Abstract
Bartonella henselae causes vasculoproliferative disorders in humans. We identified a nonfimbrial adhesin of B. henselae designated as Bartonella adhesin A (BadA). BadA is a 340-kD outer membrane protein encoded by the 9.3-kb badA gene. It has a modular structure and contains domains homologous to the Yersinia enterocolitica nonfimbrial adhesin (Yersinia adhesin A). Expression of BadA was restored in a BadA-deficient transposon mutant by complementation in trans. BadA mediates the binding of B. henselae to extracellular matrix proteins and to endothelial cells, possibly via β1 integrins, but prevents phagocytosis. Expression of BadA is crucial for activation of hypoxia-inducible factor 1 in host cells by B. henselae and secretion of proangiogenic cytokines (e.g., vascular endothelial growth factor). BadA is immunodominant in B. henselae–infected patients and rodents, indicating that it is expressed during Bartonella infections. Our results suggest that BadA, the largest characterized bacterial protein thus far, is a major pathogenicity factor of B. henselae with a potential role in the induction of vasculoproliferative disorders.
Keywords: pilus, endothelial cells, HIF-1, VEGF, angiogenesis
Introduction
Bartonella henselae is a facultative intracellular bacterium causing cat scratch disease (CSD), bacillary angiomatosis (BA), and bacillary peliosis (BP; reference 1). CSD is a self-limiting disease characterized by lymphadenopathy related to a cat scratch. In immunocompromised individuals, B. henselae causes tumorous proliferations of endothelial cells (ECs) in the skin and internal organs, referred to as BA and BP, respectively (2-4).
Stimulation of angiogenesis upon a Bartonella infection represents one fascinating feature of human pathogenic bacteria. Bartonella species may cause these vasculoproliferations by at least three different mechanisms that may act synergistically: (a) triggering EC proliferation directly (5), (b) inhibition of apoptosis of ECs (6), and (c) induction of the secretion of vasculoproliferative cytokines (e.g., vasculoendothelial growth factor [VEGF]; references 7 and 8). In vivo and in vitro infection with B. henselae results in the activation of hypoxia-inducible factor (HIF)-1 (unpublished data), the key transcription factor of angiogenesis (9). The proliferating ECs are one potential habitat of B. henselae, as the pathogen survives and replicates within these cells in vitro (10, 11).
Only few bacterial factors operating in Bartonella–host cell interactions are known. One of the most important putative pathogenicity factors of B. henselae is the “type IV pilus” (12), which mediates host cell adhesion and triggering of VEGF secretion (7). “Pilus” expression undergoes phase variation with multiple passages on agar plates (12). Additional candidates in Bartonella pathogenicity are outer membrane proteins (OMPs; references 13 and 14) and the virB type IV secretion system that is responsible for the inhibition of apoptosis in ECs (15).
Many pathogenic bacteria assemble multifunctional proteinaceous surface structures that serve as adhesins. Such nonfimbrial adhesins, e.g., Yersinia adhesin A (YadA) of enteropathogenic Yersinia species and Neisseria adhesin A (NadA) of Neisseria meningitidis, have been described as a novel class of bacterial adhesins representing important pathogenicity factors (16, 17). YadA, the best investigated representative of this protein family, mediates adherence to host cells (18) and extracellular matrix (ECM) proteins (19). YadA expression is essential for pathogenicity of Yersinia enterocolitica in a murine infection model (20). Accordingly, NadA is crucial for establishing N. meningitidis infection in an infant rat model (17).
Here, we describe the identification, cloning, and characterization of Bartonella adhesin A (BadA), formerly known as “type IV pili.” The 340-kD BadA protein, encoded by the 9.3-kb badA gene, is located in the outer membrane of B. henselae. BadA is constructed modularly and contains domains homologous to Y. enterocolitica YadA. BadA mediates the binding of B. henselae to ECM proteins and ECs, and prevents phagocytosis. It is also crucial for activation of HIF-1 and secretion of VEGF. Moreover, we provide evidence that BadA is expressed during Bartonella infections in humans and rodents with implications for serodiagnosis of Bartonella infections. Our results suggest that BadA is a major pathogenicity factor of B. henselae with a potential role in the induction of the vasculoproliferative disorders BA and BP.
Materials and Methods
Bacteria and Growth Conditions.
Bacteria are summarized in Table I. B. henselae was grown on Columbia blood agar (CBA) in a humidified atmosphere at 37°C and 5% CO2. Escherichia coli was grown in Luria-Bertani broth. Antibiotics were used at the following concentrations: B. henselae: 30 μg/ml kanamycin, 1 μg/ml chloramphenicol; E. coli: 50 μg/ml kanamycin, 50 μg/ml chloramphenicol, 100 μg/ml ampicillin.
Table I.
Bacterial Strains Used in This Study
| Strain | Characteristics | Reference or source |
|---|---|---|
| B. henselae | ||
| Wild-type (WT) | B. henselae Marseille, patient isolate, early passage | 4 |
| Pil− variant | B. henselae Marseille, extensively passaged, pilus-negative | 7 |
| BadA− | B. henselae Marseille TN <KAN-2> transposon mutant, transposon integrated in badA | This study |
| BadA−/BadA+ WT | B. henselae BadA2−/− complemented with pTR15 | This study |
| Houston-1 ATCC 49882 | published genome sequence (NC_005956) | 24 |
| E. coli | ||
| TOP 10 | host strain used for cloning | Invitrogen |
| DH5α | host strain used for cloning | Invitrogen |
| BL21 (DE3) | host strain used for protein expression | Stratagene |
For production of bacterial stock suspensions, bacteria were harvested from agar plates after 5 d and frozen in Luria-Bertani–20% glycerol at −80°C. The pilus-negative variant strain (B. henselae Pil−) was produced by extensively passaging B. henselae Marseille WT (7).
SDS-PAGE and Immunoblotting.
B. henselae were resuspended in SDS sample buffer and heated at 98°C for 3 min. SDS-PAGE was performed in 12% gels. Gels were stained with Coomassie Blue R250. For immunoblotting, proteins were transferred onto nitrocellulose membranes (Schleicher and Schuell). Blots were blocked for 1 h in 5% skim milk powder in 25 mM Tris, pH 7.5, 0.15 M NaCl, and 0.05% Tween 20 (Sigma-Aldrich), and incubated with the respective primary antibody overnight. For detection, a horseradish peroxidase–conjugated secondary antibody was used and signals were visualized either via chemiluminescence (Amersham Biosciences) or with DAB (3,3′-diaminobenzidine tetrahydrochloride; Sigma-Aldrich).
A BadA-specific rabbit antiserum was raised by immunization with a BadA stalk fragment (badA-f6-badA-r6; Table II) and purified by affinity chromatography, a rabbit anti–B. henselae Marseille serum was raised by immunization with viable bacteria (11), and a mouse anti–B. henselae Marseille serum was raised by immunization with heat-killed bacteria. Human sera were obtained from patients with the clinical diagnosis of CSD and immunoreactivity of >1:200 in an immunofluorescence test according to the recommendations of the Centers of Disease Control (21).
Table II.
Plasmids, Primers, and Probes Used in This Study
| Plasmids | Description | Reference or source |
|---|---|---|
| pBluescript II KS | cloning vector, Apr | Stratagene |
| pET30b | protein expression vector, Kmr | Novagen |
| pBBR1MCS | broad host range, Cmr | 46 |
| pTR14 | pBluescript II KS containing a 13.2-kb B. henselae Marseille EcoRI/ClaI fragment with badA, Apr |
This study |
| pTR15 | pBBR1MCS containing a 13.2-kb BamHI/ClaI fragment of pTR14 with badA, Cmr |
This study |
| pTR52 | pET30b containing badA-f6/r6-PCR fragment | This study |
| Primers | ||
| badA-f2 | CTCCAGTCTGATGATTCAGC | This study |
| badA-r2 | GCTATATTGATTTCAGTACCTGC | This study |
| badA-f6 | TGCACATATGAAAGCATTAAGGGGAATGATATCAGa | This study |
| badA-r6 | TTATCTCGAG TCAAGTACGCTTATCACTTTTGTTATTAGCb | This study |
| Probe | ||
| pilin | GNTTYTTNAARGAYGARTCNGGNGCANCNGCNATHGARTAYG GNCTNATHGCNGCNTTNATHTCNGT |
This study |
Italic, NdeI restriction site.
Italic, XhoI restriction site; underline, stop codon.
Protein Identification.
