Abstract
T cell development is dependent on the integration of multiple signaling pathways, although few links between signaling cascades and downstream nuclear factors that play a role in thymocyte differentiation have been identified. We show here that expression of the HMG box protein TOX is sufficient to induce changes in coreceptor gene expression associated with β-selection, including CD8 gene demethylation. TOX expression is also sufficient to initiate positive selection to the CD8 lineage in the absence of MHC–TCR interactions. TOX-mediated positive selection is associated with up-regulation of Runx3, implicating CD4 silencing in the process. Interestingly, a strong T cell receptor–mediated signal can modify this cell fate. We further demonstrate that up-regulation of TOX in double positive thymocytes is calcineurin dependent, linking this critical signaling pathway to nuclear changes during positive selection.
Keywords: HMG box, T cell development, TCR signaling, gene regulation, Runx
Introduction
The thymus generates a vast excess of cells that fail to complete maturation and die in situ as a result of stringent selection processes, including β-selection and positive selection (1). β-Selection refers to the requirement for productive rearrangement of TCRβ chain genes and expression of a pre-TCR to induce the survival, proliferation, and differentiation of precursor thymocytes. Positive selection operates at a subsequent stage of development, requiring productive rearrangement of TCRα chain genes and TCR-mediated recognition of self-MHC to induce cell survival and continued thymocyte maturation.
Both β-selection and positive selection elicit stable changes in gene expression. Among the best characterized of these changes is expression of the CD4 and CD8 coreceptor genes, with β-selection inducing a CD4−8− (double negative [DN]) to CD4+8+ (double positive [DP]) phenotypic change in thymocytes and positive selection inducing a DP to CD4+8− or CD4−8+ (single positive [SP]) transition. A great deal of work has focused on identifying the pre-TCR– and TCR-mediated signals that are involved in β-selection and positive selection, respectively (2). In addition, there have been significant advances in understanding the regulation of coreceptor genes during thymic selection (3). In particular, members of the Ikaros and Runt domain transcription factor families have been demonstrated to bind regulatory regions of the CD8 and CD4 gene loci, respectively. Ikaros is thought to activate CD8 gene expression by increasing the accessibility of other transcription factors to the locus (4), whereas Runx1 and Runx3 function to repress CD4 gene expression by binding to a silencer regulatory element (5). In addition, components of mammalian SWI/SNF-like BAF chromatin remodeling complexes have been implicated in the control of CD4 and CD8 gene expression at various stages of T cell development (6–8).
Although the mechanistic details are still in dispute, data are accumulating that cell fate decisions associated with positive selection are regulated to some degree by the “length and strength” of intracellular signaling, with CD4 lineage commitment associated with stronger or more prolonged signaling than CD8 lineage commitment (9–17). The mitogen-activated protein kinase (MAPK) signaling pathway has been implicated in playing a central role in this process (11, 16–19). Ultimately, however, understanding lineage commitment necessitates knowledge of how quantitatively or qualitatively different signaling networks imprint distinct patterns of gene expression in the cell. One gene target of the MAPK pathway in the thymus, encoding the Egr-1 transcription factor (20), has been further linked to regulation of Id3 (21, 22), an inhibitor of E protein transcriptional activation that is required for positive selection (23). However, few other links between signaling pathways and transcriptional regulators of positive selection are known.
Calcineurin (Cn) is a calcium-activated serine/threonine phosphatase composed of catalytic (A) and regulatory (B) subunits. Two catalytic subunits, CnAα and CnAβ, are expressed in developing thymocytes. It has been known for more than a decade that positive selection is inhibited by the immunosuppressive agent and potent Cn inhibitor cyclosporin A (24, 25). More recently, genetic approaches have confirmed that Cn activation is required for positive selection. Although T cell development proceeds normally in mice that are deficient in CnAα (26), positive selection is inhibited in mice deficient in CnAβ (27). Conversely, expression of a constitutively active form of Cn in thymocytes has been reported to enhance positive selection (28). Activated Cn is known to bind and dephosphorylate NFATs in the cytoplasm, resulting in nuclear translocation of these transcriptional regulators (29). An active form of NFATc3 has been shown to substitute for calcium-mediated signals in the differentiation of a thymocyte cell line (30). However, the relevant targets of NFATs in the context of positive selection in the thymus are not known.
We have recently described a nuclear protein of the high mobility group (HMG) box family, thymocyte selection-associated HMG box (TOX), that is up-regulated during both β-selection and positive selection in the thymus (31). Transgenic mice that express TOX (TOX-transgenic [Tg]) in the majority of thymocytes show a significant increase in TCR+ CD8SP thymocytes and a concomitant reduction in CD4 SP thymocytes. Moreover, the production of CD8SP thymocytes in these animals is class I MHC independent. In this study, we demonstrate that expression of TOX can elicit changes in coreceptor expression at both the DN and DP stages that mimic aspects of both β-selection and positive selection, respectively. Moreover, production of TCR+ CD8SP thymocytes in TOX-Tg mice is independent of both class I and class II MHC, indicating that expression of this nuclear factor is sufficient to initiate a differentiation program to the CD8 lineage. The CD8SP thymocytes in TOX-Tg/MHC–deficient (MHCo) animals have down-regulated expression of the CD4 gene and up-regulated expression of Runx3, a transcriptional regulator involved in CD4 gene silencing. We also demonstrate that regulation of the TOX gene in DP thymocytes is downstream of Cn signaling, linking this signaling pathway to changes in coreceptor gene expression. Interestingly, development to the CD8 lineage in TOX-Tg mice can be overcome by TCR-mediated signaling in vivo. These results are discussed in the context of a model where lineage commitment during positive selection is regulated by an integration of MAPK and Cn signaling, the latter pathway involving up-regulation of TOX.
Materials and Methods
Animals.
All mice were bred at the Scripps Research Institute and kept under specific pathogen-free conditions. The mice used in this study were RAG-1 deficient (32), RAG-2 deficient (33), TCRα deficient (34), MHC deficient (35), 2C TCR-Tg (36) on a B6 background, AND TCR-Tg (37) on an H-2b or H-2b/k background, DO11.10 TCR-Tg (38) on an H-2d background, and CnAβ deficient (27).
Flow Cytometry.
Thymocytes were stained as described previously (31) and analyzed on a FACScan or FACSort using CellQuest software (Becton Dickinson). Typically, 5,000–20,000 viable cells were acquired based on their forward and side light scatter, and the log fluorescence is shown. For isolation of specific thymocyte subsets, cell populations were sorted using FACStar Plus or FACS® Vantage DiVa I cell sorters (Becton Dickinson).
Fluorescein isothiocyanate-conjugated anti-CD8α (53–6.7; eBioscience), phycoerythrin-conjugated or peridinine chlorophyll protein–conjugated anti-CD4 (L3T4; eBioscience), phycoerythrin-conjugated anti-CD8β (53–5.8; BD Biosciences), phycoerythrin-conjugated anti–heat stable antigen (HSA) (30-F1; eBioscience), phycoerythrin-conjugated anti-CD44 (1M7; eBioscience), allophycocyanin-conjugated anti-CD25 (PC61; eBioscience), biotinylated or allophycocyanin-conjugated anti-TCRβ (H57−597; BD Biosciences), phycoerythrin-conjugated or biotinylated anti-CD69 (H1.2F3, BD Biosciences) mAbs were used for flow cytometry.
RT-PCR.
