Abstract
Eosinophil lineage–committed progenitors (EoPs) are phenotypically isolatable in the steady-state murine bone marrow. Purified granulocyte/monocyte progenitors (GMPs) gave rise to eosinophils as well as neutrophils and monocytes at the single cell level. Within the short-term culture of GMPs, the eosinophil potential was found exclusively in cells activating the transgenic reporter for GATA-1, a transcription factor capable of instructing eosinophil lineage commitment. These GATA-1–activating cells possessed an IL-5Rα+CD34+c-Kitlo phenotype. Normal bone marrow cells also contained IL-5Rα+CD34+c-Kitlo EoPs that gave rise exclusively to eosinophils. EoPs significantly increased in number in response to helminth infection, suggesting that the EoP stage is physiologically involved in eosinophil production in vivo. EoPs expressed eosinophil-related genes, such as the eosinophil peroxidase and the major basic protein, but did not express basophil/mast cell–related mast cell proteases. The enforced retroviral expression of IL-5Rα in GMPs did not enhance the frequency of eosinophil lineage read-outs, whereas IL-5Rα+ GMPs displayed normal neutrophil/monocyte differentiation in the presence of IL-5 alone. Thus, IL-5Rα might be expressed specifically at the EoP stage as a result of commitment into the eosinophil lineage. The newly identified EoPs could be the cellular target in the treatment of a variety of disorders mediated by eosinophils.
Eosinophils are rare hematopoietic cells that normally constitute only 1–3% of peripheral blood nucleated cells. In tissues, they are found mainly in the gastrointestinal mucosa. Upon diverse stimuli, eosinophils or their progenitors are recruited from the circulation into inflammatory sites where they may modulate immune responses by releasing cytotoxic cationic proteins and a variety of inflammatory cytokines/chemokines (1). Eosinophils play an important role in defense mechanisms against parasitic infections, but also are involved in a variety of allergic reactions and can mediate tissue damage (2). Hypereosinophilic syndrome is characterized by persistent eosinophilia with tissue infiltration (1), and sometimes is fatal mainly as a result of eosinophil-mediated cardiac damage (2). Understanding the developmental pathway of eosinophils is critical to identifying potential therapeutic targets for eosinophil-mediated disorders.
Like other blood cells, eosinophils are derived from hematopoietic stem cells (HSCs). Although eosinophils are categorized as granulocytes together with neutrophils, their origin remains controversial. For example, they were found in vitro in single colonies also containing basophils, erythrocytes, or myelomonocytic cells (3–6). However, lineage read-outs of multipotent progenitors are random, at least in vitro (7); therefore, it is difficult to define the origin of cells based on the cell components in single cell–derived colonies. Thus, it is critical to isolate and locate the eosinophil lineage–committed progenitors (EoPs) in normal hematopoiesis.
Eosinophil development is supported by a variety of cytokines, including βc-related cytokines, such as GM-CSF, IL-3, and IL-5; however, to develop eosinophilia, IL-5 signaling is especially critical (8). Eosinophilia induced by helminth infection or aeroallergen exposure was not observed in IL-5–deficient mice (9), and is blocked in mice treated with neutralizing anti–IL-5 antibodies (10). In turn, IL-5 transgenic mice displayed sustained eosinophilia (11, 12). Therefore, receptors for IL-5 (IL-5Rα) should be expressed in putative EoPs, which enables EoPs to proliferate and differentiate in response to IL-5.
Other critical molecules for eosinophil development include the GATA transcription factors (13). GATA-1 and GATA-2 are expressed in mature eosinophils as well as in cells of the megakaryocyte/erythrocyte lineage. In studies using transformed chicken cell lines, these GATA factors played a key role in eosinophil differentiation when their expression was enforced by transfection (14). GATA-1 can transactivate the major basic protein (MBP; reference 15). GATA-1–deficient mice lacked eosinophils, and the enforced expression of GATA-1 or GATA-2 stimulated the formation of eosinophil colonies at the expense of granulocyte/monocyte colonies (16). Furthermore, enforced GATA-1 expression converted granulocyte/monocyte cells into the eosinophil as well as the megakaryocyte/erythrocyte lineages (17, 18). Thus, it is likely that these GATA factors are expressed in an early stage of eosinophil progenitors to support their maturation.
In the present study, we identified EoPs from the normal murine bone marrow using a FACS. By using mice harboring a reporter for GATA-1 transcription, we found that the eosinophil potential exclusively exists in a fraction of progeny of granulocyte/monocyte progenitors (GMPs) that activated the GATA-1 reporter. These cells expressed IL-5Rα in addition to immature hematopoietic cell markers, such as CD34 and c-Kit. Given the expression pattern of these molecules, we could purify EoPs within the normal murine bone marrow prospectively.