B. henselae was lysed in 7 M urea, 2 M thiourea, and 4% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate, and centrifuged at 100,000 g for 1 h at 4°C. SDS-PAGE was followed by matrix-assisted laser desorption/ionization peptide mass fingerprinting and electrospray ionization tandem mass spectrometry. In-gel digestion and protein identification with H2 18O method was performed as described previously (22). After ZipTip purification (C18-ZipTip; Millipore), aliquots were deposited on α-cyano-4-hydroxycinnamic acid-nitrocellulose for matrix-assisted laser desorption/ionization-time of flight mass analysis (Reflex III; Bruker Daltonic). All measurements were performed in the positive ion reflection mode at an accelerating voltage of 23 kV and delayed-pulsed ion extraction. Sequence verification was performed by nanoelectrospray tandem mass spectrometry on a hybrid quadrupole orthogonal acceleration time of flight mass spectrometer (QSTAR Pulsar i; Applied Biosystems). N- versus COOH-terminal peptide sequence orientation was determined using 18O labeling of the COOH termini. MASCOT database searches (National Center for Biotechnology Information [NCBI] nonredundant protein database) used methionine oxidations as variable modifications.
DNA Techniques.
Plasmids and primers are listed in Table II. DNA manipulations were performed according to standard protocols.
Transposon Mutagenesis.
Transposon mutagenesis was performed by electroporation of the EZ::TN <KAN-2> transposon (EZ::TN <KAN-2> Tnp Transposome Kit; Epicentre) as described previously (23).
Cloning of badA.
For construction of pTR14, chromosomal DNA of B. henselae Marseille was isolated with QIAGEN Genomic-tip 100/G columns and digested with EcoRI and ClaI, yielding a 13.2-kb fragment containing badA, including its putative promoter region according to the sequence of B. henselae Houston-1. Fragments larger than 11 kb were ligated into pBluescript II KS and electroporated into E. coli TOP 10. The resulting colonies were screened for badA by colony blotting using a digoxigenin-labeled badA probe (primers: badA-f2 and badA-r2; annealing at 56°C for 30 cycles). Insertion of the 13.2-kb fragment in detected clones was confirmed by sequencing (primers: M13f and M13r; not depicted). The plasmid pTR14 was digested with BamHI and ClaI, and the insert (containing badA and the putative promoter region) was ligated into the broad host range vector pBBR1MCS. The resulting plasmid pTR15 was electroporated in B. henselae BadA−.
DNA Sequencing Procedures and DNA Sequence Analysis.
Parts of the badA sequence from B. henselae Marseille were obtained by chromosomal sequencing as described previously (23). Additional parts of the sequence were obtained by sequencing of cosmid clones. Clones harboring badA were detected from a cosmid library (SuperCos 1 Cosmid Vector Kit; Stratagene) of B. henselae Marseille by colony blotting using a badA probe (primers: badA-f2 and badA-r2).
The organization of the genomic region of badA was examined by PCR in B. henselae Marseille WT and B. henselae Pil−. Unique PCR primers were designed using the sequence of the homologous region in the Houston-1 strain (sequence data are available from GenBank/EMBL/DDBJ under accession no. NC_005956) as a template (24). PCR products were sequenced (except the highly repetitive inner parts of badA) using internal primers and ET terminator chemistry (Amersham Biosciences). Sequences were separated using a MEGABASE sequenator (Amersham Biosciences). GenBank accession numbers are as follows: B. henselae Marseille WT, AY560658 (5′ region) and AY560659 (3′ region). The BLAST program of the NCBI (blastn, blastx; http://www.ncbi.nlm.nih.gov) was used to perform sequence similarity searches.
Protein Sequence Analysis.
Sequence similarity searches were performed using the programs Blast and PSI-Blast on the nonredundant and microbial genomes databases at the NCBI (http://www.ncbi.nlm.nih.gov). Sequence alignments were made in MACAW (25). Coiled coil segments were predicted using the program COILS (26) and a consensus method based on COILS (unpublished data). Secondary structure predictions were made with PSIPRED (27).
Cloning, Expression, and Purification of a BadA Stalk Fragment.
A 480-bp fragment of badA encoding for amino acid residues 377–539 (stalk region) was amplified (primers: badA-f6 and badA-r6). The 5′ primer contained an NdeI site and a start codon, and the 3′ primer contained a stop-codon followed by an XhoI site. The fragment was cloned into the expression vector pET30b (Novagen) giving plasmid pTR52. After transformation into E. coli BL21 (DE3), expression was induced with 1 mM isopropyl β-d thiogalactopyranoside for 4 h. Cells were lysed in a French press in 30 mM Tris-HCl, pH 7.4, 50 mM NaCl, 5 mM dithiothreitol, 50 μg/ml DNase I, and 1 mM PMSF, and the protein was purified to homogeneity from the high speed centrifugation supernatant of the lysate by a combination of cation-exchange (MonoS HR; Amersham Biosciences; elution conditions are as follows: 30 mM Tris-maleate, pH 6.0, 5 mM MgCl2, 5 mM dithiothreitol, 0–1 M NaCl gradient) and gel-sizing chromatography (Superdex G-75; Amersham Biosciences; in 50 mM potassium phosphate buffer, pH 7.3, 150 mM NaCl).
Culture and Infection of ECs, J774 Macrophages, and HeLa and GD25 Cells.
Human umbilical vein ECs were cultured in EC growth medium (PromoCell). Infection experiments were performed in EC basal medium (PromoCell) as described previously (11). The mouse macrophage cell line J774A.1 (American Type Culture Collection [ATCC] TIB-67) was cultured in RPMI 1640 medium with 10% FCS, 2 mM l-glutamine, 1 mM sodium pyruvate, β-mercaptoethanol, and nonessential amino acids. Macrophages were seeded in 24-well tissue culture plates. HeLa cells were grown in RPMI 1640 with 10% FCS and for infection experiments, media were removed 2 h before infection and replaced by culture media without antibiotics and FCS to avoid unspecific HIF-1 activation. β1 integrin–deficient murine GD25 cells (28) and β1 integrin–overexpressing GD25-β1A cells (29) were cultivated in DMEM (GIBCO BRL) with 10% FCS. For GD25-β1A, 10 μg/ml puromycin (Sigma-Aldrich) was added. In some experiments, B. henselae WT and GD25-β1A were preincubated with an anti-fibronectin (Fn) antibody (DakoCytomation).
Bacteria were used at a multiplicity of infection of 100 and sedimented onto cultured cells by centrifugation for 5 min at 300 g. The actual inoculum for each experiment was determined by plating serial dilutions and calculating the number of CFUs. For J774 cells, intracellular bacteria were quantified 3 h after infection by gentamicin kill assays as described previously (11).
Determination of B. henselae Binding to ECM Proteins.
To assess binding of B. henselae to ECM proteins, coverslips were coated with 10 μg/ml human collagen type I, type III (Chemicon), and type IV (Calbiochem), and laminin (Chemicon) and Fn (Sigma-Aldrich). After washing with PBS, 107 bacteria were resuspended in 1 ml RPMI 1640 and sedimented on coverslips. After 1 h, coverslips were washed twice with RPMI, fixed with 3.75% PBS-buffered paraformaldehyde (PFA), and bacteria were stained with 1 μg/ml DAPI for 10 min. Adherence was determined via confocal laser scanning microscopy (CLSM) and by counting 20 randomly selected high power fields (1,000-fold magnification).
For detection of Fn binding by Western blotting, bacteria were harvested in PBS and the OD550 was adjusted to 1.0. Bacteria were lysed in SDS sample buffer and separated by 12% SDS-PAGE. Membranes were incubated with a monoclonal anti-Fn antibody (Becton Dickinson). Purified human plasma Fn (Chemicon) was used as a positive control (not depicted).
Detection of VEGF, IL-8, Insulin-like Growth Factor Binding Protein 3 (IGFBP-3), and Adrenomedullin (ADM) in Cell Culture Supernatants.
Determination of VEGF, IL-8, IGFBP-3, and ADM secretion upon B. henselae infection was performed without antibiotics and FCS. 25 ng/ml PMA (Sigma-Aldrich) was used as a positive control (not depicted; reference 7). VEGF concentration in culture medium was measured using a human VEGF165-ELISA kit (R&D Systems). IL-8 was determined by ELISA as described previously (30). Secreted IGFBP-3 was measured using a specific RIA (31) and secreted ADM was quantified by a commercially available RIA (Phoenix Pharmaceuticals).
Detection of HIF-1α Activation.
For reporter gene assays, a VEGF promoter luciferase reporter construct (pVEGF.4) was used (32). Transfection efficiency was normalized by cotransfection with pCMV β-galactosidase (pCMV β-gal; CLONTECH Laboratories, Inc.). 105 HeLa cells were transiently transfected with 0.5 μg pVEGF.4 Luc reporter construct and 0.25 μg pCMV β-gal using ExGen500 transfection reagent (Fermentas), and incubated for 24 h at 37°C. Transfected cells were infected with B. henselae or exposed to hypoxia. After 36 h, cells were lysed for determination of Luc activity, protein quantification, and measurement of β-gal using a Luciferase Reporter Gene Assay (Roche). Luminescence was measured with a Topcount scintillation counter (Packard Instrument Co.). Levels of Luc expression were normalized to β-gal activity and total protein concentration (30). Every experiment was performed in quadruplicate. The degree of induction was determined as the ratio of Luc activity of B. henselae–infected or hypoxia-exposed cells to that of uninfected control cells.
Immunostaining and CLSM.
