Abstract
Although stable repression of CD4 and CD8 genes is a central feature of T cell lineage commitment, we lack detailed information about the timing and mechanism of this repression. Stable gene repression has been linked to the position of genes within the nucleus. Therefore, information about the nuclear position of CD4 and CD8 genes during T cell development could provide insights into both the mechanism of regulation of CD4 and CD8 genes, and the process of lineage commitment. Here, we report that lineage-specific repression of CD4 and CD8 genes is associated with the repositioning of alleles close to heterochromatin. We also provide evidence that the relocalization of CD4 and CD8 genes to heterochromatin can occur as an early response to positive selection signals. We discuss our results in terms of our current knowledge of CD4 and CD8 gene regulation and CD4 versus CD8 lineage commitment.
Keywords: lineage commitment, heterochromatin, positive selection, gene repression, nuclear position
Introduction
The two major lineages of mature T cells are distinguished by expression of the cell surface proteins CD4 and CD8. CD4 and CD8 are coreceptors that bind to MHC class II and I, respectively, and cooperate with the TCR to recognize peptide–MHC complexes during antigen recognition in the periphery, and during T cell selection in the thymus. In the thymus, early T cell progenitors initially lack CD4 and CD8 expression (CD4− CD8−, double negative [DN] thymocytes), and then turn on expression of both coreceptors (CD4+ CD8+, double positive [DP] thymocytes). DP thymocytes then undergo a stringent selection process to ensure that their newly formed TCRs have a weak reactivity to self-peptide/MHC (positive selection), and are not strongly self-reactive (negative selection). After positive selection, DP thymocytes down-regulate either CD4 or CD8 to give rise to mature thymocytes (CD4+ CD8− or CD4− CD8+ single positive [SP] thymocytes). The decision to down-regulate CD4 or CD8 is linked to the specificity of TCR for MHC class I or II. Thymocytes that undergo positive selection via recognition of MHC class I down-regulate CD4 and give rise to mature CD8 T lineage cells. In contrast, thymocytes that undergo positive selection via MHC class II down-regulate CD8 and give rise to CD4 lineage T cells. Thus, the down-regulation of CD4 and CD8 in the thymus is tightly linked to the processes of positive selection and lineage commitment.
Because of their central role in T cell function, and their prominence as differentiation markers for T cells, considerable effort has gone into understanding the regulation of CD4 and CD8 genes (for review see references 1–3). CD4 repression in DN and CD8 SP thymocytes is largely controlled by an intronic silencer (4) and Runx transcription factors (5–7). Other transcription factors including SAF and HES have also been implicated in CD4 regulation. For CD8, a number of cis- and trans-acting factors have been identified as positive regulators of gene expression, including Ikaros (8). Other transcription factors have been implicated in CD4 and CD8 T cell development including GATA3, which is required for CD4 T cell development (9, 10), and Tox, which can promote CD8 T cell development when ectopically expressed (11). How these transcription factors work together during the development of CD4 and CD8 T cells, and how they are regulated by positive selection signals, is not yet clear. In addition to cis-acting DNA sequences, and the transcription factors that bind them, information about mechanism of gene regulation can also come from analysis of the position of alleles within the nucleus (12–15). This type of approach has not yet been applied to the study of CD4 and CD8 gene regulation during thymic development.
Progress in understanding the mechanism of CD4/CD8 lineage commitment would be advanced by assays to assess the early stages of lineage commitment. In theory, this could be done by examining the initial down-regulation of the CD4 and CD8 genes. However, attempts to define intermediate stages of positive selection by identifying thymocyte populations expressing intermediate levels of surface CD4 and CD8, have led to surprising results. For example, numerous studies have shown that CD4+ CD8low thymocytes, which were originally assumed to be in transition to the CD4 lineage, can give rise to mature CD8 SP thymocytes as well as mature CD4 SP thymocytes (16–20). In addition, negative selection signals can lead to coordinate down-regulation of CD4 and CD8 surface levels (21–23). These complexities arise in part from multiple controls on surface coreceptor expression including protein turnover and mRNA stability, as well as transcription (24, 25). Attempts to assess CD4 and CD8 commitment using pronase stripping and reexpression assays (18) are subject to many of the same confounding issues, as well as the potential artifacts introduced by the pronase treatment itself. Perhaps the most direct approach to assess the developmental potential of a thymocyte population is to isolate the population in question and follow its differentiation after transfer into the thymus or thymic organ culture (16, 17). Although this approach can be informative, the low cell recoveries make it difficult to draw firm conclusions regarding the lineage commitment of the starting population.
Previous studies have shown that stable repression of gene expression can be accompanied by repositioning of the silenced gene to heterochromatin (12–14). We reasoned that the location of the CD4 and CD8 genes relative to heterochromatin could provide information both about the mechanism of stable repression of these genes and insights into the timing and mechanism of T cell lineage commitment. We find that CD4 and CD8 genes tend to be positioned near heterochromatin in thymocyte and T cell populations in which they are not expressed. We also show that the repositioning of CD4 and CD8 near heterochromatin can be observed in DP thymocytes undergoing positive selection. We discuss our results in terms of current models for the CD4 versus CD8 lineage choice.
Materials and Methods
Mice.
C57BL/6 (B6), MHC−/− (C57BL/6 β2-microglobulin I-Ab−/− I-E null; Taconic; reference 26), F5 TCR transgenic (provided by D. Kioussis, NIMR, London UK; reference 27), P14 TCR transgenic RAG-2−/− (Taconic; reference 28), 5CC7 TCR transgenic C57BL/10.A RAG-2−/− (Taconic; reference 29), AND TCR transgenic (The Jackson Laboratory; reference 30), OT-1, and OT-2 RAG-2−/− TCR transgenic (31) mice were bred and maintained in the University of California, Berkeley mouse facility. All animal procedures were approved by the Animal Care and Use Committee of University of California, Berkeley. Mice between 4 and 12 wk of age were used for analyses (31).
Purification of Thymocyte and T Cell Subsets.
DP thymocytes from F5 and P14 TCR transgenic mice were incubated with anti-CD4 beads and thymocytes from 5CC7 and AND TCR transgenic mice were incubated with anti-CD8 beads, and the positive fractions were purified by AutoMacs magnetic bead separation according to the manufacturer's guidelines (Miltenyi Biotec). TCR transgenic CD4+ SP thymocytes were purified by negative selection of DP thymocytes with anti-CD8 beads followed by positive selection with anti-CD4 beads. CD8+ SP thymocytes were similarly purified with beads incubated in the reverse order. Whole B6 thymocytes and thymocytes complement depleted of heat stable antigen–expressing cells were FACS sorted to isolate B6 DP and SP thymocytes. LN T cells were purified by negative selection on mouse CD3+ mini T cell enrichment columns (R&D Systems). Populations were determined to be 90–99% pure based on flow cytometry. For activation of mature T cells, LN cells were cultured at a concentration of 106/ml in presence of 1 μg/ml Con A for 48 h. CD4+ T cells from cultures were isolated by positive selection using anti-CD4 beads.