Total RNA was isolated from sorted DP and CD8+CD4lo/− thymocytes from TOX-Tg/MHCo mice using the RNeasy RNA kit (QIAGEN). Primer pairs (Invitrogen) used were: CD8α 5′-TGCCATGAGGGACACGAATAATAAG-3′ and 5′-TAAATATCACAGGCGAAGTCCAATC-3′; CD8β 5′-TTCTTGGTTGGGGCAGTTGTAGGAA-3′ and 5′-TGGCTCTGGCTGGTCTTCAGTATGA-3′; CD4 5′-CTGATGTGGAAGGCAGAGAAGGATTC-3′ and 5′-CAGCACGCAAGCCAGGAACACTGTCT-3′; TCRα 5′-TGGGGCCATTGCCTGGAGCAACCAGA-3′ and 5′-CACAGCCTCAGCGTCATGAGCAGGT-3′; Thy-1 5′-CGTACCGCTCCCGCGTCACCCTCTCC-3′ and 5′-GCAGGCTTATGCCGCCACACTTGACCA-3′; and β-actin 5′-GGCAACGAGCGGTTCCGATGCCCTGA-3′ and 5′-GCCAGGATGGAGCCACCGATCCACA-3′.
DNA Methylation Assay.
Genomic DNA was isolated from sorted thymocytes using the DNeasy tissue kit (QIAGEN) and digested with HhaI or NcoI. 100 ng of DNA template was used for PCR amplification of the sequence flanking the HhaI and NcoI sites in the CD8 locus with the primer pair 5′-GCCCCAGCCTGCACACCTGGGCTACA-3′ and 5′-GATGTTCACAGGACCCTGCTGGCCAAA-3′.
In Vitro Activation and Culture of Thymocytes.
Thymocytes from TCRα chain–deficient (TCRαo) or CnAβ-deficient mice were isolated and cultured in the absence or presence of 0.2 ng/ml PMA (Calbiochem) and/or 0.05–0.2 μg/ml ionomycin (Calbiochem), with and without inhibitors cyclosporin A (5 μg/ml) (Sigma-Aldrich) or U0126 (10 μM) (Calbiochem) for 6–8 h at 37°C. Cells were analyzed by flow cytometry and/or by Western blot. In other experiments, sorted DP thymocytes from TOX-Tg/MHCo mice or whole thymocytes from MHCo mice were cultured for 44 h at 37°C and 10% CO2 in DMEM medium supplemented as described previously (18) and used for flow cytometric analysis.
Immunoblotting.
3 × 105 thymocytes were lysed in SDS sample buffer and subjected to Western blot as described previously (31). Primary antibodies used were affinity purified rabbit anti-TOX antibody (150–300 ng/ml) (31), polyclonal anti-Runx (1:3,000) (39), or anti-β-actin (1:5,000) (Ab-1; Oncogene Research Products). Identification of Runx proteins was confirmed by Western blot analysis of transfected cells expressing Runx1 or Runx3 (unpublished data).
Results
TOX Is Sufficient to Initiate Coreceptor Changes Associated with β-Selection.
DN thymocytes progress through distinct developmental stages that can be monitored by the expression of CD44 and CD25; the stages are termed DN1 (CD44+ CD25−), DN2 (CD44+CD25+), DN3 (CD44−CD25+), and DN4 (CD44−CD25−) (40). Transition of thymocytes from the DN3 to the DN4 stage during β-selection requires signaling by the pre-TCR complex (41).
We have reported previously that TOX is up-regulated in the DN3 blast population as a consequence of β-selection and subsequently down-regulated before the DP stage (31). To determine the role of TOX during β-selection, we generated TOX-Tg mice on a RAG-deficient (RAGo) background. RAGo thymocytes have a developmental block at the DN3 stage and severely reduced thymic cellularity due to the failure to undergo β-selection (33). Although the thymic cellularity of TOX-Tg/RAGo mice was similar to that of RAGo mice, the expression of the TOX transgene resulted in a complex pattern of coreceptor expression (Fig. 1 A). Four distinct cell populations, DN, CD4loCD8−, DP, and CD8SP thymocytes, were present in these mice. An identical staining pattern was obtained when cells were stained for CD4 and CD8β, indicating that the CD8+ thymocytes expressed a CD8αβ heterodimer, as do normal DP and CD8SP thymocytes (unpublished data).
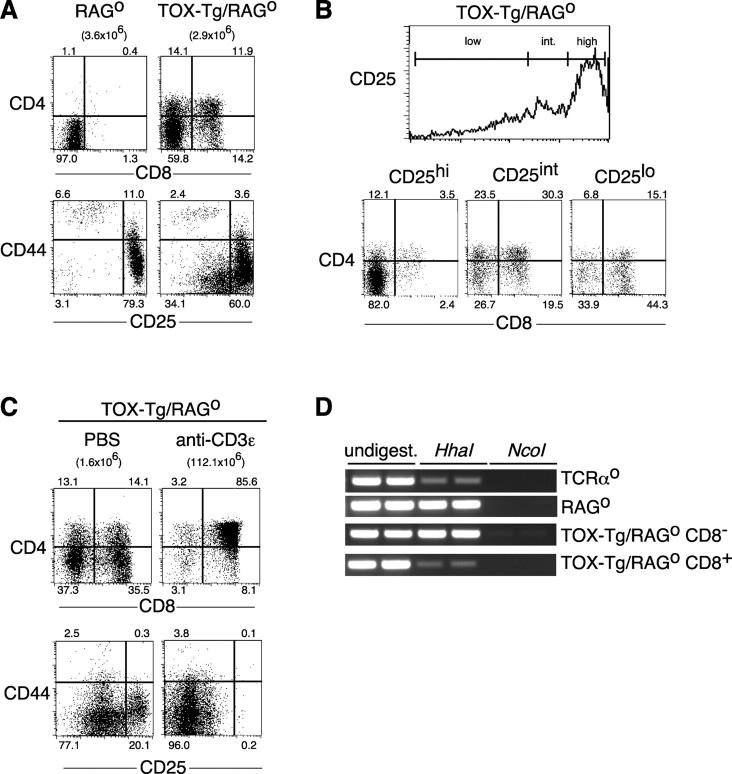
Figure 1.
TOX induces development of coreceptor positive thymocytes in RAGo mice. (A) TOX-Tg/RAGo thymocytes were stained for expression of CD4, CD8α, CD44, and CD25 and analyzed by flow cytometry. Percentages of total thymocytes in each quadrant and total thymic cellularity are indicated. (B) CD4 and CD8α expression (dot plots) on CD25 gated thymocytes (histogram) in a TOX-Tg/RAGo mouse. (C) TOX-Tg/RAGo mice were injected i.p. with PBS or anti-CD3ɛ antibody, and the thymocytes were analyzed 5 d later for expression of CD4 and CD8α or CD44 and CD25 as in A. (D) Methylation status of the CD8 locus in thymocytes from mice with the indicated genotype as analyzed by restriction digest and PCR. CD8α+ and CD8α2 thymocyte cell subpopulations were purified by cell sorting.
Expression of TOX on a RAGo background also resulted in the appearance of CD44−CD25lo thymocytes, consistent with initiation of the DN3 to DN4 transition in some cells (Fig. 1 A). To determine the developmental relationship between different cell populations, thymocytes from TOX-Tg/RAGo mice were four-color stained for CD4, CD8, CD25, and CD44. The TOX-Tg/RAGo DN and some CD4loCD8− thymocytes were CD44−CD25hi, similar to the majority DN3 population found in RAGo thymocytes (Fig. 1 A). In contrast, the majority of DP and some CD4lo cells displayed intermediate levels of CD25, consistent with a post-DN3 stage of development (Fig. 1 B). Thymocytes with a CD8SP phenotype are found both as a DN4 to DP transitional population (CD4−CD8+ immature single positive thymocyte [CD8ISP]) and as a product of DP thymocytes after positive selection. Surprisingly, the CD8SP thymocytes in TOX-Tg/RAGo mice were CD25lo when compared with DP thymocytes in these same animals, most consistent with a post-DP stage rather than a CD8ISP stage of development (Fig. 1 B).