RESULTS AND DISCUSSION
A transgenic GATA-1 reporter marks eosinophil lineage–committed progenitors downstream of granulocyte/monocyte progenitors
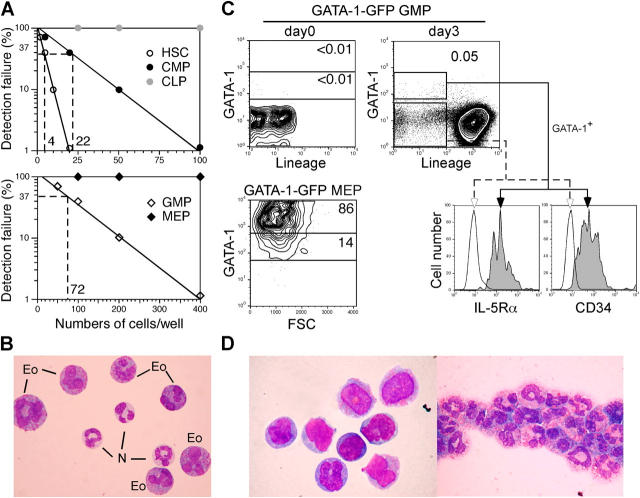
We first evaluated the distribution of the developmental potential into eosinophils in normal hematopoiesis. We previously identified the earliest lymphoid-committed progenitor (common lymphoid progenitor [CLP]; reference 19) and the earliest myeloid-committed progenitor (common myeloid progenitor [CMP]; reference 20) that represent the choice point of the lymphoid versus the myeloid lineage decision. In the myeloid pathway, the GMP and the megakaryocyte/erythrocyte progenitor (MEP; reference 20) also have been isolated downstream of CMPs. Development of eosinophils was evaluated in liquid cultures containing Slf, IL-3, GM-CSF, IL-5, and IL-9. By limiting dilution assays, 1 in 4 HSCs, 22 CMPs, and 72 GMPs could generate eosinophils in vitro (Fig. 1 A), whereas no eosinophil progeny was obtained in >10,000 MEPs, CLPs, proB or proT cells (not depicted); this indicated that the eosinophil potential exists along with the granulocyte/monocyte differentiation pathway. The higher frequencies of the eosinophil potential in HSCs and CMPs compared with GMPs may be because both populations can generate multiple GMPs in culture (20). In GMP cultures, eosinophils were found in 14 out of 1,098 single GMP-derived colonies (Table I). 6 were pure eosinophil colonies; 4 contained neutrophils and eosinophils (Fig. 1 B); and 4 consisted of eosinophils, neutrophils, and macrophages. Colonies that contained only macrophages and eosinophils were not found. These data indicate that the eosinophil potential exists along with the granulocyte/monocyte differentiation pathway from HSCs, and at least a fraction of GMPs are bipotent for the eosinophil and the neutrophil lineages.
Figure 1.
Eosinophils develop from GMPs together with neutrophils and monocytes. (A) Frequency of eosinophil read-outs from purified HSCs stem cells or from myeloid- or lymphoid-committed progenitors determined by limiting dilution assays. (B) Single GMP-derived colony contained eosinophils (Eo) and neutrophils (N). (C) Isolation of EoPs within GMP cultures was achieved by using a transgenic GFP reporter for GATA-1 transcription. GFP is expressed in MEPs, but not in GMPs. The eosinophil potential was found exclusively in the GATA-1–GFP+ fraction of day 3 GMP progeny that expressed IL-5Rα and CD34. (D) Purified day 3 GATA-1–GFP+IL-5Rα+ cells (left) exclusively generated mature eosinophils (right).
Table I.
Results of single GMP cultures
| Pure eosinophils | Neutrophils and macrophages | Neutrophils | Macrophages | Total | |
|---|---|---|---|---|---|
| No eosinophils | NA | 240 | 682 | 162 | 1,084 |
| Eosinophil-containing wells | 6 | 4 | 4 | 0 | 14 |
| Total | 6 | 244 | 686 | 162 | 1,098 |
Total 1,200 single GMPs were cultured in the presence of Slf, IL-3, GM-CSF, IL-5, and IL-9. Plating efficiency was 91% (1,098/1,200). Each colony type was determined by May-Grünwald Giemsa (MGG) staining. In case eosinophils were detected in a colony, EoPO transcripts were additionally analyzed by RT-PCR.
