Abstract
Escape mutations are believed to be important contributors to immune evasion by rapidly evolving viruses such as hepatitis C virus (HCV). We show that the majority of HCV-specific cytotoxic T lymphocyte (CTL) responses directed against viral epitopes that escaped immune recognition in HCV-infected chimpanzees displayed a reduced CDR3 amino acid diversity when compared with responses in which no CTL epitope variation was detected during chronic infection or with those associated with protective immunity. Decreased T cell receptor (TCR) CDR3 amino acid diversity in chronic infection could be detected long before the appearance of viral escape mutations in the plasma. In both chronic and resolved infection, identical T cell receptor clonotypes were present in liver and peripheral blood. These findings provide a deeper understanding of the evolution of CTL epitope variations in chronic viral infections and highlight the importance of the generation and maintenance of a diverse TCR repertoire directed against individual epitopes.
Keywords: CD8+ T lymphocytes, CD4+ T lymphocytes, hepatitis C, antigenic variation, epitopes
Introduction
Strong hepatitis C virus (HCV)–specific CD4+ and CD8+ T cell responses against multiple viral epitopes have been associated with clearance of HCV during acute infection (1–6) and are thought to be important contributors to protective immunity (7–9). Failure to eradicate the virus despite detectable CTL responses has been associated with emergence of viral escape mutations within epitopes targeted by CTLs during acute HCV (10, 11) and several other viral infections (12–15). Maturation defects or functional impairment of antigen-specific CTL responses have been observed by several investigators and are thought to be possible reasons for viral persistence despite measurable T cell responses in HCV (16–19). T cell receptor diversity against viral epitopes may be another factor that influences the outcome of viral infection.
In various chronic viral and intracellular infections, antigen-specific T cell responses are characterized by a marked structural TCR diversity (20–23). The majority of expanded CD8+ T cells during acute lymphocytic choriomeningitis virus infection are pathogen specific (24), and many of these initially expanded CD8+ T cells undergo apoptosis (25, 26), leaving behind a stable pool of memory cells with structural and functional resemblance to the primary T cell response (27, 28). This contrasts findings in some experimental models where repeated rechallenge with antigen results in a narrowing of the antigen-specific T cell receptor repertoire, probably caused by a selective expansion of high affinity T cell clones (29–32). However, after simian immunodeficiency virus infection of rhesus macaques, fluctuations within the TCR repertoire, but no evidence of a maturation of the TCR repertoire, could be observed over the course of infection (33). It has been hypothesized that in chronic viral infections characterized by high viral mutation rates, the generation and maintenance of a highly diverse epitope-specific TCR repertoire might decrease the chance of CTL escape due to cross-recognition of mutated epitopes (34–36). The benefit of TCR diversity is also supported by studies demonstrating that a diverse TCR repertoire is more likely to contain high affinity T cell clones that results in better epitope recognition and control of the pathogen (32, 33, 37). This view is also supported by rapid evolution of viral escape mutations associated with a narrow response against a single CTL epitope in HIV infection (38) and deletions of the HIVnef gene after infusion of CTL clones against a single viral epitope in HIV infection (39). However, to date, there is no convincing evidence linking the TCR diversity of virus-specific T cells with evolution of CTL escape mutations in a chronic viral infection in vivo.
Structural analyses of the TCR–pMHC complex highlight the importance of the CDR3 region as a dominant antigen recognition site within the TCR (40). Here, we tested the hypothesis that in vivo–generated T cell responses that induce CTL escape are more likely to display a narrow CDR3 repertoire as compared with T cell responses where no CTL epitope variation occurs or to those that are found in protective immunity. We took advantage of the chimpanzee HCV disease model in which all animals were infected with the same HCV-1/910 virus inoculum (10), eliminating the problem of infection with variant viruses, which is unavoidable in human studies of HCV infection. Some of the infected chimpanzees resolved the infection spontaneously (2), even after homologous rechallenge (9), whereas others developed persistent disease despite existing CTL responses (41). In chronically infected animals, detectable T cell responses were found in the liver and the peripheral blood (2, 10, 41, 42), and they were associated with the evolution of escape mutations in almost all of the targeted epitopes (for an overview of analyzed immune responses and CTL epitope variations, see Table I; references 11, 42). To assess the diversity of HCV-specific T cell responses, we used a recently published method that allows the generation of a high number of antigen-specific TCR sequences (43–45). In addition, we performed TCRα/β sequence analysis of HCV-specific T cell clones derived by limiting dilution from the liver or blood of the infected animals.
Table I.
HCV-1 Epitopes and Disease Status of Study Animals
| Animal | Patr allele | HCV region | Wild-type sequence/ escape mutations |
Month of detection | Status |
|---|---|---|---|---|---|
| CBO603 | A*0901 | NS41963–1972 | QWISSECTTPC/ ----------- (8/24) ------S---- (2/24) --L-----A—- (1/24) --L-------- (13/24) |
63a | chronic |
| CH-503 | A*0401 | E2588–596 | KHPDATYSR/ -------T- (9/9) |
21 | chronic |
| B*1601 | E1233–241 | GNASRCWVA/ --------- (15/15) |
NA | ||
| NS31446–1454 | GDFDSVIDC/ ---E----- (10/10) |
3 | |||
| NS5a1989–1997 | SDFKTWLKA/ ---R----- (11/11) |
10 | |||
| CBO572 | B*2302 | E2445–456 | HKFNSSGCPERL/ NA |
NA | resolved |
| DRB1*1001 | NS31248–1257 | GYKVLVNPSV/ NA |
NA | ||
| DRB5*0310 | NS31380–1389 | YGKAIPLEVI/ NA |
NA |
No sequencing performed between months 4 and 63 (reference 11).
Materials and Methods
Subjects.
Infection of the chimpanzees enrolled in this study with the HCV-1/910 virus stock has been described in detail previously (2, 10, 11, 46, 47). CBO572 was initially infected with 100 CID of HCV-1/910 stock and rechallenged 7 yr later with the same virus (9). CBO603 and CH-503 have been chronically infected for 7 or 9 yr, respectively. Chimpanzees were housed at the New Iberia Research Center. Animals were cared for under an experimental protocol approved by the New Iberia Research Center Animal Care and Use Committee.
Cell Lines.
CTL lines were derived from liver biopsy tissue as described previously (10). In brief, after homogenization of liver biopsies, CD8+ T cells were enriched using anti–human CD8+ dynabeads (Dynal) and cultured at 10–50 cells per well in a 96-well tissue culture plate in the presence of anti–human CD3 (Immunotech), irradiated human PBMCs, and R10 medium supplemented with 50 U/ml rIL-2. At least 300 independently derived CD8+ T cell lines were tested for recognition of autologous B cell target cells infected with recombinant vaccinia viruses expressing different regions of the HCV-1/910 polyprotein. HCV-specific CD4+ and CD8+ T cells were enriched from peripheral blood after Ficoll-Paque (Amersham Biosciences) by culturing 20–40 × 106 PBMCs with the respective HCV peptide (50 μM). Cells were restimulated on days 7 and 14 with 2 × 106 irradiated autologous PBMCs pulsed with 50 μM of the peptide. Analysis of peptide-specific IFN-γ production and purification of IFN-γ+ cells was performed between days 21 and 25.
Purification of HCV-specific Cells by IFN-γ Secretion.
IFN-γ+ HCV-specific T cells were purified using a cytokine-secretion assay (Miltenyi Biotec) as described previously (44) with minor modifications. In brief, 30–40 × 106 T cells were incubated with 20 μM peptide and 1 μg/ml of the anti-CD28 and anti-CD49d mAbs (Becton Dickinson) at 37°C, 5% CO2 for 6–8 h. Subsequently, IFN-γ+ cells were stained with monoclonal antibodies according to the manufacturer's protocol (Miltenyi Biotec) and separated by cell sorting using the FACSvantage™ cell sorter (Becton Dickinson). In all experiments, the purity of the enriched populations was >90% (range 90–99%). FACS® data on representative purifications are provided in Fig. S1, available at http://www.jem.org/cgi/content/full/jem.20040638.
cDNA Synthesis and TCR Sequencing.