Expression of TOX in transgenic animals on a wild-type background does not block β-selection, nor does it result in aberrant expression of CD25 on DP or CD8SP thymocytes (31). Thus, it seems likely that the reduction in CD25 in CD8SP compared with DP cells in TOX-Tg/RAGo mice represents a developmental progression. The failure of CD25 to be fully down-regulated in these cells may reflect the absence of proliferation and/or pre-TCR signals normally associated with β-selection. In support of this, anti-CD3ɛ treatment of TOX-Tg/RAGo mice led to increases in thymic cellularity and loss of CD25 on DP and CD8SP cells (Fig. 1 C) as it does in RAGo mice (42).
Transcriptional activation of the CD8 gene locus during the transition from the DN to DP stage of thymocyte development is associated with DNA demethylation of the gene (43). Since expression of TOX was sufficient to induce up-regulation of both CD8α and CD8β on DN thymocytes, we asked whether TOX also induced a change in the methylation status of these loci. A CpG sequence that forms part of an HhaI restriction site is found between the linked CD8β and CD8α genes and is a site of DNA methylation (43, 44). Methylation status of this site was assessed by restriction digest of genomic DNA followed by PCR. In RAGo thymocytes, the CD8 locus is transcriptionally silent and this site is methylated, as evidenced by resistance to digestion with the methylation-sensitive enzyme HhaI and successful PCR with primers that flank this site (Fig. 1 D). However, PCR fails after digestion of the DNA with the control enzyme NcoI that recognizes an adjacent sequence that does not include a CpG methylation site (Fig. 1 D). In contrast, this site is partially demethylated in DP thymocytes (total thymocytes from TCRα-deficient mice) and is therefore sensitive to HhaI digestion (Fig. 1 D).
To determine if TOX was sufficient to alter the methylation pattern of the CD8 locus, CD8+ and CD8− thymocytes were purified by cell sorting from TOX-Tg/RAGo mice and analyzed as above. Although the HhaI site was methylated in CD8− thymocytes and resistant to digestion, the CD8+ thymocytes showed partial demethylation, consistent with activation of the CD8 gene locus (Fig. 1 D).
TOX Is Sufficient to Initiate Coreceptor Changes Associated with Positive Selection to the CD8 Lineage.
We have reported previously that TOX-Tg mice have increased numbers of CD8SP thymocytes and reduced numbers of CD4SP thymocytes (31). Production of CD8SP thymocytes in these mice is not dependent on expression of class I MHC. The fact that TOX-Tg/RAGo mice also had CD8SP thymocytes and that these cells had down-regulated expression of CD25 when compared with DP thymocytes in these same animals, suggested that TOX might be sufficient to initiate positive selection and β-selection.
To test this, we bred TOX-Tg mice onto a TCRαo background. Thymocytes from TCRαo mice are unable to undergo positive selection and are blocked at the DP stage of development. However, expression of TOX resulted in appearance of a pronounced CD8SP subpopulation, even in the absence of a functional TCR complex (Fig. 2 A). In contrast, no CD4SP thymocytes were detected in these mice (Fig. 2 A). The CD8SP thymocytes from TOX-Tg/TCRαo mice were neither functionally nor phenotypically mature (not depicted). However, in the absence of the TCR as a marker it was not clear whether these cells were CD8ISP thymocytes or CD8SP thymocytes that had failed to undergo complete maturation.
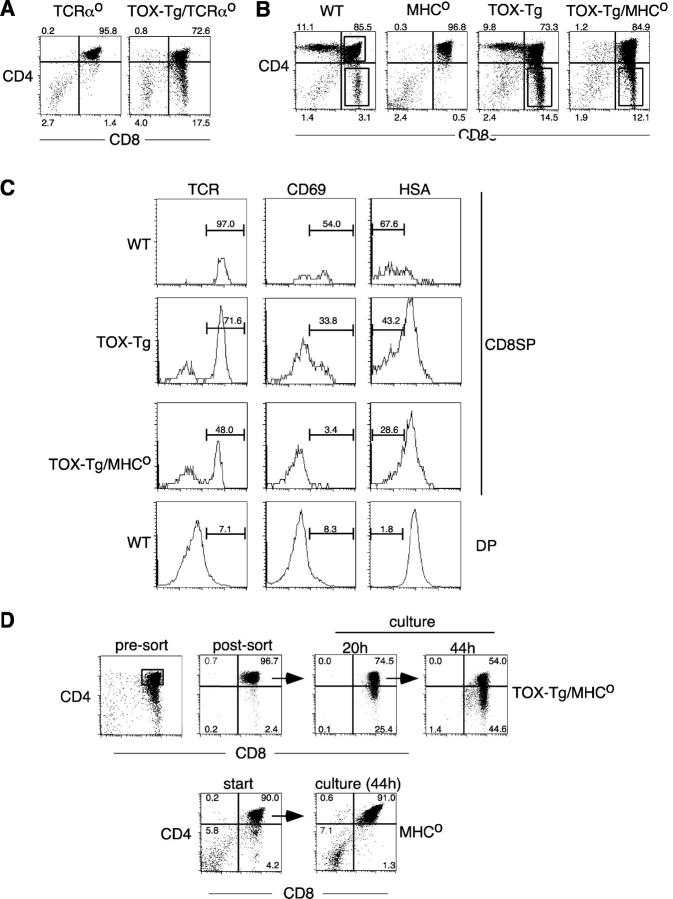
Figure 2.
TOX induces development of CD8SP thymocytes in the absence of TCR–MHC interactions. (A and B) Expression of CD4 and CD8α on thymocytes from mice with the indicated genotype. Percentages of total thymocytes in each quadrant are indicated. (C) Expression of TCRβ, CD69, and HSA on CD8SP thymocytes gated as in B from mice with the indicated genotypes. DP thymocytes from wild-type mice were used as a control to set appropriate gates. The percentage of TCRhi, CD69+, and HSAlo cells are shown. (D) Total MHCo thymocytes or sorted DP thymocytes from TOX-Tg/MHCo mice were cultured ex vivo and analyzed for CD4 and CD8α expression by flow cytometry after 20 or 44 h. Percentages of total thymocytes in each quadrant are shown. Viable yield of TOX-Tg/MHCo thymocytes was 73% after 20 h of culture.
This issue was addressed using MHC class I and II doubly deficient (MHCo) mice, since the thymocytes in these animals are unable to undergo positive selection but express the TCR. Thus, CD8ISP and CD8SP thymocytes can be distinguished by expression of the TCR on the latter. In contrast to MHCo mice, TOX-Tg/MHCo mice had a pronounced CD8SP population, similar to that seen in TOX-Tg/TCRαo mice (Fig. 2 B). A significant proportion (48.0%) of these CD8SP thymocytes also expressed the TCR, indicating that they are not CD8ISP cells (Fig. 2 C). No TCR+ CD4SP thymocytes were present in these mice (Fig. 2 C and not depicted).
CD69 is a marker of TCR-mediated signaling associated with positive selection (45). In a normal thymus, approximately half of the CD8SP thymocytes express CD69 (Fig. 2 C). TOX-Tg mice on a wild-type background also contained CD69+ CD8SP thymocytes (Fig. 2 C). However, few CD8SP thymocytes in TOX-Tg/MHCo mice expressed CD69, consistent with TCR-independent development of these cells. Down-regulation of HSA occurs during the later stages of SP thymocyte maturation (46). Approximately 70 and 40% of CD8SP thymocytes in normal animals and TOX-Tg mice, respectively, were HSAlo (Fig. 2 C). HSAlo cells were also present among TOX-Tg/MHCo CD8SP thymocytes, although the extent of down-regulation was reduced compared with wild-type cells (Fig. 2 C).
To definitively demonstrate that DP cells can give rise to CD8SP cells in the presence of TOX and absence of positive selection, DP thymocytes were purified by cell sorting and cultured in vitro (Fig. 2 D). A significant proportion of TOX-Tg/MHCo DP thymocytes spontaneously down-regulated CD4 over a 44-h culture period, giving rise to CD8SP cells (Fig. 2 D, top row). In contrast, DP thymocytes from MHCo mice maintained expression of both CD4 and CD8 (Fig. 2 D, bottom row). Together, the data demonstrate that expression of TOX in DP thymocytes is sufficient to induce down-regulation of CD4 and partial maturation in the absence of normal positive selection signals.