DPK In Vitro Differentiation.
DCEK cells, an L cell derivative transfected with I-Ek, were pretreated with mitomycin C and pulsed with PCC peptide. DPK cells (32) were then added at a concentration of 106/ml in Click's media supplemented with 10% fetal calf serum, penicillin, streptomycin, and 50 μM 2-mercaptoethanol. At the indicated times, DPK cells were removed by gentle pipetting and analyzed by flow cytometry for CD4, CD8, and CD69 expression to monitor in vitro differentiation. For fluorescence in situ hybridization (FISH) analysis, DPK cells were removed from cocultures by gentle pipetting and FACS sorted for CD4 expression to exclude DCEK cells from the analysis. Where indicated, DPK cells from 18-h stimulation cultures were sorted into CD4+ CD69+ and CD4+ CD69− fractions by FACS before analysis by FISH.
FISH.
The CD4 probe was a 10-kb insert from a λ genomic clone (provided by A. Rahemtulla, University of Oxford, Oxford, UK). The CD8α probe was a 30-kb insert from a CD8 genomic cosmid clone (provided by D. Kioussis). CD4 and CD8α probes were labeled with digoxigenin and the γ satellite probe was labeled with FITC using a nick translation kit (Roche Applied Science) according to the manufacturer's instructions. γ satellite and CD4 or CD8α genes were detected as described previously (14) with small modifications. In brief, cells fixed with NaBH4 were blocked in 1% BMB blocking reagent (Roche Applied Science) containing 50 ug/ml herring sperm DNA for 2 h at 37°C. Probes were denatured in 50% formamide/10% dextran sulfate in 2X SSC at 94°C for 4 min, and then incubated at 37°C for 10 min. Chromosomal DNA was denatured in 50% formamide/2X SSC at 94°C for 4 min. Hybridization was performed overnight at 37°C in humid chambers. Digoxigenin-labeled probes were detected with sheep anti–digoxigenin-rhodamine (Roche Applied Science) and for CD4 detection, were amplified with donkey anti–sheep IgG-rhodamine (Jackson ImmunoResearch Laboratories).
Cells were stained with DAPI and analyzed using multiwavelength wide-field, three-dimensional microscopy with computer-driven shutters, filter wheels, focus movement, and data collection. For fluorescence analysis of fixed cells, data stacks of immunofluorescent images were acquired in the FITC and rhodamine channels by moving the stage in successive 0.25-μm focal planes through the sample. Out of focus light was removed with a nearest neighbor deconvolution algorithm. Data were scored manually by identifying cells that had one or two clear signals for the signal copy probe. The proximity of each allele to γ satellite probe was assessed by examination of all focal planes. A number of samples were rescored after being identified only by letter. In each case the values for percent of alleles associated with γ satellite probe agreed within 3%.
Flow Cytometry.
Thymocytes were examined by three-color flow cytometry using combinations of anti-CD4, anti-CD5, anti-CD8, anti-CD69, and anti–TCR antibody conjugated to FITC, phycoerythrin, or phycoerythrin-Cy5 (BD Biosciences and eBioscience), and anti-CD4 and anti-CD8 conjugated to phycoerythrin Texas red (Caltag). Cells were processed on a Coulter Epics XL-MCL and analyzed using FlowJo software (Tree Star).
Results
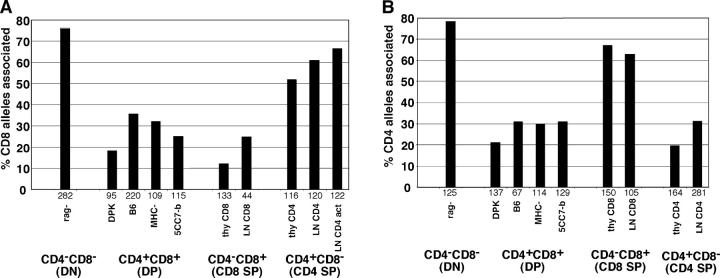
To examine the location of the CD4 and CD8 genes relative to heterochromatin, we used the technique of FISH. In this technique nuclei are stained with a gene-specific probe together with a probe for γ satellite DNA as a marker for centromeric heterochromatin, and individual alleles are scored as being associated with heterochromatin. Analysis of various thymocyte and T cell populations revealed a strong correlation between expression of CD4 and CD8 and their association with heterochromatin (Figs. 1 and 2). Both genes were strongly associated with heterochromatin in early CD4− CD8− DN thymocytes (76–78% of alleles), but only weakly associated (18–35%) in CD4+ CD8+ DP thymocytes. In addition, the lineage-specific repression of CD4 and CD8 in mature CD4+ CD8− and CD4− CD8+ SP thymocytes was accompanied by repositioning of the repressed coreceptor gene to heterochromatin. Importantly, the extent of heterochromatin association was similar between mature peripheral T cells and SP thymocytes, implying that repositioning to heterochromatin is an early event after positive selection. It is also interesting to note that the extent of heterochromatin association of the repressed coreceptor gene in mature T cells and thymocytes (52–67%) was less pronounced than the association seen in early DN thymocytes. This may reflect a difference in the mechanism of repression of a gene that has been recently expressed versus one that has not. Together, these data indicate that the induction and repression of the CD4 and CD8 genes during different stages of T cell development correlates with their association with centromeric heterochromatin.
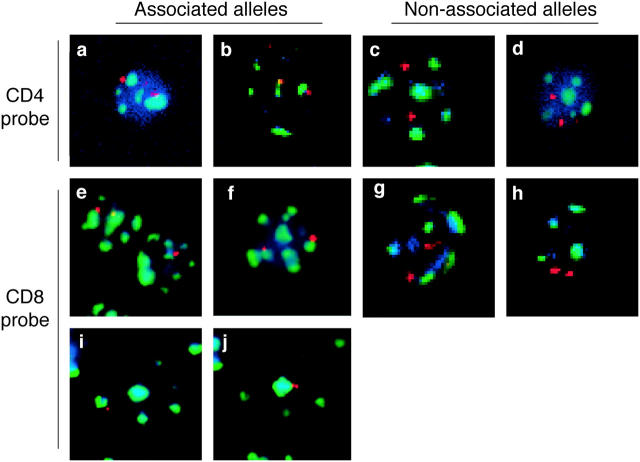
Figure 1.
