional Review Board. All individuals gave informed consent for participating in this study.
Sequencing.
Proviral DNA was extracted from fresh PBMCs using the Puregene™ DNA isolation kit. HIV-Gag sequences were amplified by nested PCR as previously described (37). Sequencing reactions were performed using the ABI Prism BigDye™ Ready Reaction and using the six primers previously described (37). The plates were analyzed on the ABI 3700 DNA Analyzer from Applied Biosystems.
ELISPOT.
HIV-1–specific CD8 T cell responses were quantified by ELISPOT assay using fresh or frozen PBMCs (0.5–1 × 105 per well) and single peptides (final concentration of 20 μg/ml) as previously described (38).
Cell Lines.
EBV-transformed B lymphoblastoid cell lines were established in R20 medium (RPMI 1640; Sigma-Aldrich) supplemented with 2 mM l-glutamine, 50 U/ml penicillin, 50 μg/ml streptomycin, 10 mM Hepes, and 20% heat-inactivated FCS (Sigma-Aldrich). CTL clones were isolated by limiting dilution and characterized and maintained as previously described (39), using the CD3-specific mAb 12F6 and irradiated feeder cells as stimulus for T cell proliferation. For this study, we used CTL clones specific for the following epitopes: B57 KAFSPEVIPMF, B57 ISPRTLNAW, and A31 RLRDLLLIVTR. Specific lysis for all clones was 70–90% in 51Cr release assay at the E/T ratio of 10:1. Primary CD4 cells were derived from PBMCs of an HIV-uninfected donor with the HLA class I genotype: HLA A2, 11; B57; Cw7, 11. CD4 T cells were expanded using the bispecifc mAb CD3/CD8.
51Cr Release Assay.
B lymphoblastoid cell line target cells were labeled with 50 μCi Na2(51CrO4) (1 Ci = 37 GBq; New England Nuclear) for 1 h at 37°C, 5% CO2. Targets were incubated with peptide either at the time of Cr labeling or after washing twice with peptide dilutions for titration assays. CTL clones were added as effectors at the E/T ratio of 10:1 or 5:1. Supernatants were harvested after an additional 4-h incubation at 37°C, 5% CO2. For assays using primary CD4 cells as targets, primary CD4 cells were infected with either WT or mutant virus at a multiplicity of infection (MOI) of 1 and used on day 4 after infection. Infection rates were comparable for both viruses: 8.6 and 8.7%, respectively, as measured by intracellular p24 staining.
Site-directed Mutagenesis.
The mutant virus A146P was constructed by substituting an alanine with a proline in position 146 of HIV-1 Gag with the GeneTailor™ Site-Directed Mutagenesis System (Invitrogen). The whole plasmid DNA was PCR amplified in a mutagenesis reaction with two overlapping primers, one of which contains the target mutation: p24/1209–1238 5′-GCA AAT GGT ACA TCA GCC CAT ATC ACC TAG-3′ and p24/1222–1195 5′-GAT GTA CCA TTT GCC CCT GGA GGT TCT G-3′. The presence of A146P was verified by DNA sequencing from nucleotide position 983 to 1656 in Gag in newly generated plasmid clones. Infectious virus was produced by cotransfection of MT-4 cells with the mutant proviral 5′ half-genome plasmid and the HIV-1NL4–3 3′ half-genome plasmid (p83-10; references 40 and 41).
Growth Competition Assays.
Infections were initiated with unequal amounts of two competing virus variants, typically 10 and 90%, based on virus infectivity titrations. Inocula of 5,000 TCID50 of each virus were used to infect 5 × 106 stimulated PBMCs (MOI = 0.001) as previously described (42, 43). PBMCs were also separately infected with the mutant A146P in the absence of WT virus to evaluate the potential for true genetic reversion over the course of the experiment. 5-ml cultures were maintained in six-well tissue culture plates at 0.5 × 106 cells/ml. At day 3 of the first passage, half of the culture was replaced with fresh medium. At day 7, fresh MT-4 cells were reinfected with a dilution of 1:10 of the supernatant and cultured at 0.5 × 106 cells/ml. Two additional passages were performed in the same way. Cell-free supernatant fluid aliquots were taken at the end of the culture and viral RNA was extracted (QIAamp™ Viral RNA kit; QIAGEN). The relative proportion of the two competing variants was determined over time based on the relative peak heights in electropherograms obtained by automated sequencing of HIV-1 p24 from culture supernatants. To further quantify the proportion of sequences expressing the A146P (GCC to CCC) mutation, allele-specific real-time PCR combining nonselective and selective primer pairs was also used. For nonselective amplification, we used primer sets ISW9-Fw (1144–1165) 5′-GCAGCTGACACAGGAAACAA-CA-3′ and ISW9NS-Rv (1227–1254) 5′-CCATGCATTTAAAGTTCTAGGTGATATG-3′ and the TaqMan MGB probe ISW9_Pr (1167–1181) 5′-(6-FAM)CCAGGTCAGCCAAAA-3′ obtained from Applied Biosystems. For preferential amplification of Ala-146, the ISW9NS-Rv primer was replaced by the primer ISW9Swt-Rv (1225–1250) 5′-GCATTTAAAGTTCTAGGTGATATCGC-3′. This primer included an intentional mismatch at the −2 position relative to the 3′ terminus of the oligonucleotide with the objective to destabilize the amplification of mutant sequences. Samples were evaluated by real-time PCR using an ABI PRISM 7000 sequence detection system (Applied Biosystems) with the following cycling parameters: 50°C for 2 min, 95°C for 10 min, followed by 40 cycles at 95°C for 15 s and 60°C for 1 min. The number of cycles required to reach threshold fluorescence (Ct) was determined, and the quantity of sequences initially present was calculated by extrapolation onto the standard curve. The same dilutions of standard (plasmid containing the Ala-146) were used for both the nonselective and the Ala-146–selective amplifications. The percentage of viral sequences containing Ala-146 was then calculated as follows: % Ala-146 sequences = [(quantity of Ala-146 sequences in the sample)/(quantity of total sequences in the sample)] × 100. Nonselective and selective amplifications were always performed at the same time. All reactions were performed in triplicate and the mean of the three values was used for calculations.
Viral Replication Inhibition Assay.
Inocula of 80,000 TCID50 of either HIV-1 NL4-3 WT or A146P mutant virus were used to infect 8 × 106 primary CD4 cells (MOI = 0.01) on day 7 after stimulation with the bispecific antibody CD3/CD8. These were cultured with HIV-1–specific CTL clones at E/T ratios 4:1, 1:1, and 1:4 in duplicate in 24-well plates in 2 ml. All assays were performed in R10 with 50 U/ml rIL-2. Half of the supernatant was harvested for quantitative HIV-1 p24 antigen capture ELISA (Dupont) and replenished with fresh medium at the indicated time points after infection (44).
ERAPI Assay.
The peptides used were >90% pure by mass spectrometry analysis. 50 nmol/ml of NH2-extended peptides were incubated at 37°C with 3.5 μg/ml human recombinant ERAPI in a buffer containing 50 mM Tris-HCl, pH 7.8. At each time point a 50-μl aliquot was removed and the reaction was stopped by adding an equal volume of 0.6% trifluoroacetic acid. Samples were loaded on a 4.6 × 50-mm TARGA 3-μm C18 column (Higgins Analytical, Inc.) in 10 mM sodium phosphate buffer, pH 6.6, containing 7% acetonitrile. Elution was performed with a linear 7–31.5% acetonitrile gradient. The amount of a trimmed peptide was calculated by the integration of peptide peaks. The identity of peaks was determined by comparing their retention time to those of pure synthetic peptides.