TOX and Coreceptor Gene Expression.
Loss of CD4 expression during the DP to CD8SP thymocyte transition is controlled by activation of a silencer element in the CD4 gene locus (47). To determine whether loss of cell surface CD4 on TOX-Tg/MHCo CD8SP thymocytes reflected a change in gene expression, RT-PCR was performed on purified cell populations. No significant difference in expression of TCRα, CD8α, or CD8β genes was observed when TOX-Tg/MHCo CD8SP and DP thymocytes were compared (Fig. 3 A). However, CD4 gene expression was markedly reduced in the CD8SP subpopulation (Fig. 3 A). Interestingly, some diminution of Thy-1 gene expression was also apparent in the CD8SP population (Fig. 3 A), another indication of partial maturation of these cells (48).
Figure 3.
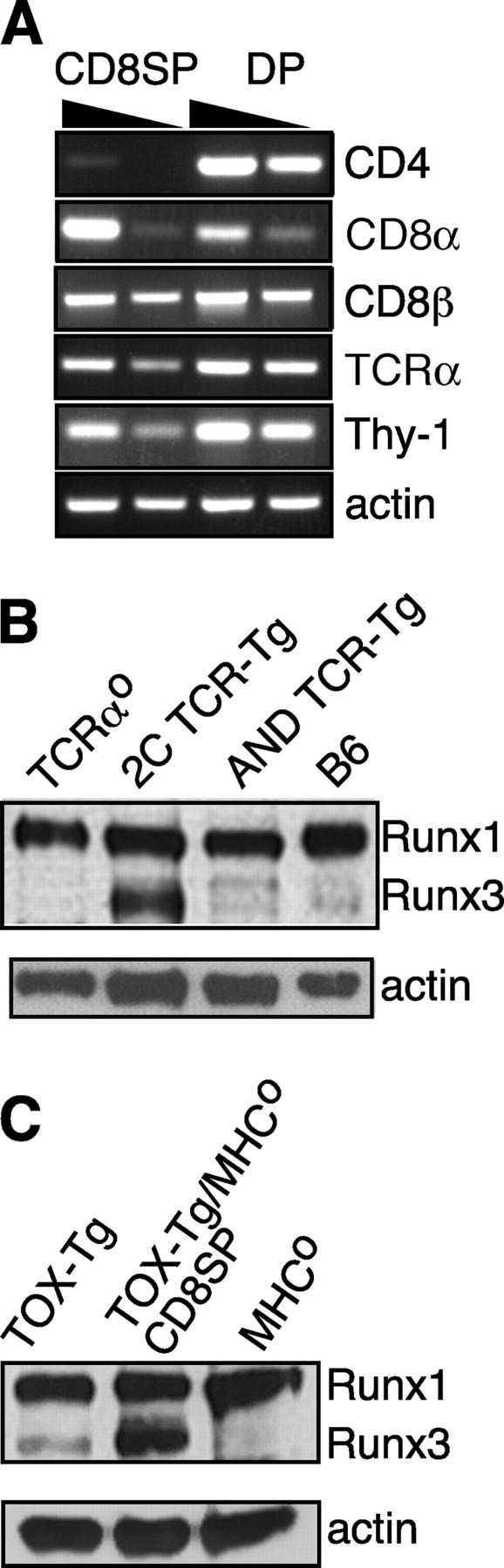
CD4 gene silencing in TOX-Tg mice. (A) RT-PCR analysis for expression of the indicated genes in purified CD8SP or DP thymocytes isolated from TOX-Tg/MHCo mice. (B and C) Western blot analysis for expression of Runx proteins in whole cell lysates prepared from total thymocytes or isolated CD8SP cells as indicated. Expression of β-actin was used as a loading control.
Recently, the Runx1 and Runx3 proteins have been demonstrated to regulate the expression of the CD4 gene by binding to the CD4 silencer element (5). Moreover, CD8SP thymocytes were absent in mice that were deficient in Runx3 and were heterozygous for a deletion mutant of Runx1 (49). Conversely, the overexpression of Runx1 in vivo resulted in an increase in the total number of CD8SP thymocytes (39).
As ectopic expression of TOX also led to an increase in CD8SP thymocytes, we assessed the levels of Runx proteins in thymocytes from different mouse strains used in this study (Fig. 3 B). Runx3 is more highly expressed by CD8SP thymocytes than CD4SP thymocytes (5, 39), consistent with its role in CD4 silencing. Thus, in circumstances where almost all the thymocytes were DP or CD4SP including TCRαo, AND TCR-Tg, and MHCo mice, no Runx3 was detectable (Fig. 3, B and C). In contrast, Runx1 was expressed at similar amounts in thymocytes from these mice (Fig. 3, B and C) as expected (5, 50). Conversely, in animals where CD8SP thymocytes were present including normal mice and especially in class I MHC-restricted 2C TCR-Tg mice, Runx3 was up-regulated (Fig. 3 B).
To assess the changes induced by expression of TOX in vivo, we then investigated the levels of Runx proteins in TOX-Tg thymocytes. TOX-Tg thymocytes expressed both Runx1 and Runx3, consistent with the production of CD8SP thymocytes in these animals (Fig. 3 C). Most interestingly, however, CD8SP thymocytes that were produced in the absence of positive selection in TOX-Tg/MHCo mice had up-regulated Runx3 and maintained expression of Runx1 (Fig. 3 C). This indicates that expression of TOX is sufficient to initiate a sequence of events in developing thymocytes, including CD4 gene silencing and the associated up-regulation of the CD4 silencer binding protein Runx3.
TOX Is Downstream of Cn Signaling.
The Tox gene was isolated from DP thymocytes which were cultured with PMA and ionomycin under conditions that induce cell differentiation (31). Both MAPK and Cn signaling are required for positive selection (as described in Introduction). To address whether these signaling pathways are also involved in regulation of the Tox gene, we asked whether one or both pharmacologic activators needed to be present to up-regulate TOX expression. Cultured TCRαo (primarily DP) thymocytes expressed little TOX as assessed by Western blotting (Fig. 4 A). As expected, PMA and ionomycin induced up-regulation of TOX in these cultured cells (Fig. 4 A). However, PMA alone was a poor activator of TOX expression, whereas ionomycin alone fully up-regulated the protein in DP thymocytes (Fig. 4 A). Consistent with these results, cyclosporin A, an inhibitor of Cn, but not U0126, a MEK antagonist, inhibited up-regulation of TOX in this system (Fig. 4 A). In contrast, induction of the MAPK-dependent activation marker CD69 (20, 51) was inhibited by U0126 (Fig. 4 A).
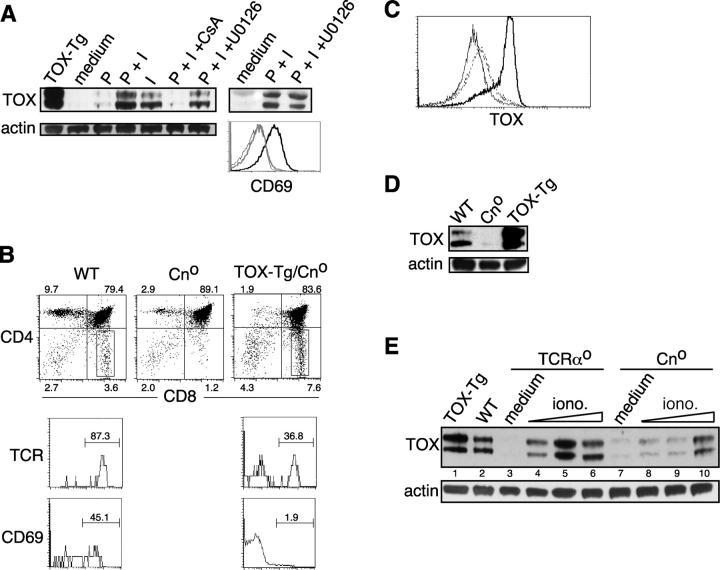
Figure 4.