Analysis of the position of CD4 and CD8 alleles relative to heterochromatin using FISH analysis. Thymocyte or T cell populations were isolated and analyzed by FISH using probes for either CD4 (a–d) or CD8α (e–j) genes (red). Samples were costained with probes for γ satellite DNA (green) as a marker for heterochromatin as described in Materials and Methods. DAPI staining (blue) is included to help locate the nucleus. Examples of associated alleles are shown on the left and examples of nonassociated alleles are shown on the right. Representative deconvolved fluorescent images are shown for the following samples: (a) CD4− CD8− thymocytes from RAG-1− mice, (b) CD4+ CD8+ thymocytes from P14 TCR transgenic mice, (c) CD4+ CD8− LN T cells from wild-type mice, (d) CD4+ CD8+ thymocyte from F5 TCR transgenic mice, (e) CD4− CD8− thymocytes from RAG-1− mice, (f) CD4+ CD8− thymocytes from 5CC7 TCR transgenic mice, (g) CD4+ CD8+ thymocytes from wild-type mice, (h) CD4+ CD8+ thymocytes from F5 TCR transgenic mice, (i and j) activated CD4+ CD8− LN T cells. (i and j) Two different focal (z) planes of the same cell taken 1.8 μm apart are shown. Compiled data for percent of alleles associated are shown in Figs. 2 and 3. Note that each image corresponds to a single focal plane, however complete z-series for each cell was recorded and individual alleles were scored as associated or nonassociated with heterochromatin based on examination of all focal planes.
Figure 2.
Dynamic repositioning of CD4 and CD8 genes relative to heterochromatin during T cell development. The indicated thymocyte and T cell populations were isolated and analyzed by FISH using probes for either CD8α (A) or CD4 (B) genes. Samples were costained with probes for γ satellite DNA as a marker for heterochromatin. Individual CD4 or CD8 alleles were scored for association with heterochromatin and the percent of alleles associated with heterochromatin is displayed. The number of alleles scored for each sample is indicated beneath each set of bars. Data are compiled from two to four independent experiments for each sample. Samples are denoted as follows: rag-, thymocytes from RAG-1–deficient mice that were >95% CD4−CD8−; DPK, a CD4+ CD8+ thymocyte cell line (reference 32); B6, isolated CD4+ CD8+ thymocytes from wild-type C57Bl/6 mice; MHC-, isolated CD4+ CD8+ thymocytes from β-2 microglobulin and Ab mutant mice; 5CC7-b, isolated CD4+ CD8+ thymocytes from 5CC7 TCR transgenic mice on a nonselecting (B10) background; thy CD8, isolated CD4− CD8+ thymocytes from F5 TCR transgenic mice; LN CD8, isolated CD4− CD8+ LN cells from wild-type mice; thy CD4, isolated CD4+ CD8− thymocytes from 5CC7 TCR transgenic mice on a positive selecting background (B10.A); LN CD4, isolated CD4+ CD8− LN cells from wild-type mice; LN CD4 act, LN CD4 cells that were activated in the presence of Con A for 2 d. Similar trends are seen when data is scored as percent of cells with one or two alleles associated with heterochromatin (not depicted).
It is interesting that the heterochromatic association of the CD4 and CD8 genes was maintained in resting mature T cells. This is in contrast to studies with B cells, in which heterochromatic association of repressed genes was observed in B cell lines and activated B cells, but not seen in resting primary B cells (13). In contrast, the percent of CD8 alleles associated with heterochromatin did not change significantly upon activation of mature CD4 T cells (Fig. 2 A, last two bars). It is also noteworthy that 20–35% of cells that lack expression of CD4 or CD8 do not score as showing heterochromatin-associated alleles in our assay. One possible explanation is that there are additional sites of heterochromatin that are not revealed by the γ satellite probe. It is also possible that events can lead to transcription silencing of some alleles independent of heterochromatin association.
Because the relocalization of CD4 and CD8 genes is likely to reflect their stable repression, information about the timing of this repositioning, and in which population it occurs, could give insights into the process of lineage commitment. In particular, evidence for repositioning of CD4 and CD8 genes to heterochromatin in DP thymocytes undergoing positive selection could provide an early indication of their lineage choice. Although we did not observe a significant increase in the percent of alleles associated with heterochromatin in DP thymocytes from wild-type mice compared with DP from mice that cannot undergo positive selection (MHC-null or the nonselecting TCR transgenic mice 5CC7-B10), this may reflect the fact that only a small fraction of thymocytes in wild-type mice can undergo positive selection.
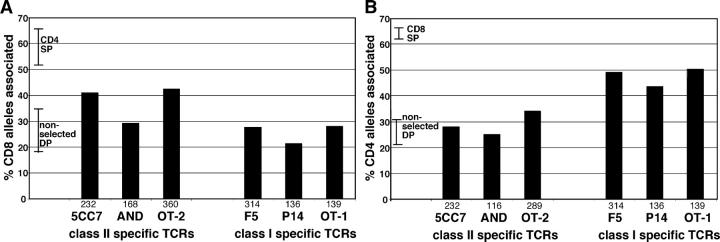
To examine whether repositioning to heterochromatin occurs in DP thymocytes undergoing positive selection, we performed FISH analysis from DP thymocytes from mice expressing rearranged TCR transgenes that can induce positive selection (Figs. 1 and 3). Interestingly, DP thymocytes from all three of the class I–restricted TCR transgenic mice examined (F5, P15, and OT1) showed significant repositioning of the CD4 gene, but not the CD8 gene, to heterochromatin. The proportion of CD4 alleles associated with heterochromatin in class I–restricted DP thymocytes (43–50%) was intermediate between the amount of association seen in nonselecting DP thymocytes (21–31%) and the amount seen in mature CD4− CD8+ cells (63–67%). This partial relocalization might be due to the fact that not all DP thymocytes in TCR transgenic mice complete positive selection (33, 34). Alternatively, it could reflect the fact that CD4+ CD8+ populations contain cells at different stages of the selection process, and the process of relocalization to heterochromatin is ongoing during the DP stage. In either case, these data indicate that repositioning of the CD4 gene to heterochromatin occurs early during class I positive selection. Moreover, the observation that the CD4 gene, but not the CD8 gene, repositioned to heterochromatin suggests that most DP thymocytes being selected via MHC class I are choosing the CD8 lineage.