Phylogenetic Analysis of Selection Pressures.
We used a maximum likelihood method that assesses the fit to the data of various models of codon evolution that differ in how d N/d S varies across the sequence alignment and takes into account the phylogenetic relationships of the sequences in question (45). We used two models of codon evolution: “M7,” which specifies ten categories of d N/d S in the alignment, none of which may be >1 so that evolution is entirely neutral, and “M8,” which only differs from M7 in that it incorporates an extra (11th) class of codons that can take on any value of d N/d S, including those supporting positive selection (d N/d S > 1). The selection pressures at individual codons can then be assessed using Bayesian methods. All analyses were undertaken using the CODEML program from the PAML package (46) with input phylogenetic trees inferred using the maximum likelihood method available in the PAUP* package (47).
Online Supplemental Material.
The Online Supplemental Material shows the sequence data of HIV transmission pairs (Table S1), the phylogenetic tree showing the relationship of the viruses of the transmission pairs (Fig. S1), the HPLC elution times for synthetic peptides used in this study (Table S2), and comparison of viral replication fitness of A146 wild-type and A146P mutant virus (Fig. S2). Supplemental Materials and Methods, Tables S1 and S2, and Figs. S1 and S2 are available at http://www.jem.org/cgi/content/full/jem.20031982/DC1.
Results
Identification of Allele-specific Mutations in Persons Expressing HLA-B57.
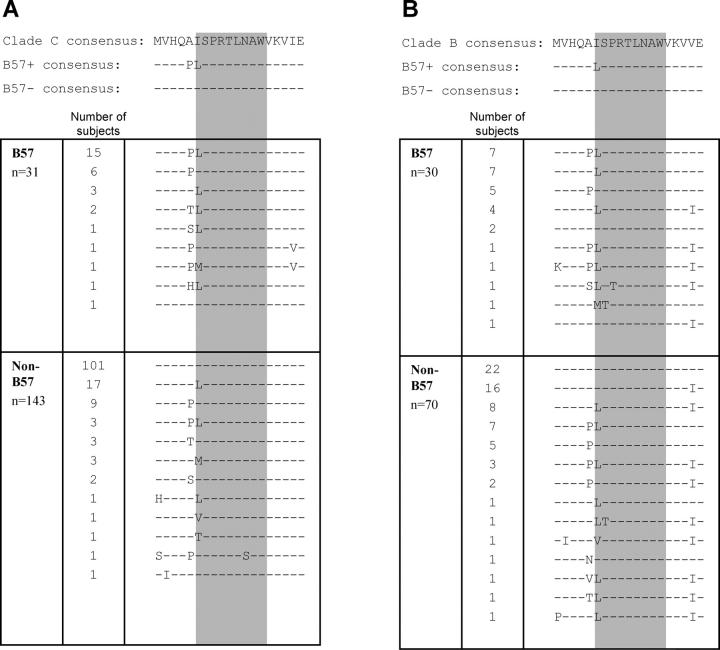
Complete Gag sequencing in 174 clade C–infected persons revealed two sites at which there were differences in consensus sequence between persons expressing HLA-B57 and those not expressing this allele. Both sites were within regions encoding defined HLA-B57–restricted epitopes: ISPRTLNAW (“ISW9,” HXB2 Gag residues 147–155; Fig. 1 A) and TSTLQEQIAW (“TW10,” residues 240–249; reference 37). For TW10, the amino acid change was situated within the previously described epitope located at position 3, believed to be a TCR contact residue (48). Of the two amino acid changes in relation to the ISW9 epitope that were specific to HLA-B57+ subjects, one was found at Gag residue 147 located at the NH2-terminal position within epitope (49). The other was an alanine to proline change at Gag residue 146 immediately preceding the NH2-terminal amino acid position (Fig. 1 A). The same analysis was performed in 100 persons infected with HIV clade B, and similar significant differences in sequences were observed in HLA-B57+ persons (Fig. 1 B).
Figure 1.
Amino acid sequences of the region spanning the ISPRTLNAW epitope (Gag residues 142–160). Consensus sequence as listed in the Los Alamos Database (top row), consensus sequence according to the sequence data obtained from B57+ subjects (middle row), and non-B57+ subjects (bottom row) for clade C (A) and clade B (B). Listed below are the actual sequence data for 174 clade C–infected subjects and 100 clade B–infected subjects. For clade C–infected subjects, variation at residues 146 and 147 was associated with expression of B57 (P < 0.0001 and P < 0.0001, respectively, Fisher's exact test), as well as in clade B–infected subjects (P = 0.03 and P = 0.0006, respectively).
Analysis of the specific polymorphisms at residues 146 and 147 revealed strong allele-specific selection for certain amino acids. When the clade B and clade C cohorts were combined and analyzed, 37 out of 61 (61%) of B57+ subjects had variation at residue 146 involving an A146P polymorphism versus 30 out of 213 (14%) of non-B57 controls (relative risk = 4.3; P < 0.0001). Similarly, the variation at position Gag 147 involved an isoleucine to leucine polymorphism (“I147L”) in 43 out of 61 (70%) of B57+ subjects versus 44 out of 213 (21%) of non-B57 controls (relative risk = 3.4; P < 0.0001). The association of these A146P and I147L mutations with HLA-B57 expression was less strong in B clade infection than in C clade infection, although remaining significant (P = 0.03 and 0.0006, respectively).
HLA-B57–specific Positive Selection Pressure Drives Variation at Position 146 and Is Found after Transmission in Persons Expressing HLA-B57.
To determine whether the variation observed at residues 146 and 147 was the result of positive selection pressure, the ratio of nonsynonymous to synonymous nucleotide change (d N/d S) was calculated for each individual codon in Gag and analyzed using the maximum likelihood of Yang (45). Initial studies were performed in clade C–infected persons, where the association between polymorphism in Gag and expression of HLA-B57 was strongest. Amino acid variation was observed at only two sites within Gag that was under positive selection (d N/d S > 1) specific to B57+ subjects (not depicted). These were at Gag residue 242 (within the TW10 epitope) and at Gag residue 146, immediately preceding the ISW9 epitope (mean d N/d S: 3.22). There was no evidence of positive selection at these sites when sequences from HLA-B57− subjects alone were analyzed, nor was there evidence of positive selection for the conservative I147L mutation.
To further demonstrate that the variation at Gag residues 146 and 147 involves immune selection pressure mediated by HLA-B57, we sought examples of known transmission of virus from an individual expressing neither of these alleles to an HLA-B57+ individual. Three such mother-child pairs and a pair of hemophiliacs were identified, in which the WT virus was present in the HLA-B57− mother/donor. The A146P and/or I147L variants arose subsequent to transmission in each B57+ child/recipient (Table S1, available at http://www.jem.org/cgi/content/full/jem.20031982/DC1). In each of the pairs evaluated, the common ancestry of the viruses was confirmed by phylogenetic analysis (Fig. S1, available at http://www.jem.org/cgi/content/full/jem.20031982/DC1).