A modified anchored RT-PCR was performed with Powerscript Reverse transcriptase (CLONTECH Laboratories, Inc.) from total RNA as described previously (43, 44) using gene-specific primers for the TCRα constant region or the β constant region with a modified cDNA anchor primer (CLONTECH Laboratories, Inc.). Amplification of the cDNA by PCR was performed using the TCR constant region based primers and an anchor-specific primer 5′-AATCCTTTCTCTTGACCATG-3′. PCR products of 500–600 base pairs were gel purified and cloned using the TOPO TA cloning kit (Invitrogen). Selected colonies were sequenced using the Taq DyeDeoxy Terminator cycle sequencing kit (Applied Biosystems) and capillary electrophoresis on a PRISM automated sequencer (Applied Biosystems). Full-length sequences of all TCRβ variable regions have been published previously (44) and have been submitted to GenBank/EMBL/DDBJ (accession nos. AY168667 and AY168708).
CTL Assays.
Autologous lymphoblastoid B cell lines were used as targets and incubated with 50 μCi of 51Cr and varying concentrations of the respective peptide epitopes. After washing 5 × 103 target, cells were cocultured with effector CTL lines at different effector/target ratios for 4 h. 50 μl of supernatant was harvested into Lumaplates (Packard Instrument Co.) and radioactivity was counted in a Microbeta scintillation instrument (Wallac). To determine the binding kinetics of variant and wild-type peptides, we analyzed CTL clone recognition of peptide-pulsed target cells over a 48-h period. B cell lines were first pulsed with 100 μM of the variant or the wild-type peptide. CTL recognition was measured with the standard chromium release assay as aforementioned at hours 0, 16, and 48.
Entropy Calculations and Statistical Analysis.
Genetic analysis was performed as described previously (44). In brief, after alignment of sequences, amino acid variability was determined using the Shannon entropy (H; reference 48) calculation for protein sites as described previously (49) by the formula: H = −Σ pilog2 p i where p i is the fraction of residues at a site that is amino acid type i. For the 20 amino acids, H can range from 0 (site contains only one amino acid in all sequences) to 4.32 (all amino acids are represented equally at this site). Positions that contained >50% gaps were excluded from analysis. Statistical analyses were performed with Prism 3.0 (GraphPad Software Inc.), Version 11.0. (SPSS Inc.), and Version 8.02 (SAS Institute). Mann-Whitney U tests were used to compare distributions of entropy values between responses, and the Wilcoxon Sign test was used to compare within epitope difference between memory and rechallenged entropy values of the E2445 peptide. A linear mixed regression was used to compare entropy values by escape from immune recognition. The use of a mixed regression model controls for dependency among entropy values within a response.
Online Supplemental Material.
IFN-γ production of expanded HCV-specific T cells derived from CH-503 and purity after enrichment using FACS® are shown in Fig. S1. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20040638.
Results
TCRα/β Repertoire of CD8+ T Cell Clones Directed against HCV Epitopes Undergoing Escape Mutations.
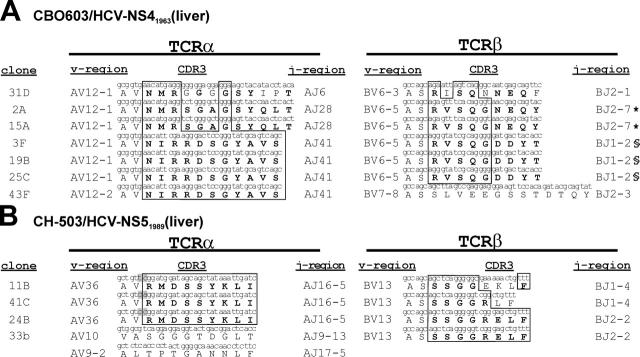
Our initial studies identified a narrow spectrum of TCR usage from CTL clones isolated from the liver of infected chimpanzees. In two chronically infected chimpanzees (CBO603 and CH-503), we were able to isolate multiple T cell clones from the liver after 5 yr of persistent viremia that targeted identical CTL epitopes (Fig. 1). These clones targeted the NS41963 and NS51989 epitopes in CBO603 and CH-503, respectively (Table I). At the time of isolation of these CTL clones, viral escape mutations within these epitopes were found in the plasma of both chimpanzees as described previously (Table I and reference 11), and none of these CTL clones was able to recognize the variant epitope sequences (unpublished data). TCRα/β analysis demonstrated common variable and joining region usage by most of the clones as well as highly homogenous CDR3 amino acid motifs present in most of the TCRα and TCRβ chain sequences (Fig. 1). These data indicate that CTL escape mutations may be associated with the presence of a highly uniform TCRα/β repertoire.
Figure 1.
TCRα/β repertoire of liver-derived HCV-specific CD8+ T cells targeting CTL epitopes undergoing escape mutations in chronically infected chimpanzees. (A) TCRα/β rearrangements of HCV-specific, liver-derived CD8+ T cell clones in chimpanzee CBO603 directed against NS41963. (B) TCR α/β rearrangements of HCV-specific, liver-derived CD8+ T cell clones in chimpanzee CH-503 directed against NS51989. T cell clones with identical nucleotide sequences in TCRα and TCRβ are identified with symbols (*, §) at the end of the sequence.
Longitudinal Analysis of the TCR Repertoire Associated with CTL Escape.
CH-503 recognized another viral epitope located within the HCV envelope protein 2 (E2588/′588-KHPDATYSR-596′) early after primary infection (11). CD8+ T cell clones targeting that epitope were isolated from the liver of CH-503 at multiple time points after infection. A single viral escape mutation (S595T) within this epitope was found as early as 21 mo after infection and remained present as the dominant clone in the viral population thereafter (Table I and reference 11). All liver-derived E2588-specific CTL clones isolated at 7 mo after infection, 14 mo before detection of the escape mutation in the plasma, used similar variable and joining regions, and, more importantly, displayed a highly homologous amino acid motif within their CDR3 region in both TCRα and TCRβ chains (Fig. 2 A). 7 yr after primary infection and 5 yr after the viral escape occurred, nearly identical amino acid motifs were detectable in the CDR3 regions of TCRα/β chains from liver-derived E2588-specific CTL clones (Fig. 2 B). Interestingly, we observed that clonotypes with amino acid motifs identical to those observed 7 mo after primary infection displayed different nucleotide sequences, indicating that those T cell clones were derived from different T cell progenitors (Fig. 2, gray areas).
Figure 2.
Longitudinal analysis of the TCRα/β repertoire of HCV-E2588–specific CD8+ T cells before and after the detection of CTL epitope variations. (A) E2588-specific T cells were derived from the liver of chimpanzee CH-503 7 mo after primary infection, but before the appearance of escape mutations. (B) TCRα/β rearrangement of liver-derived T cell clones generated by limiting dilution 7 yr after primary infection. (C) PBMC-derived TCRβ clonotypes 9 yr after primary infection. 67 sequences were evaluated at month 110, and 70 sequences were evaluated at month 112 after infection. Amino acid motifs within the CDR3 antigen recognition site are shown (bold). Homogenous areas within the CDR3 repertoire are indicated (boxed). Gray areas indicate nucleotide differences of clonotypes with identical amino acid motifs. T cell clones with identical nucleotide sequences in TCRα and TCRβ are identfied with symbols (*, **, §) at the end of the sequence.
Because this response was also detectable in the peripheral blood, we analyzed a total of 137 TCRβ rearrangements from PBMC-derived CD8+ T cells specific for E2588 at two independent time points 9 yr after primary infection (Fig. 2 C). The TCR repertoire at both time points was nearly identical, with >90% of sequences present on both occasions and with a similar hierarchy of the individual clonotypes, demonstrating the reproducibility of the results (Fig. 2 C). The majority of clonotypes from peripheral blood had highly similar amino acid motifs within their CDR3 region as compared with clonotypes of liver-derived T cell clones derived from earlier time points, some with identical nucleotide sequences (Fig. 2). We observed predominant recombination of T cell receptor β variable (TRBV) region 30 with T cell receptor β joining (TRBJ) region 1-1 in most of the sequences. Despite the high similarity on the amino acid level, the T cell response detected 9 yr after primary infection contained many clonotypes that were not previously detected. Interestingly, one clonotype displayed a CDR3 region amino acid sequence that was highly similar to the predominant CDR3 region sequence that was found in clonotypes rearranging TRBV30 with TRBJ1-1, but this clonotype used the TRBV7-2 variable region (Fig. 2 C). Together, these data indicate that a restricted TCR repertoire can be detected long before viral escape mutations become evident and that new clonotypes with identical or highly similar amino acid motifs within the antigen recognition site can be detected over the course of infection.