Figure 3.
Repositioning of CD8 and CD4 genes to heterochromatin can begin during the DP stage. DP thymocyte populations from the indicated TCR transgenic mice were isolated and analyzed by FISH for CD8 (A) and CD4 (B). For comparison, the ranges for percent of centromeric alleles for SP and nonselecting DP thymocytes from Fig. 2 are indicated as bars. For nonselecting DP, this range includes values for the DP cell line DPK, DP thymocytes from wild-type mice, which contain a low proportion of cells undergoing positive selection, DP thymocytes from MHC mice, and DP thymocytes from mice expressing a nonselectable TCR transgene. Individual CD4 or CD8 alleles were scored as being associated or not with heterochromatin. The number of alleles scored for each sample is indicated beneath each set of bars.
In contrast, the pattern of coreceptor gene relocalization for three class II–restricted TCR transgenic thymocytes (5CC7-B10A, AND, and OT2) was less clear cut (Fig. 3). DP thymocytes from 5CC7-B10A mice displayed relocalization of CD8 and not CD4. This is in line with results from class I–restricted TCRs and fits with the idea that these DP thymocytes are becoming committed to the CD4 lineage. However, DP thymocytes from AND TCR transgenic mice did not display significant repositioning of either coreceptor gene. This might indicate that a relatively low proportion of DP thymocytes in these mice have begun the process of lineage commitment. DP thymocytes from OT2 TCR transgenic mice showed significant repositioning of the CD8 gene to heterochromatin. In addition, there was also a slight increase in the percent of CD4 alleles associated with heterochromatin on OT2 DP thymocytes relative to nonselected DP thymocytes. The different patterns of CD4 and CD8 gene relocalization seen with class II–restricted TCR, and particularly indications of repositioning of the CD4 gene to heterochromatin in DP thymocytes from OT2 TCR transgenic mice, suggests that lineage commitment of class II–restricted thymocytes may involve a more complex series of events than those that occur for class I–restricted thymocytes. These differences may reflect differences in the timing, levels, and avidity of particular transgenic TCRs examined.
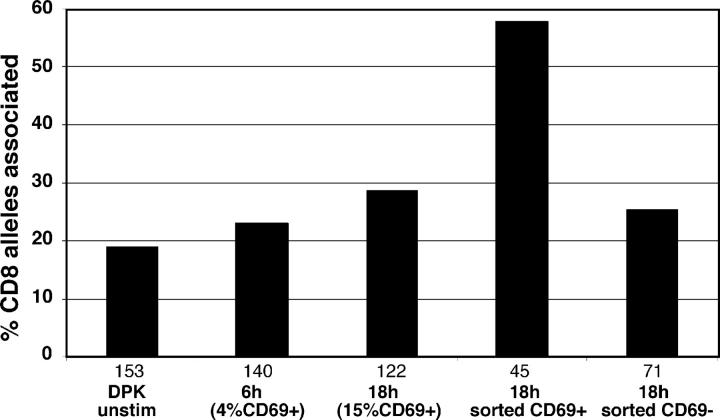
The observation that relocalization of the CD4 and CD8 genes to heterochromatin was detectable in DP from TCR transgenic mice suggests that this repositioning might represent an early response to lineage commitment signals. In fact, given the estimated lifespan of 1–3 d for DP thymocytes from TCR transgenic mice (33, 34), it seems likely that the repositioning begins within 1 d of initiating positive selection. To gain further information about the timing of repositioning to heterochromatin, we turned to a cell line model for positive selection, the CD4+ CD8+ thymocyte cell line, DPK (32). DPK cells respond to TCR stimulation by down-regulating CD8 surface expression, along with other phenotypic changes that mimic the DP to CD4SP transition. Thus, this cell line provides the opportunity to examine the repression of the CD8 gene by positive selection in a synchronized system. We examined location of the CD8 gene at 6 and 18 h after TCR stimulation, well before the down-regulation of surface CD8 is detectable. We found that stimulation of DPK cells for 18 h led to a slight increase in association of the CD8 gene with heterochromatin from 24 to 28% associated (Fig. 4). This modest relocalization of CD8 could reflect the fact that not all DPK cells in the culture respond to TCR engagement. Indeed, in our hands only ∼30% of DPK cells showed CD8 surface down-regulation 2 d after TCR stimulation and only 15% of DPK cells up-regulated CD69 18 h after TCR stimulation (not depicted). Therefore, we sorted the DPK cells from the 18-h stimulation cultures into CD69+ and CD69− fractions, and subjected them to FISH analysis. CD69+ DPK cells displayed 58% of CD8 alleles associated with heterochromatin, compared with 25% for CD69− cells. Together, these data indicate that CD8 locus undergoes substantial relocalization to heterochromatin as early as 18 h after TCR engagement, and well before the down-regulation of surface CD8 is detectable.
Figure 4.
Rapid relocalization of a CD8 gene after TCR stimulation of DPK cells. DPK cells were stimulated for 6 or 18 h by coculture with DCEK cells bearing MHC class II I-EK and cytochrome C peptide, and analyzed by FISH. The number of alleles scored for each sample is indicated beneath each set of bars. The percent of CD69+ cells within each population is indicated. DPK cells from the 18-h time point were also sorted into CD69+ and CD69− fractions before FISH analysis. After 48 h of stimulation, a substantial fraction of DPK cells down-regulate surface CD8 expression; however, at 18 h, surface levels of CD4 and CD8 remain unchanged (reference 32 and not depicted). No increase in the percent of CD4 alleles associated with centromeric heterochromatin was observed at any time point (not depicted).
Association of genes with heterochromatin often correlates with transcriptional repression. If this correlation holds for the CD4 and CD8 genes, then steady-state mRNA levels might correlate with repositioning of CD4 and CD8 genes to heterochromatin, and might provide equivalent information regarding lineage commitment. However, gene repression can occur in the absence of heterochromatic association when repression is a transient event (13). Moreover, previous analysis of cell surface levels of CD4 and CD8 indicated that expression of both coreceptors is repressed by positive selection via MHC class I and II, as well as by negative selection (19, 21–23). This down-regulation appears to result in part from destabilization of CD4 and CD8 mRNA, in addition to transcriptional repression (24). These observations suggest that CD4 and CD8 mRNA levels in DP thymocytes might not reflect stable repression of the CD4 and CD8 genes, and would likely not correspond to the degree of association with heterochromatin.