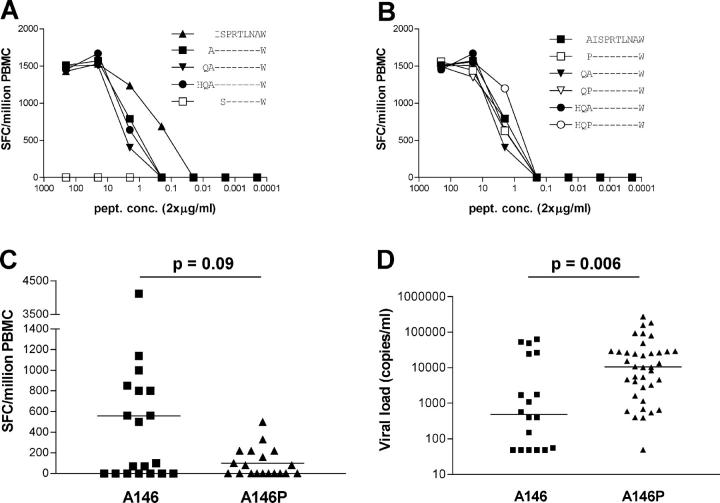
The A146P Mutation Is in the Epitope's Flanking Region and Is Associated with Low Frequency ISW9-specific CTL Activity and High HIV Viral Load.
The finding of an HLA-B57–specific difference in the consensus sequence at Gag residue 146 was unexpected as it did not lie within the defined CTL epitope. Because previous studies have shown that HLA-B57 can accommodate peptides of different lengths and that one HLA-B57 epitope can lie entirely within another (50), we examined the possibility that residue 146 might indeed lie within a targeted epitope. Data from the study of seven HLA-B57+ individuals showed that ISPRTLNAW (ISW9) is the only HLA-B57–restricted epitope in this region (not depicted) and confirmed that it is the best-recognized epitope at low peptide concentrations (Fig. 2 A). There was no difference in recognition of N-extended WT or A146P-mutant synthetic peptides when using PBMCs in an ex vivo ELISPOT assay (Fig. 2 B). In addition, ISW9-specific CTL clones recognize both the WT and mutant peptides using peptide-pulsed HLA-B57+ primary CD4 T cells as targets (not depicted). However, in spite of the preserved recognition of mutant peptides, a trend toward stronger responses was seen in persons expressing alanine at position 146 (P = 0.09; Mann-Whitney test), and for those with a positive response, the magnitude was significantly greater in persons with the A146 WT autologous sequence (P = 0.02; Mann-Whitney test; Fig. 2 C).
Figure 2.
Impact of A146P mutation using synthetic peptides. (A and B) Peptide titration ELISPOT using ISW9 and WT or mutant N-extended synthetic peptides. Results are representative for assays using ex vivo PBMCs of seven different individuals recognizing ISW9. (C) Ex vivo ELISPOT response toward ISW9 in 38 subjects according to the sequence of the autologous virus at residue Gag 146. P = 0.09, Mann-Whitney test. Magnitude of ISW9 response for subjects with A146, median 560 (range 0–4,120) spot-forming cells/million PBMCs; for subjects with A146P, median 100 (range 0–500) spot-forming cells/million PBMCs. (D) Association of A146P expression with higher viremia. All HAART-naive HLA-B57+ subjects from these cohorts and those for whom published data are available (reference 41) were included for analysis. P = 0.006, Mann-Whitney test.
To address the question of whether this A146P flanking mutation might be associated with less effective control of HIV infection in HLA-B57+ subjects, we compared viral load in B57+ subjects whose autologous virus encoded the A146P mutant with that in B57+ subjects whose autologous virus encoded the A146 WT. Despite the considerable diversity in our own and in the other B57+ cohorts (51) for which these data are available, this single amino acid change is associated with a 22-fold increase in viral load (median: 483 HIV RNA copies/ml vs. 10,521 HIV RNA copies/ml; P = 0.006; Fig. 2 D). This compares with a 5.1-fold difference between the 75th and 25th centiles for viral load in the MACS cohort (52). Because viral load is a strong predictor of disease progression (53), this A146P mutation is associated with a substantially increased risk of disease progression.
These results confirm that although the A146P mutation is associated with HLA-B57 and is the result of HLA-B57–associated positive selection, it does not lie within the region targeted by the TCR but instead in the flanking region of the ISW9 epitope, thus providing evidence for an escape mechanism involving antigen processing. In those subjects targeting the ISW9 epitope, the magnitude of the response is lower in association with the A146P mutation. Furthermore, this A146P mutation appears to have significance for HLA-B57+ subjects because this mutation is associated with high viremia and therefore higher risk of progression to disease.
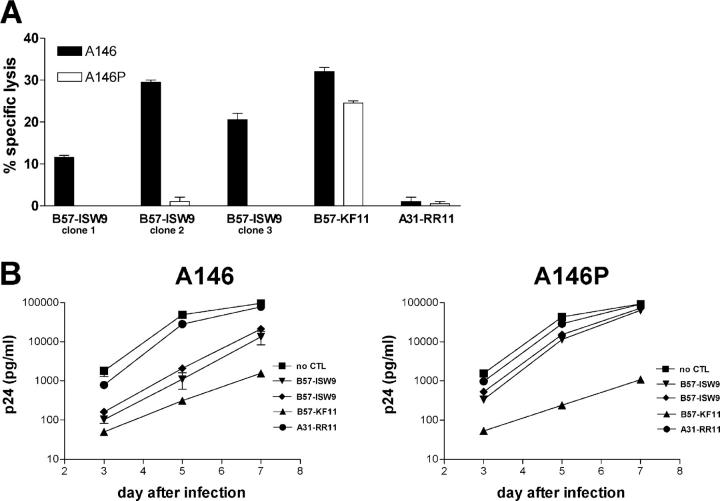
The A146P Mutation Leads to Impaired Recognition of Virus-infected Cells by ISW9-specific CTL Clones.
Because the use of synthetic peptides to pulse target cells does not involve intracytoplasmic generation of peptides and therefore does not assess potential alterations in antigen processing that might be induced by the flanking residue mutation, we next constructed viruses differing only at position Gag 146, the NL4-3 WT virus and a mutant virus isogenic with NL4-3 except in expression of proline at Gag residue 146. Cytotoxicity assays using CTL clones as effectors and virus-infected primary CD4 T cells from an uninfected HLA-B57+ donor as targets showed that WT infected target cells were lysed by ISW9-specific clones but mutant infected cells were not (Fig. 3 A). In contrast, HLA-B57–restricted KAFSPEVIPMF (“KF11”)-specific CTL (targeting a p24 Gag epitope conserved in both viruses) lysed cells infected with either virus, and, as expected, an HLA-mismatched HLA-A31–restricted CTL clone did not lyse either of the target cells. The proportion of targets infected with WT and mutant virus as assessed using intracellular cytokine staining of targets by anti-p24 antibody was identical (9% in each case).
Figure 3.
Impact of the A146P mutations using single point mutation viruses. (A) 51Cr release assay using CTL clones as effectors (E/T ratio, 100:1) and primary CD4 T cells as targets infected either with A146 WT virus or A146P mutant virus (MOI = 1). Infection rates were 8.6% for A146 WT virus and 8.7% for A146P mutant virus. Representative results for three assays using ISW9 clones of two different donors. (B) Replication inhibition assay using A146 WT or A146P mutant–infected primary CD4 cells (MOI = 0.01) and CTL clones as effectors. Representative results for five assays and ISW9 clones of two different subjects. ISW9, ISPRTLNAW; KF11, KAFSPEVIPMF; RR11, RLRDLLLIVTR.
