Abstract
Mycolic acids represent a major component of the unique cell wall of mycobacteria. Mycolic acid biosynthesis is inhibited by isoniazid, a key frontline antitubercular drug that is inactivated by mycobacterial and human arylamine N-acetyltransferase (NAT). We show that an in-frame deletion of Mycobacterium bovis BCG nat results in delayed entry into log phase, altered morphology, altered cell wall lipid composition, and increased intracellular killing by macrophages. In particular, deletion of nat perturbs biosynthesis of mycolic acids and their derivatives and increases susceptibility of M. bovis BCG to antibiotics that permeate the cell wall. Phenotypic traits are fully complemented by introduction of Mycobacterium tuberculosis nat. We infer from our findings that NAT is critical to normal mycolic acid synthesis and hence other derivative cell wall components and represents a novel target for antituberculosis therapy. In addition, this is the first report of an endogenous role for NAT in mycobacteria.
Keywords: isoniazid, macrophage, Mycobacterium tuberculosis, cell wall, metabolism
Introduction
Mycobacterium tuberculosis, the etiological agent of tuberculosis, is responsible for two to three million deaths per year, worldwide (1, 2). Chemotherapy is available for tuberculosis, but requires an extremely long, complex multiple therapy regimen to resolve infection (3). The length of this course leads to high rates of patient noncompliance, suspected to account for the increasing number of drug-resistant clinical isolates of M. tuberculosis now observed (4). Additional antituberculosis drugs are now urgently required both to treat organisms already resistant to existing therapeutics, and to limit the emergence of drug resistance by use with current treatment regimens to shorten the course of therapy.
M. tuberculosis belongs to the genus Mycobacterium, characterized by a unique cell wall rich in unusual glycolipids, polysaccharides, and lipids, including mycolic acids. Biosynthetic pathways of cell wall components have proved to be effective targets for several major antitubercular drugs, including the frontline drug isoniazid (INH), which inhibits mycolic acid biosynthesis. Mycolic acids are large, α alkyl β hydroxy fatty acids with the general structure of R1-CH(OH)-CH(R2)-COOH, where R1 is a meromycolate chain typically containing 50–56 carbons and R2 is a shorter α branch containing 22–26 carbons. They constitute the inner leaflet of the lipid bilayer of the mycobacterial cell wall and form an effective barrier to the penetration of antibiotics and chemotherapeutic agents (5). The effect of INH on cell wall and mycolic acid synthesis is long established (6–13), although there has been controversy over the molecular targets of INH. We recently reported arylamine N-acetyltransferase (NAT) of M. tuberculosis is a modifier of INH involved in mediating INH resistance (14,15).
Genes encoding NAT are present in a range of bacterial genomes (15, 16). This observation first provoked intrigue because human NATs have long been identified as drug metabolizing enzymes (17). NAT represents one of the first examples of pharmacogenetic variation and its study revealed the role of acetyl-CoA as an acetyl donor. In particular, NAT2 in humans is known to be responsible for the inactivation of INH through acetylation (18–20). We have studied mycobacterial NATs in this laboratory as potential contributors to the variation in INH resistance among M. tuberculosis clinical isolates. It is now known that nat in M. tuberculosis is polymorphic (14). The expression product acetylates and inactivates INH in vitro, and it has been suggested that this activity and polymorphism might be a contributory factor to INH resistance (14, 15, 21). We know that nat is expressed in M. tuberculosis and Mycobacterium bovis BCG and the gene product is active (14). The genomes of M. tuberculosis (22) and M. bovis (23) have been sequenced. M. bovis is a member of the M. tuberculosis complex. M. bovis BCG is an attenuated M. bovis strain in use as a vaccine. The nat gene is maintained in M. bovis BCG and is identical in sequence to that of M. tuberculosis (14–16, 22, 23).
An endogenous role for the N-acetylation activity of NAT has not been reported for any mycobacterial species and to this end we have generated an in-frame deletion of nat in M. bovis BCG. We examined the growth, cell morphology, and extractable cell wall lipid composition of the resulting knockout strain. The most important finding is that NAT is essential for normal mycolic acid synthesis in M. bovis BCG, suggesting that mycolic acid biosynthesis involves an as yet unidentified pathway involving NAT. In addition, loss of NAT activity resulted in increased intracellular killing of M. bovis BCG by macrophages. We were able to restore M. bovis BCG wild-type phenotype via functional complementation with M. tuberculosis nat, indicating that NAT has an endogenous role within mycobacteria. We propose that NAT, with its crucial role in mycolic acid biosynthesis, represents a novel antituberculosis drug target.
Materials and Methods
Bacterial Strains and Culture Conditions.
M. bovis BCG Pasteur and genetically modified strains, including the Δnat complemented with nat in pACE1 (24), were cultured at 37°C in roller bottles with rotation at two revolutions per minute in Middlebrook 7H9 liquid medium containing 10% (vol/vol) albumin-dextrose-catalase (ADC; Difco) and 0.05% (vol/vol) Tween 80 (Sigma-Aldrich), and on Middlebrook 7H10 agar plus 10% (vol/vol) oleic acid–ADC (OADC; Difco), unless otherwise stated. Cultures were harvested from log phase at an OD600 of 0.6–1.2. NAT activity in cell lysates was determined after HPLC analysis as previously described (14).
Generation of nat Knockout.
M. bovis BCG Pasteur with an in-frame unmarked deletion of the nat open reading frame (ORF) was generated by homologous recombination, using plasmids and methods previously described (24, 25). The suicide construct comprised p2NIL, homology arms of ∼1 Kb flanking the M. bovis BCG nat ORF, and a selectable marker cassette from pGOAL19. Preparation and transformation of M. bovis BCG electrocompetent cells and selection of the knockout strain were performed exactly as described (25). For the resultant strain, the nat ORF (Rv3566c) and the five upstream ORFs (Rv3566A, Rv3567c, Rv3568c, Rv3569c, and Rv3570c) in the putative nat operon (16) were amplified using pFU DNA polymerase (Promega) with gene-specific primers (Rv3566c nat: 5′-GAC GAG GTC AGA ATG GCA AC-3′ and 5′-GGG GTT CGT TTG TTC GGA TA-3′; Rv3566A: 5′-GTGTCCGGCGCCGAT-3′ and 5′-TCAGATCCAGTGCCATGTTGC-3′; Rv3567c: 5′-ATGTCGGCTCAGATCGATCC-3′ and 5′-CTAGAGCCAGGTGTCCTGG-3′; Rv3568c: 5′-TGAGCATCCGGTCGCTG-3′ and 5′-CTAGCCGCGAGCGCCTAC-3′; Rv3569c: 5′-ATGACAGCTACCGAGGAATTG-3′ and 5′-TCATCTGCCACCTCCCAG-3′; Rv3570c: 5′-GTGACGTCCCATTCAACAGCG-3′ and 5′-TAGACCATGGTGTCGCCG-3′). DMSO at 6% (vol/vol) was added to all reactions. Mycobacterial cells were denatured at 95°C for 10 min as an extra cycle before the addition of the enzyme mix. The PCR cycle was repeated 30 times. The DNA sequences of the PCR products were confirmed by automated sequencing (Biochemistry Department, DNA Sequencing Facility, Oxford University).
Complementation of M. bovis BCG Δnat with M. tuberculosis nat.