Bacteria were resuspended in PBS, dried on glass slides, and fixed in 3.75% PBS-buffered PFA. 105 ECs were seeded onto coverslips and infection was stopped by 3.75% PFA. Immunostaining of B. henselae was performed as described previously (11) using a BadA-specific rabbit antiserum or mouse polyclonal antibodies raised against B. henselae Marseille. FITC-conjugated secondary antibodies and TRITC-labeled phalloidin were purchased from Dianova and Sigma-Aldrich. Bacteria were stained with DAPI. Cellular fluorescence was evaluated using a Leica DM IRE 2 CLSM. Three different fluorochromes were detected representing the green (FITC), red (TRITC), and blue (DAPI) channels. Images were digitally processed with Photoshop 6.0 (Adobe Systems). Adherence to ECs and GD25 cells was quantified by counting adherent bacteria from 20 randomly selected cells.
Transmission electron microscopy (TEM) and immunoelectronmicroscopy (IEM).
TEM was performed as described previously (11). In brief, B. henselae cell pellets were fixed and after embedding in glycide ether, the blocks were cut using an ultra microtome (Ultracut; Reichert). 80-nm ultra-thin sections were stained (Ultrastainer; Leica) with 0.5% uranyl acetate for 10 min at 30°C and 2.7% lead citrate for 5 min at 20°C. For IEM of BadA, post-embedding immunogold labeling was performed. Cells were fixed and after centrifugation, the sediment was embedded in 3% agarose at 37°C and then cooled on ice. Small parts of the agarose blocks were embedded in Lowicryl (Polysciences Ltd.). 50-nm ultra-thin sections were mounted on formvar-coated nickel grids and incubated with anti-BadA rabbit serum, followed by 10 nm gold-conjugated goat anti–rabbit IgG (Auroprobe EM; Amersham Biosciences). In control samples, the primary antibody was omitted. Grids were counterstained with uranyl acetate and lead citrate and examined using a transmission electron microscope (Zeiss EM 109; Carl Zeiss MicroImaging, Inc.).
Statistical Analysis.
All experiments were performed at least three times and revealed comparable results. Differences between mean values of experimental and control groups were analyzed by student's t test. A p-value of <0.05 was considered statistically significant.
Online Supplemental Material.
In Fig. S1, BadA is shown in more detail by electron microscopy (negative staining). In Fig. S2, BadA immunoreactivity of sera from patients suffering from CSD is evaluated by Western blotting of whole cell bacterial lysates. Figs. S1 and S2 are available at http://www.jem.org/cgi/content/full/jem.20040500/DC1.
Results
B. henselae WT, But Not B. henselae Pil− Phase Variant, Binds Fn.
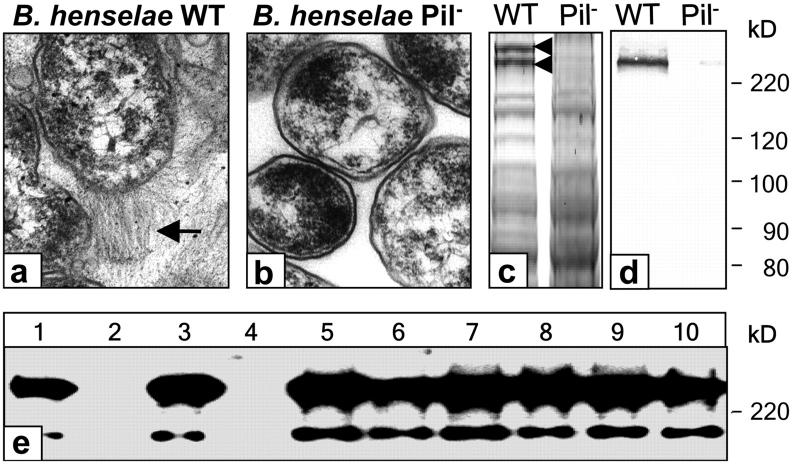
To analyze the protein composition of the “pilus”-like structures of B. henselae, a comparative protein analysis of both B. henselae WT and a spontaneous B. henselae Pil− variant was performed. TEM revealed no pilus expression of the variant strain (Fig. 1, a and b; reference 7). Expression of “pili” correlated with the observation that bacterial colonies of the WT stuck more tightly to the surface of CBA than Pil−. Protein patterns of whole bacterial lysates (WT and Pil−) showed two high molecular weight (HMW) proteins in B. henselae WT (>220 kD) not present in Pil− (Fig. 1 c). Using electrospray ionization-mass spectrometry/mass spectrometry, the lower band was identified as Fn (not depicted). Analysis of the upper band revealed four peptide fragments (ATN(I/L)(I/L)T(I/L)GK; (I/L)TY (I/L)(I/L)F; AAVT(I/L); and (I/L)YS(I/L)NE(Q/K)(I/L) ATYFG), which matched no entry in the database (blastp; http://www.ncbi.nlm.nih.gov). These were later identified as fragments of BadA (see below). We hypothesized that B. henselae WT, but not Pil−, might bind to Fn present in CBA. To further elucidate this hypothesis, Fn binding of B. henselae was assessed by Western blotting of whole bacterial lysates and detection of Fn using anti-Fn antibodies. Fn bound to bacteria was detectable in WT, but not in Pil− lysates (Fig. 1, d and e, lanes 1 and 2). Taken together, the results provide evidence that expression of pilus-like structures on the surface of B. henselae is associated with binding to Fn.
Figure 1.
Phase variation of B. henselae. (a and b) Expression of so-called type IV-like pili (BadA, arrow; see also Fig. S1, available at http://www.jem.org/cgi/content/full/jem.20040500/DC1) of B. henselae Marseille WT not detectable on the surface of B. henselae Pil−. (c) SDS-PAGE of B. henselae WT and Pil− showing two differential HMW bands. The lower band is Fn (240 kD), the upper is BadA (calculated mass: 340 kD). (d) Detection of Fn binding of B. henselae WT and Pil− by Western blotting. Bacteria-bound Fn was detected using an anti-Fn antibody. (e) Screening of a B. henselae transposon library for Fn binding. B. henselae WT (lane 1), Pil− (lane 2), and transposon mutants (lanes 3–10). Fn binding is not detectable in B. henselae Pil− and in a transposon mutant (lane 4), suggesting loss of BadA (pilus) expression.
Transposon Insertion in badA Results in Loss of Fn Binding.
To identify the gene(s) coding for the putative type IV pilus, a transposon library of B. henselae Marseille (23) was screened, revealing one transposon mutant (Fig. 1 e, lane 4) deficient in Fn binding, similar to the Pil− variant described above. By chromosomal sequencing, the site of transposon insertion (nucleotide position 3438) was identified as a 9.3-kb gene of unknown function, based on the genomic sequence of B. henselae Houston-1 (24). Database searches revealed protein sequence similarities to hypothetical genes of yet unknown functions in Brucella melitensis, Sinorhizobium meliloti, and Mesorhizobium loti (not depicted). Analysis of the deduced amino acid sequence revealed a protein with a calculated mass of 340 kD, consistent with the size of the differentially expressed upper HMW protein of B. henselae WT (Fig. 1 c). The four unidentified peptide fragments mentioned above were present in this translated protein sequence. Protein sequence analysis indicated that this protein belongs to a novel class of nonfimbrial adhesins exemplified by Yersinia adhesin A (YadA; reference 16) and Neisseria adhesin A (NadA; reference 17). Therefore, we designated the protein Bartonella adhesin A (BadA).
BadA Has a Modular Structure and Belongs to the Class of Nonfimbrial Adhesins.
BadA shows a high degree of modularity in its domain structure. After a putative signal sequence and a region not similar to any other currently known protein (Fig. 2 a), BadA contains 11 degenerate 14-residue repeats resembling those found in the YadA head sequence (16), which form a novel-type of left-handed β-helix (33). Two of these repeats are interrupted by sequence inserts. At the COOH terminus, after a motif that is highly conserved (often in multiple copies in most nonfimbrial adhesins [16], termed a “neck sequence”), BadA again resembles YadA in the succession of a right-handed coiled coil segment with pentadecad periodicity, a left-handed coiled coil segment with heptad periodicity, and a membrane anchor domain containing four transmembrane β strands. The membrane anchor is thought to form a 12-stranded pore upon trimerization and allow the autotransport of the adhesin across the outer membrane (34). It is conserved in all nonfimbrial adhesins and may represent their defining feature. In between these two N- and COOH-terminal YadA-like regions, BadA contains a single occurrence of a sequence found repetitively in proteins of Xylella spp., followed by 21 occasionally truncated copies of a sequence also seen in putative adhesins from uropathogenic and enterohaemorrhagic E. coli, Shigella flexneri, and Salmonella spp. (Fig. 2 b). These central repeats of BadA contain left-handed coiled coil segments and are separated by neck sequences (24 in total). This is the highest number observed in any protein so far. Structurally, the fairly regular alternation of coiled coil and globular sequences suggests an extended, rod-like shape with periodically recurring bulkier and thinner parts, rather like a segmented rope. This conjecture fits well with the great estimated length of BadA (∼100–300 nm), as well as its hair-like, flexible appearance in electron micrographs (Fig. 1 a and Fig. S2, available at http://www.jem.org/cgi/content/full/jem.20040500/DC1).