Analysis of the Entropy of HCV-specific TCR Repertoires.
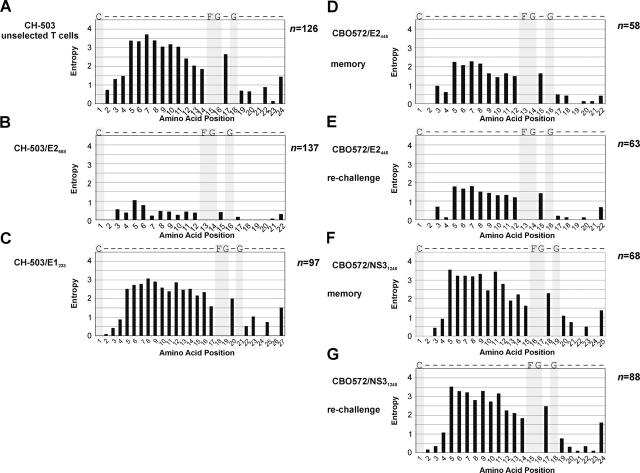
To further evaluate and compare the degree of TCR diversity of different HCV-specific T cell responses, we computed the Shannon entropy (48) for all amino acids sites within each response (Fig. 3). This method has been shown to be useful in the quantification of amino acid diversity in distinct regions of antigen receptors such as immunoglobulins and TCRs (49). A highly diverse TCR repertoire was observed within an unselected T cell population from CH-503 (Fig. 3 A). In marked contrast, liver-derived CTL clones from this animal specific for the NS51989 epitope (Fig. 1 B), which mutated 10 mo after infection, displayed a low CDR3 entropy value of 0.31 ± 0.54. Similarly, the entropy of the TCR sequences generated from six liver-derived CTL clones specific for the E2588 epitope (isolated before escape from immune recognition) was very low (0.17 ± 0.37), due to the presence of a dominant amino acid motif within the CDR3 region (Fig. 2 A). The entropy of a cell line specific for this same epitope generated from PBMCs in month 110 (9 yr after viral escape) was still low (0.45 ± 0.28; P < 0.02; Fig. 3 B) and far lower than most responses associated with lack of escape. In this response, 85% of the TCR sequences were comprised of a single TCR clonotype, the majority of the remaining sequences used the BV30 gene and highly similar CDR3 region sequences, and 94% of the sequences contained identical amino acid motifs (despite different nucleotide sequences) to those from liver-derived CTL clones (Fig. 2 C). A follow-up analysis of PBMCs from month 112 after infection showed nearly identical results. There was no difference in entropy compared with the month 110 result, and in this case 90% of sequences showed the same CDR3 amino acid motif associated with this immune response (Fig. 2 C).
Figure 3.
Analysis of amino acid diversity of the CDR3/joining region in HCV-specific T cell responses. Shannon entropies per amino acid site for unselected T cells (A) and CD8+ T cells specific for E2588 (B) and E1233 (C) in CH-503 are shown. (D–G) CD8+ T cells specific for E2445 in CBO572 6 yr after resolution of primary infection (D) and 14 d after rechallenge with a homologous virus at the peak of the immune response (E), and for CD4+ T cells specific for NS31248 6 yr after resolution of primary infection (F) and 14 d after rechallenge with a homologous virus at the peak of the immune response (G). Bar graphs indicate the entropy at the respective amino acid site. Conserved amino acid residues are shown (gray bars). Amino acid residues are numbered starting with the conserved NH2-terminal cysteine of the TCR variable region.
TCR Repertoire of HCV-specific Responses Directed against an Epitope without CTL Epitope Variation.
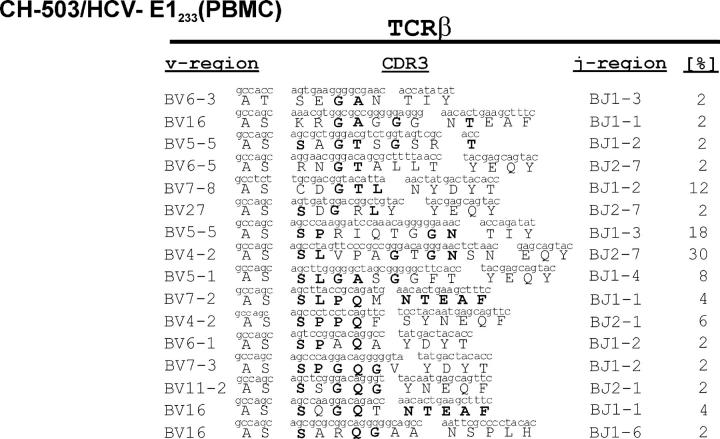
The entropy of the E2588-specific CD8+ T cell response was compared with a CD8+ T cell response derived from CH-503 that was directed against the envelope protein 1 (Table I, E1233/′GNASRCWVA′). This was the only epitope targeted by the CD8+ T cell response in this animal that showed no evidence of viral escape throughout infection (11). Of 97 TCR sequences generated from PBMC-derived E1233-specific CD8+ T cells specific for this epitope, we identified at least 16 distinct TCR clonotypes with diverse TRBV- and TRBJ-region usage (Fig. 4). The mean CDR3 entropy value of this response was 2.14 ± 0.91, which was significantly higher than the entropy observed for the E2588-specific CD8+ T cell response (P = 0.002; Fig. 3 C). Thus, we were able to identify and quantify differences between the TCR repertoire diversity of CD8+ T cell responses directed against epitopes that underwent escape and those that did not show CTL epitope variations during chronic infection. These results also demonstrate that in this chronically infected animal, a diverse repertoire could be generated after in vitro expansion for the one response not associated with escape from immune recognition. For each of these immune responses the level of circulating CTL was low, yet epitope-specific CTLs were easily expanded from peripheral blood, and made up 10–30% of cell lines as assessed by intracellular IFN-γ staining. There was no correlation between the baseline levels of immune responses ex vivo, the degree of in vitro expansion, and the CDR3 amino acid diversity for these responses (Fig. S1).
Figure 4.
TCRβ clonotypes in chronic infection without CTL epitope variants. TCRβ clonotypes of E1233-specific CD8+ cells derived from the peripheral blood of CH-503, where no escape was detectable during chronic infection.
TCR Repertoire Associated with Resolved HCV Infection.
To determine whether our observations might have any relevance for the outcome of HCV infection, we evaluated the TCR diversity of T cell responses associated with spontaneous clearance of hepatitis C. We characterized the TCR repertoires of HCV-specific T cell responses in CBO572, where we observed successful termination of the primary HCV infection and accelerated viral clearance after homologous rechallenge. A detailed description of these immune responses has been recently published (Table I and reference 9). The TCR repertoire of a dominant CD8+ T cell response (E2445/′445-HKFNSSGCPERL-456′; reference 9) from chimpanzee CBO572 at 6 mo before rechallenge (6 yr after primary infection) and at day 14 after rechallenge (at the peak of the immune response, when the virus was cleared) was found to be highly diverse. Fig. 5 A demonstrates the sequences and frequencies of the TCRβ clonotypes of E2445–specific CD8+ T cells that were observed during the memory phase of infection and after rechallenge. Similar to previous findings in different infectious models, the majority of E2445-specific CD8+ T cell sequences evaluated at the peak of the immune response were identical to clonotypes detected within the prechallenge memory repertoire (28). Moreover, clonotypes that were dominant during the memory phase of the infection were also dominant at the peak of the immune response after rechallenge. TCRβ sequences derived from E2445-specific CTL clones (Fig. 5 B), which were independently isolated from the liver and peripheral blood of this animal 5 yr after primary infection by limiting dilution, corresponded to sequences within the TCR repertoire derived from T cells expanded from PBMCs. HCV specificity of these T cell clones was confirmed in a chromium release cytotoxicity assay using E2445 peptide–pulsed autologous target cells (Fig. 5 C). After rechallenge, the magnitude of this response expanded dramatically (9). Within the population of TCR sequences, the dominant TCR sequences from the memory phase of infection showed a preferential expansion after rechallenge. This was associated with a small but statistically significant decrease in the entropy value at each amino acid position of the CDR3 region (P = 0.002), and with a similar pattern of entropy values at each amino acid position (Fig. 3, D and E). Despite this decrease, the entropy value after rechallenge was still higher than the majority of entropies observed in T cell responses associated with CTL escape.