GATA-1 is expressed in mature eosinophils, instructs eosinophil commitment at a myeloid progenitor stage (14, 17), and transactivates MBP (15). Thus, it is possible that EoPs exist within cells activating GATA-1 transcription. To separate cells committed to the eosinophil lineage within GMP cultures by transcriptional activation of GATA-1, we established mice harboring the transgenic GATA-1 reporter tagged with GFP. The promoter construct encompasses all three DNase hypersensitive regions and all six GATA-1 exons. In other transgenic models, this construct accurately reproduced endogenous GATA-1 transcriptional control (21). Consistent with our previous RT-PCR data (18), GATA-1–GFP was not detectable in GMPs or CLPs, but increased at a high level in MEPs (Fig. 1 C). When culturing GMPs with Slf, IL-3, GM-CSF, IL-5, and IL-9, a small fraction of Lin− cells expressing an intermediate level of GFP appeared on day 3 (Fig. 1 C). Most GATA-1–GFP+ cells in the immature Lin− fraction expressed IL-5Rα, CD34 (Fig. 1 C), and a low level of c-Kit, but not Sca-1 (not depicted). Purified GATA-1–GFP+IL-5Rα+ GMP progeny differentiated only into eosinophils (Fig. 1 D) in liquid cultures, whereas day 3 GATA–1-GFP− GMP progeny gave rise only to neutrophils and macrophages (not depicted) in the presence of the cytokine cocktail. These data strongly suggest that commitment into the eosinophil lineage completes within 3 d during the GMP culture, and that the GATA-1–GFP expression marks the vast majority of cells capable of differentiation into eosinophils within day 3 GMP cultures. Because Lin−GATA-1–GFP− cells did not express IL-5Rα (Fig. 1 C), IL-5Rα was likely to be a useful marker for EoPs in normal mice. We tried to isolate EoPs in the normal bone marrow by using this phenotypic definition.
Isolation of IL-5R–expressing eosinophil lineage–committed progenitors in murine bone marrow
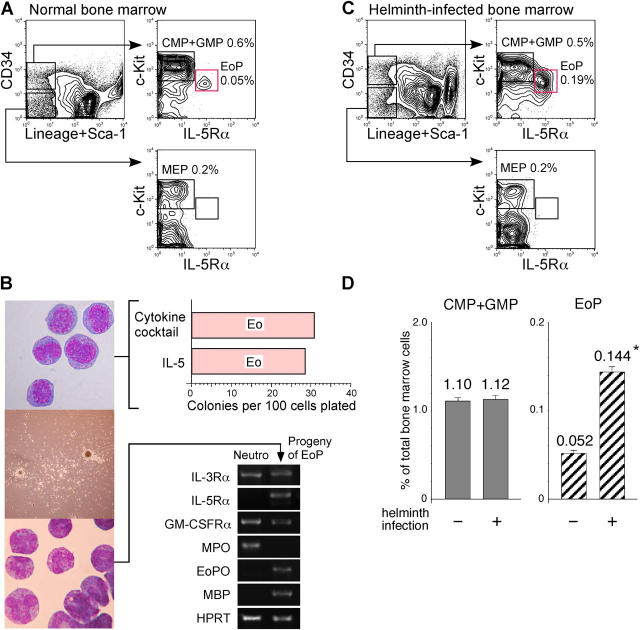
In the normal bone marrow, the Lin−Sca-1−CD34+ fraction contained a small number of cells (∼0.05% of total) expressing IL-5Rα and a low level of c-Kit (Fig. 2 A). Similar to GATA-1–GFP+ GMP progeny, purified IL-5Rα+Lin−Sca-1−CD34+c-Kitlo EoPs were blastic cells with scattered eosinophilic granules (Fig. 2 B, left). As expected, purified EoPs differentiated exclusively into eosinophils in liquid and methylcellulose cultures. Single EoPs formed eosinophil colonies at ∼30% of plating efficiency in methylcellulose containing Slf, IL-3, IL-5, IL-9, GM-CSF, Epo, and Tpo, or IL-5 alone (Fig. 2 B). Progeny of EoPs were all eosinophils with IL-5Rα+Gr-1+ phenotype by FACS (not depicted), and possessed eosinophil peroxidase (EoPO) and MBP, but not myeloperoxidase transcripts (Fig. 2 B). Thus, EoPs have the eosinophil lineage–restricted differentiation capacity, and their proliferation and maturation can be supported solely by IL-5.
Figure 2.
EoPs are prospectively isolatable in the murine bone marrow. (A) FACS analysis of the normal bone marrow demonstrated the existence of IL-5Rα+Lin−Sca-1−CD34+c-Kitlo EoPs. (B) Purified EoPs were blastic cells (left, top), and formed homogenous compact colonies (left, middle) that contained only eosinophils (bottom left). The results of the methylcellulose colony assay also are shown (top right). Cytokine cocktails used in this study contained Slf, IL-3, IL-5, IL-9, GM-CSF, Epo, and Tpo. RT-PCR analyses of purified Gr-1+CD11b+ neutrophils and progeny of EoPs are shown (bottom right). HPRT, hypoxanthine phosphoribosyltransferase; MPO, myeloperoxidase. (C) FACS analysis of the bone marrow from T. spiralis–infected mice. (D) Percentages of EoPs and CMPs plus GMPs in the bone marrow of mice with or without T. spiralis infection.