Expression of TOX is regulated by Cn activation. (A) Expression of TOX in TCRαo thymocytes activated with PMA and ionomycin in the absence or presence of inhibitors of Cn (cyclosporin A, CsA) or MEK (U0126). Also shown is CD69 expression on cells cultured in medium (thin line), with PMA and ionomycin (thick black line), or with PMA/ionomycin and U0126 (thick gray line). Expression of TOX in thymocytes derived from a TOX-Tg mouse is shown as a control. (B) Expression of CD4 and CD8α on thymocytes derived from various animals as indicated (dot plots). Percentage of total thymocytes in each quadrant is shown. Expression of TCRβ and CD69 was also analyzed on gated CD8SP thymocytes, and the percentage of TCRhi or CD69+ cells is indicated. Expression of TOX was determined by intracellular staining and flow cytometry (C) in WT (dashed line), Cno (thin line), and TOX-Tg (thick line) thymocytes or by Western blot of whole cell lysates (D). (E) Thymocytes from TCRαo and Cno mice were cultured in medium (lanes 3 and 7) or 0.2 ng/ml of PMA and 0.05 (lanes 4 and 8), 0.1 (lanes 5 and 9), or 0.2 (lanes 6 and 10) μg/ml of ionomycin. TOX-Tg thymocytes were analyzed as a control (lane 1).
We also took a genetic approach to analyze the dependence of TOX up-regulation on Cn. The reduction in Cn signaling in CnAβ-deficient (Cno) mice results in severe inhibition of positive selection evidenced by a decrease in TCR+ CD4SP and CD8SP thymocytes (27) (Fig. 4 B). The incomplete block in positive selection in these animals is likely due to compensation by CnAα. Cno thymocytes express lower amounts of TOX than wild-type thymocytes when analyzed by flow cytometry (Fig. 4 C) or by Western blot (Fig. 4 D). Moreover, the efficacy of induction of TOX by ionomycin is severely reduced in Cno thymocytes compared with TCRαo thymocytes (Fig. 4 E). The residual TOX induction is presumably mediated by CnAα, consistent with a reduction but not complete inhibition of positive selection in these animals (Fig. 4 B).
If TOX is downstream of Cn signaling one would predict that loss of the CnAβ gene would have no effect on the production of CD8SP thymocytes in TOX-Tg mice. To test this, we bred TOX-Tg mice onto a Cno background. Although expression of TOX did not rescue CD4SP thymocyte development in Cno mice, there was a fivefold increase in the frequency of TCR+ CD8SP thymocytes in TOX-Tg/Cno mice (Fig. 4 B). In addition, few CD8SP thymocytes in TOX-Tg/Cno mice expressed CD69, consistent with their production in the absence of TCR signaling normally associated with positive selection (Fig. 4 B).
These results demonstrate that expression of TOX induces development of a post-DP CD8SP thymocyte population in the absence of MHC-mediated positive selection. The question arises as to why expression of TOX skews development to the CD8 lineage. CD4SP thymocytes are present in TOX-Tg mice, although somewhat reduced in number (31). As shown here, the development of these CD4SP thymocytes is dependent on expression of TCR and MHC and on Cn (Fig. 2, A and B, and Fig. 4 B). In addition, CD4SP thymocytes in TOX-Tg mice are indistinguishable phenotypically from CD4SP in wild-type mice and show normal up-regulation of TCR, CD5, and CD69 (31). Thus, TOX is not sufficient to initiate CD4 lineage commitment, and development of the CD4 lineage maintains a requirement for positive selection even in TOX-Tg mice. Therefore, we reasoned that CD4SP cells would develop as a result of TCR-mediated signals that could overcome the ability of TOX to initiate CD8 lineage development.
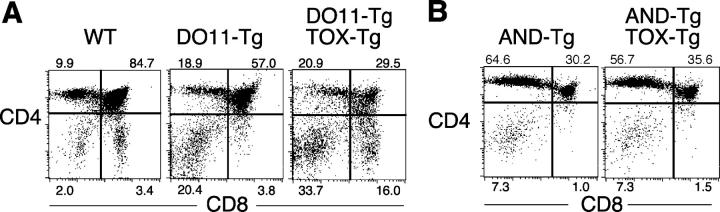
We have reported previously that expression of TOX promoted the appearance of CD8SP cells in class II MHC-restricted DO11 TCR Tg mice (31) (Fig. 5 A). However, CD4SP thymocytes were also present in these double transgenic animals, raising the possibility that DO11 TCR signaling was only partially effective in competing with TOX. To test this, we bred TOX-Tg mice to AND TCR-Tg mice on a selecting background. AND TCR-Tg mice exhibit more pronounced skewing to the CD4 lineage than DO11 TCR-Tg mice, presumably because of increased affinity of the AND TCR for class II MHC expressed in the thymus. Importantly, when the TOX transgene was expressed in conjunction with the AND TCR transgene, few CD8 SP thymocytes were observed (Fig. 5 B).
Figure 5.
TCR signaling in vivo competes with TOX activity. (A and B) Expression of CD4 and CD8α on thymocytes from indicated mice was analyzed by flow cytometry. Percentage of total thymocytes in each quadrant is shown.
Discussion
Expression of TOX is transiently up-regulated during both β-selection and positive selection of thymocytes. In this study, we demonstrated that enforced TOX expression was sufficient to induce the differentiation of cells through these developmental checkpoints as assessed by changes in coreceptor expression but failed to induce cellular proliferation normally associated with the DN to DP transition. This shows a clear split in the downstream effectors that are sufficient for changes in coreceptor gene expression and those that initiate cell proliferation during β-selection. The failure of TOX to induce proliferation of DN thymocytes also indicates that the vast majority of DP thymocytes that develop in TOX-Tg mice on a wild-type background are the product of β-selection. Although expression of TOX was sufficient to induce subpopulations of DP and CD8SP thymocytes in RAGo mice, these cells maintained intermediate levels of CD25 on the cell surface. This is an additional indication that TOX is an incomplete mimic of pre-TCR signaling. Similarly, the expression of a stable form of β-catenin results in development of DP thymocytes in RAGo mice, with limited proliferation and only partial down-regulation of CD25 (52).
The CD8 locus is demethylated in TOX-Tg/RAGo thymocytes that express CD8 at the cell surface (Fig. 1 D). In some contexts, demethylation appears to be causative for CD8 gene expression. Thus, mice lacking the maintenance DNA methyltransferase Dnmt1 abnormally express CD8αβ heterodimer in the γδ T cell subset (53). Demethylation may also play an important role in regulation of CD8 gene expression at the DN to DP transition, although remethylation is not required for loss of CD8 expression in CD4 T cells (43, 53). In addition to the activation of the CD8 gene, we also observed CD4lo-expressing thymocytes in TOX-Tg/RAGo mice (Fig. 1 A). Based on the expression of CD44 and CD25 (Fig. 1 B), it seems most likely that these cells arise from derepression of CD4 gene expression in the DN3 population. The fact that ectopic expression of a single nuclear factor can elicit such a complex collection of cell phenotypes suggests that timing and cellular context are critical for the function of TOX.