Viral replication inhibition assays using the same effectors and targets further confirmed preferential targeting of the WT virus over the A146P mutant, whereas control CTL specific for an epitope conserved in both viruses inhibited both viruses and the HLA-mismatched clone did not inhibit either virus (Fig. 3 B). These data suggest that the A146P mutation acts as a processing mutant by which the virus can evade the ISW9-specific response.
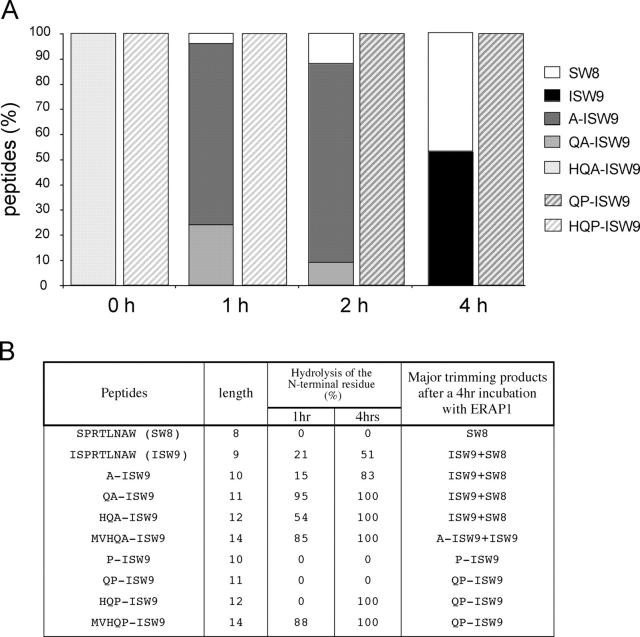
The A146P Mutation Results in Altered Trimming by ERAPI.
Next, we evaluated the effects of the A146P mutation on epitope processing. ERAPI is the first ER-resident aminopeptidase shown to be involved in antigen presentation (25–27). Reported studies were undertaken in the mouse model and although it is likely that they apply also to the human system, direct evidence for the involvement of ERAPI in the trimming of HLA-restricted N-extended epitopes has not yet been presented. Like other aminopeptidases, ERAPI cannot hydrolyze an X-P bond (23, 25) and we therefore investigated whether ERAPI might be involved in the NH2-terminal trimming of the N-extended ISW9 epitope. We focused our study on four N-extended WT or mutant ISW9 peptides (MVHQA/P-ISW9, HQA/P-ISW9, QA/P-ISW9, and A/P-ISW9). These 9–14 mers fall into the range of peptide lengths translocated by TAP into the ER (54).
The unique HPLC profiles and elution times were determined for each synthetic peptide to serve as standards (Table S2, available at http://www.jem.org/cgi/content/full/jem.20031982/DC1). We then incubated ERAPI with the WT and variant peptides for 0, 1, 2, and 4 h and performed reverse phase HPLC profile analysis of the digestion products (Fig. 4 A). In 1 h the HQA-ISW9 peptide is trimmed to a mix of QA-ISW9 and A-ISW9, showing that this N-extended HLA-B57–restricted epitope is a substrate for ERAPI, and by 4 h the major product is the optimal ISW9 peptide (Fig. 4 A). In contrast, HQP-ISW9 is trimmed to QP-ISW9 in 2 h and no other products are produced even at 4 h.
Figure 4.
Trimming of N-extended WT and mutant ISW9 by ERAPI. (A) HQA-ISW9 or HQP-ISW9 were incubated with human recombinant ERAPI. At each time point an aliquot was removed and fractionated by reverse phase HPLC. 100% correspond to 50 nmol/ml. (B) Hydrolysis rates and digestion products obtained after a 4-h incubation of each intermediate A146 or A146P peptide with ERAPI.
Next, we studied the digestion products obtained after incubation of each intermediate peptide as well as ISW9 and SW8 (Fig. 4 B). All WT N-extended peptides are trimmed to a mix of ∼50% ISW9 and 50% SW8 (except QA-ISW9, for which SW8 represents 75% of the final product), whereas the mutants are never fully trimmed to the optimal epitope. These results demonstrate that if these 10–14-mer peptides indeed reach the ER lumen, ERAPI is able to process only those peptides containing an alanine at residue 146, but cannot trim any of the 10–14-mer N-extended ISW9 peptides containing a proline at position 146 down to the optimal epitope. Thus, these results are consistent with the lack of CD8 T cell recognition of cells infected with HIV containing proline at Gag residue 146.
Accumulation of A146P Variants over Time in the Population.
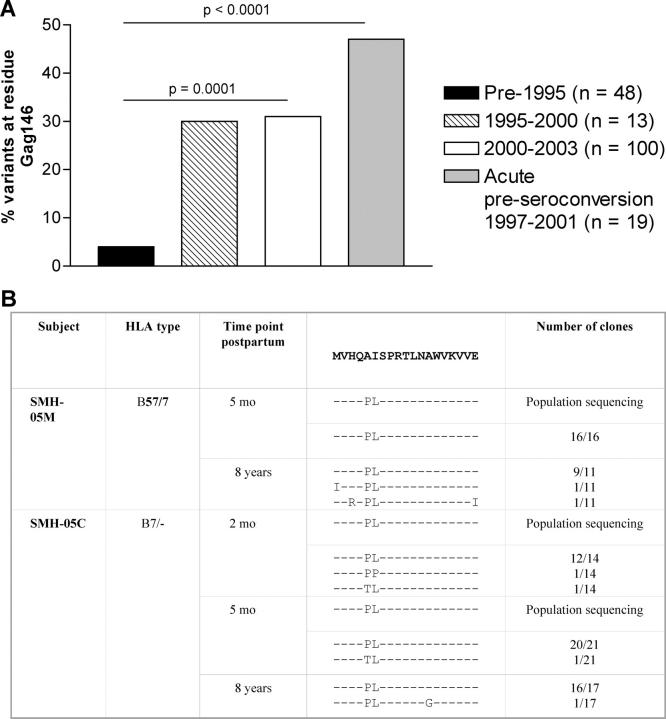
To determine whether the A146P mutation affected viral fitness, replication of the WT NL4-3 virus was compared in culture to that of the A146P mutant virus that had been generated. Both in these experiments (Fig. 3 B) and in competition experiments in which relative replication fitness was analyzed (Fig. S2, available at http://www.jem.org/cgi/content/full/jem.20031982/DC1), there was no evidence that viral fitness was compromised by the A146P mutation. It was therefore hypothesized that transmission of this variant to an individual lacking HLA-B57 would not result in its disappearance, and that persistence of this variant would be reflected in an accumulation of this variant over time through the epidemic. Sequences in the Los Alamos database with a known sampling date show that of the B clade sequences obtained from subjects early in the epidemic, for example pre-1995, only 2 out of 48 (4%) showed variation at Gag residue 146, compared with 29–31% of sequences obtained from subjects sampled later in the epidemic (between 1995 and 1999 or since 2000 using the clade B–infected samples used in Fig. 1; P = 0.0001; Fig. 5 A). Out of 19 acutely infected subjects in Boston (B clade) whose virus was sampled before seroconversion (1997–2001; reference 55), 47% showed virus-encoding mutants at A146 (P < 0.0001). Transmission of virus from an HLA-B57+ mother to an HLA-B57− child showed that over 8 yr there was no reversion in this A146P mutation (Fig. 5 B), in contrast to a mutation in the B57-restricted TW10 epitope (37). The percentage of mutations at position Gag 146 is lower in the B57− clade C–infected individuals than in the B57− clade B–infected cohort (13 vs. 29%) sequenced for this study. These C clade–infected subjects were recruited in South Africa where the HIV epidemic is much younger, which may explain why the accumulation is not visible to date. One may therefore hypothesize that this process of transmission of the A146P mutant and maintenance in the absence of the evolutionary force that originally drove its selection would gradually lead to the loss of ISW9 as an HLA-B57–restricted epitope as the epidemic progresses.