We next considered whether the IL-5Rα+Lin−CD34+c-Kitlo EoP is involved in the physiological eosinophil development in response to inflammation. Mice infected with Trichinella spiralis display accumulation of mature eosinophils in the intestine within 5 d after infection (22). Therefore, we analyzed the bone marrow of infected mice. On day 5, in the bone marrow, EoPs expanded by approximately threefold in number, whereas numbers of GMPs and CMPs were not affected (Fig. 2 C); this suggested that the EoP population represents a critical stage for in vivo eosinophil development. Purified EoPs from infected mice again differentiated exclusively to eosinophils (unpublished data). Because we could not find IL-5Rα+Lin−Sca-1−CD34+c-Kitlo EoPs in the spleen or the intestine of normal or helminth-infected mice (unpublished data), these data suggest that eosinophils are produced mainly in the bone marrow via the EoP stage.
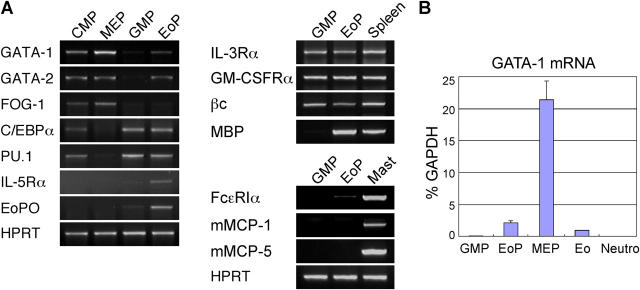
We then analyzed the expression of eosinophil lineage–related genes in purified EoPs (Fig. 3 A). EoPs expressed transcription factors critical for eosinophil development, including GATA-1, GATA-2, PU.1, and C/EBPα, whereas GMPs only expressed PU.1 and C/EBPα. The expression level of GATA-1 in purified EoPs, GMPs, MEPs, Gr-1+CD11c+ neutrophils, and Gr-1+IL-5Rα+ eosinophils was evaluated by a real-time PCR assay (Fig. 3 B). Consistent with the GFP level of each progenitor in GATA-1–GFP reporter mice (Fig. 1 C), EoPs possessed a low amount of GATA-1 transcripts, whose level was ∼10-fold less than that in MEPs. Mature eosinophils expressed a further two- to threefold lower amount of GATA-1 transcripts as compared with EoPs. The expression pattern of GATA-1 in these purified cells was consistent with the fact that in a chicken myeloblast cell line, the ectopic GATA-1–induced reprogramming into the eosinophil and the megakaryocyte lineages was associated with intermediate and high levels of GATA-1, respectively (14). Friend of GATA-1, a transcription factor that promotes erythroid, but suppresses eosinophil, development (23) was expressed in MEPs but not EoPs. EoPs also expressed genes related to eosinophil functions, such as MBP and EoPO. FcɛRIα transcripts were barely detectable in EoPs. EoPs did not express basophil/mast cell–related genes, such as murine mast cell protease–1 or –5. Although previous studies demonstrated the presence of granulocytes with a hybrid eosinophil/basophil phenotype in patients who had chronic or acute myelogenous leukemia (24, 25), these data suggest that the developmental pathway for eosinophils is independent of that for basophils or mast cells at least after the EoP stage in normal murine hematopoiesis.
Figure 3.
RT-PCR analyses of lineage-affiliated genes in purified EoPs and other myeloid progenitors. (A) Conventional RT-PCR analyses of lineage-affiliated genes. Mast, peritoneal mast cells. HPRT, hypoxanthine phosphoribosyltransferase. (B) A quantitative real-time PCR assay for GATA-1 mRNA. Eo, purified IL-5Rα+Gr-1+ eosinophils; Neutro, Gr-1hiCD11bhi neutrophils.