M. tuberculosis nat cloned into the Escherichia coli mycobacterial shuttle expression vector pACE1, under control of the inducible acetamidase promoter (15, 24), was used to complement M. bovis BCG Δnat. Preparation and transformation of electrocompetent M. bovis BCG cells with the construct were as previously described, with selection of transformants on 50 μg/ml 7H10 OADC agar-containing hygromycin. Cultures of M. bovis BCG Δnat complemented with nat in pACE1 were initially maintained in minimal medium induced with 2 mg/ml acetamide (15) and the log phase was reached after 7 d. For experiments in which growth characteristics were compared with other strains, the complemented strain was grown in liquid culture in 7H9 Middlebrook medium, under which conditions the acetamide promoter is known to drive basal level expression (26).
Western Blotting.
We performed SDS-PAGE and Western blotting as previously described using rabbit antiserum raised against recombinant M. tuberculosis NAT as first antibody (21) at a dilution of 1:10,000.
Electron Microscopy.
Transmission electron microscopy (TEM) was performed as previously described (27) and viewed on a Philips EM410 transmission electron microscope. Digital images were taken using the Gatan multiscan Camera, model 791.
Scanning electron microscopy (SEM) was performed on polylysine-coated sterile coverslips kept under PBS buffer within a 24-well plate. A drop of concentrated bacterial culture was placed on the surface, washed three times after 1 h with PBS, and then fixed with 3% glutaraldehyde and stained with 1% osmium tetroxide. Critical point drying was performed after a serial wash with 75, 85, and 95%, and twice in absolute alcohol and dry alcohol (kept under CuSO4). The sample was coated with platinum vapor and observed by SEM (28).
Preparation and Analysis of Polar and Nonpolar Lipids, and Mycolic Acids.
100 ml roller cultures were harvested at mid-exponential phase (OD600 = 1.0) for each strain. The CFU values for each culture were determined separately to confirm equivalence of biomass. The complex lipids and mycolic acids were extracted and the same proportion of each culture was loaded onto thin layer chromatography (TLC) plates as previously described (29) to allow direct visual comparison between the different strains. Identification of components was by comparison with authenticated standards.
Drug Susceptibility/Sensitivity Assay.
M. bovis BCG Pasteur and Δnat strains were grown to mid-exponential phase (OD600 = 1.0) and serially diluted using fresh liquid medium (7H9 ADC Tween 80) and spotted at 102 cells/well in six-well plates with or without antibiotics in 5 ml 7H10 OADC agar. Growth was observed after 14 d of incubation at 37°C. Mid-exponential phase cultures were dispensed and grown in 7H9 Middlebrook medium containing ADC and INH from 0 to 0.3 μg/ml in 96-well plates until cultures without INH reached the late exponential phase (4 d). The growth rate was followed by measuring the turbidity at 600 nm. The relative growth rate is expressed as a fraction of the OD of the untreated wild-type cells (21).
Infectivity and Killing Assay with Macrophages.
Monolayers of the mouse macrophage cell line RAW 264.7 were grown in RPMI 1640 with 10% FBS and plated on coverslips in multiwell plates for 3–4 h before infection (30). Cells were washed three times with OPTIMEM and labeled with TOPRO3 (Molecular Probe) as previously described (31). Elicited macrophage were prepared as previously described (32). M. bovis BCG Pasteur or Δnat cells were harvested at mid-log phase, washed, and resuspended in PBS. One half of the mycobacterial cells were washed and resuspended in OPTIMEM for infection and killing assays, and the remainder were labeled with 500 μg/ml FITC in NaHCO3, pH 8.5. For opsonization, mycobacteria were incubated at 37°C for 30 min with human serum (4:1 vol/vol), and used immediately. All solutions contained 0.01% Tween 80 (Sigma-Aldrich).
Infection Assay.
Bacteria were added to macrophages in a ratio of 10:1 for fluorimetry, FACS® analysis, and microscopy. Cultures were incubated for 2 h at 37°C, washed three times in PBS, and fixed with 4% paraformaldehyde. Cells were either stained with ZN (Tb-color kit; Bund Deutscher Hebammen Laboratory) for light microscopy or with TOPRO 3 for 1 h at room temperature after permeabilization with 1% Triton X-100 (31) and washing, before FACS® analysis. For FACS® and fluorimetry, cells were recovered by scraping. Protein was measured using the Bradford Colorimetric Assay (Sigma-Aldrich).
Killing Assay.
After infection with M. bovis BCG Pasteur and the corresponding Δnat mutant in 16-well plates for 2 h, macrophage were washed thoroughly with OPTIMEM with 0.01% Tween, incubated at 37°C for 2 h, 3 d, or 7 d, and then washed with PBS with 0.01% Tween 80. After discarding the medium, cells were lysed in distilled water at room temperature for 10 min, and dilutions were incubated on Middlebrook 7H10 OADC agar at 37°C for 28 d to determine the CFUs.
Results
Generation of M. bovis BCG Pasteur Δnat by Allelic Exchange.
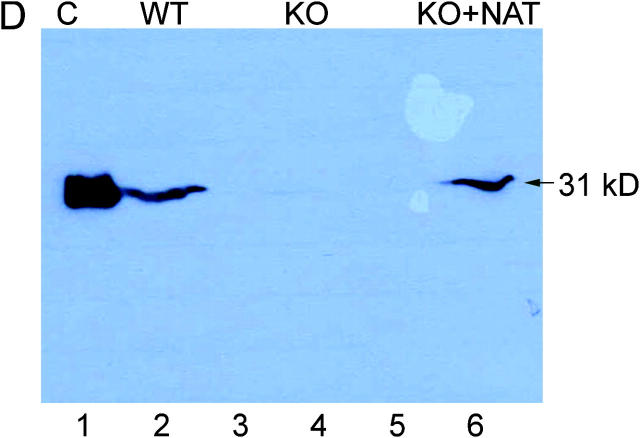
M. bovis BCG with an in-frame unmarked deletion of the nat gene (M. bovis BCG Δnat) was generated using homologous recombination (25). Deletion of nat in M. bovis BCG Δnat was confirmed by PCR, Southern blotting, and sequencing of the vestigial nat sequence. We also confirmed the integrity of the adjacent genes by sequencing and PCR. The effect of deleting nat was confirmed by NAT activity, which fell from 17.2 pmoles INH acetylated/min/mg protein to an undetectable level in lysates of the M. bovis BCG Δnat strain. Western blotting using an antibody that specifically recognizes NAT in cell lysates of M. tuberculosis and M. bovis BCG (fig. 1 D) showed NAT to be clearly present in the parental strain, but we do not detect NAT protein in the lysate of M. bovis BCG Δnat. We introduced the E. coli mycobacterial shuttle expression vector pACE1 (24) containing M. tuberculosis nat (14) into M. bovis BCG Δnat to generate a complemented M. bovis BCG Δnat strain. The level of expression of NAT in the complemented strain as determined by Western blotting appears to be similar to the wild-type strain (Fig. 1 D).