Figure 2.
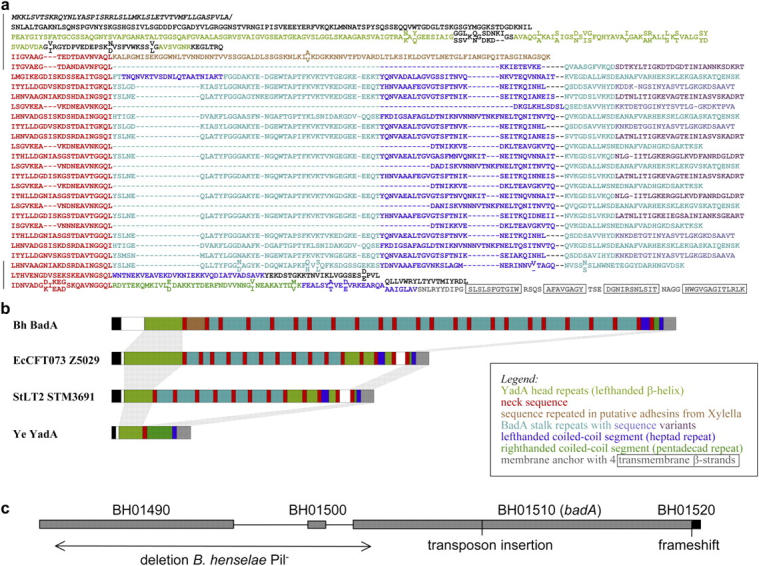
Protein domain structure of BadA and genomic organization of badA region. (a) Sequence representation and (b) schematic diagram of the BadA domain structure. Individual domains are color coded. For simplicity, the colors are explained in the figure itself. Regions that could not be assigned by sequence similarity are left black in (a) and shown as open boxes in (b). The sequence is BadA of B. henselae Houston-1. Regions also sequenced in B. henselae Marseille are marked by black bars in (a) and residues different in the two strains are shown staggered, with the Houston-1 sequence above and the Marseille sequence below. In (b), structurally equivalent regions are connected by gray bars. The organisms are as follows: Bh, B. henselae; EcCFT073, E. coli strain CFT073; StLT2, Salmonella typhimurium strain LT2; Ye, Y. enterocolitica. (c) Schematic diagram of the arrangement of genes in a 17.7-kb genomic region of B. henselae Houston-1. Details are given in Results. Site of transposon insertion in B. henselae Marseille BadA− and position of the frameshift in the membrane anchor region of B. henselae Houston-1 are indicated. Loss of BadA (pilus) expression in a spontaneous B. henselae Marseille variant (B. henselae Pil−) is correlated with an 8.5-kb deletion (arrow).
Genomic Organization of the badA Region.
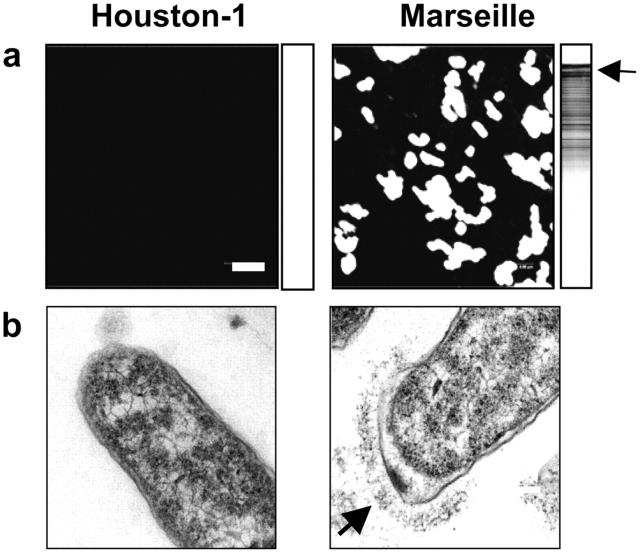
The upstream region of the badA gene (B. henselae Houston-1; gene BH01510) contains three short open reading frames, one of which (BH01500) shows sequence similarity to the 3′ end of badA, thought to encode the membrane anchor region (Fig. 2 c). Further upstream is another open reading frame (BH01490) that displays sequence similarity with badA (47% over aligned amino acid regions) and also encodes an anchor region. A PCR-based survey revealed an identical gene organization pattern in B. henselae Marseille WT. A similar survey of B. henselae Marseille Pil− revealed a large deletion of 8.5 kb spanning from the 5′ terminal end of BH01490 to the 5′ terminal end of badA. The deletion seems to have been mediated by repeated sequences present in the beginning of BH01490 and badA, resulting in the loss of most of BH01490, the 5′ terminal end of badA, and the intergenic segment flanked by these two genes. As a consequence, the 5′ terminal end of BH01490 has fused with a truncated version of badA in the Marseille Pil− strain. Additionally, in the Houston-1 strain there is a 1-bp deletion at the anchor region of badA that is not detected in the Marseille strain. As a consequence, the anchor region is not located inside BH01510, but in a separate small gene (BH01520) that is partially overlapping with the 3′ terminal end of BH01510 in the Houston-1 strain. In contrast, BH01510 and BH01520 are merged into a single gene in the Marseille strain. BadA is expressed in B. henselae Marseille, but not in Houston-1 (Fig. 3). From this data we conclude that the B. henselae Houston-1 strain we investigated (ATCC 49882) lacks BadA expression.
Figure 3.
BadA expression of B. henselae Houston-1 (ATCC 49882) and B. henselae Marseille. (a) BadA expression was analyzed by immunofluorescence and Western blotting using an anti-BadA rabbit serum (arrow). Bar, 2 μm. (b) Detection of BadA expression by TEM. BadA is expressed on the surface of B. henselae Marseille (arrow), but missing in the Houston-1 strain (ATCC 49882).
Complementation of B. henselae BadA− Reveals That BadA Forms the Pilus-like Structure.
Complementation of the BadA− mutant was performed with plasmid pTR15 containing a 13.2-kb chromosomal fragment carrying WT badA, including its putative promoter region. TEM revealed that expression of BadA in B. henselae BadA−/BadA+ WT leads to the formation of the pilus-like structure (Fig. 4 a). An anti-BadA rabbit serum generated by immunization with a BadA stalk fragment revealed a strong surface staining of the WT and the complemented mutant not detectable in Pil− and BadA− (Fig. 4 b). Consistent data were obtained by IEM showing clearly that BadA is stained (Fig. 4 c). In accordance, this serum was reactive against BadA in OMP preparations of B. henselae WT not present in Pil− (not depicted). These results suggest strongly that the formerly described type IV pilus (12) is in fact the nonfimbrial adhesin BadA.
Figure 4.
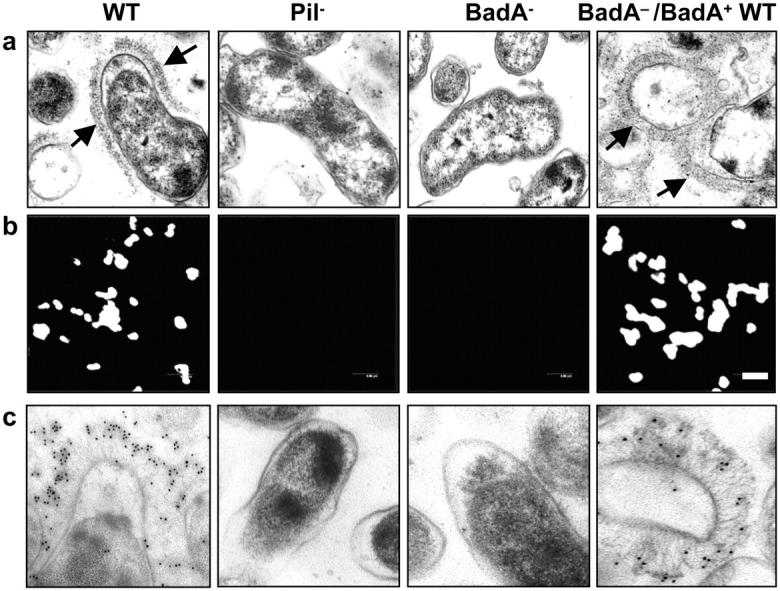
Complementation of B. henselae BadA−. (a) Detection of BadA expression in B. henselae WT, Pil−, BadA−, and BadA−/BadA+ WT by TEM. Note the brush-like arrangement of BadA on the surface of the WT and the complemented mutant (arrows) missing in the Pil− variant and the BadA− mutant. (b) Detection of BadA expression by immunofluorescence using an anti-BadA rabbit serum. Bar, 2 μm. (c) Detection of BadA expression by IEM using 10 nm gold-conjugated goat anti–rabbit IgG.