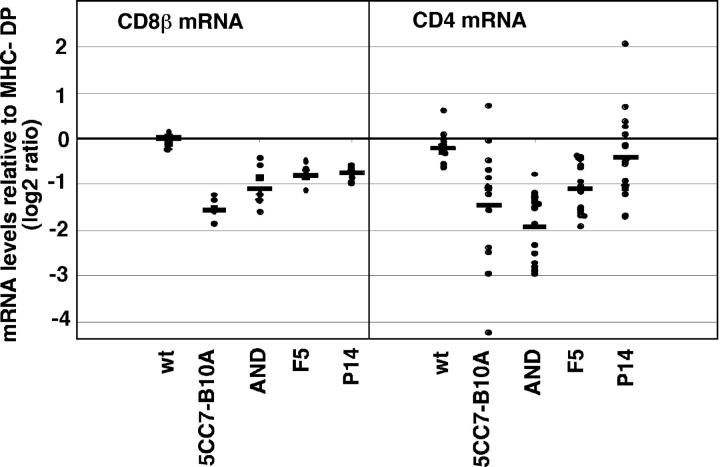
To examine this issue more directly, we analyzed CD4 and CD8 mRNA levels in DP thymocytes from TCR transgenic mice using DNA microarrays. Our previous DNA microarray analysis indicated that CD4 and CD8 steady-state mRNA levels are coordinately repressed during positive selection (35). However, because this work was focused on global changes in gene expression, our earlier analysis did not permit careful quantitative comparisons between samples or the statistical analysis of the differences in individual genes observed. Therefore, we reanalyzed our microarray data to focus on differences in CD4 and CD8 gene expression (Fig. 5). Consistent with other studies, we found that down-regulation of steady-state CD4 and CD8 mRNA was a general feature of positive selection, regardless of TCR specificity. In addition, in some cases we observed a decrease in steady-state mRNA levels in samples that did not show any detectable increase in heterochromatin localization (e.g., AND DP for CD4 and CD8, 5CC7-B10A DP for CD4, and P14 and F5 DP for CD8). The lack of correlation between steady-state mRNA and CD4/CD8 gene position implies that the early down-regulation of coreceptor mRNA in response to TCR engagement probably does not involve relocalization to heterochromatin, and might be distinct mechanistically from stable lineage-specific repression.
Figure 5.
CD4 and CD8 mRNA levels from DP thymocytes. Values for CD4 and CD8 mRNA levels are from published DNA microarray analysis (reference 35). Data are replotted here to show values for each individual measurement. Data are presented as the log2 ratio of signal from the indicated CD4+ CD8+ thymocyte sample relative to CD4+ CD8+ thymocytes from MHC-deficient mice. Each dot represents the value from an individual spot and the average fold change for all measurements for each sample is shown as a line. Average fold changes for CD8β mRNA were as follows: MHC v B6 0.012(0.15), 5CC7 TCR −1.57(0.26), AND TCR −1.10(0.44), F5 −0.81(0.23), and P14 −0.75(0.16). Average fold changes for CD4 mRNA were as follows: MHC v B6 −0.22(0.33), 5CC7 TCR −1.46(1.30), AND TCR −1.94(0.71), F5 −1.09(0.51), and P14 −0.42(0.880). Standard errors of means are given in parenthesis.
Discussion
One obstacle to a mechanistic understanding of CD4 versus CD8 lineage commitment is the lack of early markers for the lineage commitment process. Lineage commitment is associated with stable repression of CD4 or CD8 genes, an event that can be accompanied by positioning of loci near regions of heterochromatin. In this work we have examined the position of CD4 and CD8 loci relative to heterochromatin at different stages of thymic development to gain insight into the timing and mechanism of stable gene repression and lineage commitment. We found that CD4 and CD8 loci tended to be located near centromeric heterochromatin in early thymocytes that have not yet expressed CD4 or CD8, and in mature thymocytes and T cells that have repressed either CD4 or CD8. We also provided evidence that repositioning of CD4 or CD8 genes to heterochromatin can occur as an early response to positive selection, being detectable in CD4+ CD8+ thymocytes that are receiving positive selection signals. This implies that relocalization of either CD4 or CD8 genes to heterochromatin represents an early event during CD4/CD8 lineage commitment.
Lineage commitment might best be thought of as a series of gradual steps, rather than a single discrete event. At the beginning of the process, a precursor cell has an equal probability of adopting alternative fates. At an intermediate stage in the process, changes accumulate that increase the likelihood that a precursor will adopt one fate over another. An example of such an intermediate phase in lineage commitment has been described during vulval development in Caenorhabditis elegans (36). In this example, changes in expression levels of the Notch family member, lin-12, predict the ultimate fate of a precursor cell, even while the cell remains plastic. Finally late in the lineage commitment process, additional steps reinforce the cell fate decision. An example of a late, reinforcing event is the expression of Pax5 in developing B cells that represses alternative blood cell fates (37). The association of the CD4 and CD8 genes with heterochromatin is likely to represent one of several steps that occur as an uncommitted CD4+ CD8+ thymocyte differentiates into a peripheral CD4+ or CD8+ T cell.
With this in mind, it is worth considering how repositioning of CD4 and CD8 genes to heterochromatin relates to other events that occur as part of lineage commitment. With regard to silencing of the CD4 locus in CD8 lineage cells, two distinct phases have been identified (38). The initiation of silencing is controlled by a silencer element in the first intron of the CD4 gene (4) and is mediated by Runx transcription factors (5–7). This phase occurs in the thymus, and is likely to be closely linked to the repositioning of the locus near heterochromatin described here. Perhaps the positioning of the CD4 gene near heterochromatin facilitates silencing by bringing the locus into proximity with machinery that represses transcription, such as histone-modifying enzymes, or by preventing access to factors that would promote transcription. Runx protein binding to the silencer could lead to the recruitment of the CD4 locus to heterochromatin, or alternatively, recruitment of the locus to heterochromatin by other factors could facilitate the binding or activity of Runx proteins.
In contrast, the maintenance of CD4 silencing, which is not dependent on the intronic silencer, occurs after mature CD8 SP T cells leave the thymus (38). It appears that this later maintenance phase of CD4 silencing is not accompanied by increased association of the CD4 gene with heterochromatin because we find a similar percent of CD4 alleles associated with heterochromatin in thymic CD8 SP and peripheral CD8 SP. Thus, a picture emerges in which positive selection via MHC class I leads to initial CD4 repression via the silencer and the repositioning of CD4 gene near heterochromatin as an early step in lineage commitment. Some time later, after the mature CD8 cell leaves the thymus, the repression of the CD4 gene is reinforced by a distinct mechanism that locks the CD4 gene into its silent state without increasing the percent of alleles associated with heterochromatin.