Figure 5.
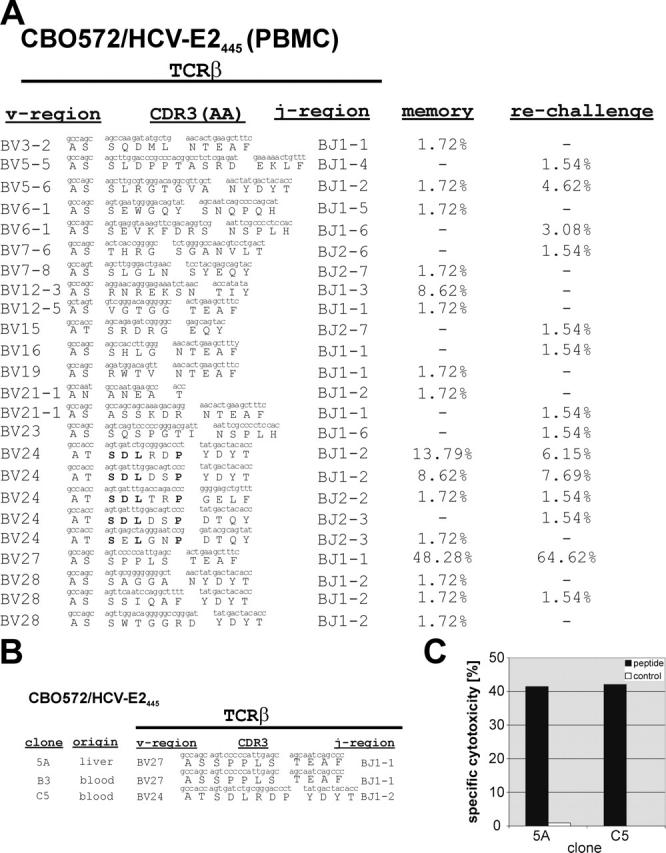
TCR clonotypes after resolved infection, and after rechallenge with homologous virus. (A) The majority of TCRβ clonotypes of E2445-specific CD8+ T cells from CBO572 are identical before and after rechallenge. The frequencies of individual clonotypes 6 yr after resolved primary infection (memory) and 14 d after rechallenge with homologous virus are shown. (B) TCRβ rearrangement and CDR3 sequences of three CD8+ T cell clones derived from liver or PBMCs derived from CBO572 5 yr after primary infection. (C) T cell clones show E2445-specific cytotoxic activity as tested in chromium release assay against autologous target cells at an effector/target cell ratio of 5:1.
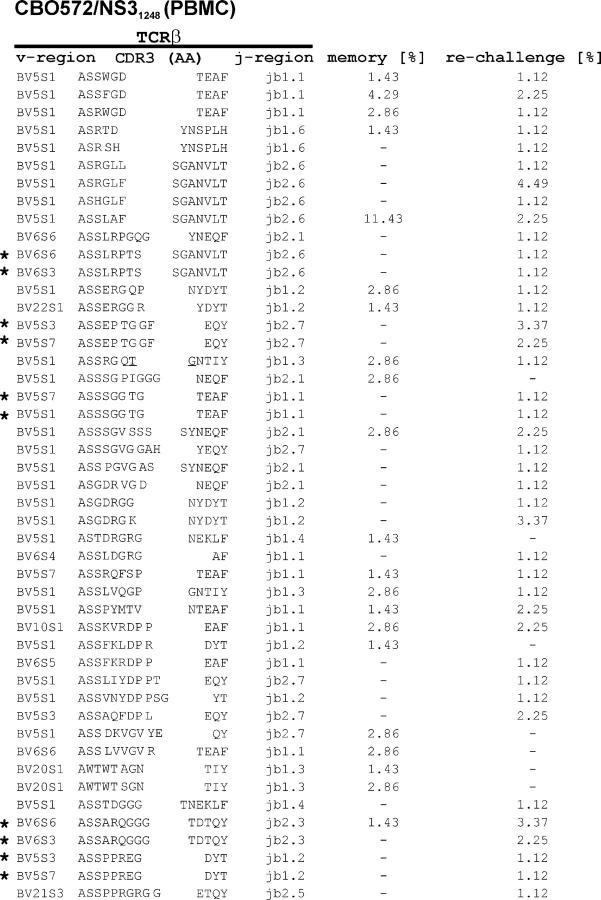
The TCR repertoires of two dominant CD4+ T cell responses directed against the HCV-nonstructural protein 3 (NS31248/′1248-GYKVLVNPSV-1257′ and NS31380/′1380-YGKAIPLEVI-1389′) also displayed a high degree of CDR3 diversity. The highest level of clonal diversity in CBO572 could be detected within the NS31248-specific CD4+ T cell response, where we analyzed the TCR repertoire of the memory response (before virus rechallenge; entropy 2.3 ± 1.14) and at the peak of the immune response 14 d after rechallenge (entropy 2.29 ± 1.10; Fig. 3, F and G, and Fig. 6). This polyclonal response was characterized by rearrangement of genes from the TRBV5 family, but a highly diverse CDR3 and joining region repertoire. After rechallenge, there was no significant change in entropy, despite a 10-fold increase in the magnitude of this response (9). However, several new but closely related sequences were detectable after rechallenge (Fig. 6). Again, we observed the rearrangement of identical CDR3 sequences with different yet closely related TRBV sequences, but no evidence of somatic mutations within the antigen-specific T cell receptor repertoire.
Figure 6.
TCR clonotypes after resolved infection, and after rechallenge with homologous virus. TCRβ clonotypes of NS31248-specific CD4+ T cells from CBO572 before and after rechallenge. The frequencies of individual clonotypes 6 yr after resolved primary infection (memory) and 14 d after rechallenge with homologous virus are shown. *, sequences with identical CDR3 amino acid sequences, but rearrangement of different variable regions.
Low Entropies of HCV-specific T Cell Responses Are Associated with Immune Escape.
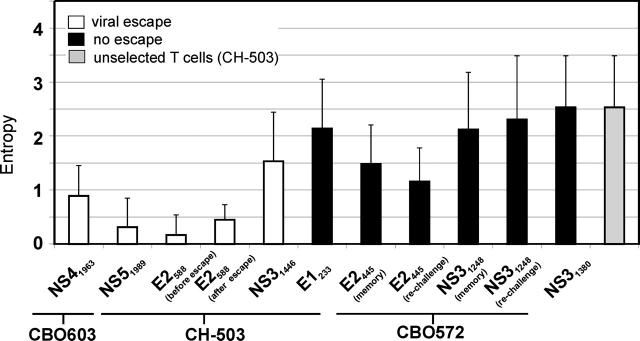
Together, we observed striking differences in TCR diversity between the majority of HCV-specific responses directed against epitopes that underwent CTL escape and those that were associated either with no escape during chronic infection or with protective immunity. Fig. 7 summarizes the hierarchies of the CDR3 entropy values analyzed in this paper. The highest entropy values were present in the unselected T cells (2.53 ± 0.96), the two CD4+ T cell responses, NS31380 (2.12 ± 1.06) and NS31248 (2.29 ± 1.14), and the E1233 CD8+ T cell response (2.14 ± 0.91). Each of these values was significantly higher than three out of the four responses (NS41963, NS51989, and E2588) associated with escape from immune recognition (p-values range from <0.001 to 0.012 for each comparison). In addition, the dominant CD8+ T cell response in chimpanzee CBO572, which was directed against the E2445 epitope and had an entropy value of 1.49 ± 0.72, showed greater diversity than each of the escape-associated CD8+ T cell responses (p-values range from 0.006–0.03 for each comparison), except for NS31446.