BadA Is Crucial for Interaction of B. henselae with ECM and Host Cells.
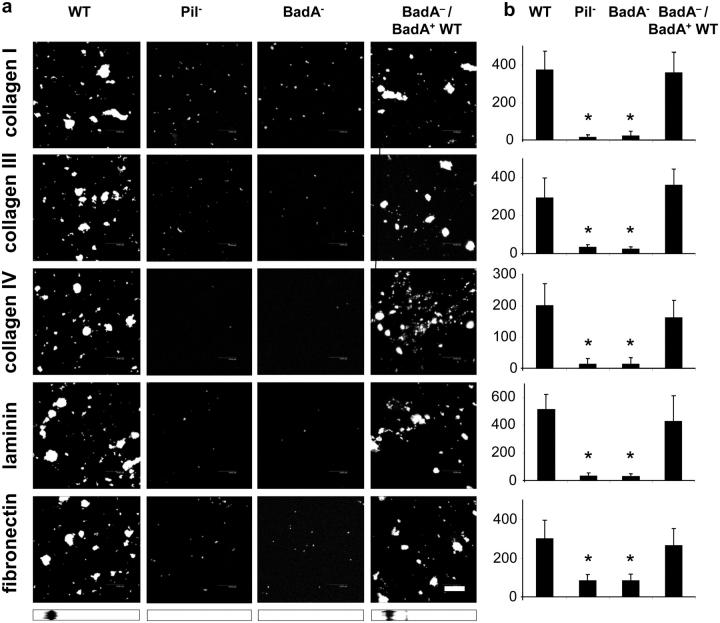
To investigate whether BadA is involved in binding to ECM proteins, B. henselae was exposed to collagen type I, III, and IV, and laminin- and Fn-coated coverslips (Fig. 5). BadA-expressing B. henselae WT and BadA−/BadA+ WT bound to a higher degree to these ECM protein–coated surfaces than Pil− or BadA−. The ability to bind Fn was reconstituted in BadA−/BadA+ WT as additionally shown by Western blotting. From this data we conclude that expression of BadA is crucial for adherence of B. henselae to ECM proteins.
Figure 5.
BadA-dependent interaction of B. henselae with ECM proteins. Collagen type I, III, and IV, and laminin and Fn binding of B. henselae WT, Pil−, BadA−, and BadA−/BadA+ WT. (a) Bacteria were seeded onto coated coverslips and DAPI stained. Bar, 20 μm. Fn binding was additionally assessed by Western blotting. (b) Microscopic quantification of bacterial binding to ECM proteins (adherent bacteria per high power field). *, significant difference compared with B. henselae WT (P < 0.01).
Nonfimbrial adhesins mediate adherence of bacteria to host cells (16). As one potential target in B. henselae infections is ECs (6, 10, 11), the role of BadA in adherence to these cells was investigated. ECs were infected with B. henselae WT, Pil−, BadA−, and BadA−/BadA+ WT and adherence was assessed 30 min after infection by CLSM (Fig. 6, a and b). Both B. henselae Pil− and BadA− hardly adhered to ECs, showing that BadA expression is crucial for host cell adherence. Consistently, TEM suggests that BadA mediates adherence of B. henselae to ECs (Fig. 6 c). Next, we analyzed the role of β1 integrins in the binding of B. henselae to host cells, as they mediate YadA-dependent adherence of Yersinia (35). For this purpose, the fibroblast cell lines GD25 (lacking β1 integrins; reference 28) and GD25-β1A (overexpressing β1 integrins; reference 29) were infected with B. henselae WT. Data revealed a significantly decreased adherence of B. henselae WT to GD25, suggesting that β1 integrins act as a cellular binding partner of BadA. This binding might be mediated by Fn, as pretreatment of cells and bacteria with anti-Fn antibodies abolished bacterial adherence to the background level (Fig. 6, d and e). Furthermore, like YadA (36), expression of BadA prevents B. henselae WT from phagocytosis in J774 murine macrophages (Fig. 6 f).
Figure 6.
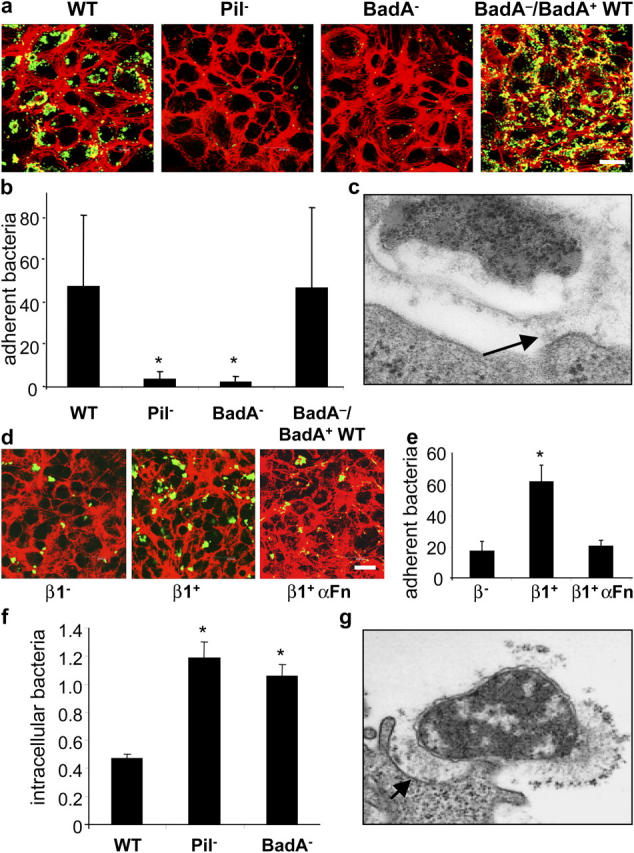
BadA mediates cell adherence and shares antiphagocytic properties. (a) Adherence of B. henselae WT, Pil−, BadA−, and BadA−/BadA+ WT to ECs shown by CLSM. Bacteria were labeled by FITC-conjugated antibodies (green signal) and filamentous actin was stained with TRITC-labeled phalloidin (red signal). Bar, 20 μm. (b) Microscopic quantification (bacteria/cell) of EC adherence. (c) Adherence of B. henselae WT to ECs shown by TEM. Note that adherence of B. henselae WT to the cell surface is mediated via BadA (arrow). (d) Adherence of B. henselae WT to GD25 (β1−) and GD25 β1 integrin–overexpressing (β1+) fibroblasts shown by CLSM. For modulation, bacteria and β1+ cells were preincubated with anti-Fn antibodies (β1+ αFN). (e) Microscopic quantification of bacterial adherence to GD25 and GD25 β1 integrin+ fibroblasts. (f) Intracellular B. henselae WT, Pil−, and BadA− 3 h after infection of J774 macrophages. Bacteria (percent of inoculum) were calculated from gentamicin protection assays. (g) Interaction of B. henselae WT with J774 macrophages shown by TEM. Note that adherence of B. henselae WT to the cell surface is mediated via BadA (arrow). *, Significant difference compared with B. henselae WT (P < 0.01).
Induction of a Proangiogenic Host Cell Response Depends on BadA Expression.
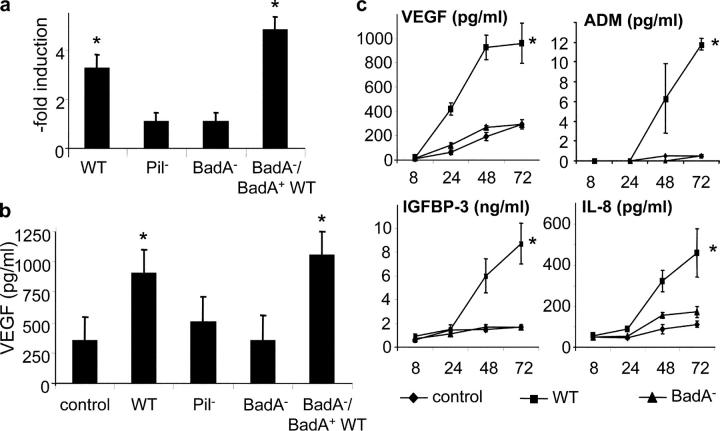
HIF-1 is the key transcription factor of angiogenesis regulating, e.g., VEGF, IGFBP-3, and ADM (9). As a B. henselae infection activates HIF-1 in host cells (epithelial and endothelial) in vitro and in BA lesions in vivo (unpublished data), we elucidated the role of BadA in this process (Fig. 7 a). HeLa cells were transiently transfected with a VEGF promoter luciferase reporter construct (pVEGF.4; reference 32) specifically regulated by HIF-1. Infection with B. henselae WT resulted in a 3.3-fold increase in HIF-1–regulated VEGF gene transcription (positive control: hypoxia, 13.2-fold stimulation). Both B. henselae Pil− (1.1-fold) and BadA− (1.1-fold) did not activate the VEGF promoter in contrast to BadA−/BadA+ WT (4.9-fold). Consistently, secretion of VEGF was fully induced by BadA−/BadA+ WT (Fig. 7 b). Next, the role of BadA in mediating an angiogenic host cell response was investigated. HeLa cells were infected with B. henselae WT and BadA−, and secretion of VEGF, ADM, IGFBP-3, and IL-8 was assessed over 72 h (Fig. 7 c). Infection with B. henselae WT led to a strong induction of these cytokines, whereas BadA− failed to induce any of these. From these data we conclude that expression of BadA by B. henselae is crucial for the activation of HIF-1 and has major importance in the proangiogenic reprogramming of host cells.