IL-5R is expressed as a result of commitment into the eosinophil lineage
The fact that IL-5Rα is expressed in EoPs, but not in the vast majority of GMPs, suggests that IL-5 targets EoPs to stimulate eosinophil production in vivo, but it is unclear whether IL-5 signaling exerts an instructive or permissive effect on eosinophil development. To test the effect of IL-5 signaling on eosinophil lineage commitment, we retrovirally transduced the IL-5Rα gene into GMPs. GMPs were infected with GFP-tagged IL-5Rα retroviruses (Fig. 4 A) or control GFP retroviruses, and GFP+ cells were purified on day 2. Only purified GFP+ GMPs that were infected with the GFP–IL-5Rα retrovirus expressed IL-5Rα (Fig. 4 B). Although control GMPs could not respond to IL-5 to form colonies, IL-5Rα+ GMPs gave rise to a variety of GM-related colonies in the presence of IL-5 alone, as efficiently as control GMPs cultured with GM-CSF, IL-3, and IL-5 (Fig. 4 C). Most progeny from IL-5Rα+ GMPs cultured with IL-5 alone were neutrophils and macrophages (Fig. 4 D). There was no increase in frequency of eosinophil development between control and IL-5Rα+ GMPs in these cultures (Fig. 4 E). These data indicate that IL-5 signaling does not instruct eosinophil lineage commitment at the GMP stage, but can support proliferation and maturation of myelomonocytic cells as well as eosinophils. Thus, IL-5Rα expression in EoPs might occur as a result of eosinophil lineage commitment, and IL-5 might regulate eosinophil production in vivo after the completion of the eosinophil lineage fate decision. These results are consistent with a previous report that the bone marrow cells of mice that constitutively expressed transgenic IL-5Rα could form megakaryocyte and GM colonies in the presence of IL-5 alone (26). In this context, in myelopoiesis, a high level of serum IL-5 mainly can induce eosinophilia (11) simply because IL-5Rα is expressed only in EoPs but not in other myeloid progenitors. Therefore, the treatment of eosinophilia or hypereosinophilic syndrome with neutralizing IL-5 antibodies (27–29) might be effective through inhibiting proliferation and maturation of EoPs.
Figure 4.
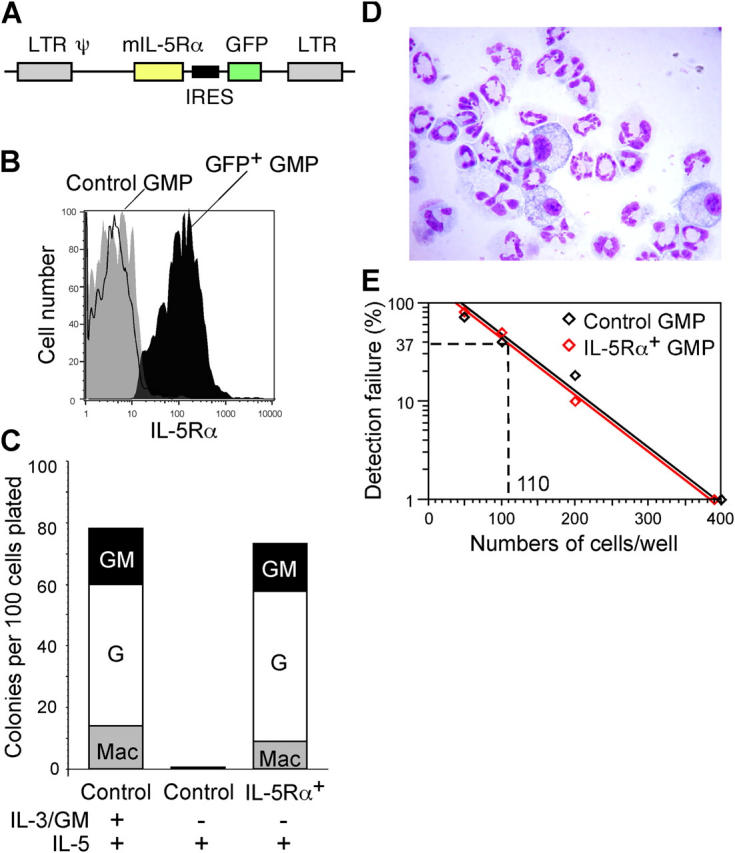
The enforcement of IL-5 signaling did not affect eosinophil lineage read-outs at the GMP stage. (A) The construct of MSCV-mIL-5Rα-ires-GFP retrovirus. LTR, long terminal repeat. (B) GMP infected with GFP-tagged mIL-5Rα retroviruses expressed IL-5Rα protein detected by anti–mIL-5Rα monoclonal antibodies (H7). (C) The effect of IL-5 or other cytokines on myeloid differentiation of control-GFP+ and IL-5Rα-GFP+ GMPs. Mac, macrophage. (D) Cells derived from IL-5Rα-GFP+ GMPs in the presence of IL-5 alone on day 6. IL-5Rα-GFP+ GMPs generate mainly neutrophils and macrophages. (E) Frequency of eosinophil read-outs from control-GFP+ and IL-5Rα-GFP+ GMPs determined by limiting dilution assays.
In summary, we have delineated the eosinophil developmental pathway in normal murine hematopoiesis. Eosinophils developed with neutrophils and monocytes from single GMPs. EoPs were cells activating GATA-1 at a low level as compared with MEPs, and were isolatable prospectively downstream of GMPs as a distinct population with the IL-5Rα+Lin−Sca-1−CD34+c-Kitlo phenotype. Thus, the eosinophil developmental pathway might diverge from neutrophils and monocytes at the GMP stage, and result in the formation of EoPs. The newly identified EoPs and other purified progenitor populations (19, 20) might be useful in investigating the mechanism of commitment and differentiation of the eosinophil lineage. EoPs also could be a therapeutic target to control a variety of eosinophil-related disorders, including allergic diseases and the hypereosinophilic syndrome.