Figure 7.
CDR3 entropy of HCV-specific responses from chronically infected chimpanzees CBO603 and CH-503 and chimpanzee CBO572 after resolved HCV infection. Bars represent mean CDR3 entropy (±SD) of unselected T cells (gray bar), HCV-specific T cell responses associated with CTL escape (white bars), and those not associated with CTL escape (black bars).
There was no significant difference between the entropy value of each of these responses and the NS31446 response (1.53 ± 9.1), which was the only T cell response with a high entropy value that was associated with viral escape (Fig. 7 and Table I). Overall, the four responses associated with escape from immune recognition were significantly less diverse than those associated with resolution of infection, or that did not lead to escape from immune recognition (P = 0.011).
Discussion
Although there is increasing evidence for the importance of HCV-specific T cell responses in the resolution of HCV infection, reasons for the failure of the immune system to eradicate the virus and to prevent viral escape mutations are less clear (17). The aim of this paper was to characterize the TCR repertoire of immunodominant HCV-specific T cell responses in chimpanzees with chronic HCV infection or after successfully resolved primary infection and subsequent homologous viral rechallenge. One of our goals was to understand why HCV-specific CTL responses that are easily detectable during chronic HCV infection in both humans (1, 50) and chimpanzees (2, 11) are not capable of clearing viremia.
We observed striking differences between the T cell receptor repertoire diversity of the majority of responses that were associated with viral escape in chronic infection and those where no escape occurred, either in chronic or resolved infection. Marked diversity within the antigen-specific TCR repertoire has been noted for a variety of chronic viral infections (20–23, 33, 43, 45, 51) including HCV (52). However, despite the general descriptions of TCR repertoire fluctuations over the course of viral infections (33, 43, 45) or TCR usage of individual CTL clones (52), there has been no comprehensive analysis of the diversity of the antigen-specific TCR repertoire in relation to clearance of viremia or the generation of viral escape variants. In our analysis, three out of four T cell responses directed against T cell epitopes in which viral escape mutations developed during chronic HCV infection displayed a highly homogeneous amino acid motif within their antigen recognition site, and this held true for both the TCRα and TCRβ chains. A Shannon entropy analysis confirmed this limited TCR diversity, showing decreased entropy values at the antigen recognition sites within these TCR populations. In a longitudinal analysis, the highly homogenous CDR3 repertoire was present long before the viral escape mutation could be detected, and ruled out the possibility this was an oligoclonal T cell expansion that occurred in response to a mutation within the recognized epitope. It is important to note that in each of these responses, the dominant T cell clonotypes were unable or at least substantially reduced in their capacity to recognize the variant peptides in vitro. Furthermore, it was not possible to expand T cell responses to these epitope variants despite repetitive peptide stimulations in vitro.
Our results are unlikely to be due to a selection bias in CD8+ T cell responses with restricted TCR repertoires (e.g., due to low frequency of epitope-specific T cells). For example, the NS31446 response generated quite a diverse TCR repertoire after in vitro expansion, despite the low magnitude of the response as measured directly via ELISPOT (unpublished data). There was also no correlation between the magnitude of in vitro expansion of IFN-γ+ cells and the TCR amino diversity of epitope-specific T cell responses in chronic infection. Although the low entropy repertoire generated after expansion with the E2588 peptide (Fig. 2 C) was dominated by a single clonotype, there were several clonotypes present in this response that conformed to a particular motif that directly corresponded to that of liver-derived CTL clones derived several years earlier. Therefore, these responses were comprised of many distinct clonotypes as defined by differences in the encoding nucleotides (indicated by the gray areas within the nucleotide sequences in Figs. 1 and 2) and yet displayed a restricted TCR repertoire at the amino acid level. This may reflect the inability of new clonotypes with different CDR3 amino acid motifs to emerge. Importantly, the restricted amino acid variability seemed to be stable over an extended period of time (at least 9 yr), although the emergence of different clonotypes (with the same restricted amino acid repertoire) could be observed over time. The observation of long-term clonal stability in association with persistent viral replication extends our earlier findings in human HIV infection (53, 54). However, our results indicate an inability of the immune system to directly respond to the emergence of viral mutations, and in this case a less effective immune response is maintained over a prolonged period of time, a situation that has been termed “original antigenic sin” (55).
In resolved infection, there was a substantial increase in the magnitude of HCV-specific T cell responses after viral rechallenge (∼10-fold for both the CD8+ [E2445] and CD4+ T cell responses ([NS31248] evaluated here; reference 9) and, whereas the entropy of the CD8 response declined slightly at the peak of the response, the CDR3 region entropy of the CD4+ T cell response evaluated after rechallenge before remained diverse, despite rather restricted usage of the BV5 TCR gene. This association between the ability to expand diverse TCR repertoires after T cell stimulation and a favorable disease outcome may be related to prior observations that T cells with enhanced proliferative potential are associated with better control of viremia in murine models (56, 57) and after HIV infection (58).
Although our paper describes the association between TCR diversity and the emergence of viral escape mutations during chronic HCV infection, the reasons why limited TCR repertoires are generated in the first place cannot be answered with our study design. The antigen-specific TCR repertoire could be shaped by positive and negative selection during thymopoesis (59, 60) or by overrepresentation of certain nucleotides within the CDR3 region TCR germline sequences (61). Furthermore, differences in TCR affinity might influence a selective expansion of certain clonotypes (36, 62) as described in a previous paper where TCR diversity was found to be associated with the binding affinity of the peptide to its MHC class I complex (21). Whatever the reasons may be for a reduced diversity of antigen-specific TCR clonotypes against particular epitopes, we suggest that this inflexibility to substantially change the TCR repertoire at the amino acid level might be crucial for successful immune evasion by the virus.
But how can reduced amino acid variability within the TCR repertoire facilitate immune evasion by the virus? We hypothesize that reduced amino acid sequence variability of the antigen recognition site within an HCV-specific T cell response might result in a reduced flexibility to recognize viral sequence variations within the epitope. This might result in better chances to evade immune recognition and thereby provide a survival benefit for the virus by two mechanisms. First, survival might be more likely with lack of T cell recognition itself. Evidence for this mechanism has been demonstrated in chronic viral infections such as HIV infection, where a single amino acid change in an epitope can abrogate recognition by an individual CTL clone, but where efficient cross-recognition of epitope variations by diverse T cell clonotypes can be observed (34, 63). Second, we hypothesize that CD8+ T cell responses with a limited TCR repertoire might be more vulnerable to the induction of clonal unresponsiveness due to antagonistic properties of the epitope variation, which has been observed in various chronic viral infections (12–14).
For the robust, successful CD8+-mediated immune response directed against E2445 in animal CBO572, we observed a narrowing of TCR diversity after rechallenge with HCV. This response was diverse in the memory phase and after rechallenge; however, at the peak of the immune response after rechallenge, the dominant TCR clonotypes showed preferential expansion. This led to an overall decrease in CDR3 entropy within this population of TCR sequences, and is consistent with a previous paper showing TCR repertoire narrowing after rechallenge infection (21). This would be consistent with the hypothesis that the initial generation of a diverse TCR repertoire, followed by clearance of the pathogen, leaves behind a diverse memory pool of T cells. After rechallenge those T cells with the highest avidity and highest antiviral activity will preferentially expand and result in more rapid viral clearance upon rechallenge. For a polyclonal CD4+ T cell response, we observed the emergence of previously undetected clonotypes that displayed identical CDR3 amino acid sequences but rearranged different yet closely related variable regions (Fig. 6). This finding also supports the idea of a preferential expansion of T cell clones with optimal antigen recognition that is related to the structure of the CDR3 antigen recognition site. Interestingly, this diverse response was characterized by preferential rearrangement of variable regions belonging to the TRBV5 family, indicating that restriction of TRBV rearrangement does not predict the degree of TCR diversity at the antigen recognition site.