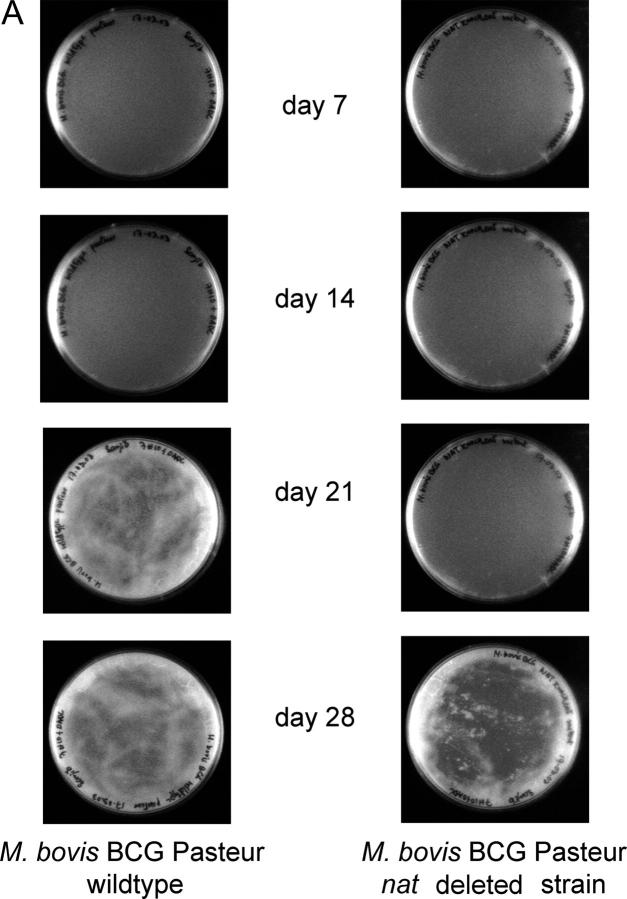
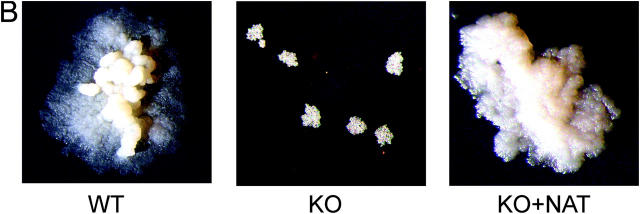
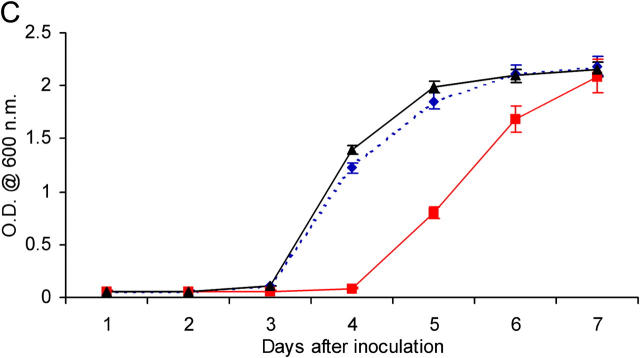
Figure 1.
Deleting the nat gene affects the growth of M. bovis BCG Pasteur. (A) Growth of M. bovis BCG and M. bovis BCG Δnat over a 28-d period on solid medium. Colonies of M. bovis BCG are visible by day 21 compared with day 28 for the Δnat strain (magnification, 0.2-fold). (B) Colonies of the Δnat strain (KO) are smaller than the corresponding colonies of the M. bovis BCG (WT) and complemented strain (KO+NAT; magnification, 10-fold). (C) When M. bovis BCG (▴), M. bovis BCG Pasteur Δnat (▪), and nat complemented strains (♦) are grown in liquid culture (7H9 ADC and Tween 80), growth of the M. bovis BCG Δnat strain is altered such that the lag phase is extended and it is restored to the wild-type phenotype when the nat gene is reintroduced. (D) Lysates of cells harvested at mid-log phase were run on SDS-PAGE. In each lane, lysate corresponding to 10 ml culture was loaded. Western blots were developed with specific antibodies against recombinant M. tuberculosis NAT (reference 21) used at a 1:10,000 dilution. Lane 1, pure recombinant NAT (reference 14) as standard (C); lane 2, M. bovis BCG (WT); lanes 3 and 5, Rainbow Molecular Weight Markers (Amersham Biosciences); lane 4, M. bovis BCG Δnat (KO); lane 6, M. bovis BCG Δnat complemented with nat (KO+NAT).
M. bovis BCG Pasteur Δnat Is Defective for Entry into Exponential Growth Phase.
The growth of M. bovis BCG is slower when nat is deleted. This is observed both on solid agar and in liquid cultures. On agar, colonies in which the nat gene is deleted appear at day 28, as opposed to day 21 for the parental strain (Fig. 1 A). Growth curves in liquid culture reveal that the strain with the nat gene deleted has an increased lag phase (Fig. 1 C), whereas its exponential growth rate mirrors that of the parental strain. In the complemented strain, the growth is restored to that of the wild-type, consistent with the restoration of NAT protein.
M. bovis BCG Pasteur Δnat Has Altered Cellular Morphologies.
We observed, initially by eye, that colonies of M. bovis BCG Δnat appear smaller than their parental strain counterparts (Fig. 1, A and B). This was confirmed by light microscopy; the knockout colonies appear smaller. We attempted to quantify this morphological change using SEM and TEM. From SEM, the individual cells of M. bovis BCG Δnat are significantly (P < 0.05) smaller (Fig. 2 C) than those of the parental cells (M. bovis BCG, total area 5.47 ± 0.84 μ2; M. bovis BCG Δnat, total area 3.97 ± 0.65; average ± SD, n = 40).
Figure 2.
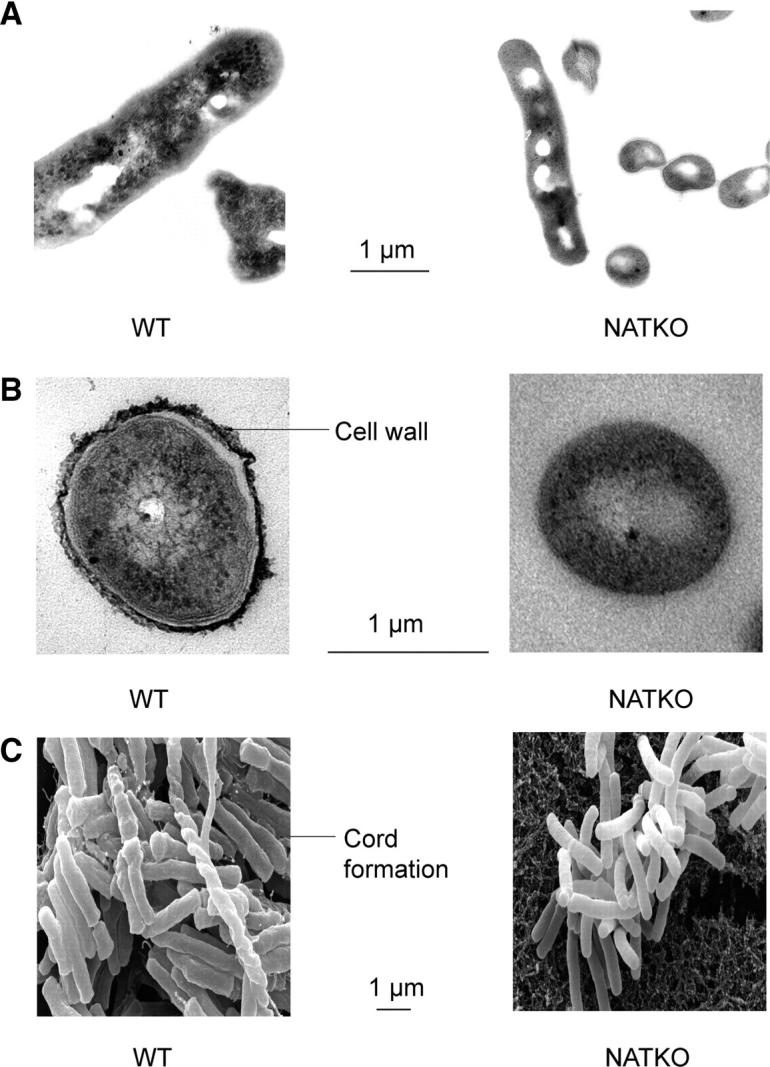
Morphology and ultrastructure of individual M. bovis BCG Pasteur cells are modified when the nat gene is deleted. Longitudinal (A) and transverse (B) TEM images show that the size of M. bovis BCG is altered on deleting the nat gene. The outer cell wall (arrow), present in M. bovis BCG, is absent in the Δnat strain. SEM (C) also shows the difference in size of the bacilli. Cord formation (arrow) in M. bovis BCG is missing when the nat gene is deleted. Bar is 1 μm for all frames.