MATERIALS AND METHODS
Mice.
C57BL6 mice were purchased from Jackson ImmunoResearch Laboratories. GATA-1–GFP transgenic reporter mice have been developed by using the promoter construct that encompasses all three DNase hypersensitive regions and all six GATA-1 exons (21). GATA-1–GFP mice were crossed with C57BL6 mice for eight generations. Mice were bred and maintained in the Research Animal Facilities at the Dana-Farber Cancer Institute or Harvard Medical School, in accordance with institutional guidelines.
Antibodies, cell staining, and sorting.
HSCs, CLPs, CMPs, MEPs, and GMPs were purified as reported elsewhere (19, 20). PE-Cy5–conjugated rat antibodies specific for the following lineage markers were used: CD3 (CT-CD3), CD4 (RM4-5), CD8 (5H10), B220 (6B2), Gr-1 (8C5), and CD19 (6D5; Caltag). To sort EoPs, bone marrow cells were stained with biotinylated anti–IL-5Rα chain (H7; reference 30), FITC-conjugated anti-CD34 (RAM34), APC-conjugated anti–c-Kit (2B8; BD Biosciences), and PE-Cy5–conjugated anti–Sca-1 (D7; eBioscience) monoclonal antibodies and with a lineage cocktail, followed by avidin-PE (Caltag). EoPs were purified as Lin−Sca-1−CD34+IL-5Rα+c-Kitlo cells with a low level of side scatter profile. Mature eosinophils were purified as IL-5Rα+Gr-1+ cells. Dead cells were excluded by propidium iodide staining in a PE-Cy5 channel. All progenitor populations were sorted or analyzed using a double laser (488 nm/350 nm Enterprise II + 647nm Spectrum) high-speed cell sorter (Moflo-MLS, DakoCytomation). For single cell and limiting dilution assays, cells were sorted directly into 60-well Terasaki plates or 96-well plates using an automatic cell deposition unit. Data were analyzed with the FlowJo software (Treestar, Inc.).
Cultures.
For limiting dilution and single GMP culture assays, progenitor cells were cultured in 96-well or 60-well Terasaki plates with the following liquid medium; Iscove's Modified Dulbecco's Medium (GIBCO BRL) supplemented with 20% FCS, 5 × 10−5 M 2-mercaptoethanol, sodium pyruvate, and antibiotics. Cells in each well were split into two preparations; one is cytospun with May-Grünwald Giemsa staining, and the other is for preparing total RNA in Trizol reagent (Invitrogen). Eosinophil-containing wells were determined by the presence of eosinophilic granules on May-Grünwald Giemsa and of EoPO transcripts. For clonogenic analyses of EoPs, cells were cultured in a methylcellulose medium (Methocult H4100, Stem Cell Technologies) supplemented with 30% FCS, 1% BSA, and 2 mM L-glutamine (Stem Cell Technologies). Each EoP-derived colony was picked up, and its transcripts were analyzed by RT-PCR. The cytokines used were: murine Slf (20 ng/ml), IL-3 (20 ng/ml), IL-5 (50 ng/ml), IL-7 (10 ng/ml), IL-9 (50 ng/ml), IL-11 (10 ng/ml), GM-CSF (10 ng/ml), Epo (2 U/ml), and Tpo (10 ng/ml; R&D Systems). All cultures were incubated at 37°C in a humidified chamber under 5% CO2.
Retroviral transduction of GMPs.
A murine IL-5Rα cDNA was subcloned into the XhoI site of MSCV-ires-EGFP vector. An empty vector was used as control. In brief, FACS-purified GMPs were plated onto a recombinant fibronectin fragment-coated culture dish (RetroNectin dish; Takara), and were cultured for 48 h with 1 ml of the virus supernatant containing Slf (20 ng/ml) and IL-11 (10 ng/ml). GFP+ GMPs were purified by FACS, and were subjected to further analyses.
Analysis of gene expression from total RNA.
Total RNA extracted from 200 cells of each population was subjected to RT-PCR analyses. Primer sequences and PCR protocols for each specific gene are listed in Table S1 (available at http://www.jem.org/cgi/content/full/jem.20050548/DC1). A quantitative real-time PCR assay for GATA-1 was performed with ABI PRISM 7700 Sequence Detector. The forward primer was 5′-CAGAACCGGCCTCTCATCC-3′, the reverse primer was 5′-TAGTGCATTGGGTGCCTGC-3′, and the probe was 5′-FAM-CCCAAGAAGCGAATGATTGTCAGCAAA-TAMRA-3′. Rodent GAPDH control reagents (Applied Biosystems) were used as an internal control.
Infection with T. spiralis.