Figure 5.
Accumulation of the A146P mutation in the population. (A) Percentage of sequences with amino acid variation at Gag residue 146 by date of sampling in clade B–infected individuals. Sequences from subjects with a known sampling date were taken from the Los Alamos HIV Sequence Database (http://hiv-web.lanl.gov) and included 100 B clade sequences for the 2000–2003 time period from the subjects whose virus was sequenced for this paper, of whom 70 were HLA-B57− (see Fig. 1 B). The acute preseroconversion group was recruited at the Massachusetts General Hospital as part of the “acute infection cohort” (reference 55). Statistical analysis was performed using the Fisher's exact test. (B) Population sequencing and sequencing of multiple clones in an HLA-B57+ mother who transmitted the virus to a HLA-B57− child at different time points.
Discussion
Here we present evidence of immune escape due to viral mutation in the flanking region of an epitope that is induced by CTL selection pressure. Variant peptide-pulsed target cells were well recognized but variant virus-infected cells were not well seen either in cytotoxicity assays or in viral inhibition assays in vitro. The processing variant, occurring in the NH2-terminal flanking position of the epitope, involved a substitution of proline for alanine and, if the N-extended peptide were to reach the ER lumen, this substitution would lead to the inability of ERAPI to cleave the peptide to the optimal size. This is the first study to address the role of ERAPI in trimming of the NH2 terminally extended peptide in the human system. The functional importance of this processing mutation is evidenced by the demonstration that it arises as a consequence of positive selection pressure in association with HLA-B57 expression. Subjects who targeted the ISW9 epitope and in whom the A146P mutation was present showed weaker recognition of the ISW9 peptide affected by the processing mutation. Furthermore, the A146P mutation was associated with a 22-fold higher viral load in HAART-naive subjects with B57. Finally, viral replication capacity was not reduced by this A146P mutation in vitro, consistent with evidence presented that this mutation is accumulating in the viral population over time.
Although proposed for many years, evidence that intrahost mutations in the flanking region of a CTL epitope can lead to immune escape in a natural human viral infection has been absent. It has been shown that either inserting a CTL epitope at different positions into unrelated carrier proteins or mutating the flanking region of an epitope can affect epitope presentation (29, 31–34). A natural mutation in the C terminus of a murine leukemia virus CTL epitope led to the abolition of proper epitope presentation (35). Yellen-Shaw et al. (30), using a mouse model of influenza infection, experimentally substituted proline for alanine immediately upstream of the H-2Kd influenza NP147–155 epitope, and demonstrated that the mutation blocks presentation of this epitope. The data presented here indicate that a similar alanine to proline substitution can occur as a consequence of natural selection pressure, and thus provides evidence of a mechanism for immune escape in chronic human viral infections.
It is unclear why the adjacent intra-epitopic mutation I147L is associated with HLA-B57 expression and yet phylogenetic analysis failed to demonstrate positive selection at this site. Two possibilities seem likely. First, that amino acid changes at site 147 occurred less often and at deeper branches of the tree than those at site 146, so that there was insufficient variability to detect positive selection under the analytical methods used here. Alternatively, it might be that the I147L mutation in HLA-B57+ patients is not selected in itself, but rather is in linkage with the selected mutation at site 146. To resolve these theories will evidently require the analysis of this epitope in a larger sample of patients.
The processing mutant we characterized may result from a failure of ERAPI to trim NH2-terminal residues flanking the optimal peptide. In common with other aminopeptidases, ERAPI fails to cleave the NH2-terminal residue when it is followed by a proline (X-P-epitope), at least in the case of an artificial mutation introduced upstream from a mouse epitope (23, 25). This characteristic of the enzyme may explain why the mutation described in our study leads to loss of presentation. The full specificity of ERAPI has yet to be defined, however, and our data additionally show failure of the QP-ISW9 peptide to be cleaved to ISW9. Our studies do not exclude the possibility that other steps involved in antigen processing may have been affected by the A146P mutation. Potential sites for processing escape mutations include the cleavage of proteins by proteasomes (56) and the translocation of the peptide into the ER by TAP (21, 22, 54, 57). It has been shown that a proline at position 2 or 3 of a peptide decreases its translocation rate via TAP (58, 59). Yellen-Shaw et al. (30), examining an experimentally induced A→P substitution flanking the NH2 terminus of an epitope, could not explain the loss of epitope presentation by interference with proteasome cleavage or poor transport by TAP. Failure of ERAPI to trim the NH2-terminal flanking residues may therefore contribute substantially to the processing escape mechanism in the example described here.
The extent to which processing mutation is an immune escape mechanism used by pathogens is unknown. There are obstacles to identifying processing mutations, the first of which is defining the CTL epitopes themselves. Although no virus has received more intensive research attention than HIV, only a small minority of all the HIV-specific CTL epitopes that are targeted have been defined to date (60, 61). A second difficulty in detecting potential processing mutants is that variation occurring outside defined CTL epitopes might be the consequence of selection pressures other than those driving processing escape mutations. This would include compensatory mutations enabling the virus to achieve improved fitness after a deleterious mutation arising elsewhere (13, 37), and mutations occurring within other epitopes than the one under study. However, there are indications that processing escape is a mechanism more widely used than is currently apparent. First, perusal of the defined HIV-specific CTL epitopes shows that the occurrence of proline at the position immediately preceding the NH2 terminus of CTL epitopes is rare: only 6 out of 187 defined HIV-specific epitopes have a proline residue at position −1. In at least one case, the Nef epitope QVPLRPMTYK, turns out to be dependent not upon ERAPI for processing but upon TPPII (24). Second, Moore et al. (62) described associations between viral sequence mutations and certain HLA alleles, a substantial proportion of which are not within defined epitopes and a number of which were present in the immediate flanking region of defined epitopes. Finally, a second processing mutation adjacent to the C terminus of the HLA-A3–restricted p17 Gag epitope KIRLRPGGK has recently been identified (63). This time it is not a mutation impairing ERAPI activity, but potentially a proteasomal or other endoproteolytic enzyme. Thus, we believe that processing escape mutations such as the one we have described in this paper might be substantially more frequent than hitherto recognized.
In conclusion, this is the first report not only to define intrahost viral escape via mutation in the flanking region of a CTL epitope in a natural human infection, but also to identify a potential mechanism of this processing failure. Failure of ERAPI to process the mutant peptide resulted in an inability of the optimal epitope to be generated. It will be important to evaluate whether this mutation also affects other steps of the processing pathway and to define further the role of viral escape as a consequence of antigen processing failure. The A146P mutation itself does not influence the replicative capacity of the virus and the accumulation over time of this processing escape mutation in the population of viruses circulating through the epidemic may therefore ultimately lead to the extinction of this epitope as a CTL target, even though the epitope itself may remain invariant. These data extend the knowledge about viral escape mechanisms and have implications for vaccine design, both in terms of vaccine construction and also to take into account HIV-1 evolution as the epidemic proceeds.