Other transcription factors have been implicated in the development of CD4 or CD8 lineage T cells, including GATA3, Tox, and SAF (9–11, 39). For example, loss of function of GATA3 prevents CD4 T cell development, and GATA3 expression is up-regulated early after CD4-inducing positive selection signals (9, 10). This suggests the possibility that GATA3 up-regulation may lead, directly or indirectly, to the repositioning of the CD8 locus close to heterochromatin in CD4 lineage cells. It will be interesting to examine the relationship between the activity of factors such as GATA3 and the nuclear position of the CD4 and CD8 genes during positive selection.
Our results have implications for the question of how TCR specificity for MHC class I or II influences the CD4/CD8 lineage choice. If recognition of MHC class I or II directs the lineage choice at the CD4+ CD8+ stage (instructive model), we would expect that DP thymocytes undergoing selection via MHC class I recognition would exclusively relocalize the CD4 gene, whereas DP thymocytes undergoing selection via class II would relocalize only the CD8 gene. We find that this prediction holds for three different class I–restricted TCRs. Thus, for positive selection of class I–restricted thymocytes, our data fit well with a simple instructive model for lineage commitment.
In contrast, positive selection of thymocytes bearing class II–restricted TCRs provides a more complex picture. DP thymocytes from 5CC7 TCR transgenic mice showed repositioning of CD8, and not CD4, as predicted by instructive models. In contrast, we did not observe significant repositioning of either CD4 or CD8 genes in DP thymocytes from AND TCR transgenic mice, a result that is likely a reflection of the low proportion of DP thymocytes in these mice that are engaged in the process of positive selection. In support of this idea, an independent study found evidence for CD8 repositioning in a selected subset of surface CD8low DP thymocytes from AND TCR transgenic mice (40). However, this population made up only 20% of DP thymocytes, and thus repositioning might be difficult to detect within the entire DP population.
Most surprisingly, we found that DP thymocytes bearing the class II–restricted OT-2 TCR showed some evidence for repositioning of CD4 as well as CD8 genes. There are several possible interpretations of this result. One possibility is that the relocalization of CD4 to heterochromatin in DP thymocytes from OT-2 TCR transgenic mice occurs in response to negative selection, rather than positive selection signals. Although these mice are generally thought of as models for positive selection, there is evidence that some negative selection can also occur in positively selecting TCR transgenic mice (41, 42). Thus, negative selection signals could induce the repositioning of both CD4 and CD8 genes to heterochromatin, a possibility that could contribute to the coordinate down-regulation of surface CD4 and CD8 that is observed in thymocytes undergoing negative selection (21–23). It is also possible that relocalization of the CD4 gene to heterochromatin occurs early in the response to class II positive selection, but can be reversed as cells continue through the positive selection process. Both of these explanations predict that CD4 and CD8 genes might reposition to heterochromatin in the same cells. We cannot at present determine whether CD4 and CD8 repositioning occurs in the same or different populations of thymocytes due to the technical difficulty of detecting two single copy genes simultaneously.
Perhaps the most interesting possibility is that DP thymocytes expressing the OT-2 TCR consist of mixtures of thymocytes choosing the CD4 or CD8 lineages. Indeed, numerous studies suggest that some class II–restricted thymocytes may down-regulate CD4 instead of CD8, but that most of these mismatched thymocytes fail to complete positive selection because they lose the ability to recognize MHC class II as they mature (stochastic/selection model; references 43–46). In fact, mixed models incorporating elements of both instructive and stochastic models have been proposed in which MHC recognition imposes an instructive bias on lineage choice, and the continued requirement for expression of the appropriate coreceptor later during selection serves to reinforce this bias (47).
Our results are compatible with the widely held quantitative model for CD4/CD8 lineage commitment. This model was originally proposed based on the differential ability of CD4 and CD8 cytoplasmic domains to recruit the tyrosine kinase, Lck, and promote CD4 or CD8 development (41, 48). Subsequently, a number of approaches that have altered the strength or duration of signaling during positive selection have provided additional support for this model (for review see references 49 and 50). We propose that the weak or transient signals generated during positive selection via MHC class I recognition are sufficient to induce relocalization of the CD4 gene, but not the CD8 gene, to heterochromatin. In contrast, stronger, or more prolonged signaling generated during positive selection via MHC class II would be required to induce relocalization of the CD8 gene. For certain TCRs, positive selection could lead to mistakes in the initial lineage choice. In these cases, the requirement for prolonged MHC recognition would serve as a check to ensure that thymocytes that down-regulate the wrong coreceptor do not complete positive selection.
In summary, our data indicate that changes in the nuclear position of CD4 and CD8 genes can be used to monitor lineage commitment during thymic development. When used in combination with other assays, such as those that monitor the developmental potential of cells and the activity of transcription factors, this approach may provide a new tool to help to unravel the complex, multi-step process of lineage commitment.
Acknowledgments
We would like to thank Jane Grogan for help with setting up the FISH assay; Amanda Fischer, Nigel Killeen, and Amin Rahemtulla for providing FISH probes; B.J. Fowlkes for helpful discussion; and Matthias Merkenschlager and Amanda Fischer for communication of unpublished data.
This work was supported by National Institutes of Health grants AI32985 and AI053039 to E.A. Robey.
The authors have no conflicting financial interests.
S. Delaire and Y.H. Huang contributed equally to this work.
Y.H. Huang's present address is Genomics Institute of the Novartis Research Foundation, 10675 John Jay Hopkins Dr., San Diego, CA 92121.
Abbreviations used in this paper: DN, double negative; DP, double positive; FISH, fluorescence in situ hybridization; SP, single positive.