Infectious larvae (L1) were isolated from the skeletal muscle of infected mice. The larvae were washed by low-speed centrifugation (50 g for 5 min), and adjusted to ∼2,000 larvae per ml. Mice were infected with ∼400 larvae each by gavage.
Online supplemental material.
Table S1 provides PCR primer sequences and protocols that were used in Figs. 2 and 3. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20050548/DC1.
Acknowledgments
We thank M.A. Handley for technical assistance with flow cytometric analysis and sorting, and D. Dalma-Weiszhausz for critically reviewing the manuscript.
This work was supported in part by grants from National Institutes of Health (nos. DK050654 and AI031599), the Mitsubishi Pharma Research Foundation (to K. Akashi), and the Leukemia and Lymphoma Society (to S.-i. Mizuno).
The authors have no conflicting financial interests.
Abbreviations used: CLP, common lymphoid progenitor; CMP, common myeloid progenitor; EoP, eosinophil lineage–committed progenitor; EoPO, eosinophil peroxidase; GMP, granulocyte/monocyte progenitor; HSC, hematopoietic stem cell; MBP, major basic proteins; MEP, megakaryocyte/erythrocyte progenitor.
References
- 1.Rothenberg, M.E., A. Mishra, E.B. Brandt, and S.P. Hogan. 2001. Gastrointestinal eosinophils. Immunol. Rev. 179:139–155. [DOI] [PubMed] [Google Scholar]
- 2.Rothenberg, M.E. 1998. Eosinophilia. N. Engl. J. Med. 338:1592–1600. [DOI] [PubMed] [Google Scholar]
- 3.Denburg, J.A., S. Telizyn, H. Messner, B. Lim, N. Jamal, S.J. Ackerman, G.J. Gleich, and J. Bienenstock. 1985. Heterogeneity of human peripheral blood eosinophil-type colonies: evidence for a common basophil-eosinophil progenitor. Blood. 66:312–318. [PubMed] [Google Scholar]
- 4.Nakahata, T., S.S. Spicer, and M. Ogawa. 1982. Clonal origin of human erythro-eosinophilic colonies in culture. Blood. 59:857–864. [PubMed] [Google Scholar]
- 5.Clutterbuck, E.J., E.M. Hirst, C.J. Sanderson, J.M. Wang, A. Rambaldi, A. Biondi, Z.G. Chen, and A. Mantovani. 1989. Human interleukin-5 (IL-5) regulates the production of eosinophils in human bone marrow cultures: comparison and interaction with IL-1, IL-3, IL-6, and GMCSF. Blood. 73:1504–1512. [PubMed] [Google Scholar]
- 6.Nakahata, T., K. Tsuji, A. Ishiguro, O. Ando, N. Norose, K. Koike, and T. Akabane. 1985. Single-cell origin of human mixed hemopoietic colonies expressing various combinations of cell lineages. Blood. 65:1010–1016. [PubMed] [Google Scholar]
- 7.Suda, T., J. Suda, and M. Ogawa. 1984. Disparate differentiation in mouse hemopoietic colonies derived from paired progenitors. Proc. Natl. Acad. Sci. USA. 81:2520–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanderson, C.J. 1992. Interleukin-5, eosinophils, and disease. Blood. 79:3101–3109. [PubMed] [Google Scholar]
- 9.Foster, P.S., S.P. Hogan, A.J. Ramsay, K.I. Matthaei, and I.G. Young. 1996. Interleukin 5 deficiency abolishes eosinophilia, airways hyperreactivity, and lung damage in a mouse asthma model. J. Exp. Med. 183:195–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coffman, R.L., B.W. Seymour, S. Hudak, J. Jackson, and D. Rennick. 1989. Antibody to interleukin-5 inhibits helminth-induced eosinophilia in mice. Science. 245:308–310. [DOI] [PubMed] [Google Scholar]
- 11.Dent, L.A., M. Strath, A.L. Mellor, and C.J. Sanderson. 1990. Eosinophilia in transgenic mice expressing interleukin 5. J. Exp. Med. 172:1425–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tominaga, A., S. Takaki, N. Koyama, S. Katoh, R. Matsumoto, M. Migita, Y. Hitoshi, Y. Hosoya, S. Yamauchi, Y. Kanai, et al. 1991. Transgenic mice expressing a B cell growth and differentiation factor gene (interleukin 5) develop eosinophilia and autoantibody production. J. Exp. Med. 173:429–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McNagny, K., and T. Graf. 2002. Making eosinophils through subtle shifts in transcription factor expression. J. Exp. Med. 195:F43–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kulessa, H., J. Frampton, and T. Graf. 1995. GATA-1 reprograms avian myelomonocytic cell lines into eosinophils, thromboblasts, and erythroblasts. Genes Dev. 9:1250–1262. [DOI] [PubMed] [Google Scholar]