Acknowledgments
We want to thank the following people for their superb efforts providing samples: B. Kupfer (Universitatsklinik Bonn, Germany), G. Taylor, H. Lyall, G. Tudor-Williams (St. Mary's Hospital, London, UK), Tim Roach, A. St. John (Queen Elizabeth II Hospital, Barbados), V. Novelli (GOS, London, UK), A. Edwards (The Harrison Clinic, Radcliffe Infirmary Hospital, Oxford, UK), G. Luzzi (Department of Genitourinary Medicine, High Wycombe General Hospital, High Wycombe, UK), and J. Berz (New England Medical Center, Tufts University, Boston, MA). In addition, we gratefully acknowledge Alfred Goldberg's contribution in facilitating this work and for critical reading of the manuscript, and Bette Korber and Werner Abfalter for assistance in accessing HIV sequences from the Los Alamos database of a known sampling date.
This work was supported by the Wellcome Trust (to P.J.R. Goulder and A.J. Leslie), the Elizabeth Glaser Pediatric AIDS Foundation (to P.J.R. Goulder and M.E. Feeney), the National Institutes of Health (Contract nos. NO1-A1-15442, RO1 AI46995-01A1, and RO1 AI28568), the Doris Duke Charitable Foundation, a FIS 01/1122, a BEFI grants from the Spanish Health Department (Red G03/173; to J. Martinez-Picado), the Deutsche Forschungsgemeinschaft (DR424/1-1), and the Howard Hughes Medical Institute (to R. Draenert and B.D. Walker). P.J.R. Goulder is an Elizabeth Glaser Scientist and B.D. Walker is a Doris Duke Distinguished Clinical Science Professor.
R. Draenert, S. Le Gall, and K.J. Pfafferott contributed equally to this work.
The online version of this article contains supplemental material.
Abbreviations used in this paper: ERAPI, ER aminopeptidase I; MOI, multiplicity of infection; TAP, transporter associated with antigen processing.
References
- 1.Hislop, A.D., N.E. Annels, N.H. Gudgeon, A.M. Leese, and A.B. Rickinson. 2002. Epitope-specific evolution of human CD8+ T cell responses from primary to persistent phases of Epstein-Barr virus infection. J. Exp. Med. 195:893–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dunn, H.S., D.J. Haney, S.A. Ghanekar, P. Stepick-Biek, D.B. Lewis, and H.T. Maecker. 2002. Dynamics of CD4 and CD8 T cell responses to cytomegalovirus in healthy human donors. J. Infect. Dis. 186:15–22. [DOI] [PubMed] [Google Scholar]
- 3.Deliyannis, G., D.C. Jackson, N.J. Ede, W. Zeng, I. Hourdakis, E. Sakabetis, and L.E. Brown. 2002. Induction of long-term memory CD8(+) T cells for recall of viral clearing responses against influenza virus. J. Virol. 76:4212–4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmitz, J.E., M.J. Kuroda, S. Santra, V.G. Sasseville, M.A. Simon, M.A. Lifton, P. Racz, K. Tenner-Racz, M. Dalesandro, B.J. Scallon, et al. 1999. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science. 283:857–860. [DOI] [PubMed] [Google Scholar]
- 5.Koup, R.A., J.T. Safrit, Y. Cao, C.A. Andrews, G. McLeod, W. Borkowsky, C. Farthing, and D.D. Ho. 1994. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J. Virol. 68:4650–4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Phillips, R.E. 2002. Immunology taught by Darwin. Nat. Immunol. 3:987–989. [DOI] [PubMed] [Google Scholar]
- 7.Allen, T.M., D.H. O'Connor, P. Jing, J.L. Dzuris, B.R. Mothe, T.U. Vogel, E. Dunphy, M.E. Liebl, C. Emerson, N. Wilson, et al. 2000. Tat-specific cytotoxic T lymphocytes select for SIV escape variants during resolution of primary viraemia. Nature. 407:386–390. [DOI] [PubMed] [Google Scholar]
- 8.O'Connor, D.H., T.M. Allen, T.U. Vogel, P. Jing, I.P. DeSouza, E. Dodds, E.J. Dunphy, C. Melsaether, B. Mothe, H. Yamamoto, et al. 2002. Acute phase cytotoxic T lymphocyte escape is a hallmark of simian immunodeficiency virus infection. Nat. Med. 8:493–499. [DOI] [PubMed] [Google Scholar]
- 9.Borrow, P., H. Lewicki, X. Wei, M.S. Horwitz, N. Peffer, H. Meyers, J.A. Nelson, J.E. Gairin, B.H. Hahn, M.B. Oldstone, et al. 1997. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat. Med. 3:205–211. [DOI] [PubMed] [Google Scholar]
- 10.Evans, D.T., D.H. O'Connor, P. Jing, J.L. Dzuris, J. Sidney, J. da Silva, T.M. Allen, H. Horton, J.E. Venham, R.A. Rudersdorf, et al. 1999. Virus-specific cytotoxic T-lymphocyte responses select for amino-acid variation in simian immunodeficiency virus Env and Nef. Nat. Med. 5:1270–1276. [DOI] [PubMed] [Google Scholar]
- 11.Barouch, D.H., J. Kunstman, M.J. Kuroda, J.E. Schmitz, S. Santra, F.W. Peyerl, G.R. Krivulka, K. Beaudry, M.A. Lifton, D.A. Gorgone, et al. 2002. Eventual AIDS vaccine failure in a rhesus monkey by viral escape from cytotoxic T lymphocytes. Nature. 415:335–339. [DOI] [PubMed] [Google Scholar]
- 12.Goulder, P.J., R.E. Phillips, R.A. Colbert, S. McAdam, G. Ogg, M.A. Nowak, P. Giangrande, G. Luzzi, B. Morgan, A. Edwards, et al. 1997. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat. Med. 3:212–217. [DOI] [PubMed] [Google Scholar]
- 13.Kelleher, A.D., C. Long, E.C. Holmes, R.L. Allen, J. Wilson, C. Conlon, C. Workman, S. Shaunak, K. Olson, P. Goulder, et al. 2001. Clustered mutations in HIV-1 gag are consistently required for escape from HLA-B27–restricted cytotoxic T lymphocyte responses. J. Exp. Med. 193:375–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goulder, P.J., C. Brander, Y. Tang, C. Tremblay, R.A. Colbert, M.M. Addo, E.S. Rosenberg, T. Nguyen, R. Allen, A. Trocha, et al. 2001. Evolution and transmission of stable CTL escape mutations in HIV infection. Nature. 412:334–338. [DOI] [PubMed] [Google Scholar]
- 15.Klenerman, P., S. Rowland-Jones, S. McAdam, J. Edwards, S. Daenke, D. Lalloo, B. Koppe, W. Rosenberg, D. Boyd, A. Edwards, et al. 1994. Cytotoxic T-cell activity antagonized by naturally occurring HIV-1 Gag variants. Nature. 369:403–407. [DOI] [PubMed] [Google Scholar]
- 16.Cascio, P., C. Hilton, A.F. Kisselev, K.L. Rock, and A.L. Goldberg. 2001. 26S proteasomes and immunoproteasomes produce mainly N-extended versions of an antigenic peptide. EMBO J. 20:2357–2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rock, K.L., C. Gramm, L. Rothstein, K. Clark, R. Stein, L. Dick, D. Hwang, and A.L. Goldberg. 1994. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell. 78:761–771. [DOI] [PubMed] [Google Scholar]
- 18.York, I.A., A.X. Mo, K. Lemerise, W. Zeng, Y. Shen, C.R. Abraham, T. Saric, A.L. Goldberg, and K.L. Rock. 2003. The cytosolic endopeptidase, thimet oligopeptidase, destroys antigenic peptides and limits the extent of MHC class I antigen presentation. Immunity. 18:429–440. [DOI] [PubMed] [Google Scholar]
- 19.Saric, T., J. Beninga, C.I. Graef, T.N. Akopian, K.L. Rock, and A.L. Goldberg. 2001. Major histocompatibility complex class I-presented antigenic peptides are degraded in cytosolic extracts primarily by thimet oligopeptidase. J. Biol. Chem. 276:36474–36481. [DOI] [PubMed] [Google Scholar]
- 20.Serwold, T., and N. Shastri. 1999. Specific proteolytic cleavages limit the diversity of the pool of peptides available to MHC class I molecules in living cells. J. Immunol. 162:4712–4719. [PubMed] [Google Scholar]