References
- 1.Kioussis, D., and W. Ellmeier. 2002. Chromatin and CD4, CD8A and CD8B gene expression during thymic differentiation. Nat. Rev. Immunol. 2:909–919. [DOI] [PubMed] [Google Scholar]
- 2.Ellmeier, W., S. Sawada, and D.R. Littman. 1999. The regulation of CD4 and CD8 coreceptor gene expression during T cell development. Annu. Rev. Immunol. 17:523–554. [DOI] [PubMed] [Google Scholar]
- 3.Siu, G. 2002. Controlling CD4 gene expression during T cell lineage commitment. Semin. Immunol. 14:441–451. [DOI] [PubMed] [Google Scholar]
- 4.Sawada, S., J. Scarborough, N. Killeen, and D. Littman. 1994. A lineage-specific transcriptional silencer regulates CD4 gene expression during T lymphocyte development. Cell. 77:917–929. [DOI] [PubMed] [Google Scholar]
- 5.Ehlers, M., K. Laule-Kilian, M. Petter, C.J. Aldrian, B. Grueter, A. Wurch, N. Yoshida, T. Watanabe, M. Satake, and V. Steimle. 2003. Morpholino antisense oligonucleotide-mediated gene knockdown during thymocyte development reveals role for Runx3 transcription factor in CD4 silencing during development of CD4−/CD8+ thymocytes. J. Immunol. 171:3594–3604. [DOI] [PubMed] [Google Scholar]
- 6.Woolf, E., C. Xiao, O. Fainaru, J. Lotem, D. Rosen, V. Negreanu, Y. Bernstein, D. Goldenberg, O. Brenner, G. Berke, et al. 2003. Runx3 and Runx1 are required for CD8 T cell development during thymopoiesis. Proc. Natl. Acad. Sci. USA. 100:7731–7736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taniuchi, I., M. Osato, T. Egawa, M.J. Sunshine, S.C. Bae, T. Komori, Y. Ito, and D.R. Littman. 2002. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell. 111:621–633. [DOI] [PubMed] [Google Scholar]
- 8.Harker, N., T. Naito, M. Cortes, A. Hostert, S. Hirschberg, M. Tolaini, K. Roderick, K. Georgopoulos, and D. Kioussis. 2002. The CD8alpha gene locus is regulated by the Ikaros family of proteins. Mol. Cell. 10:1403–1415. [DOI] [PubMed] [Google Scholar]
- 9.Hernandez-Hoyos, G., M.K. Anderson, C. Wang, E.V. Rothenberg, and J. Alberola-Ila. 2003. GATA-3 expression is controlled by TCR signals and regulates CD4/CD8 differentiation. Immunity. 19:83–94. [DOI] [PubMed] [Google Scholar]
- 10.Pai, S.Y., M.L. Truitt, C.N. Ting, J.M. Leiden, L.H. Glimcher, and I.C. Ho. 2003. Critical roles for transcription factor GATA-3 in thymocyte development. Immunity. 19:863–875. [DOI] [PubMed] [Google Scholar]
- 11.Wilkinson, B., J.Y. Chen, P. Han, K.M. Rufner, O.D. Goularte, and J. Kaye. 2002. TOX: an HMG box protein implicated in the regulation of thymocyte selection. Nat. Immunol. 3:272–280. [DOI] [PubMed] [Google Scholar]
- 12.Brown, K.E., S.S. Guest, S.T. Smale, K. Hahm, M. Merkenschlager, and A.G. Fisher. 1997. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell. 91:845–854. [DOI] [PubMed] [Google Scholar]
- 13.Brown, K.E., J. Baxter, D. Graf, M. Merkenschlager, and A.G. Fisher. 1999. Dynamic repositioning of genes in the nucleus of lymphocytes preparing for cell division. Mol. Cell. 3:207–217. [DOI] [PubMed] [Google Scholar]
- 14.Grogan, J.L., M. Mohrs, B. Harmon, D.A. Lacy, J.W. Sedat, and R.M. Locksley. 2001. Early transcription and silencing of cytokine genes underlie polarization of T helper cell subsets. Immunity. 14:205–215. [DOI] [PubMed] [Google Scholar]
- 15.Kosak, S.T., J.A. Skok, K.L. Medina, R. Riblet, M.M. Le Beau, A.G. Fisher, and H. Singh. 2002. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science. 296:158–162. [DOI] [PubMed] [Google Scholar]
- 16.Kydd, R., K. Lundberg, D. Vremec, A.W. Harris, and K. Shortman. 1995. Intermediate steps in thymic positive selection. Generation of CD4−8+ T cells in culture from CD4+8+, CD4int8+, and CD4+8int thymocytes with up-regulated levels of TCR-CD3. J. Immunol. 155:3806–3814. [PubMed] [Google Scholar]
- 17.Lundberg, K., W. Heath, F. Kontgen, F. Carbone, and K. Shortman. 1995. Intermediate steps in positive selection: differentiation of CD4+ CD8int TCRint thymocytes into CD4− CD8+ TCRhi thymocytes. J. Exp. Med. 181:1643–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suzuki, H., J.A. Punt, L.G. Granger, and A. Singer. 1995. Asymmetric signaling requirements for thymocyte commitment to the CD4+ versus CD8+ T cell lineages: a new perspective on thymic commitment and selection. Immunity. 2:413–425. [DOI] [PubMed] [Google Scholar]
- 19.Lucas, B., and R. Germain. 1996. Unexpectedly complex regulation of CD4/CD8 coreceptor expression supports a revised model for CD4+CD8+ thymocyte differentiation. Immunity. 5:461–477. [DOI] [PubMed] [Google Scholar]
- 20.Correia-Neves, M., D. Mathis, and C. Benoist. 2001. A molecular chart of thymocyte positive selection. Eur. J. Immunol. 31:2583–2592. [DOI] [PubMed] [Google Scholar]
- 21.Swat, W., L. Ignatowicz, H. von Boehmer, and P. Kisielow. 1991. Clonal deletion of immature CD4+8+ thymocytes in suspension culture by extrathymic antigen-presenting cells. Nature. 351:150–153. [DOI] [PubMed] [Google Scholar]
- 22.Page, D., L. Kane, J. Allison, and S. Hedrick. 1993. Two signals are required for negative selection of CD4+CD8+ thymocytes. J. Immunol. 151:1868–1880. [PubMed] [Google Scholar]
- 23.McGargill, M.A., and K.A. Hogquist. 1999. Antigen-induced coreceptor down-regulation on thymocytes is not a result of apoptosis. J. Immunol. 162:1237–1245. [PubMed] [Google Scholar]