Changes in CD4 gene expression during T cell development are regulated by a silencer element located in the first intron of the CD4 gene (47). The loss of CD4 gene expression in TOX-Tg thymocytes subsequent to the DP stage is consistent with activation of this silencer. TOX is highly expressed in CD4CD8 double dull thymocytes in normal mice (31), a post-DP stage at which silencer activity is also observed (54). Moreover, Runx3 is up-regulated in CD8SP thymocytes from TOX-Tg/MHCo mice (Fig. 5 C). Runx3 binds the CD4 silencer element and plays a role in extinguishing CD4 expression in thymocytes committed to the CD8 lineage (5, 49), further implicating TOX in CD4 silencing. TOX contains an HMG box, a protein domain that bends and unwinds DNA by primarily contacting the minor groove (55). The crystal structure of Runx1, a closely related protein to Runx3, revealed that DNA bending is required in order for this transcription factor to bind to its recognition sequence (56). Thus, TOX could be involved in both induction of Runx3 and in changing the conformation of DNA targets to allow Runx factors to bind and exert their function.
Regulation of coreceptor expression in the thymus is at least partly regulated by changes in chromatin. Ikaros, a nuclear factor that is essential for fetal lymphoid development and that plays multiple roles during adult T cell development (57, 58), has been reported to act as both a transcriptional activator and repressor through interactions with proteins that can alter chromatin structure (59). The repressor activity of Ikaros is associated with packaging of genes into transcriptionally silent heterochromatin (60). Interestingly, the effect of loss of Ikaros on DN thymocytes has some similarities with the effects of expression of TOX. In both instances, there is pre-TCR–independent production of DP thymocytes in the absence of proliferation (58). However, Ikaros has been demonstrated to be required for the full activation of the CD8α locus (4). BAF57, a component of a SWI/SNF-like chromatin remodeling complex, is an HMG box protein with a single HMG box motif that is predicted to bind DNA in a sequence-independent fashion (61) as is TOX (62). Thymocytes from mice lacking wild-type BAF57, or its associated ATPase Brg-1, also exhibited changes in coreceptor expression including derepression of CD4 in DN thymocytes and impairment of CD8 gene activation (6–8). Thus, the common thread in the ability of TOX to induce activation and epigenetic modification of the CD8 locus in DN cells and activation or silencing of the CD4 locus in DN or DP cells, respectively, could be the induction of architectural changes in chromatin, as reported for Ikaros and members of the SWI/SNF-like complex. This is certainly consistent with the known and proposed functions for HMG box proteins in transcriptional regulation and nucleosome remodeling (55, 63).
The loss of CD4 gene expression on a subpopulation of thymocytes in TOX-Tg mice was independent of normal TCR and MHC-mediated positive selection. This argues against the possibility that ectopic expression of TOX alters T cell development as a result of changes in expression of the TCR or associated signaling proteins (3). We had reported previously a decrease in the CD4SP to CD8SP thymocyte ratio in TOX-Tg mice (31). This phenotype is now understood by the ability of TOX to promote CD8SP development in the absence of positive selection. The reduction of CD4SP thymocytes in these mice is likely due to a preemption of normal positive selection by early expression of the TOX transgene in DP thymocytes.
Positive selection requires TCR-mediated activation of MAPK and Cn signaling pathways. How these pathways are integrated by the cell and how they regulate lineage commitment remains unknown, although a number of lines of evidence suggest that the MAPK pathway can have an overriding deterministic effect on the CD4 T cell fate (11, 16–19). We have shown here that up-regulation of TOX in DP thymocytes is Cn dependent, providing a link between this signaling pathway and CD8 lineage commitment. Expression of TOX induced differentiation of CD8SP thymocytes independent of expression of MHC or TCR or activation of Cn. However, TOX is not sufficient to induce CD4SP development under these same conditions, suggesting a requirement for additional downstream effectors for CD4 T cell development. One such factor, GATA-3, has been shown recently to promote differentiation of thymocytes to the CD4 fate while inhibiting CD8SP development (64). Moreover, GATA-3 expression has been linked with “strong” TCR signaling (64), correlating with previous findings that the strength of TCR signaling can bias lineage commitment decisions.
These data lead to a model where development of CD4 and CD8 lineages each require both MAPK- and Cn-mediated signals. The strength and balance between these signaling pathways determines cell fate. Cn-mediated signaling during the initiation of positive selection to either T cell lineage results in up-regulation of TOX. Unopposed, TOX plays a role in CD4 silencing and CD8 lineage commitment, possibly via up-regulation of Runx3 and/or changes in chromatin structure. However, in the presence of sufficient MAPK signaling and possibly additional Cn-mediated signals (17) the effect of TOX is negated, leading to CD4 lineage commitment. This model explains why CD4SP development is not affected by expression of TOX in circumstances where there is a potent TCR signal and where timing of TCR and TOX expression are coincident, such as in TOX/AND TCR double Tg mice. We predict that one function of increased/prolonged signaling during positive selection to the CD4 lineage is induction or activation of an inhibitor of TOX. One intriguing possibility is a role for GATA-3 in this activity. However, since sustained expression of GATA-3 is not sufficient to divert class I MHC-restricted developing T cells into the CD4 lineage (64), it seems unlikely that GATA-3 alone would function in this regard. Nevertheless, one might predict that the threshold of signaling that is necessary to compete with TOX will also be the set point for CD4 lineage commitment (16).
Acknowledgments
This work was supported by grants AI44110 and AI054977 from the National Institutes of Health to J. Kaye. This is manuscript no. 16120-IMM from the Scripps Research Institute.
P. Aliahmad and E. O'Flaherty contributed equally to this work.
Abbreviations used in this paper: CD8ISP, CD4−CD8+ immature single positive thymocyte; Cn, calcineurin; DN, CD4−8− double negative thymocyte; DP, CD4+8+ double positive thymocyte; HMG, high mobility group; MAPK, mitogen-activated protein kinase; SP, CD4+8− or CD4−8+ single positive thymocyte; Tg, transgenic; TOX, thymocyte selection-associated HMG box protein.