The emergence of viral escape mutations within a CTL epitope might occur within contact residues interacting with the TCR or through mutations that affect antigen presentation. The latter could be located within the epitope, especially within MHC anchor residues, or even outside the epitope at positions that affect antigen processing. With highly diverse antigen-specific TCR repertoires, there is an increased likelihood that viral mutations that affect only the peptide–TCR interaction site, and do not completely interrupt peptide presentation or processing, will be recognized by clonotypes within the repertoire. Furthermore, in this case it would be unlikely that a particular virus mutation could function as an antagonist to the entire TCR repertoire. Alternately, it has to be emphasized that escape mutations triggered by narrow TCR repertoires may also occur within residues critical for binding to the MHC class I molecule. At which amino acid position the mutation finally occurs is the result of the balance between the impact of amino acid variation on the function/structure of the respective viral protein (viral fitness) and the efficiency of immune evasion (survival benefit). In conclusion, a narrow TCR repertoire might thus be more beneficial for the virus because it is less adaptive (cross-reactive) to viral variations within the epitopes and, thus, allows the virus to select for the mutations that are most beneficial in terms of viral fitness.
For one CD8+ T cell response (NS31446), we observed an amino acid mutation that led to decreased recognition despite a diverse TCR repertoire. This could be explained by a mutation interfering with successful peptide presentation on the MHC class I molecule. If there are constraints on particular viral epitopes that prevent the generation of variants that completely abrogate MHC class I binding or interfere with antigen processing, it may still be advantageous for the host to generate a diverse TCR repertoire. In this case, the generation of a broad array of T cell clones may include T cell receptors with high affinity for the peptide MHC complex, and this may to some extent compensate for reduced binding to MHC class I.
The finding of highly similar TCR repertoires of antigen-specific T cells in different compartments, liver and peripheral blood, is in accordance with recent findings in a murine model of a localized viral infection (64). Our findings of similar TCR motifs as well as similar degrees of TCR diversity between T cell clones isolated from the liver and T cells specific for the same antigen isolated from peripheral blood also suggest that virus selection pressure is mediated by T cells homing to the site of infected cells and then entering the peripheral circulation. This argues against views that peripheral blood and liver might act as completely different immunological compartments, at least for immune responses characterized by an oligoclonal TCR repertoire. However, future studies will be needed to address whether there are quantitative differences between the two compartments.
In summary, we provide evidence that the amino acid diversity within the T cell receptor CDR3 antigen recognition site may have direct effects on the evolution of escape mutations within viral epitopes, and, therefore, might be a crucial factor for the outcome of chronic viral infections such as HCV or HIV, where viral escape mutations contribute to immune evasion. Further studies such as these will be critical to our understanding of the molecular basis of viral immunity.
Acknowledgments
We thank Drs. D. Hasselschwert and S. Roy of the New Iberia Research Center for the excellent care of the animals cited in this paper. We would also like to extend our thanks to Dr. R. O'Neill at the National Institutes of Health (NIH) National Center for Research Resources for support of our analyses.
This work was supported by Public Health Service grants AI-RO1-A1-47367 and U19AI48231 (to C.M. Walker and S.A. Kalams) and NIH grant R01 AI39966 (to S.A. Kalams). S.A. Kalams is an Elizabeth Glaser Scientist of the Elizabeth Glaser Pediatric AIDS Foundation. D. Meyer-Olson is supported by a postdoctoral fellowship from the Deutsche Forschungsgemeinschaft (Me 1891-1), and N.H. Shoukry is supported by postdoctoral fellowships from the Canadian Institute for Health Research and the American Liver Foundation. This research was supported by the Vanderbilt-Meharry Center for AIDS Research, an NIH-funded program (no. P30 AI 54999).
The authors have no conflicting financial interests.
Abbreviations used in this paper: HCV, hepatitis C virus; TRBJ, T cell receptor β joining; TRBV, T cell receptor β variable.
References
- 1.Lauer, G.M., and B.D. Walker. 2001. Hepatitis C virus infection. N. Engl. J. Med. 345:41–52. [DOI] [PubMed] [Google Scholar]
- 2.Cooper, S., A.L. Erickson, E.J. Adams, J. Kansopon, A.J. Weiner, D.Y. Chien, M. Houghton, P. Parham, and C.M. Walker. 1999. Analysis of a successful immune response against hepatitis C virus. Immunity. 10:439–449. [DOI] [PubMed] [Google Scholar]
- 3.Lechner, F., D.K. Wong, P.R. Dunbar, R. Chapman, R.T. Chung, P. Dohrenwend, G. Robbins, R. Phillips, P. Klenerman, and B.D. Walker. 2000. Analysis of successful immune responses in persons infected with hepatitis C virus. J. Exp. Med. 191:1499–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thimme, R., D. Oldach, K.M. Chang, C. Steiger, S.C. Ray, and F.V. Chisari. 2001. Determinants of viral clearance and persistence during acute hepatitis C virus infection. J. Exp. Med. 194:1395–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerlach, J.T., H.M. Diepolder, M.C. Jung, N.H. Gruener, W.W. Schraut, R. Zachoval, R. Hoffmann, C.A. Schirren, T. Santantonio, and G.R. Pape. 1999. Recurrence of hepatitis C virus after loss of virus-specific CD4(+) T-cell response in acute hepatitis C. Gastroenterology. 117:933–941. [DOI] [PubMed] [Google Scholar]
- 6.Diepolder, H.M., J.T. Gerlach, R. Zachoval, R.M. Hoffmann, M.C. Jung, E.A. Wierenga, S. Scholz, T. Santantonio, M. Houghton, S. Southwood, et al. 1997. Immunodominant CD4+ T-cell epitope within nonstructural protein 3 in acute hepatitis C virus infection. J. Virol. 71:6011–6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takaki, A., M. Wiese, G. Maertens, E. Depla, U. Seifert, A. Liebetrau, J.L. Miller, M.P. Manns, and B. Rehermann. 2000. Cellular immune responses persist and humoral responses decrease two decades after recovery from a single-source outbreak of hepatitis C. Nat. Med. 6:578–582. [DOI] [PubMed] [Google Scholar]
- 8.Mehta, S.H., A. Cox, D.R. Hoover, X.H. Wang, Q. Mao, S. Ray, S.A. Strathdee, D. Vlahov, and D.L. Thomas. 2002. Protection against persistence of hepatitis C. Lancet. 359:1478–1483. [DOI] [PubMed] [Google Scholar]
- 9.Shoukry, N.H., A. Grakoui, M. Houghton, D.Y. Chien, J. Ghrayeb, K.A. Reimann, and C.M. Walker. 2003. Memory CD8+ T cells are required for protection from persistent hepatitis C virus infection. J. Exp. Med. 197:1645–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erickson, A.L., M. Houghton, Q.L. Choo, A.J. Weiner, R. Ralston, E. Muchmore, and C.M. Walker. 1993. Hepatitis C virus-specific CTL responses in the liver of chimpanzees with acute and chronic hepatitis C. J. Immunol. 151:4189–4199. [PubMed] [Google Scholar]