TEM also reveals the much smoother surface of the M. bovis BCG Δnat cells compared with those of the parental strain (Fig. 2, A and B). Moreover, it is clear from the scanning electron micrographs that the M. bovis BCG Δnat strain lacks cord formation, which is clearly visible in the parental cells (Fig. 2 C).
NAT Is Required for Normal Synthesis of Mycolic Acids, Complex Lipids, and Glycolipids in M. bovis BCG Pasteur.
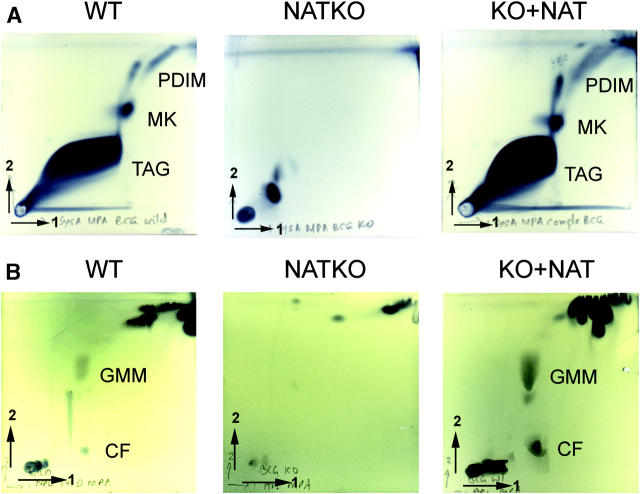
The effects of deleting nat on the morphology of the individual cells suggest that the cell wall might be altered. Therefore, we compared the lipid composition of the parental and M. bovis BCG Δnat cells at the same stage of the growth cycle. These comparisons show a distinct difference in the total lipid composition of the M. bovis BCG Δnat strain (Fig. 3, A–C). The mutant appeared to have very reduced quantities of a number of complex lipids and glycolipids, which were identified in the wild-type as phthiocerol dimycocerosate (PDIM), menaquinone (MK), glucose monomycolate (GMM), and trehalose dimycolate or cord factor (CF) using two-dimensional TLC (Fig. 3, A and B). We further analyzed the delipidated cell walls to compare bound mycolic acids (Fig. 3 C). There appears to be very little mycolic acid present within M. bovis BCG Δnat, either in extractable lipids or bound to the cell wall (Fig. 3, A and B). However, the presence of normal mycolic acid production and complex lipid patterns is restored on complementation with M. tuberculosis nat. The cultures used for mycolic acid analyses were plated out to ensure no contamination by nonmycobacterial organisms had occurred. It is these cultures that are illustrated in Fig. 1 A. We see no visible difference between the Δnat and parental strain among other extractable components of the cell wall, particularly the phospholipids (Fig. 3 E), although the amount of extractable fatty acid is slightly diminished in the M. bovis BCG Δnat strain (Fig. 3 D).
Figure 3.
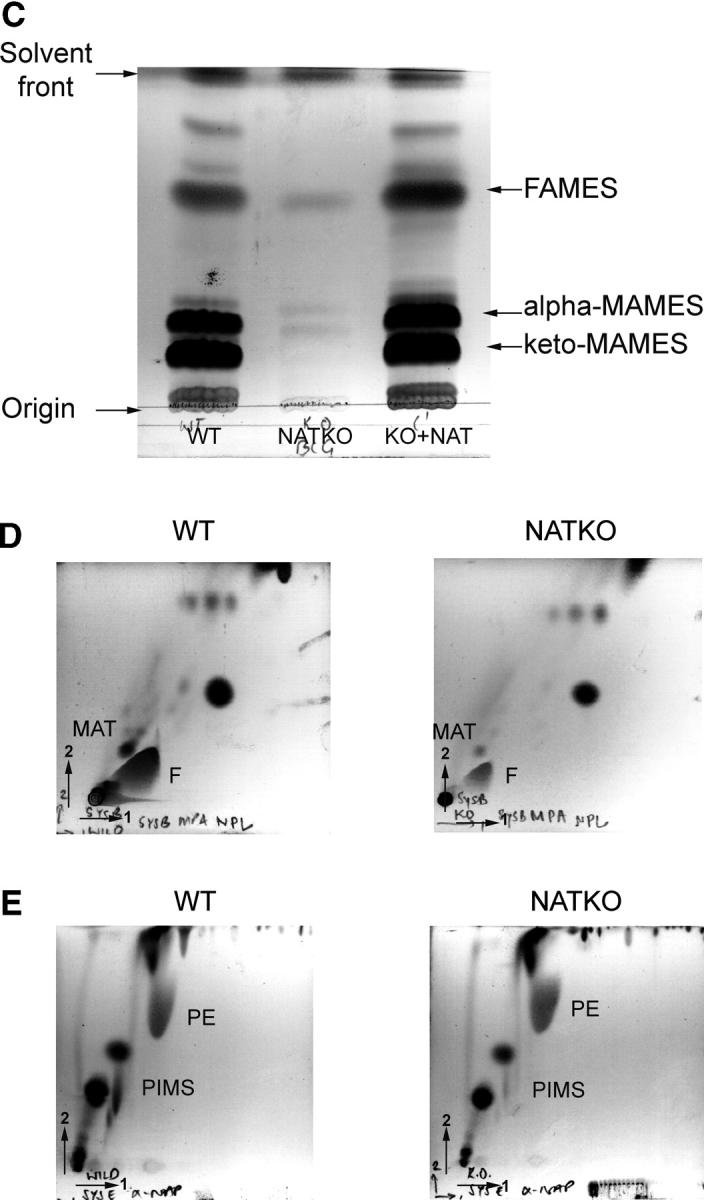
Total lipid and mycolate profile of M. bovis BCG Pasteur is changed when the nat gene is deleted. Analysis of total lipids by two-dimensional TLC shows that tri-acyl glycerol (TAG), MK, and PDIM (A), as well as CF and GMM (B), are present in M. bovis BCG (WT), but missing from the corresponding Δnat BCG strain (NAT KO). All of these complex lipids are restored by complementation with nat (KO+NAT). Separation of mycolates from the same dry weight of delipidated cells by one-dimensional TLC from WT, NATKO, and KO+NAT, shows that the synthesis of mycolate in WT is perturbed by deletion of the nat gene and is fully restored when the M. tuberculosis nat gene is introduced (C). Analyses of fatty acids and multi-acetylated trehaloses (D) and of phospholipids (E) shows very little change between the M. bovis BCG (WT) and Δnat BCG strain (NAT KO). Analyses were performed using the same biomass of the parental and mutant cultures. TAG, tri-acyl glycerol; MK, menaquinone; PDIM, pthiocerol dimycocerosate; CF, cord factor; GMM, glucose monomycolate; MAMES, mycolic acid methyl esters; FAMES, fatty acid methyl esters; MAT, multi-acylated trehaloses; F, Fatty acids; PIMs, phosphatidyl-inositol mannosides; PE, phosphatidyl ethanolamines.