References
- 1.Starr, T.K., S.C. Jameson, and K.A. Hogquist. 2003. Positive and negative selection of T cells. Annu. Rev. Immunol. 21:139–176. [DOI] [PubMed] [Google Scholar]
- 2.van Oers, N.S. 1999. T cell receptor-mediated signs and signals governing T cell development. Semin. Immunol. 11:227–237. [DOI] [PubMed] [Google Scholar]
- 3.Kioussis, D., and W. Ellmeier. 2002. Chromatin and CD4, CD8A and CD8B gene expression during thymic differentiation. Nat. Rev. Immunol. 2:909–919. [DOI] [PubMed] [Google Scholar]
- 4.Harker, N., T. Naito, M. Cortes, A. Hostert, S. Hirschberg, M. Tolaini, K. Roderick, K. Georgopoulos, and D. Kioussis. 2002. The CD8alpha gene locus is regulated by the Ikaros family of proteins. Mol. Cell. 10:1403–1415. [DOI] [PubMed] [Google Scholar]
- 5.Taniuchi, I., M. Osato, T. Egawa, M.J. Sunshine, S.C. Bae, T. Komori, Y. Ito, and D.R. Littman. 2002. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell. 111:621–633. [DOI] [PubMed] [Google Scholar]
- 6.Chi, T.H., M. Wan, K. Zhao, I. Taniuchi, L. Chen, D.R. Littman, and G.R. Crabtree. 2002. Reciprocal regulation of CD4/CD8 expression by SWI/SNF-like BAF complexes. Nature. 418:195–199. [DOI] [PubMed] [Google Scholar]
- 7.Chi, T.H., M. Wan, P.P. Lee, K. Akashi, D. Metzger, P. Chambon, C.B. Wilson, and G.R. Crabtree. 2003. Sequential roles of Brg, the ATPase subunit of BAF chromatin remodeling complexes, in thymocyte development. Immunity. 19:169–182. [DOI] [PubMed] [Google Scholar]
- 8.Gebuhr, T.C., G.I. Kovalev, S. Bultman, V. Godfrey, L. Su, and T. Magnuson. 2003. The role of Brg1, a catalytic subunit of mammalian chromatin-remodeling complexes, in T cell development. J. Exp. Med. 198:1937–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Itano, A., P. Salmon, D. Kioussis, M. Tolaini, P. Corbella, and E. Robey. 1996. The cytoplasmic domain of CD4 promotes the development of CD4 lineage T cells. J. Exp. Med. 183:731–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matechak, E.O., N. Killeen, S.M. Hedrick, and B.J. Fowlkes. 1996. MHC class II-specific T cells can develop in the CD8 lineage when CD4 is absent. Immunity. 4:337–347. [DOI] [PubMed] [Google Scholar]
- 11.Sharp, L.L., D.A. Schwarz, C.M. Bott, C.J. Marshall, and S.M. Hedrick. 1997. The influence of the MAPK pathway on T cell lineage commitment. Immunity. 7:609–618. [DOI] [PubMed] [Google Scholar]
- 12.Basson, M.A., U. Bommhardt, M.S. Cole, J.Y. Tso, and R. Zamoyska. 1998. CD3 ligation on immature thymocytes generates antagonist-like signals appropriate for CD8 lineage commitment, independently of T cell receptor specificity. J. Exp. Med. 187:1249–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yasutomo, K., C. Doyle, L. Miele, C. Fuchs, and R.N. Germain. 2000. The duration of antigen receptor signalling determines CD4+ versus CD8+ T-cell lineage fate. Nature. 404:506–510. [DOI] [PubMed] [Google Scholar]
- 14.Hernandez-Hoyos, G., S.J. Sohn, E.V. Rothenberg, and J. Alberola-Ila. 2000. Lck activity controls CD4/CD8 T cell lineage commitment. Immunity. 12:313–322. [DOI] [PubMed] [Google Scholar]
- 15.Watanabe, N., H. Arase, M. Onodera, P.S. Ohashi, and T. Saito. 2000. The quantity of TCR signal determines positive selection and lineage commitment of T cells. J. Immunol. 165:6252–6261. [DOI] [PubMed] [Google Scholar]
- 16.Wilkinson, B., and J. Kaye. 2001. Requirement for sustained MAPK signaling in both CD4 and CD8 lineage commitment: a threshold model. Cell. Immunol. 211:86–95. [DOI] [PubMed] [Google Scholar]
- 17.Adachi, S., and M. Iwata. 2002. Duration of calcineurin and Erk signals regulates CD4/CD8 lineage commitment of thymocytes. Cell. Immunol. 215:45–53. [DOI] [PubMed] [Google Scholar]
- 18.Shao, H., B. Wilkinson, B. Lee, P.C. Han, and J. Kaye. 1999. Slow accumulation of active mitogen-activated protein kinase during thymocyte differentiation regulates the temporal pattern of transcription factor gene expression. J. Immunol. 163:603–610. [PubMed] [Google Scholar]
- 19.Bommhardt, U., M.A. Basson, U. Krummrei, and R. Zamoyska. 1999. Activation of the extracellular signal-related kinase/mitogen-activated protein kinase pathway discriminates CD4 versus CD8 lineage commitment in the thymus. J. Immunol. 163:715–722. [PubMed] [Google Scholar]
- 20.Shao, H., D.H. Kono, L.Y. Chen, E.M. Rubin, and J. Kaye. 1997. Induction of the early growth response (Egr) family of transcription factors during thymic selection. J. Exp. Med. 185:731–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bain, G., C.B. Cravatt, C. Loomans, J. Alberola-Ila, S.M. Hedrick, and C. Murre. 2001. Regulation of the helix-loop-helix proteins, E2A and Id3, by the Ras-ERK MAPK cascade. Nat. Immunol. 2:165–171. [DOI] [PubMed] [Google Scholar]
- 22.Bettini, M., H. Xi, J. Milbrandt, and G.J. Kersh. 2002. Thymocyte development in early growth response gene 1-deficient mice. J. Immunol. 169:1713–1720. [DOI] [PubMed] [Google Scholar]
- 23.Rivera, R.R., C.P. Johns, J. Quan, R.S. Johnson, and C. Murre. 2000. Thymocyte selection is regulated by the helix-loop-helix inhibitor protein, Id3. Immunity. 12:17–26. [DOI] [PubMed] [Google Scholar]
- 24.Gao, E.K., D. Lo, R. Cheney, O. Kanagawa, and J. Sprent. 1988. Abnormal differentiation of thymocytes in mice treated with cyclosporin A. Nature. 336:176–179. [DOI] [PubMed] [Google Scholar]
- 25.Jenkins, M.K., R.H. Schwartz, and D.M. Pardoll. 1988. Effects of cyclosporine A on T cell development and clonal deletion. Science. 241:1655–1658. [DOI] [PubMed] [Google Scholar]
- 26.Chan, V.S., C. Wong, and P.S. Ohashi. 2002. Calcineurin Aα plays an exclusive role in TCR signaling in mature but not in immature T cells. Eur. J. Immunol. 32:1223–1229. [DOI] [PubMed] [Google Scholar]
- 27.Bueno, O.F., E.B. Brandt, M.E. Rothenberg, and J.D. Molkentin. 2002. Defective T cell development and function in calcineurin Aβ-deficient mice. Proc. Natl. Acad. Sci. USA. 99:9398–9403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hayden-Martinez, K., L.P. Kane, and S.M. Hedrick. 2000. Effects of a constitutively active form of calcineurin on T cell activation and thymic selection. J. Immunol. 165:3713–3721. [DOI] [PubMed] [Google Scholar]
- 29.Crabtree, G.R., and E.N. Olson. 2002. NFAT signaling: choreographing the social lives of cells. Cell. 109:S67–S79. [DOI] [PubMed] [Google Scholar]
- 30.Amasaki, Y., S. Adachi, Y. Ishida, M. Iwata, N. Arai, K. Arai, and S. Miyatake. 2002. A constitutively nuclear form of NFATx shows efficient transactivation activity and induces differentiation of CD4(+)CD8(+) T cells. J. Biol. Chem. 277:25640–25648. [DOI] [PubMed] [Google Scholar]
- 31.Wilkinson, B., J.Y. Chen, P. Han, K.M. Rufner, O.D. Goularte, and J. Kaye. 2002. TOX: an HMG box protein implicated in the regulation of thymocyte selection. Nat. Immunol. 3:272–280. [DOI] [PubMed] [Google Scholar]
- 32.Mombaerts, P., J. Iacomini, R.S. Johnson, K. Herrup, S. Tonegawa, and V.E. Papaioannou. 1992. RAG-1-deficient mice have no mature B and T lymphocytes. Cell. 68:869–877. [DOI] [PubMed] [Google Scholar]
- 33.Shinkai, Y., G. Rathbun, K.P. Lam, E.M. Oltz, V. Stewart, M. Mendelsohn, J. Charron, M. Datta, F. Young, A.M. Stall, et al. 1992. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell. 68:855–867. [DOI] [PubMed] [Google Scholar]