- 21.Lauvau, G., K. Kakimi, G. Niedermann, M. Ostankovitch, P. Yotnda, H. Firat, F.V. Chisari, and P.M. van Endert. 1999. Human transporters associated with antigen processing (TAPs) select epitope precursor peptides for processing in the endoplasmic reticulum and presentation to T cells. J. Exp. Med. 190:1227–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neefjes, J., E. Gottfried, J. Roelse, M. Gromme, R. Obst, G.J. Hammerling, and F. Momburg. 1995. Analysis of the fine specificity of rat, mouse and human TAP peptide transporters. Eur. J. Immunol. 25:1133–1136. [DOI] [PubMed] [Google Scholar]
- 23.Serwold, T., S. Gaw, and N. Shastri. 2001. ER aminopeptidases generate a unique pool of peptides for MHC class I molecules. Nat. Immunol. 2:644–651. [DOI] [PubMed] [Google Scholar]
- 24.Seifert, U., C. Maranon, A. Shmueli, J.F. Desoutter, L. Wesoloski, K. Janek, P. Henklein, S. Diescher, M. Andrieu, H. de la Salle, et al. 2003. An essential role for tripeptidyl peptidase in the generation of an MHC class I epitope. Nat. Immunol. 4:375–379. [DOI] [PubMed] [Google Scholar]
- 25.Serwold, T., F. Gonzalez, J. Kim, R. Jacob, and N. Shastri. 2002. ERAAP customizes peptides for MHC class I molecules in the endoplasmic reticulum. Nature. 419:480–483. [DOI] [PubMed] [Google Scholar]
- 26.Saric, T., S.C. Chang, A. Hattori, I.A. York, S. Markant, K.L. Rock, M. Tsujimoto, and A.L. Goldberg. 2002. An IFN-gamma-induced aminopeptidase in the ER, ERAP1, trims precursors to MHC class I-presented peptides. Nat. Immunol. 3:1169–1176. [DOI] [PubMed] [Google Scholar]
- 27.York, I.A., S.C. Chang, T. Saric, J.A. Keys, J.M. Favreau, A.L. Goldberg, and K.L. Rock. 2002. The ER aminopeptidase ERAP1 enhances or limits antigen presentation by trimming epitopes to 8-9 residues. Nat. Immunol. 3:1177–1184. [DOI] [PubMed] [Google Scholar]
- 28.Furman, M.H., and H.L. Ploegh. 2002. Lessons from viral manipulation of protein disposal pathways. J. Clin. Invest. 110:875–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Del Val, M., H.J. Schlicht, T. Ruppert, M.J. Reddehase, and U.H. Koszinowski. 1991. Efficient processing of an antigenic sequence for presentation by MHC class I molecules depends on its neighboring residues in the protein. Cell. 66:1145–1153. [DOI] [PubMed] [Google Scholar]
- 30.Yellen-Shaw, A.J., E.J. Wherry, G.C. Dubois, and L.C. Eisenlohr. 1997. Point mutation flanking a CTL epitope ablates in vitro and in vivo recognition of a full-length viral protein. J. Immunol. 158:3227–3234. [PubMed] [Google Scholar]
- 31.Shastri, N., T. Serwold, and F. Gonzalez. 1995. Presentation of endogenous peptide/MHC class I complexes is profoundly influenced by specific C-terminal flanking residues. J. Immunol. 155:4339–4346. [PubMed] [Google Scholar]
- 32.Gileadi, U., A. Gallimore, P. Van der Bruggen, and V. Cerundolo. 1999. Effect of epitope flanking residues on the presentation of N-terminal cytotoxic T lymphocyte epitopes. Eur. J. Immunol. 29:2213–2222. [DOI] [PubMed] [Google Scholar]
- 33.Bergmann, C.C., L. Tong, R. Cua, J. Sensintaffar, and S. Stohlman. 1994. Differential effects of flanking residues on presentation of epitopes from chimeric peptides. J. Virol. 68:5306–5310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mo, A.X., S.F. van Lelyveld, A. Craiu, and K.L. Rock. 2000. Sequences that flank subdominant and cryptic epitopes influence the proteolytic generation of MHC class I-presented peptides. J. Immunol. 164:4003–4010. [DOI] [PubMed] [Google Scholar]
- 35.Beekman, N.J., P.A. van Veelen, T. van Hall, A. Neisig, A. Sijts, M. Camps, P.M. Kloetzel, J.J. Neefjes, C.J. Melief, and F. Ossendorp. 2000. Abrogation of CTL epitope processing by single amino acid substitution flanking the C-terminal proteasome cleavage site. J. Immunol. 164:1898–1905. [DOI] [PubMed] [Google Scholar]
- 36.Brander, C., O.O. Yang, N.G. Jones, Y. Lee, P. Goulder, R.P. Johnson, A. Trocha, D. Colbert, C. Hay, S. Buchbinder, et al. 1999. Efficient processing of the immunodominant, HLA-A*0201-restricted human immunodeficiency virus type 1 cytotoxic T-lymphocyte epitope despite multiple variations in the epitope flanking sequences. J. Virol. 73:10191–10198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leslie, A.J., K.J. Pfafferott, P. Chetty, R. Draenert, M.M. Addo, M.E. Feeney, Y. Tang, E.C. Holmes, T.M. Allen, J.G. Prado, et al. 2004. HIV evolution: CTL escape mutation and reversion following transmission. Nat. Med. 10:282–289. [DOI] [PubMed] [Google Scholar]
- 38.Altfeld, M.A., A. Trocha, R.L. Eldridge, E.S. Rosenberg, M.N. Phillips, M.M. Addo, R.P. Sekaly, S.A. Kalams, S.A. Burchett, K. McIntosh, et al. 2000. Identification of dominant optimal HLA-B60- and HLA-B61-restricted cytotoxic T-lymphocyte (CTL) epitopes: rapid characterization of CTL responses by enzyme-linked immunospot assay. J. Virol. 74:8541–8549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walker, B.D., C. Flexner, K. Birch-Limberger, L. Fisher, T.J. Paradis, A. Aldovini, R. Young, B. Moss, and R.T. Schooley. 1989. Long-term culture and fine specificity of human cytotoxic T-lymphocyte clones reactive with human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. USA. 86:9514–9518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gibbs, J.S., D.A. Regier, and R.C. Desrosiers. 1994. Construction and in vitro properties of HIV-1 mutants with deletions in “nonessential” genes. AIDS Res. Hum. Retroviruses. 10:343–350. [DOI] [PubMed] [Google Scholar]
- 41.Martinez-Picado, J., A.V. Savara, L. Sutton, and R.T. D'Aquila. 1999. Replicative fitness of protease inhibitor-resistant mutants of human immunodeficiency virus type 1. J. Virol. 73:3744–3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martinez-Picado, J., A.V. Savara, L. Shi, L. Sutton, and R.T. D'Aquila. 2000. Fitness of human immunodeficiency virus type 1 protease inhibitor-selected single mutants. Virology. 275:318–322. [DOI] [PubMed] [Google Scholar]
- 43.Prado, J.G., T. Wrin, J. Beauchaine, L. Ruiz, C.J. Petropoulos, S.D. Frost, B. Clotet, R.T. D'Aquila, and J. Martinez-Picado. 2002. Amprenavir-resistant HIV-1 exhibits lopinavir cross-resistance and reduced replication capacity. AIDS. 16:1009–1017. [DOI] [PubMed] [Google Scholar]
- 44.Yang, O.O., A.C. Tran, S.A. Kalams, R.P. Johnson, M.R. Roberts, and B.D. Walker. 1997. Lysis of HIV-1-infected cells and inhibition of viral replication by universal receptor T cells. Proc. Natl. Acad. Sci. USA. 94:11478–11483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang, Z. 2000. Maximum likelihood estimation on large phylogenies and analysis of adaptive evolution in human influenza virus A. J. Mol. Evol. 51:423–432. [DOI] [PubMed] [Google Scholar]
- 46.Yang, Z. 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13:555–556. [DOI] [PubMed] [Google Scholar]
- 47.Swofford, D.L. 2002. PAUP: version 4: phylogenetic analysis using parsimony. Sunderland, MA, Sinauer.