Figure 7.
BadA-dependent HIF-1 activation and secretion of proangiogenic cytokines. (a) HIF-1 activation detected by the use of a VEGF promoter luciferase reporter construct (pVEGF.4). HeLa cells were transfected before infection with B. henselae WT, Pil−, BadA−, and BadA−/BadA+ WT. Activation was determined by chemiluminescence 36 h after infection. (b) Induction of VEGF secretion upon infection of HeLa cells. Supernatants were taken 48 h after infection and analyzed by ELISA. (c) BadA-dependent secretion of proangiogenic cytokines in Hela cells. Cells were infected with B. henselae WT and BadA−, and the amount of secreted VEGF, IGFBP-3, ADM, and IL-8 was determined 8, 24, 48, and 72 h after infection by ELISA or RIA. *, significant difference compared with uninfected control (P < 0.01).
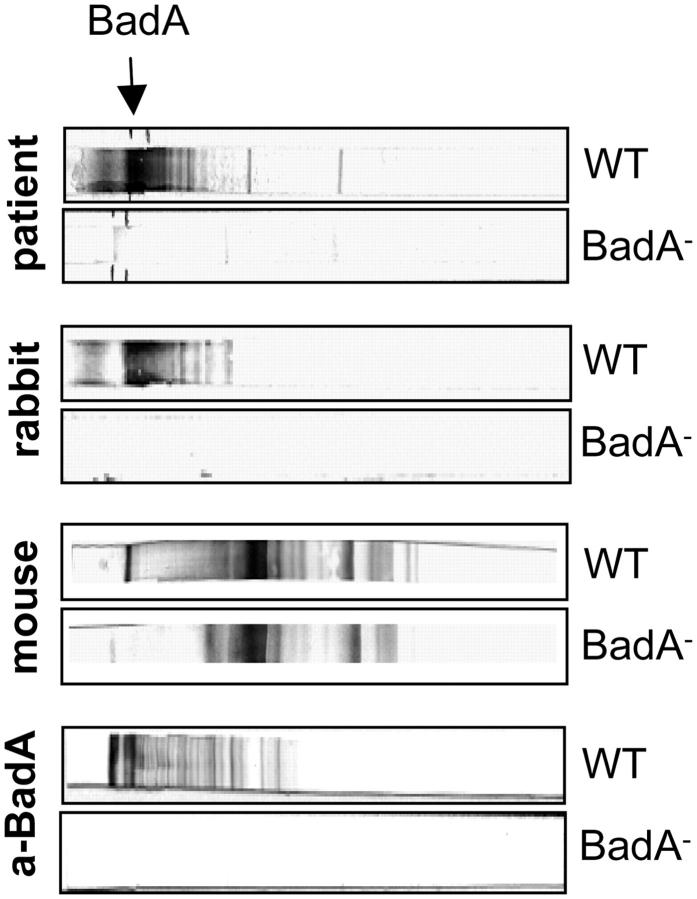
BadA Is an Immunodominant Surface Molecule in B. henselae–infected Patients and Rodents.
It has been shown that NadA leads to an anti-NadA immune response in mice (17). Therefore, we were interested in the immunological properties of BadA in B. henselae infections (Fig. 8 and Fig. S2). Serum of 7 out of 11 patients suffering from a clinically diagnosed and serologically confirmed CSD (IgG reactivity in immunofluorescence test: 1:200) showed a strong reactivity to BadA identified by Western blots. The BadA-specific bands were absent in B. henselae Pil− (not depicted) and BadA−. The same results were obtained with the serum of a rabbit infected with viable B. henselae WT. Anti-BadA reactivity was also seen in sera of mice immunized with heat-killed B. henselae. These results suggest that (a) BadA is expressed in vivo in patients suffering from Bartonella infections, (b) BadA-specific antibodies are produced in the course of CSD, and (c) that BadA might be a diagnostic marker for the serological diagnosis of B. henselae infections.
Figure 8.
Immunodominance of BadA in humans, rabbits, and mice. Western blots of whole cell lysates of B. henselae WT and BadA− were incubated with serum of a patient suffering from CSD (see also Fig. S2), serum of a rabbit infected with viable B. henselae, and serum of a mouse immunized with heat-killed B. henselae. The arrow indicates BadA. Note that antibodies against BadA are detected in each of the sera. Immunoreactivity of a specific anti-BadA rabbit serum is shown for control.
Discussion
Besides pili, nonpilus-associated adhesins, which are monomeric or oligomeric proteins anchored to the outer membrane, are important bacterial pathogenicity factors. The nonfimbrial adhesins, e.g., YadA of enteropathogenic Yersinia species and NadA of N. meningitidis represent a novel class of adhesins widely distributed in proteobacteria (16, 17).
BadA Represents a Novel Nonfimbrial Adhesin of B. henselae.
The large, filamentous surface structures of B. henselae were formerly called type IV pili based on phenotypic and functional properties (adhesion to host cells, autoagglutination; reference 12). Serial passage in vitro leads to a loss of pilus expression in B. henselae. However, we have not been able to detect pilin genes in B. henselae Marseille using a degenerated probe (Table II) for Southern blotting deduced from pilA of Caulobacter crescentus (sequence data are available from GenBank/EMBL/DDBJ under accession no. AF_229646), Agrobacterium tumefaciens (accession no. AE_007963), S. meliloti (accession no. AL_591782), and M. loti (accession no. NP_104554; not depicted). Moreover, no pilin genes were found in the B. henselae Houston-1 genome sequence (24).
We show that a transposon mutant deficient in expression of the 340-kD BadA was complemented by badA (9.3 kb) cloned with its putative promoter region. Our investigations revealed that the so-called type IV pilus of B. henselae belongs, like YadA and NadA, to a novel family of nonfimbrial adhesins (16).
B. henselae OMPs have been suggested to be relevant for attachment to ECs (13), induction of a proinflammatory cell response (14), and induction of EC proliferation (5). However, the presence of BadA was not indicated in any of these reports. Reasons for the lack of BadA in the published OMP patterns might be explained by difficulties in gel electrophoresis due to protein size (340 kD) or loss of BadA expression due to extensive passaging and phase variation (7, 12).
Most data on the pathogenicity of B. henselae have been obtained using bacteria of variable and unstated passage number. The identification of a deletion mutation in the published genome sequence of the Houston-1 strain and the high density of repeated sequences within the badA gene and BH01490, which provides targets for recombination and deletion events, suggests that a high level of variability in sequence and expression among strains is to be expected. Therefore, we recommend that expression of BadA should be evaluated when performing infection experiments with B. henselae.
BadA Is an Unusual Representative of the Nonfimbrial Adhesins.
Nonfimbrial adhesins share a common architecture, consisting of a head, a stalk, and a membrane anchor (16). This architecture is reflected in a number of broadly occurring sequence motifs, most notably the degenerate 14-residue head repeats, which have been shown in YadA to form a left-handed β-helix (33), the neck sequence, which in YadA separates the head from the stalk and forms a novel trimerization motif (33), and the membrane anchor region, consisting of four transmembrane β strands with potential pore-forming and autotransporter properties (34). All of these motifs are also found in BadA, showing that this protein is a canonical nonfimbrial adhesin. BadA, however, contains additional sequence motifs that have not, so far, been described. Strikingly, these motifs have their closest matches in proteins from γ proteobacteria (Xylella, Escherichia, Shigella, and Salmonella), whereas B. henselae belongs to the α proteobacteria, suggesting an active “trade” in adhesion domains between phylogenetically distant bacteria. Most conspicuous are segments of ∼100 residues, which occur in 21 partly truncated copies in the center of the molecule and are separated from each other by neck sequences. These segments, which contain interspersed coiled coil sequences, account for the surprising size of BadA and probably represent a novel type of stalk architecture judging from electron micrographs. Sequence similarity leads us to expect that similar adhesins will be found on the surface of uropathogenic and enterohaemorrhagic E. coli as well as of Salmonella spp.
The Role of BadA in the Infection Process.
Collagen binding of Y. enterocolitica depends on the head repeats of YadA (37). Similar repeats are also present in the BadA head domain and it might be speculated that these motifs mediate adhesion of B. henselae to ECM proteins of the basal membrane of blood vessels, which mainly consist of collagen IV and laminin (38), facilitating subsequent infection of ECs. In accordance, adherence of B. henselae to collagen type I, III, and IV depends on BadA expression. The molecular basis of the laminin- and Fn-binding capacity of BadA remains unclear. Like YadA (36), BadA shares antiphagocytic capacities, as expression of BadA prevents B. henselae from phagocytosis in J774 murine macrophages.