M. bovis BCG Pasteur Δnat Is More Sensitive toward Intracellular Killing within Macrophages.
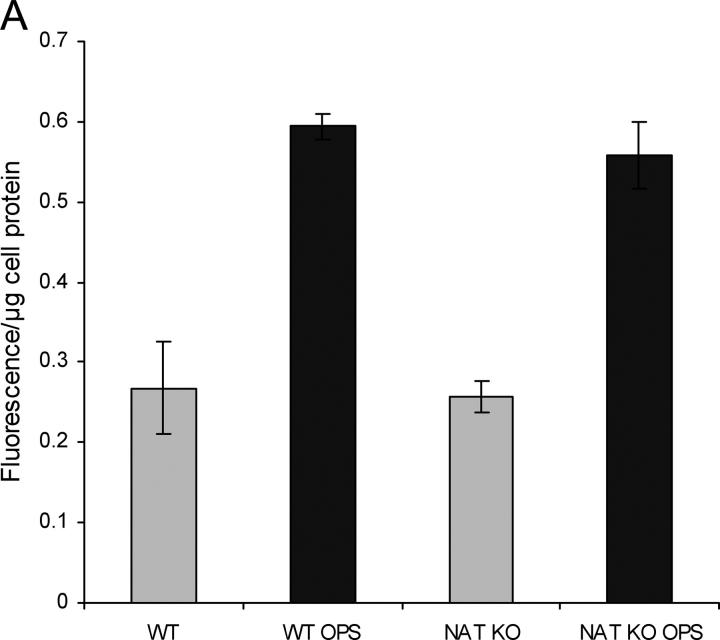
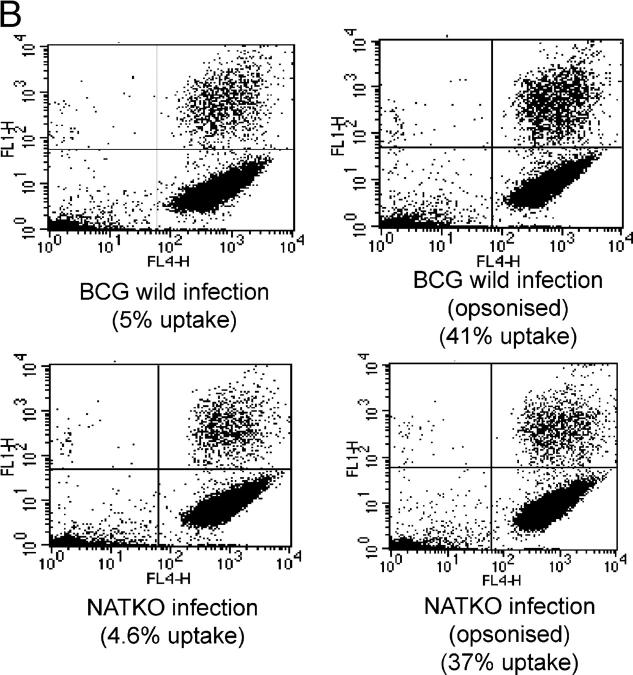
When unopsonized M. bovis BCG or M. bovis BCG Δnat bacilli are incubated with the mouse macrophage cell line (RAW 264.7), we observed no difference in cellular uptake (Fig. 4, A and B) into macrophages. Experiments with elicited macrophages showed the same results. However, intracellular M. bovis BCG Δnat bacilli are killed after 3 d, whereas the wild-type strain is not (Fig. 4 C). Opsonization does not appear to alter the marked difference in intracellular killing, although after opsonization there is a fivefold increase in the number of M. bovis BCG cells taken up for both the M. bovis BCG and M. bovis BCG Δnat bacilli.
Figure 4.
Deleting the nat gene affects the intracellular killing of M. bovis BCG Pasteur in macrophages. Infection of mouse macrophage cell line RAW showing that the uptake of M. bovis BCG (WT) and M. bovis BCG Δnat (NATKO) into RAW cells is the same, and there is an equal increase in uptake for both M. bovis BCG and the corresponding Δnat mutant after opsonization (OPS). (A) Fluorimetric assay. (B) FACS®. Upper right quadrants of B show RAW cells that have taken up FITC-BCG. (C) Intracellular killing assay. The mouse macrophage cell line RAW was infected and samples were taken at the times indicated, plated on agar, and CFUs were counted, showing that M. bovis BCG with and without opsonization can survive and is able to grow within macrophages after 7 d, whereas M. bovis BCG Δnat with and without opsonization are killed between 2 and 72 h after infection. ▴, wild-type strain; ▪, Δnat strain. Solid lines are opsonized and dotted lines are unopsonized.
M. bovis BCG Pasteur Δnat Is More Susceptible to Antibiotics.
We reasoned that the change in the composition of the cell wall of M. bovis BCG Δnat strain would result in greater accessibility of antibiotics. We show this is the case using the antibiotics hygromycin and gentamycin. These antibiotics are approximately one order of magnitude more effective in the strains with the nat gene deleted (Table I). Studies with β lactam antibiotics show that deletion of nat increases susceptibility only marginally.
Table I.
Knocking out nat Affects Sensitivity of M. bovis BCG Pasteur to a Variety of Antibiotics
| Ampicillin
|
Cloxacillin
|
Carbenicillin
|
Gentamycin
|
Hygromycin
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Concentration | WT | KO | WT | KO | WT | KO | WT | KO | WT | KO |
| Control | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| 5 μg/ml | +++ | +++ | +++ | +++ | +++ | +++ | +++ | + | +++ | + |
| 10 μg/ml | +++ | +++ | +++ | +++ | +++ | +++ | +++ | − | +++ | − |
| 20 μg/ml | +++ | ++ | +++ | +++ | +++ | +++ | + | − | +++ | − |
| 30 μg/ml | +++ | ++ | +++ | ++ | +++ | +++ | + | − | + | − |
| 40 μg/ml | ++ | − | ++ | − | +++ | + | + | − | + | − |
| 50 μg/ml | − | − | − | − | + | − | + | − | − | − |
M. bovis BCG (WT) and M. bovis BCG with nat deleted (KO) were plated at 102 cells/well on 7H10 OADC agar-containing antibiotics as indicated, in six-well plates. Cell growth was recorded after 14 d at 37°C. Good growth (+++), poor growth (+), and no visible growth (−) are indicated.