- 48.Barber, L.D., L. Percival, K.L. Arnett, J.E. Gumperz, L. Chen, and P. Parham. 1997. Polymorphism in the alpha 1 helix of the HLA-B heavy chain can have an overriding influence on peptide-binding specificity. J. Immunol. 158:1660–1669. [PubMed] [Google Scholar]
- 49.Goulder, P.J., M. Bunce, P. Krausa, K. McIntyre, S. Crowley, B. Morgan, A. Edwards, P. Giangrande, R.E. Phillips, and A.J. McMichael. 1996. Novel, cross-restricted, conserved, and immunodominant cytotoxic T lymphocyte epitopes in slow progressors in HIV type 1 infection. AIDS Res. Hum. Retroviruses. 12:1691–1698. [DOI] [PubMed] [Google Scholar]
- 50.Goulder, P.J., Y. Tang, S.I. Pelton, and B.D. Walker. 2000. HLA-B57-restricted cytotoxic T-lymphocyte activity in a single infected subject toward two optimal epitopes, one of which is entirely contained within the other. J. Virol. 74:5291–5299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Migueles, S.A., A.C. Laborico, H. Imamichi, W.L. Shupert, C. Royce, M. McLaughlin, L. Ehler, J. Metcalf, S. Liu, C.W. Hallahan, et al. 2003. The differential ability of HLA B(*)5701(+) long-term nonprogressors and progressors to restrict human immunodeficiency virus replication is not caused by loss of recognition of autologous viral gag sequences. J. Virol. 77:6889–6898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lyles, R.H., A. Munoz, T.E. Yamashita, H. Bazmi, R. Detels, C.R. Rinaldo, J.B. Margolick, J.P. Phair, and J.W. Mellors. 2000. Natural history of human immunodeficiency virus type 1 viremia after seroconversion and proximal to AIDS in a large cohort of homosexual men. Multicenter AIDS cohort study. J. Infect. Dis. 181:872–880. [DOI] [PubMed] [Google Scholar]
- 53.Mellors, J.W., C.R. Rinaldo, Jr., P. Gupta, R.M. White, J.A. Todd, and L.A. Kingsley. 1996. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science. 272:1167–1170. [DOI] [PubMed] [Google Scholar]
- 54.Koopmann, J.O., M. Post, J.J. Neefjes, G.J. Hammerling, and F. Momburg. 1996. Translocation of long peptides by transporters associated with antigen processing (TAP). Eur. J. Immunol. 26:1720–1728. [DOI] [PubMed] [Google Scholar]
- 55.Rosenberg, E.S., M. Altfeld, S.H. Poon, M.N. Phillips, B.M. Wilkes, R.L. Eldridge, G.K. Robbins, R.T. D'Aquila, P.J. Goulder, and B.D. Walker. 2000. Immune control of HIV-1 after early treatment of acute infection. Nature. 407:523–526. [DOI] [PubMed] [Google Scholar]
- 56.Goldberg, A.L., P. Cascio, T. Saric, and K.L. Rock. 2002. The importance of the proteasome and subsequent proteolytic steps in the generation of antigenic peptides. Mol. Immunol. 39:147–164. [DOI] [PubMed] [Google Scholar]
- 57.Shepherd, J.C., T.N. Schumacher, P.G. Ashton-Rickardt, S. Imaeda, H.L. Ploegh, C.A. Janeway, Jr., and S. Tonegawa. 1993. TAP1-dependent peptide translocation in vitro is ATP dependent and peptide selective. Cell. 74:577–584. [DOI] [PubMed] [Google Scholar]
- 58.Neisig, A., J. Roelse, A.J. Sijts, F. Ossendorp, M.C. Feltkamp, W.M. Kast, C.J. Melief, and J.J. Neefjes. 1995. Major differences in transporter associated with antigen presentation (TAP)-dependent translocation of MHC class I-presentable peptides and the effect of flanking sequences. J. Immunol. 154:1273–1279. [PubMed] [Google Scholar]
- 59.van Endert, P.M., D. Riganelli, G. Greco, K. Fleischhauer, J. Sidney, A. Sette, and J.F. Bach. 1995. The peptide-binding motif for the human transporter associated with antigen processing. J. Exp. Med. 182:1883–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu, X.G., M.M. Addo, E.S. Rosenberg, W.R. Rodriguez, P.K. Lee, C.A. Fitzpatrick, M.N. Johnston, D. Strick, P.J. Goulder, B.D. Walker, et al. 2002. Consistent patterns in the development and immunodominance of human immunodeficiency virus type 1 (HIV-1)-specific CD8+ T-cell responses following acute HIV-1 infection. J. Virol. 76:8690–8701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Altfeld, M., M.M. Addo, R. Shankarappa, P.K. Lee, T.M. Allen, X.G. Yu, A. Rathod, J. Harlow, K. O'Sullivan, M.N. Johnston, et al. 2003. Enhanced detection of human immunodeficiency virus type 1-specific T-cell responses to highly variable regions by using peptides based on autologous virus sequences. J. Virol. 77:7330–7340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moore, C.B., M. John, I.R. James, F.T. Christiansen, C.S. Witt, and S.A. Mallal. 2002. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science. 296:1439–1443. [DOI] [PubMed] [Google Scholar]
- 63.Allen, T.M., M. Altfeld, X.G. Yu, K.M. O'Sullivan, M. Lichterfeld, P.K. Lee, S. Legall, B. Mothe, D.E. Cohen, A.K. Freedberg, et al. 2004. Selection and transmission of an antigen-processing CTL escape mutation in HIV-1. J. Virol. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]