- 34.Mombaerts, P., A.R. Clarke, M.A. Rudnicki, J. Iacomini, S. Itohara, J.J. Lafaille, L. Wang, Y. Ichikawa, R. Jaenisch, M.L. Hooper, et al. 1992. Mutations in T-cell antigen receptor genes alpha and beta block thymocyte development at different stages. Nature. 360:225–231. [DOI] [PubMed] [Google Scholar]
- 35.Grusby, M.J., H. Auchincloss, Jr., R. Lee, R.S. Johnson, J.P. Spencer, M. Zijlstra, R. Jaenisch, V.E. Papaioannou, and L.H. Glimcher. 1993. Mice lacking major histocompatibility complex class I and class II molecules. Proc. Natl. Acad. Sci. USA. 90:3913–3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sha, W.C., C.A. Nelson, R.D. Newberry, D.M. Kranz, J.H. Russell, and D.Y. Loh. 1988. Positive and negative selection of an antigen receptor on T cells in transgenic mice. Nature. 336:73–76. [DOI] [PubMed] [Google Scholar]
- 37.Kaye, J., M.L. Hsu, M.E. Sauron, S.C. Jameson, N.R. Gascoigne, and S.M. Hedrick. 1989. Selective development of CD4+ T cells in transgenic mice expressing a class II MHC-restricted antigen receptor. Nature. 341:746–749. [DOI] [PubMed] [Google Scholar]
- 38.Murphy, K.M., A.B. Heimberger, and D.Y. Loh. 1990. Induction by antigen of intrathymic apoptosis of CD4+ CD8+TCRlo thymocytes in vivo. Science. 250:1720–1723. [DOI] [PubMed] [Google Scholar]
- 39.Hayashi, K., N. Abe, T. Watanabe, M. Obinata, M. Ito, T. Sato, S. Habu, and M. Satake. 2001. Overexpression of AML1 transcription factor drives thymocytes into the CD8 single-positive lineage. J. Immunol. 167:4957–4965. [DOI] [PubMed] [Google Scholar]
- 40.Godfrey, D.I., J. Kennedy, T. Suda, and A. Zlotnik. 1993. A developmental pathway involving four phenotypically and functionally distinct subsets of CD3−CD4−CD8− triple-negative adult mouse thymocytes defined by CD44 and CD25 expression. J. Immunol. 150:4244–4252. [PubMed] [Google Scholar]
- 41.Fehling, H.J., A. Krotkova, C. Saint-Ruf, and H. von Boehmer. 1995. Crucial role of the pre-T-cell receptor alpha gene in development of alpha beta but not gamma delta T cells. Nature. 375:795–798. [DOI] [PubMed] [Google Scholar]
- 42.Shinkai, Y., and F.W. Alt. 1994. CD3 epsilon-mediated signals rescue the development of CD4+CD8+ thymocytes in RAG-2−/− mice in the absence of TCR beta chain expression. Int. Immunol. 6:995–1001. [DOI] [PubMed] [Google Scholar]
- 43.Carbone, A.M., P. Marrack, and J.W. Kappler. 1988. Demethylated CD8 gene in CD4+ T cells suggests that CD4+ cells develop from CD8+ precursors. Science. 242:1174–1176. [DOI] [PubMed] [Google Scholar]
- 44.Pestano, G.A., Y. Zhou, L.A. Trimble, J. Daley, G.F. Weber, and H. Cantor. 1999. Inactivation of misselected CD8 T cells by CD8 gene methylation and cell death. Science. 284:1187–1191. [DOI] [PubMed] [Google Scholar]
- 45.Swat, W., M. Dessing, H. von Boehmer, and P. Kisielow. 1993. CD69 expression during selection and maturation of CD4+8+ thymocytes. Eur. J. Immunol. 23:739–746. [DOI] [PubMed] [Google Scholar]
- 46.Ramsdell, F., M. Jenkins, Q. Dinh, and B.J. Fowlkes. 1991. The majority of CD4+8− thymocytes are functionally immature. J. Immunol. 147:1779–1785. [PubMed] [Google Scholar]
- 47.Sawada, S., J.D. Scarborough, N. Killeen, and D.R. Littman. 1994. A lineage-specific transcriptional silencer regulates CD4 gene expression during T lymphocyte development. Cell. 77:917–929. [DOI] [PubMed] [Google Scholar]
- 48.Groves, T., M. Parsons, N.G. Miyamoto, and C.J. Guidos. 1997. TCR engagement of CD4+CD8+ thymocytes in vitro induces early aspects of positive selection, but not apoptosis. J. Immunol. 158:65–75. [PubMed] [Google Scholar]
- 49.Woolf, E., C. Xiao, O. Fainaru, J. Lotem, D. Rosen, V. Negreanu, Y. Bernstein, D. Goldenberg, O. Brenner, G. Berke, et al. 2003. Runx3 and Runx1 are required for CD8 T cell development during thymopoiesis. Proc. Natl. Acad. Sci. USA. 100:7731–7736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hayashi, K., W. Natsume, T. Watanabe, N. Abe, N. Iwai, H. Okada, Y. Ito, M. Asano, Y. Iwakura, S. Habu, et al. 2000. Diminution of the AML1 transcription factor function causes differential effects on the fates of CD4 and CD8 single-positive T cells. J. Immunol. 165:6816–6824. [DOI] [PubMed] [Google Scholar]
- 51.D'Ambrosio, D., D.A. Cantrell, L. Frati, A. Santoni, and R. Testi. 1994. Involvement of p21ras activation in T cell CD69 expression. Eur. J. Immunol. 24:616–620. [DOI] [PubMed] [Google Scholar]
- 52.Gounari, F., I. Aifantis, K. Khazaie, S. Hoeflinger, N. Harada, M.M. Taketo, and H. von Boehmer. 2001. Somatic activation of beta-catenin bypasses pre-TCR signaling and TCR selection in thymocyte development. Nat. Immunol. 2:863–869. [DOI] [PubMed] [Google Scholar]
- 53.Lee, P.P., D.R. Fitzpatrick, C. Beard, H.K. Jessup, S. Lehar, K.W. Makar, M. Perez-Melgosa, M.T. Sweetser, M.S. Schlissel, S. Nguyen, et al. 2001. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 15:763–774. [DOI] [PubMed] [Google Scholar]
- 54.Adlam, M., and G. Siu. 2003. Hierarchical interactions control CD4 gene expression during thymocyte development. Immunity. 18:173–184. [DOI] [PubMed] [Google Scholar]
- 55.Thomas, J.O., and A.A. Travers. 2001. HMG1 and 2, and related ‘architectural’ DNA-binding proteins. Trends Biochem. Sci. 26:167–174. [DOI] [PubMed] [Google Scholar]
- 56.Bartfeld, D., L. Shimon, G.C. Couture, D. Rabinovich, F. Frolow, D. Levanon, Y. Groner, and Z. Shakked. 2002. DNA recognition by the RUNX1 transcription factor is mediated by an allosteric transition in the RUNT domain and by DNA bending. Structure (Camb). 10:1395–1407. [DOI] [PubMed] [Google Scholar]
- 57.Wang, J.H., A. Nichogiannopoulou, L. Wu, L. Sun, A.H. Sharpe, M. Bigby, and K. Georgopoulos. 1996. Selective defects in the development of the fetal and adult lymphoid system in mice with an Ikaros null mutation. Immunity. 5:537–549. [DOI] [PubMed] [Google Scholar]
- 58.Winandy, S., L. Wu, J.H. Wang, and K. Georgopoulos. 1999. Pre-T cell receptor (TCR) and TCR-controlled checkpoints in T cell differentiation are set by Ikaros. J. Exp. Med. 190:1039–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim, J., S. Sif, B. Jones, A. Jackson, J. Koipally, E. Heller, S. Winandy, A. Viel, A. Sawyer, T. Ikeda, et al. 1999. Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity. 10:345–355. [DOI] [PubMed] [Google Scholar]
- 60.Brown, K.E., S.S. Guest, S.T. Smale, K. Hahm, M. Merkenschlager, and A.G. Fisher. 1997. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell. 91:845–854. [DOI] [PubMed] [Google Scholar]
- 61.Wang, W., T. Chi, Y. Xue, S. Zhou, A. Kuo, and G.R. Crabtree. 1998. Architectural DNA binding by a high-mobility-group/kinesin-like subunit in mammalian SWI/SNF-related complexes. Proc. Natl. Acad. Sci. USA. 95:492–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.O'Flaherty, E., and J. Kaye. 2003. TOX defines a conserved subfamily of HMG-box proteins. BMC Genomics. 4:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Travers, A.A. 2003. Priming the nucleosome: a role for HMGB proteins? EMBO Rep. 4:131–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hernandez-Hoyos, G., M.K. Anderson, C. Wang, E.V. Rothenberg, and J. Alberola-Ila. 2003. GATA-3 expression is controlled by TCR signals and regulates CD4/CD8 differentiation. Immunity. 19:83–94. [DOI] [PubMed] [Google Scholar]