- 24.Cibotti, R., A. Bhandoola, T.I. Guinter, S.O. Sharrow, and A. Singer. 2000. CD8 coreceptor extinction in signaled CD4(+)CD8(+) thymocytes: coordinate roles for both transcriptional and posttranscriptional regulatory mechanisms in developing thymocytes. Mol. Cell. Biol. 20:3852–3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barthlott, T., H. Kohler, and K. Eichmann. 1997. Asynchronous coreceptor downregulation after positive thymic selection: prolonged maintenance of the double positive state in CD8 lineage differentiation due to sustained biosynthesis of the CD4 coreceptor. J. Exp. Med. 185:357–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grusby, M., H.J. Auchincloss, R. Lee, R. Johnson, J. Spencer, M. Zijlstra, R. Jaenisch, V. Papaioannou, and L. Glimcher. 1993. Mice lacking major histocompatibility complex class I and class II molecules. Proc. Natl. Acad. Sci. USA. 90:3913–3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mamalaki, C., J. Elliott, T. Norton, N. Yannoutsos, A.R. Townsend, P. Chandler, E. Simpson, and D. Kioussis. 1993. Positive and negative selection in transgenic mice expressing a T-cell receptor specific for influenza nucleoprotein and endogenous superantigen. Dev. Immunol. 3:159–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pircher, H., K. Burki, R. Lang, H. Hengartner, and R.M. Zinkernagel. 1989. Tolerance induction in double specific T-cell receptor transgenic mice varies with antigen. Nature. 342:559–561. [DOI] [PubMed] [Google Scholar]
- 29.Seder, R.A., W.E. Paul, M.M. Davis, and B. Fazekas de St. Groth. 1992. The presence of interleukin 4 during in vitro priming determines the lymphokine-producing potential of CD4+ T cells from T cell receptor transgenic mice. J. Exp. Med. 176:1091–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaye, J., M.L. Hsu, M.E. Sauron, S.C. Jameson, N.R. Gascoigne, and S.M. Hedrick. 1989. Selective development of CD4+ T cells in transgenic mice expressing a class II MHC-restricted antigen receptor. Nature. 341:746–749. [DOI] [PubMed] [Google Scholar]
- 31.Hogquist, K.A., S.C. Jameson, W.R. Heath, J.L. Howard, M.J. Bevan, and F.R. Carbone. 1994. T cell receptor antagonist peptides induce positive selection. Cell. 76:17–27. [DOI] [PubMed] [Google Scholar]
- 32.Kaye, J., and D.L. Ellenberger. 1992. Differentiation of an immature T cell line: a model of thymic positive selection. Cell. 71:423–435. [DOI] [PubMed] [Google Scholar]
- 33.Huesmann, M., B. Scott, P. Kisielow, and H. von Boehmer. 1991. Kinetic and efficacy of positive selection in the thymus of normal and T cell receptor transgenic mice. Cell. 66:533–540. [DOI] [PubMed] [Google Scholar]
- 34.Itano, A., and E. Robey. 2000. Highly efficient selection of CD4 and CD8 lineage thymocytes supports an instructive model of lineage commitment. Immunity. 12:383–389. [DOI] [PubMed] [Google Scholar]
- 35.Huang, Y.H., D.L. Li, A. Winoto, and E.A. Robey. 2004. Distinct transcriptional programs in thymocytes responding to T cell receptor, Notch, and positive selection signals. Proc. Natl. Acad. Sci. USA. 101:4936–4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilkinson, H., K. Fitzgerald, and I. Greenwald. 1994. Reciprocal changes in expression of the receptor lin-12 and its ligand prior to commitment in a C. elegans cell fate decision. Cell. 79:1187–1198. [DOI] [PubMed] [Google Scholar]
- 37.Mikkola, I., B. Heavey, M. Horcher, and M. Busslinger. 2002. Reversion of B cell commitment upon loss of Pax5 expression. Science. 297:110–113. [DOI] [PubMed] [Google Scholar]
- 38.Zou, Y.R., M.J. Sunshine, I. Taniuchi, F. Hatam, N. Killeen, and D.R. Littman. 2001. Epigenetic silencing of CD4 in T cells committed to the cytotoxic lineage. Nat. Genet. 29:332–336. [DOI] [PubMed] [Google Scholar]
- 39.Kim, W.W., and G. Siu. 1999. Subclass-specific nuclear localization of a novel CD4 silencer binding factor. J. Exp. Med. 190:281–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Merkenschlager, M., S. Amoils, E. Roldan, A. Rahemtulla, E. O'Connor, A.G. Fisher, and K.E. Brown. 2004. Centromeric repositioning of coreceptor loci predicts their stable silencing and the CD4/CD8 lineage choice. J. Exp. Med. 200:1437–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matechak, E., N. Killeen, S. Hedrick, and B. Fowlkes. 1996. MHC class II-specific T cells can develop in the CD8 lineage when CD4 is absent. Immunity. 4:337–347. [DOI] [PubMed] [Google Scholar]
- 42.Robey, E.A., F. Ramsdell, D. Kioussis, W. Sha, D. Loh, R.A. Axel, and B.J. Fowlkes. 1992. The level of CD8 expression can determine the outcome of thymic selection. Cell. 69:1089–1096. [DOI] [PubMed] [Google Scholar]
- 43.Davis, C.B., N. Killeen, M.E.C. Crooks, D. Raulet, and D.R. Littman. 1993. Evidence for a stochastic mechanism in the differentiation of mature subsets of T lymphocytes. Cell. 73:237–247. [DOI] [PubMed] [Google Scholar]
- 44.Baron, A., K. Hafen, and H. von Boehmer. 1994. A human CD4 transgene rescues CD4−CD8+ cells in beta 2-microglobulin-deficient mice. Eur. J. Immunol. 24:1933–1936. [DOI] [PubMed] [Google Scholar]
- 45.Paterson, R., L. Burkly, D. Kurahara, A. Dunlap, R. Flavell, and T. Finkel. 1994. Thymic development in human CD4 transgenic mice. Positive selection occurs after commitment to the CD8 lineage. J. Immunol. 153:3491–3503. [PubMed] [Google Scholar]
- 46.Leung, R.K., K. Thomson, A. Gallimore, E. Jones, M. Van den Broek, S. Sierro, A.R. Alsheikhly, A. McMichael, and A. Rahemtulla. 2001. Deletion of the CD4 silencer element supports a stochastic mechanism of thymocyte lineage commitment. Nat. Immunol. 2:1167–1173. [DOI] [PubMed] [Google Scholar]
- 47.Robey, E.A., and B.J. Fowlkes. 1994. Selective events in T cell development. Annu. Rev. Immunol. 12:675–705. [DOI] [PubMed] [Google Scholar]
- 48.Itano, A., P. Salmon, D. Kioussis, M. Tolaini, P. Corbella, and E. Robey. 1996. The cytoplasmic domain of CD4 promotes the development of CD4 lineage T cells. J. Exp. Med. 183:731–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Basson, M.A., and R. Zamoyska. 2000. The CD4/CD8 lineage decision: integration of signalling pathways. Immunol. Today. 21:509–514. [DOI] [PubMed] [Google Scholar]
- 50.Germain, R.N. 2002. T-cell development and the CD4-CD8 lineage decision. Nat. Rev. Immunol. 2:309–322. [DOI] [PubMed] [Google Scholar]