- 15.Yamaguchi, Y., S.J. Ackerman, N. Minegishi, M. Takiguchi, M. Yamamoto, and T. Suda. 1998. Mechanisms of transcription in eosinophils: GATA-1, but not GATA-2, transactivates the promoter of the eosinophil granule major basic protein gene. Blood. 91:3447–3458. [PubMed] [Google Scholar]
- 16.Hirasawa, R., R. Shimizu, S. Takahashi, M. Osawa, S. Takayanagi, Y. Kato, M. Onodera, N. Minegishi, M. Yamamoto, K. Fukao, et al. 2002. Essential and instructive roles of GATA factors in eosinophil development. J. Exp. Med. 195:1379–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heyworth, C., S. Pearson, G. May, and T. Enver. 2002. Transcription factor-mediated lineage switching reveals plasticity in primary committed progenitor cells. EMBO J. 21:3770–3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iwasaki, H., S. Mizuno, R.A. Wells, A.B. Cantor, S. Watanabe, and K. Akashi. 2003. GATA-1 converts lymphoid and myelomonocytic progenitors into the megakaryocyte/erythrocyte lineages. Immunity. 19:451–462. [DOI] [PubMed] [Google Scholar]
- 19.Kondo, M., I.L. Weissman, and K. Akashi. 1997. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 91:661–672. [DOI] [PubMed] [Google Scholar]
- 20.Akashi, K., D. Traver, T. Miyamoto, and I.L. Weissman. 2000. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 404:193–197. [DOI] [PubMed] [Google Scholar]
- 21.Jasinski, M., P. Keller, Y. Fujiwara, S.H. Orkin, and M. Bessler. 2001. GATA1-Cre mediates Piga gene inactivation in the erythroid/megakaryocytic lineage and leads to circulating red cells with a partial deficiency in glycosyl phosphatidylinositol-linked proteins (paroxysmal nocturnal hemoglobinuria type II cells). Blood. 98:2248–2255. [DOI] [PubMed] [Google Scholar]
- 22.Basten, A., M.H. Boyer, and P.B. Beeson. 1970. Mechanism of eosinophilia. I. Factors affecting the eosinophil response of rats to Trichinella spiralis. J. Exp. Med. 131:1271–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Querfurth, E., M. Schuster, H. Kulessa, J.D. Crispino, G. Doderlein, S.H. Orkin, T. Graf, and C. Nerlov. 2000. Antagonism between C/EBPbeta and FOG in eosinophil lineage commitment of multipotent hematopoietic progenitors. Genes Dev. 14:2515–2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boyce, J.A., D. Friend, R. Matsumoto, K.F. Austen, and W.F. Owen. 1995. Differentiation in vitro of hybrid eosinophil/basophil granulocytes: autocrine function of an eosinophil developmental intermediate. J. Exp. Med. 182:49–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weil, S.C., and M.A. Hrisinko. 1987. A hybrid eosinophilic-basophilic granulocyte in chronic granulocytic leukemia. Am. J. Clin. Pathol. 87:66–70. [DOI] [PubMed] [Google Scholar]
- 26.Takagi, M., T. Hara, M. Ichihara, K. Takatsu, and A. Miyajima. 1995. Multi-colony stimulating activity of interleukin 5 (IL-5) on hematopoietic progenitors from transgenic mice that express IL-5 receptor alpha subunit constitutively. J. Exp. Med. 181:889–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamaguchi, Y., T. Matsui, T. Kasahara, S. Etoh, A. Tominaga, K. Takatsu, Y. Miura, and T. Suda. 1990. In vivo changes of hemopoietic progenitors and the expression of the interleukin 5 gene in eosinophilic mice infected with Toxocara canis. Exp. Hematol. 18:1152–1157. [PubMed] [Google Scholar]
- 28.Shardonofsky, F.R., J. Venzor, III, R. Barrios, K.P. Leong, and D.P. Huston. 1999. Therapeutic efficacy of an anti-IL-5 monoclonal antibody delivered into the respiratory tract in a murine model of asthma. J. Allergy Clin. Immunol. 104:215–221. [DOI] [PubMed] [Google Scholar]
- 29.Garrett, J.K., S.C. Jameson, B. Thomson, M.H. Collins, L.E. Wagoner, D.K. Freese, L.A. Beck, J.A. Boyce, A.H. Filipovich, J.M. Villanueva, et al. 2004. Anti-interleukin-5 (mepolizumab) therapy for hypereosinophilic syndromes. J. Allergy Clin. Immunol. 113:115–119. [DOI] [PubMed] [Google Scholar]
- 30.Yamaguchi, N., Y. Hitoshi, S. Mita, Y. Hosoya, Y. Murata, Y. Kikuchi, A. Tominaga, and K. Takatsu. 1990. Characterization of the murine interleukin 5 receptor by using a monoclonal antibody. Int. Immunol. 2:181–187. [DOI] [PubMed] [Google Scholar]