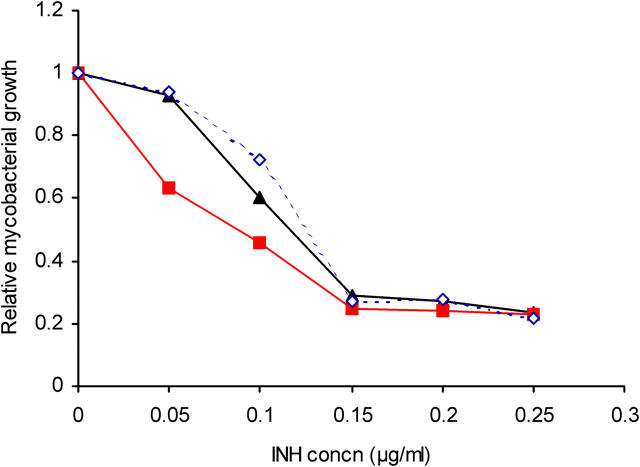
The M. bovis BCG Δnat strain is twofold more sensitive to INH than the parental strain of M. bovis BCG Pasteur (Fig. 5). The growth of the Δnat strain was inhibited at a twofold lower INH concentration than the wild-type, whereas the complemented strain was indistinguishable from the wild-type in its INH sensitivity.
Figure 5.
Deleting nat affects sensitivity of M. bovis BCG Pasteur to INH. Mid-log phase cultures of M. bovis BCG (▴), M. bovis BCG Δnat (▪), and M. bovis BCG Δnat complemented with nat (♦) were grown in the presence of differing concentrations of INH. The growth was determined after 4 d and is expressed relative to the corresponding strain cultured without INH. Mean values of three determinations are shown and the SD is within the symbol.
Discussion
To investigate the endogenous role of NAT in mycobacteria, we deleted the nat gene from M. bovis BCG Pasteur and observed phenotypic changes on (a) growth, (b) ultrastructure and cell morphology, (c) cell wall lipid composition, and (d) intracellular killing of M. bovis BCG Pasteur by mouse macrophages. Deletion of nat affects growth of M. bovis BCG Pasteur on plates and retards the growth in liquid culture by increasing the duration of the lag phase. It has been demonstrated that the nat gene in certain clinical isolates of M. tuberculosis (from amongst a related group of isolates that share a particular IS1160 genotype and are known as family 28) harbor a mutation (33) in the nat gene that renders the NAT protein less active (14). Observations on the growth of these clinical isolates have repeatedly demonstrated that they grow exceptionally slowly on agar and in liquid culture. Although the members of family 28 harbor other mutations, these data indicate that NAT activity may influence growth of M. tuberculosis as well as of M. bovis BCG. We have recently generated a strain of M. tuberculosis H37Rv in which the nat gene has been deleted and have observed that the effect on colony morphology mirrors our results on the effect of deleting the nat gene in M. bovis BCG.
The ultrastructure of M. bovis BCG Pasteur is affected when nat is deleted and the cell wall is greatly diminished as observed in TEM. Biochemical analysis of the cell wall lipids clearly shows that the M. bovis BCG Δnat mutant has less mycolic acids and other mycolic acid derivatives like GMM and CF as well as other complex lipids, such as PDIM and MK. Consistent with the lack of CF, we have found cord formation is absent in M. bovis BCG Δnat mutant from ultrastructural analysis by SEM. Studies of these cell wall components of mycobacteria are also consistent with the biochemical findings. PDIM, which is found only in pathogenic mycobacteria, is required for growth in the lungs of mice and is associated with virulence (34). Mycobacterial GMM is a target of the human immune response to mycobacteria (35) and CF is able to stimulate innate, early adaptive, and both humoral and cellular adaptive immunity (36), and induces prolonged mycobacterial survival (37). MK is a component of the respiratory chain of mycobacteria. The effects of environmental conditions on the structure and function of the respiratory chain are beginning to be understood (38). MK appears to be essential (39) and its level in the Δnat strain might be below the level of detection in the experiments presented here. We have found that the changes in mycolates and associated complex lipids from the cell wall is associated with increased sensitivity of mycobacteria to intracellular killing by mouse macrophages, regardless of opsonization. Dilapidation of M. tuberculosis similarly affects survival in bone marrow–derived macrophages (40). It is not certain how a decrease in the content of complex lipids contributes to the observed phenotype or whether the pattern of lipid changes is a primary defect or a secondary effect as a result of interference of normal mycolic acid synthesis, for example through shedding. It is possible that NAT has a role in maintaining homeostasis of acetyl-CoA, a central metabolite in lipid synthesis and hence its lack would affect different synthetic pathways. In this context, it has been observed that a truncation mutant of NAT that catalyses hydrolysis of acetyl-CoA is toxic to E. coli (41). The effects of deleting the nat gene on lipid biosynthesis in M. bovis BCG is not universal. Although mycolic acids and their methyl esters are greatly affected, fatty acids are only diminished slightly. These data suggest that NAT may play a role in extension of the chain length of fatty acids before esterification. The biochemical mechanisms of extension of fatty acyl chains is dependent on chain length (11) and in this context it might be significant that in preliminary experiments, we have found NAT associated with the cell membrane fraction under certain growth conditions.
INH is a substrate of NAT and hence INH is indisputably a NAT ligand (42). We have obtained a three-dimensional crystallographic structure of NAT with INH bound (unpublished data). The precise target of INH in mycobacteria leading to inhibition of mycolic acid synthesis has been the subject of continued debate (43, 44). Studies that we have performed with pure proteins suggest that KatG and NAT compete for INH, supporting the interpretation that NAT acts to control the amount of active INH available and hence modulates INH sensitivity. The interest in mycolic acids stems from their exclusivity to mycobacteria, making these biosynthetic pathways obvious targets for antimycobacterial drugs. Routes leading to mycolic acid biosynthesis are not fully established (45, 46), although studies with a viable strain of Mycobacterium smegmatis defective in mycolate biosynthesis (5) as well as Mycobacterium aurum treated with INH (47) suggest that mycobacterial cells can survive with severely reduced mycolic acid content of the cell wall. A reduction in mycolates makes these organisms more permeable and is in agreement with the results presented here on increased sensitivity to antibiotics in the Δnat strain of M. bovis BCG Pasteur.
Although INH is a substrate for M. tuberculosis NAT (14), NAT is unlikely to be an additional target for INH. Nevertheless, a reduction in mycolic acid as well as decreasing the effectiveness of the cell wall as a barrier to protein extrusion (47), mirrors the ultrastructural effects we have observed on deleting the nat gene. These data, together with the increased intracellular killing by macrophages of M. bovis BCG Pasteur Δnat, indicate that specifically targeting the NAT protein may serve both to increase the effectiveness of combination therapy by an order of magnitude and shorten the treatment time for active infection through inhibiting cell wall biosynthesis.
To target the NAT protein, we have developed a high throughput assay (48) and are using this with combinatorial chemistry and modeling on the NAT 3-D structure (42) to generate compounds to test both on mycobacterial growth in culture and mycobacterial killing in macrophages.
Acknowledgments
We thank Mimi Mo and Drs. F. Pompeo, J. Harris, and P. Deepalakshmi for assistance.
S. Bhakta is a Wellcome Travelling Research Fellow and G.S. Besra is a Lister Institute-Jenner Research Fellow. We acknowledge support from the Wellcome Trust and Medical Research Council, Biotechnology and Biological Sciences Research Council, and GlaxoSmithKline for a studentship for A.M. Upton.