Our data clearly show that expression of BadA is important for adherence of B. henselae to ECs that represent one potential habitat (10, 11). It is known that Fn-binding proteins, such as those of N. meningitidis, promote the infection process of host cells, possibly via Fn bridging to α5β1 integrins (39). Using β1 integrin–overexpressing GD25 cells and anti-Fn antibodies, we demonstrated that β1 integrins are crucial for cell adhesion of B. henselae, possibly via Fn bridging. Our data are also in line with previous reports (35) that show that β1 integrins are required for YadA binding to host cells.
The Role of BadA in the Induction of a Proangiogenic Host Cell Response.
HIF-1 is the key transcription factor in angiogenesis (9). Of the many genes induced by HIF, VEGF represents the major mitogen for ECs (40). We and others demonstrated that a B. henselae infection results in host cell VEGF secretion in vitro and in BA and BP in vivo (7, 8). Moreover, HIF-1 is activated in host cells by B. henselae in vitro and in BA lesions in vivo, and VEGF, IGFBP-3, ADM, and IL-8 (all sharing angiogenic capacities; references 41–43) are secreted in vitro (unpublished data). Our data show that expression of BadA is crucial for both HIF-1 activation and secretion of proangiogenic compounds. Therefore, BadA appears to play a crucial role in the induction of a proangiogenic host cell response. Whether BadA directly triggers expression of proangiogenic factors or mediates adhesion of B. henselae to host cells followed by subsequent pathogen–host cell interactions, is not yet clear. Binding of Fn to α5β1 integrins results in activation of a proangiogenic gene expression program (44). One could speculate that BadA might be involved in triggering a proangiogenic host cell response via Fn bridging to α5β1 integrins.
Immunodominance of BadA.
Sera of patients suffering from Bartonella infection and of rabbits infected with viable B. henselae reacted with BadA in immunoblotting (Fig. 8 and Fig. S2), suggesting that BadA is expressed in B. henselae infections. The biological functions of BadA expression in vivo might be to avoid phagocytosis similar to YadA (36) and to adhere to ECs. Antibodies against YadA and NadA mediate protection in Y. enterocolitica and N. meningitidis infections (17, 45). For vaccination strategies against zoonotic Bartonella in their mammalian reservoirs, BadA could therefore be a promising vaccine candidate. The immunodominance of BadA in the sera of B. henselae–infected patients suggests also that it might be a suitable marker for serodiagnosis of B. henselae infections.
In conclusion, the nonfimbrial adhesin BadA is (a) an unusual modularly constructed, surface-exposed HMW protein, (b) highly important for pathogen–host cell interactions, (c) involved in the induction of a proangiogenic host cell response, and (d) an immunodominant antigen. Further investigations will elucidate the role of this multifunctional molecule in Bartonella infections.
Acknowledgments
We thank B. Schütt for performing IGFBP-3 and ADM RIAs. We also thank A. Roggenkamp, J. Heesemann, K. Alitalo, and C. Wolz for helpful discussions and D. Neumann, E. Januschke, B. Fehrenbacher, L. Nirell, K. Strijbis, A.-S. Eriksson, and A. Ursinus for excellent technical assistance.
The work of V.A.J. Kempf was supported by grants from the Deutsche Forschungsgemeinschaft (DFG), the “Landesforschungsschwerpunktprogramm” of the Ministry of Science, Research and Arts Baden-Württemberg, and from the University of Tübingen (Fortüne-Programm). The work of S.G.E. Andersson was supported from the Wallenberg Foundation, the Foundation for Strategic Research, and the Swedish Research Council. A. Nordheim is supported by the DFG, the MWK Stuttgart, and the Fonds der Chemischen Industrie.
The authors have no conflicting financial interests.
P. Kyme's present address is Centre for Infectious Diseases and Microbiology, University of Sydney, Westmead Hospital, New South Wales 2145, Australia.
Abbreviations used in this paper: ADM, adrenomedullin; BA, bacillary angiomatosis; BadA, Bartonella adhesin A; BP, bacillary peliosis; CBA, Columbia blood agar; CLSM, confocal laser scanning microscopy; CSD, cat scratch disease; EC, endothelial cell; ECM, extracellular matrix; Fn, fibronectin; HIF, hypoxia-inducible factor; HMW, high molecular weight; IEM, immunoelectronmicroscopy; IGFBP-3, insulin-like growth factor binding protein 3; NadA, Neisseria adhesin A; OMP, outer membrane protein; PFA, paraformaldehyde; TEM, transmission electron microscopy; VEGF, vasculoendothelial growth factor; YadA, Yersinia adhesin A.
References
- 1.Anderson, B.E., and M.A. Neuman. 1997. Bartonella spp. as emerging human pathogens. Clin. Microbiol. Rev. 10:203–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Relman, D.A., J.S. Loutit, T.M. Schmidt, S. Falkow, and L.S. Tompkins. 1990. The agent of bacillary angiomatosis. An approach to the identification of uncultured pathogens. N. Engl. J. Med. 323:1573–1580. [DOI] [PubMed] [Google Scholar]
- 3.Koehler, J.E., F.D. Quinn, T.G. Berger, P.E. LeBoit, and J.W. Tappero. 1992. Isolation of Rochalimaea species from cutaneous and osseous lesions of bacillary angiomatosis. N. Engl. J. Med. 327:1625–1631. [DOI] [PubMed] [Google Scholar]
- 4.Drancourt, M., R. Birtles, G. Chaumentin, F. Vandenesch, J. Etienne, and D. Raoult. 1996. New serotype of Bartonella henselae in endocarditis and cat-scratch disease. Lancet. 347:441–443. [DOI] [PubMed] [Google Scholar]
- 5.Conley, T., L. Slater, and K. Hamilton. 1994. Rochalimaea species stimulate human endothelial cell proliferation and migration in vitro. J. Lab. Clin. Med. 124:521–528. [PubMed] [Google Scholar]
- 6.Kirby, J.E., and D.M. Nekorchuk. 2002. Bartonella-associated endothelial proliferation depends on inhibition of apoptosis. Proc. Natl. Acad. Sci. USA. 99:4656–4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kempf, V.A., B. Volkmann, M. Schaller, C.A. Sander, K. Alitalo, T. Riess, and I.B. Autenrieth. 2001. Evidence of a leading role for VEGF in Bartonella henselae-induced endothelial cell proliferations. Cell. Microbiol. 3:623–632. [DOI] [PubMed] [Google Scholar]
- 8.Resto-Ruiz, S.I., M. Schmiederer, D. Sweger, C. Newton, T.W. Klein, H. Friedman, and B.E. Anderson. 2002. Induction of a potential paracrine angiogenic loop between human THP-1 macrophages and human microvascular endothelial cells during Bartonella henselae infection. Infect. Immun. 70:4564–4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pugh, C.W., and P.J. Ratcliffe. 2003. Regulation of angiogenesis by hypoxia: role of the HIF system. Nat. Med. 9:677–684. [DOI] [PubMed] [Google Scholar]
- 10.Brouqui, P., and D. Raoult. 1996. Bartonella quintana invades and multiplies within endothelial cells in vitro and in vivo and forms intracellular blebs. Res. Microbiol. 147:719–731. [DOI] [PubMed] [Google Scholar]
- 11.Kempf, V.A., M. Schaller, S. Behrendt, B. Volkmann, M. Aepfelbacher, I. Cakman, and I.B. Autenrieth. 2000. Interaction of Bartonella henselae with endothelial cells results in rapid bacterial rRNA synthesis and replication. Cell. Microbiol. 2:431–441. [DOI] [PubMed] [Google Scholar]
- 12.Batterman, H.J., J.A. Peek, J.S. Loutit, S. Falkow, and L.S. Tompkins. 1995. Bartonella henselae and Bartonella quintana adherence to and entry into cultured human epithelial cells. Infect. Immun. 63:4553–4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burgess, A.W., and B.E. Anderson. 1998. Outer membrane proteins of Bartonella henselae and their interaction with human endothelial cells. Microb. Pathog. 25:157–164. [DOI] [PubMed] [Google Scholar]
- 14.Fuhrmann, O., M. Arvand, A. Gohler, M. Schmid, M. Krull, S. Hippenstiel, J. Seybold, C. Dehio, and N. Suttorp. 2001. Bartonella henselae induces NF-kappaB-dependent upregulation of adhesion molecules in cultured human endothelial cells: possible role of outer membrane proteins as pathogenic factors. Infect. Immun. 69:5088–5097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmid, M.C., R. Schulein, M. Dehio, G. Denecker, I. Carena, and C. Dehio. 2004. The VirB type IV secretion system of Bartonella henselae mediates invasion, proinflammatory activation and antiapoptotic protection of endothelial cells. Mol. Microbiol. 52:81–92. [DOI] [PubMed] [Google Scholar]
- 16.Hoiczyk, E., A. Roggenkamp, M. Reichenbecher, A. Lupas, and J. Heesemann. 2000. Structure and sequence analysis of Yersinia YadA and Moraxella UspAs reveal a novel class of adhesins. EMBO J. 19:5989–5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Comanducci, M., S. Bambini, B. Brunelli, J. Adu-Bobie, B. Arico, B. Capecchi, M.M. Giuliani, V. Masignani, L. Santini, S. Savino, et al. 2002. NadA, a novel vaccine candidate of Neisseria meningitidis. J. Exp. Med. 195:1445–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roggenkamp, A., K. Ruckdeschel, L. Leitritz, R. Schmitt, and J. Heesemann. 1996. Deletion of amino acids 29 to 81 in adhesion protein YadA of Yersinia enterocolitica serotype O:8 results in selective abrogation of adherence to neutrophils. Infect. Immun. 64:2506–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schulze-Koops, H., H. Burkhardt, J. Heesemann, K. von der Mark, and F. Emmrich. 1992. Plasmid-encoded outer membrane protein YadA mediates specific binding of enteropathogenic yersiniae to various types of collagen. Infect. Immun. 60:2153–2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pepe, J.C., M.R. Wachtel, E. Wagar, and V.L. Miller. 1995. Pathogenesis of defined invasion mutants of Yersinia enterocolitica in a BALB/c mouse model of infection. Infect. Immun. 63:4837–4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Serodiagnosis of Emerging Infectious Diseases. Bartonella and Ehrlichia Infections (course manual). 1999. Centers of Disease Control and Prevention. Atlanta, USA.