- 11.Erickson, A.L., Y. Kimura, S. Igarashi, J. Eichelberger, M. Houghton, J. Sidney, D. McKinney, A. Sette, A.L. Hughes, and C.M. Walker. 2001. The outcome of hepatitis C virus infection is predicted by escape mutations in epitopes targeted by cytotoxic T lymphocytes. Immunity. 15:883–895. [DOI] [PubMed] [Google Scholar]
- 12.Niewiesk, S., S. Daenke, C.E. Parker, G. Taylor, J. Weber, S. Nightingale, and C.R. Bangham. 1995. Naturally occurring variants of human T-cell leukemia virus type I Tax protein impair its recognition by cytotoxic T lymphocytes and the transactivation function of Tax. J. Virol. 69:2649–2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klenerman, P., S. Rowland-Jones, S. McAdam, J. Edwards, S. Daenke, D. Lalloo, B. Koppe, W. Rosenberg, D. Boyd, A. Edwards, et al. 1994. Cytotoxic T-cell activity antagonized by naturally occurring HIV-1 Gag variants. Nature. 369:403–407. [DOI] [PubMed] [Google Scholar]
- 14.Bertoletti, A., A. Sette, F.V. Chisari, A. Penna, M. Levrero, M. De Carli, F. Fiaccadori, and C. Ferrari. 1994. Natural variants of cytotoxic epitopes are T-cell receptor antagonists for antiviral cytotoxic T cells. Nature. 369:407–410. [DOI] [PubMed] [Google Scholar]
- 15.Meier, U.C., P. Klenerman, P. Griffin, W. James, B. Koppe, B. Larder, A. McMichael, and R. Phillips. 1995. Cytotoxic T lymphocyte lysis inhibited by viable HIV mutants. Science. 270:1360–1362. [DOI] [PubMed] [Google Scholar]
- 16.Wedemeyer, H., X.S. He, M. Nascimbeni, A.R. Davis, H.B. Greenberg, J.H. Hoofnagle, T.J. Liang, H. Alter, and B. Rehermann. 2002. Impaired effector function of hepatitis C virus-specific CD8+ T cells in chronic hepatitis C virus infection. J. Immunol. 169:3447–3458. [DOI] [PubMed] [Google Scholar]
- 17.Willberg, C., E. Barnes, and P. Klenerman. 2003. HCV immunology-death and the maiden T cell. Cell Death Differ. 10:S39–S47. [DOI] [PubMed] [Google Scholar]
- 18.Appay, V., P.R. Dunbar, M. Callan, P. Klenerman, G.M. Gillespie, L. Papagno, G.S. Ogg, A. King, F. Lechner, C.A. Spina, et al. 2002. Memory CD8+ T cells vary in differentiation phenotype in different persistent virus infections. Nat. Med. 8:379–385. [DOI] [PubMed] [Google Scholar]
- 19.Crispe, I.N. 2003. Hepatic T cells and liver tolerance. Nat. Rev. Immunol. 3:51–62. [DOI] [PubMed] [Google Scholar]
- 20.Cose, S.C., J.M. Kelly, and F.R. Carbone. 1995. Characterization of diverse primary herpes simplex virus type 1 gB-specific cytotoxic T-cell response showing a preferential V beta bias. J. Virol. 69:5849–5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Busch, D.H., and E.G.P. Am. 1998. MHC class I/peptide stability: implications for immunodominance, in vitro proliferation, and diversity of responding CTL. J. Immunol. 160:4441–4448. [PubMed] [Google Scholar]
- 22.Cole, G.A., T.L. Hogg, and D.L. Woodland. 1994. The MHC class I-restricted T cell response to Sendai virus infection in C57BL/6 mice: a single immunodominant epitope elicits an extremely diverse repertoire of T cells. Int. Immunol. 6:1767–1775. [DOI] [PubMed] [Google Scholar]
- 23.Horwitz, M.S., Y. Yanagi, and M.B. Oldstone. 1994. T-cell receptors from virus-specific cytotoxic T lymphocytes recognizing a single immunodominant nine-amino-acid viral epitope show marked diversity. J. Virol. 68:352–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murali-Krishna, K., J.D. Altman, M. Suresh, D.J. Sourdive, A.J. Zajac, J.D. Miller, J. Slansky, and R. Ahmed. 1998. Counting antigen-specific CD8 T cells: a reevaluation of bystander activation during viral infection. Immunity. 8:177–187. [DOI] [PubMed] [Google Scholar]
- 25.Razvi, E.S., and R.M. Welsh. 1993. Programmed cell death of T lymphocytes during acute viral infection: a mechanism for virus-induced immune deficiency. J. Virol. 67:5754–5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Razvi, E.S., and R.M. Welsh. 1995. Apoptosis in viral infections. Adv. Virus Res. 45:1–60. [DOI] [PubMed] [Google Scholar]
- 27.Sourdive, D.J., K. Murali-Krishna, J.D. Altman, A.J. Zajac, J.K. Whitmire, C. Pannetier, P. Kourilsky, B. Evavold, A. Sette, and R. Ahmed. 1998. Conserved T cell receptor repertoire in primary and memory CD8 T cell responses to an acute viral infection. J. Exp. Med. 188:71–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blattman, J.N., D.J. Sourdive, K. Murali-Krishna, R. Ahmed, and J.D. Altman. 2000. Evolution of the T cell repertoire during primary, memory, and recall responses to viral infection. J. Immunol. 165:6081–6090. [DOI] [PubMed] [Google Scholar]
- 29.McHeyzer-Williams, M.G., and M.M. Davis. 1995. Antigen-specific development of primary and memory T cells in vivo. Science. 268:106–111. [DOI] [PubMed] [Google Scholar]
- 30.Savage, P.A., J.J. Boniface, and M.M. Davis. 1999. A kinetic basis for T cell receptor repertoire selection during an immune response. Immunity. 10:485–492. [DOI] [PubMed] [Google Scholar]
- 31.Busch, D.H., I. Pilip, and E.G.P. Am. 1998. Evolution of a complex T cell receptor repertoire during primary and recall bacterial infection. J. Exp. Med. 188:61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Busch, D.H., and E.G.P. Am. 1999. T cell affinity maturation by selective expansion during infection. J. Exp. Med. 189:701–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen, Z.W., Y. Li, X. Zeng, M.J. Kuroda, J.E. Schmitz, Y. Shen, X. Lai, L. Shen, and N.L. Letvin. 2001. The TCR repertoire of an immunodominant CD8+ T lymphocyte population. J. Immunol. 166:4525–4533. [DOI] [PubMed] [Google Scholar]
- 34.Kalams, S.A., R.P. Johnson, M.J. Dynan, K.E. Hartman, T. Harrer, E. Harrer, A.K. Trocha, W.A. Blattner, S.P. Buchbinder, and B.D. Walker. 1996. T cell receptor usage and fine specificity of human immunodeficiency virus 1–specific cytotoxic T lymphocyte clones: analysis of quasispecies recognition reveals a dominant response directed against a minor in vivo variant. J. Exp. Med. 183:1669–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mason, D. 1998. A very high level of crossreactivity is an essential feature of the T-cell receptor. Immunol. Today. 19:395–404. [DOI] [PubMed] [Google Scholar]
- 36.Sloan-Lancaster, J., and P.M. Allen. 1996. Altered peptide ligand-induced partial T cell activation: molecular mechanisms and role in T cell biology. Annu. Rev. Immunol. 14:1–27. [DOI] [PubMed] [Google Scholar]
- 37.Messaoudi, I., J.A. Guevara Patino, R. Dyall, J. LeMaoult, and J. Nikolich-Zugich. 2002. Direct link between mhc polymorphism, T cell avidity, and diversity in immune defense. Science. 298:1797–1800. [DOI] [PubMed] [Google Scholar]
- 38.Borrow, P., H. Lewicki, X. Wei, M.S. Horwitz, N. Peffer, H. Meyers, J.A. Nelson, J.E. Gairin, B.H. Hahn, M.B. Oldstone, and G.M. Shaw. 1997. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat. Med. 3:205–211. [DOI] [PubMed] [Google Scholar]