A. Upton's present address is The Rockefeller University, 1230 York Avenue, New York, NY 10021.
Abbreviations used in this paper: ADC, albumin-dextrose-catalase; CF, cord factor; GMM, glucose monomycolate; INH, isoniazid; MK, menaquinone; NAT, arylamine N-acetyltransferase; OADC, oleic acid–ADC; ORF, open reading frame; PDIM, phthiocerol dimycocerosate; SEM, scanning electron microscopy; TEM, transmission electron microscopy; TLC, thin layer chromatography.
References
- 1.Dye, C., S. Scheele, P. Dolin, V. Pathania, and M.C. Raviglione. 1999. Consensus statement. Global burden of tuberculosis: estimated incidence, prevalence, and mortality by country. WHO Global Surveillance and Monitoring Project. JAMA. 282:677–686. [DOI] [PubMed] [Google Scholar]
- 2.Bloom, B.R., and C.J. Murray. 1992. Tuberculosis: commentary on a reemergent killer. Science. 257:1055–1064. [DOI] [PubMed] [Google Scholar]
- 3.Bass, J.B., Jr., L.S. Farer, P.C. Hopewell, R. O'Brien, R.F. Jacobs, F. Ruben, D.E. Snider, Jr., and G. Thornton. 1994. Treatment of tuberculosis and tuberculosis infection in adults and children. American Thoracic Society and The Centers for Disease Control and Prevention. Am. J. Respir. Crit. Care Med. 149:1359–1374. [DOI] [PubMed] [Google Scholar]
- 4.Mitchison, D.A. 1998. How drug resistance emerges as a result of poor compliance during short course chemotherapy for tuberculosis. Int. J. Tuberc. Lung Dis. 2:10–15. [PubMed] [Google Scholar]
- 5.Liu, J., and H. Nikaido. 1999. A mutant of Mycobacterium smegmatis defective in the biosynthesis of mycolic acids accumulates meromycolates. Proc. Natl. Acad. Sci. USA. 96:4011–4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Winder, F.G., and P.B. Collins. 1970. Inhibition by isoniazid of synthesis of mycolic acids in Mycobacterium tuberculosis. J. Gen. Microbiol. 63:41–48. [DOI] [PubMed] [Google Scholar]
- 7.Wang, L., and K. Takayama. 1972. Relationship between the uptake of isoniazid and its action on in vivo mycolic acid synthesis in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2:438–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takayama, K., H.K. Schnoes, E.L. Armstrong, and R.W. Boyle. 1975. Site of inhibitory action of isoniazid in the synthesis of mycolic acids in Mycobacterium tuberculosis. J. Lipid Res. 16:308–317. [PubMed] [Google Scholar]
- 9.Wheeler, P.R., and P.M. Anderson. 1996. Determination of the primary target for isoniazid in mycobacterial mycolic acid biosynthesis with Mycobacterium aurum A+. Biochem. J. 318:451–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sacchettini, J.C., and J.S. Blanchard. 1996. The structure and function of the isoniazid target in M. tuberculosis. Res. Microbiol. 147:36–43. [DOI] [PubMed] [Google Scholar]
- 11.Kremer, L., L.G. Dover, H.R. Morbidoni, C. Vilcheze, W.N. Maughan, A. Baulard, S.C. Tu, N. Honore, V. Deretic, J.C. Sacchettini, et al. 2003. Inhibition of InhA activity, but not KasA activity, induces formation of a KasA-containing complex in mycobacteria. J. Biol. Chem. 278:20547–20554. [DOI] [PubMed] [Google Scholar]
- 12.Quemard, A., C. Lacave, and G. Laneelle. 1991. Isoniazid inhibition of mycolic acid synthesis by cell extracts of sensitive and resistant strains of Mycobacterium aurum. Antimicrob. Agents Chemother. 35:1035–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Slayden, R.A., and C.E. Barry III. 2000. The genetics and biochemistry of isoniazid resistance in Mycobacterium tuberculosis. Microbes Infect. 2:659–669. [DOI] [PubMed] [Google Scholar]
- 14.Upton, A.M., A. Mushtaq, T.C. Victor, S.L. Sampson, J. Sandy, D.M. Smith, P.V. van Helden, and E. Sim. 2001. Arylamine N-acetyltransferase of Mycobacterium tuberculosis is a polymorphic enzyme and a site of isoniazid metabolism. Mol. Microbiol. 42:309–317. [DOI] [PubMed] [Google Scholar]
- 15.Payton, M., R. Auty, R. Delgoda, M. Everett, and E. Sim. 1999. Cloning and characterization of arylamine N-acetyltransferase genes from Mycobacterium smegmatis and Mycobacterium tuberculosis: increased expression results in isoniazid resistance. J. Bacteriol. 181:1343–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Payton, M., A. Mushtaq, T.W. Yu, L.J. Wu, J. Sinclair, and E. Sim. 2001. Eubacterial arylamine N-acetyltransferases-identification and comparison of 18 members of the protein family with conserved active site cysteine, histidine and aspartate residues. Microbiol. 147:1137–1147. [DOI] [PubMed] [Google Scholar]
- 17.Sim, E., M. Payton, M. Noble, and R. Minchin. 2000. An update on genetic, structural and functional studies of arylamine N-acetyltransferases in eucaryotes and procaryotes. Hum. Mol. Genet. 9:2435–2441. [DOI] [PubMed] [Google Scholar]
- 18.Price-Evans, D.A., K.A. Manley, and V.A. McKusick. 1960. Genetic control of isoniazid metabolism in man. Br. Med. J. 2:485–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deguchi, T. 1992. Physiology and molecular biology of arylamine N-acetyltransferases. Biomed. Res. 13:231–242. [Google Scholar]
- 20.Bernstein, J., W.A. Lott, B.A. Steinberg, and H.L. Yale. 1952. Chemotherapy of experimental tuberculosis. V. Isonicotinic acid hydrazide (nydrazid) and related compounds. Am. Rev. Tuberc. 65:357–364. [DOI] [PubMed] [Google Scholar]
- 21.Payton, M., C. Gifford, P. Schartau, C. Hagemeier, A. Mushtaq, S. Lucas, K. Pinter, and E. Sim. 2001. Evidence towards the role of arylamine N-acetyltransferase in Mycobacterium smegmatis and development of a specific antiserum against the homologous enzyme of Mycobacterium tuberculosis Microbiol. 147:3295–3302. [DOI] [PubMed] [Google Scholar]
- 22.Cole, S.T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S.V. Gordon, K. Eiglmeier, S. Gas, C.E. Barry III, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 393:537–544. [DOI] [PubMed] [Google Scholar]
- 23.Garnier, T., K. Eiglmeier, J.C. Camus, N. Medina, H. Mansoor, M. Pryor, S. Duthoy, S. Grondin, C. Lacroix, C. Monsempe, et al. 2003. The complete genome sequence of Mycobacterium bovis. Proc. Natl. Acad. Sci. USA. 100:7877–7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Smet, K.A., K.E. Kempsell, A. Gallagher, K. Duncan, and D.B. Young. 1999. Alteration of a single amino acid residue reverses fosfomycin resistance of recombinant MurA from Mycobacterium tuberculosis Microbiol. 145:3177–3184. [DOI] [PubMed] [Google Scholar]
- 25.Parish, T., and N.G. Stoker. 2000. Use of a flexible cassette method to generate a double unmarked Mycobacterium tuberculosis tlyA plcABC mutant by gene replacement. Microbiol. 146:1969–1975. [DOI] [PubMed] [Google Scholar]
- 26.Roberts, G., D.G. Muttucumaru, and T. Parish. 2003. Control of the acetamidase gene of Mycobacterium smegmatis by multiple regulators. FEMS Microbiol. Lett. 221:131–136. [DOI] [PubMed] [Google Scholar]
- 27.Etienne, G., C. Villeneuve, H. Billman-Jacobe, C. Astarie-Dequeker, M.A. Dupont, and M. Daffe. 2002. The impact of the absence of glycopeptidolipids on the ultrastructure, cell surface and cell wall properties, and phagocytosis of Mycobacterium smegmatis Microbiol. 148:3089–3100. [DOI] [PubMed] [Google Scholar]
- 28.Klegerman, M.E., P.O. Devadoss, J.L. Garrido, H.R. Reyes, and M.J. Groves. 1996. Chemical and ultrastructural investigations of Mycobacterium bovis BCG: implications for the molecular structure of the mycobacterial cell envelope. FEMS Immunol. Med. Microbiol. 15:213–222. [DOI] [PubMed] [Google Scholar]
- 29.Besra, G.S. 1998. Preparation of cell-wall fractions from mycobacteria. Methods Mol. Biol. 101:91–107. [DOI] [PubMed] [Google Scholar]
- 30.Gordon, S. 1996. The myeloid system. Weir's Handbook of Experimental Immunology Volume IV. L.A. Herzenberg, D.M. Weir, and C. Blackwell, editors. Blackwell Science, Oxford. 153.1–153.9.