- 22.Shevchenko, A., I. Chernushevich, W. Ens, K.G. Standing, B. Thomson, M. Wilm, and M. Mann. 1997. Rapid ‘de novo’ peptide sequencing by a combination of nanoelectrospray, isotopic labeling and a quadrupole/time-of-flight mass spectrometer. Rapid Commun. Mass Spectrom. 11:1015–1024. [DOI] [PubMed] [Google Scholar]
- 23.Riess, T., B. Anderson, A. Fackelmayer, I.B. Autenrieth, and V.A. Kempf. 2003. Rapid and efficient transposon mutagenesis of Bartonella henselae by transposome technology. Gene. 313:103–109. [DOI] [PubMed] [Google Scholar]
- 24.Alsmark, C.M., A.C. Frank, E.O. Karlberg, B.A. Legault, D.H. Ardell, B. Canback, A.S. Eriksson, A.K. Naslund, S.A. Handley, M. Huvet, et al. 2004. The louse-borne human pathogen Bartonella quintana is a genomic derivative of the zoonotic agent Bartonella henselae. Proc. Natl. Acad. Sci. USA. 101:9716–9721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schuler, G.D., S.F. Altschul, and D.J. Lipman. 1991. A workbench for multiple alignment construction and analysis. Proteins. 9:180–190. [DOI] [PubMed] [Google Scholar]
- 26.Lupas, A., M. Van Dyke, and J. Stock. 1991. Predicting coiled coils from protein sequences. Science. 252:1162–1164. [DOI] [PubMed] [Google Scholar]
- 27.McGuffin, L.J., K. Bryson, and D.T. Jones. 2000. The PSIPRED protein structure prediction server. Bioinformatics. 16:404–405. [DOI] [PubMed] [Google Scholar]
- 28.Fassler, R., M. Pfaff, J. Murphy, A.A. Noegel, S. Johansson, R. Timpl, and R. Albrecht. 1995. Lack of beta 1 integrin gene in embryonic stem cells affects morphology, adhesion, and migration but not integration into the inner cell mass of blastocysts. J. Cell Biol. 128:979–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wennerberg, K., L. Lohikangas, D. Gullberg, M. Pfaff, S. Johansson, and R. Fassler. 1996. Beta 1 integrin-dependent and -independent polymerization of fibronectin. J. Cell Biol. 132:227–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schulte, R., G.A. Grassl, S. Preger, S. Fessele, C.A. Jacobi, M. Schaller, P.J. Nelson, and I.B. Autenrieth. 2000. Yersinia enterocolitica invasin protein triggers IL-8 production in epithelial cells via activation of Rel p65-p65 homodimers. FASEB J. 14:1471–1484. [DOI] [PubMed] [Google Scholar]
- 31.Blum, W.F., M.B. Ranke, K. Kietzmann, E. Gauggel, H.J. Zeisel, and J.R. Bierich. 1990. A specific radioimmunoassay for the growth hormone (GH)-dependent somatomedin-binding protein: its use for diagnosis of GH deficiency. J. Clin. Endocrinol. Metab. 70:1292–1298. [DOI] [PubMed] [Google Scholar]
- 32.Ikeda, E., M.G. Achen, G. Breier, and W. Risau. 1995. Hypoxia-induced transcriptional activation and increased mRNA stability of vascular endothelial growth factor in C6 glioma cells. J. Biol. Chem. 270:19761–19766. [DOI] [PubMed] [Google Scholar]
- 33.Nummelin, H., M.C. Merckel, J.C. Leo, H. Lankinen, M. Skurnik, and A. Goldman. 2004. The Yersinia adhesin YadA collagen-binding domain structure is a novel left-handed parallel beta-roll. EMBO J. 23:701–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roggenkamp, A., N. Ackermann, C.A. Jacobi, K. Truelzsch, H. Hoffmann, and J. Heesemann. 2003. Molecular analysis of transport and oligomerization of the Yersinia enterocolitica adhesin YadA. J. Bacteriol. 185:3735–3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bliska, J.B., M.C. Copass, and S. Falkow. 1993. The Yersinia pseudotuberculosis adhesin YadA mediates intimate bacterial attachment to and entry into HEp-2 cells. Infect. Immun. 61:3914–3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.China, B., B.T. N'Guyen, M. de Bruyere, and G.R. Cornelis. 1994. Role of YadA in resistance of Yersinia enterocolitica to phagocytosis by human polymorphonuclear leukocytes. Infect. Immun. 62:1275–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.El Tahir, Y., and M. Skurnik. 2001. YadA, the multifaceted Yersinia adhesin. Int. J. Med. Microbiol. 291:209–218. [DOI] [PubMed] [Google Scholar]
- 38.Yurchenco, P.D., and J.C. Schittny. 1990. Molecular architecture of basement membranes. FASEB J. 4:1577–1590. [DOI] [PubMed] [Google Scholar]
- 39.Unkmeir, A., K. Latsch, G. Dietrich, E. Wintermeyer, B. Schinke, S. Schwender, K.S. Kim, M. Eigenthaler, and M. Frosch. 2002. Fibronectin mediates Opc-dependent internalization of Neisseria meningitidis in human brain microvascular endothelial cells. Mol. Microbiol. 46:933–946. [DOI] [PubMed] [Google Scholar]
- 40.Yancopoulos, G.D., S. Davis, N.W. Gale, J.S. Rudge, S.J. Wiegand, and J. Holash. 2000. Vascular-specific growth factors and blood vessel formation. Nature. 407:242–248. [DOI] [PubMed] [Google Scholar]
- 41.Oehler, M.K., S. Hague, M.C. Rees, and R. Bicknell. 2002. Adrenomedullin promotes formation of xenografted endometrial tumors by stimulation of autocrine growth and angiogenesis. Oncogene. 21:2815–2821. [DOI] [PubMed] [Google Scholar]
- 42.Schmid, M.C., M. Bisoffi, A. Wetterwald, E. Gautschi, G.N. Thalmann, S. Mitola, F. Bussolino, and M.G. Cecchini. 2003. Insulin-like growth factor binding protein-3 is overexpressed in endothelial cells of mouse breast tumor vessels. Int. J. Cancer. 103:577–586. [DOI] [PubMed] [Google Scholar]
- 43.Koch, A.E., P.J. Polverini, S.L. Kunkel, L.A. Harlow, L.A. DiPietro, V.M. Elner, S.G. Elner, and R.M. Strieter. 1992. Interleukin-8 as a macrophage-derived mediator of angiogenesis. Science. 258:1798–1801. [DOI] [PubMed] [Google Scholar]
- 44.Klein, S., A.R. de Fougerolles, P. Blaikie, L. Khan, A. Pepe, C.D. Green, V. Koteliansky, and F.G. Giancotti. 2002. Alpha 5 beta 1 integrin activates an NF-kappa B-dependent program of gene expression important for angiogenesis and inflammation. Mol. Cell. Biol. 22:5912–5922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vogel, U., I.B. Autenrieth, R. Berner, and J. Heesemann. 1993. Role of plasmid-encoded antigens of Yersinia enterocolitica in humoral immunity against secondary Y. enterocolitica infection in mice. Microb. Pathog. 15:23–36. [DOI] [PubMed] [Google Scholar]
- 46.Kovach, M.E., R.W. Phillips, P.H. Elzer, R.M. Roop, and K.M. Peterson. 1994. pBBR1MCS: a broad-host-range cloning vector. Biotechniques. 16:800–802. [PubMed] [Google Scholar]