- 39.Koenig, S., A.J. Conley, Y.A. Brewah, G.M. Jones, S. Leath, L.J. Boots, V. Davey, G. Pantaleo, J.F. Demarest, C. Carter, et al. 1995. Transfer of HIV-1-specific cytotoxic T lymphocytes to an AIDS patient leads to selection for mutant HIV variants and subsequent disease progression. Nat. Med. 1:330–336. [DOI] [PubMed] [Google Scholar]
- 40.Rudolph, M.G., J.G. Luz, and I.A. Wilson. 2002. Structural and thermodynamic correlates of T cell signaling. Annu. Rev. Biophys. Biomol. Struct. 31:121–149. [DOI] [PubMed] [Google Scholar]
- 41.Weiner, A., A.L. Erickson, J. Kansopon, K. Crawford, E. Muchmore, A.L. Hughes, M. Houghton, and C.M. Walker. 1995. Persistent hepatitis C virus infection in a chimpanzee is associated with emergence of a cytotoxic T lymphocyte escape variant. Proc. Natl. Acad. Sci. USA. 92:2755–2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weiner, A.J., H.M. Geysen, C. Christopherson, J.E. Hall, T.J. Mason, G. Saracco, F. Bonino, K. Crawford, C.D. Marion, K.A. Crawford, et al. 1992. Evidence for immune selection of hepatitis C virus (HCV) putative envelope glycoprotein variants: potential role in chronic HCV infections. Proc. Natl. Acad. Sci. USA. 89:3468–3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Douek, D.C., M.R. Betts, J.M. Brenchley, B.J. Hill, D.R. Ambrozak, K.L. Ngai, N.J. Karandikar, J.P. Casazza, and R.A. Koup. 2002. A novel approach to the analysis of specificity, clonality, and frequency of HIV-specific T cell responses reveals a potential mechanism for control of viral escape. J. Immunol. 168:3099–3104. [DOI] [PubMed] [Google Scholar]
- 44.Meyer-Olson, D., K.W. Brady, J.T. Blackard, T.M. Allen, S. Islam, N.H. Shoukry, K. Hartman, C.M. Walker, and S.A. Kalams. 2003. Analysis of the TCR {beta} variable gene repertoire in chimpanzees: identification of functional homologs to human pseudogenes. J. Immunol. 170:4161–4169. [DOI] [PubMed] [Google Scholar]
- 45.Cohen, G.B., S.A. Islam, M.S. Noble, C. Lau, C. Brander, M.A. Altfeld, E.S. Rosenberg, J.E. Schmitz, T.O. Cameron, and S.A. Kalams. 2002. Clonotype tracking of TCR repertoires during chronic virus infections. Virology. 304:474–484. [DOI] [PubMed] [Google Scholar]
- 46.Kowalski, H., A.L. Erickson, S. Cooper, J.D. Domena, P. Parham, and C.M. Walker. 1996. Patr-A and B, the orthologues of HLA-A and B, present hepatitis C virus epitopes to CD8+ cytotoxic T cells from two chronically infected chimpanzees. J. Exp. Med. 183:1761–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Choo, Q.L., G. Kuo, R. Ralston, A. Weiner, D. Chien, G. Van Nest, J. Han, K. Berger, K. Thudium, C. Kuo, et al. 1994. Vaccination of chimpanzees against infection by the hepatitis C virus. Proc. Natl. Acad. Sci. USA. 91:1294–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shannon, C.E. 1948. A mathematical theory of communication. The Bell System Technical Journal. 27:379–423. [Google Scholar]
- 49.Stewart, J.J., C.Y. Lee, S. Ibrahim, P. Watts, M. Shlomchik, M. Weigert, and S. Litwin. 1997. A Shannon entropy analysis of immunoglobulin and T cell receptor. Mol. Immunol. 34:1067–1082. [DOI] [PubMed] [Google Scholar]
- 50.Lechner, F., N.H. Gruener, S. Urbani, J. Uggeri, T. Santantonio, A.R. Kammer, A. Cerny, R. Phillips, C. Ferrari, G.R. Pape, and P. Klenerman. 2000. CD8+ T lymphocyte responses are induced during acute hepatitis C virus infection but are not sustained. Eur. J. Immunol. 30:2479–2487. [DOI] [PubMed] [Google Scholar]
- 51.Casanova, J.L., P. Romero, C. Widmann, P. Kourilsky, and J.L. Maryanski. 1991. T cell receptor genes in a series of class I major histocompatibility complex–restricted cytotoxic T lymphocyte clones specific for a Plasmodium berghei nonapeptide: implications for T cell allelic exclusion and antigen-specific repertoire. J. Exp. Med. 174:1371–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tsai, S.L., Y.M. Chen, M.H. Chen, C.Y. Huang, I.S. Sheen, C.T. Yeh, J.H. Huang, G.C. Kuo, and Y.F. Liaw. 1998. Hepatitis C virus variants circumventing cytotoxic T lymphocyte activity as a mechanism of chronicity. Gastroenterology. 115:954–965. [DOI] [PubMed] [Google Scholar]
- 53.Brander, C., P.J. Goulder, K. Luzuriaga, O.O. Yang, K.E. Hartman, N.G. Jones, B.D. Walker, and S.A. Kalams. 1999. Persistent HIV-1-specific CTL clonal expansion despite high viral burden post in utero HIV-1 infection. J. Immunol. 162:4796–4800. [PubMed] [Google Scholar]
- 54.Islam, S.A., C.M. Hay, K.E. Hartman, S. He, A.K. Shea, A.K. Trocha, M.J. Dynan, N. Reshamwala, S.P. Buchbinder, N.O. Basgoz, and S.A. Kalams. 2001. Persistence of human immunodeficiency virus type 1-specific cytotoxic T-lymphocyte clones in a subject with rapid disease progression. J. Virol. 75:4907–4911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Klenerman, P., and R.M. Zinkernagel. 1998. Original antigenic sin impairs cytotoxic T lymphocyte responses to viruses bearing variant epitopes. Nature. 394:482–485. [DOI] [PubMed] [Google Scholar]
- 56.Kaech, S.M., and R. Ahmed. 2001. Memory CD8+ T cell differentiation: initial antigen encounter triggers a developmental program in naive cells. Nat. Immunol. 2:415–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kaech, S.M., E.J. Wherry, and R. Ahmed. 2002. Effector and memory T-cell differentiation: implications for vaccine development. Nat. Rev. Immunol. 2:251–262. [DOI] [PubMed] [Google Scholar]
- 58.Migueles, S.A., A.C. Laborico, W.L. Shupert, M.S. Sabbaghian, R. Rabin, C.W. Hallahan, D. Van Baarle, S. Kostense, F. Miedema, M. McLaughlin, et al. 2002. HIV-specific CD8+ T cell proliferation is coupled to perforin expression and is maintained in nonprogressors. Nat. Immunol. 3:1061–1068. [DOI] [PubMed] [Google Scholar]
- 59.Burrows, S.R., R. Khanna, J.M. Burrows, and D.J. Moss. 1994. An alloresponse in humans is dominated by cytotoxic T lymphocytes (CTLs) cross-reactive with a single Epstein-Barr virus CTL epitope: implications for graft-versus-host disease. J. Exp. Med. 179:1155–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Burrows, S.R., S.L. Silins, D.J. Moss, R. Khanna, I.S. Misko, and V.P. Argaet. 1995. T cell receptor repertoire for a viral epitope in humans is diversified by tolerance to a background major histocompatibility complex antigen. J. Exp. Med. 182:1703–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Callan, M.F., H.T. Reyburn, P. Bowness, S. Rowland-Jones, J.I. Bell, and A.J. McMichael. 1995. Selection of T cell receptor variable gene-encoded amino acids on the third binding site loop: a factor influencing variable chain selection in a T cell response. Eur. J. Immunol. 25:1529–1534. [DOI] [PubMed] [Google Scholar]
- 62.Kedl, R.M., B.C. Schaefer, J.W. Kappler, and P. Marrack. 2002. T cells down-modulate peptide-MHC complexes on APCs in vivo. Nat. Immunol. 3:27–32. [DOI] [PubMed] [Google Scholar]
- 63.Buseyne, F., and Y. Riviere. 2001. The flexibility of the TCR allows recognition of a large set of naturally occurring epitope variants by HIV-specific cytotoxic T lymphocytes. Int. Immunol. 13:941–950. [DOI] [PubMed] [Google Scholar]
- 64.Turner, S.J., G. Diaz, R. Cross, and P.C. Doherty. 2003. Analysis of clonotype distribution and persistence for an influenza virus-specific CD8(+) T cell response. Immunity. 18:549–559. [DOI] [PubMed] [Google Scholar]