- 31.Van Hooijdonk, C.A., C.P. Glade, and P.E. Van Erp. 1994. TO-PRO-3 iodide: a novel HeNe laser-excitable DNA stain as an alternative for propidium iodide in multiparameter flow cytometry. Cytometry. 17:185–189. [DOI] [PubMed] [Google Scholar]
- 32.Martinez-Pomares, L., J.A. Mahoney, R. Kaposzta, S.A. Linehan, P.D. Stahl, and S. Gordon. 1998. A functional soluble form of the murine mannose receptor is produced by macrophages in vitro and is present in mouse serum. J. Biol. Chem. 273:23376–23380. [DOI] [PubMed] [Google Scholar]
- 33.Warren, R.M., S.L. Sampson, M. Richardson, G.D. Van Der Spuy, C.J. Lombard, T.C. Victor, and P.D. van Helden. 2000. Mapping of IS6110 flanking regions in clinical isolates of Mycobacterium tuberculosis demonstrates genome plasticity. Mol. Microbiol. 37:1405–1416. [DOI] [PubMed] [Google Scholar]
- 34.Cox, J.S., B. Chen, M. McNeil, and W.R. Jacobs, Jr. 1999. Complex lipid determines tissue-specific replication of Mycobacterium tuberculosis in mice. Nature. 402:79–83. [DOI] [PubMed] [Google Scholar]
- 35.Moody, D.B., B.B. Reinhold, M.R. Guy, E.M. Beckman, D.E. Frederique, S.T. Furlong, S. Ye, V.N. Reinhold, P.A. Sieling, R.L. Modlin, et al. 1997. Structural requirements for glycolipid antigen recognition by CD1b-restricted T cells. Science. 278:283–286. [DOI] [PubMed] [Google Scholar]
- 36.Ryll, R., Y. Kumazawa, and I. Yano. 2001. Immunological properties of trehalose dimycolate (cord factor) and other mycolic acid-containing glycolipids–a review. Microbiol. Immunol. 45:801–811. [DOI] [PubMed] [Google Scholar]
- 37.Nuzzo, I., M. Galdiero, C. Bentivoglio, R. Galdiero, and C. Romano Carratelli. 2002. Apoptosis modulation by mycolic acid, tuberculostearic acid and trehalose 6,6′-dimycolate. J. Infect. 44:229–235. [DOI] [PubMed] [Google Scholar]
- 38.Kana, B.D., E.A. Weinstein, D. Avarbock, S.S. Dawes, H. Rubin, and V. Mizrahi. 2001. Characterization of the cydAB-encoded cytochrome bd oxidase from Mycobacterium smegmatis. J. Bacteriol. 183:7076–7086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Truglio, J.J., K. Theis, Y. Feng, R. Gajda, C. Machutta, P.J. Tonge, and C. Kisker. 2003. Crystal structure of Mycobacterium tuberculosis MenB, a key enzyme in vitamin K2 biosynthesis. J. Biol. Chem. 278:42352–42360. [DOI] [PubMed] [Google Scholar]
- 40.Indrigo, J., R.L. Hunter, Jr., and J.K. Actor. 2002. Influence of trehalose 6,6′-dimycolate (TDM) during mycobacterial infection of bone marrow macrophages. Microbiol. 148:1991–1998. [DOI] [PubMed] [Google Scholar]
- 41.Mushtaq, A., M. Payton, and E. Sim. 2002. The COOH terminus of arylamine N-acetyltransferase from Salmonella typhimurium controls enzymic activity. J. Biol. Chem. 277:12175–12181. [DOI] [PubMed] [Google Scholar]
- 42.Sandy, J., A. Mushtaq, A. Kawamura, J. Sinclair, E. Sim, and M. Noble. 2002. The structure of arylamine N-acetyltransferase from Mycobacterium smegmatis–an enzyme which inactivates the anti-tubercular drug, isoniazid. J. Mol. Biol. 318:1071–1083. [DOI] [PubMed] [Google Scholar]
- 43.Slayden, R.A., R.E. Lee, and C.E. Barry III. 2000. Isoniazid affects multiple components of the type II fatty acid synthase system of Mycobacterium tuberculosis. Mol. Microbiol. 38:514–525. [DOI] [PubMed] [Google Scholar]
- 44.Larsen, M.H., C. Vilcheze, L. Kremer, G.S. Besra, L. Parsons, M. Salfinger, L. Heifets, M.H. Hazbon, D. Alland, J.C. Sacchettini, et al. 2002. Overexpression of inhA, but not kasA, confers resistance to isoniazid and ethionamide in Mycobacterium smegmatis, M. bovis BCG and M. tuberculosis. Mol. Microbiol. 46:453–466. [DOI] [PubMed] [Google Scholar]
- 45.Asselineau, C., J. Asselineau, G. Laneelle, and M.A. Laneelle. 2002. The biosynthesis of mycolic acids by Mycobacteria: current and alternative hypotheses. Prog. Lipid Res. 41:501–523. [DOI] [PubMed] [Google Scholar]
- 46.Barry, C.E., III, R.E. Lee, K. Mdluli, A.E. Sampson, B.G. Schroeder, R.A. Slayden, and Y. Yuan. 1998. Mycolic acids: structure, biosynthesis and physiological functions. Prog. Lipid Res. 37:143–179. [DOI] [PubMed] [Google Scholar]
- 47.Bardou, F., A. Quemard, M.A. Dupont, C. Horn, G. Marchal, and M. Daffe. 1996. Effects of isoniazid on ultrastructure of Mycobacterium aurum and Mycobacterium tuberculosis and on production of secreted proteins. Antimicrob. Agents Chemother. 40:2459–2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brooke, E.W., S.G. Davies, A.W. Mulvaney, F. Pompeo, E. Sim, and R.J. Vickers. 2003. An approach to identifying novel substrates of bacterial arylamine N-acetyltransferases. Bioorg. Med. Chem. 11:1227–1234. [DOI] [PubMed] [Google Scholar]