Abstract
The mechanisms that target class switch recombination (CSR) to antibody gene switch (S) regions are unknown. Analyses of switch site locations in wild-type mice and in mice that lack the Sμ tandem repeats show shifts indicating that a 4–5-kb DNA domain (bounded upstream by the Iμ promoter) is accessible for switching independent of Sμ sequences. This CSR-accessible domain is reminiscent of the promoter-defined domains that target somatic hypermutation. Within the 4–5-kb CSR domain, the targeting of S site locations also depends on the Msh2 mismatch repair protein because Msh2-deficient mice show an increased focus of sites to the Sμ tandem repeat region. We propose that Msh2 affects S site location because sequences with few activation-induced cytidine deaminase targets generate mostly switch DNA cleavages that require Msh2-directed processing to allow CSR joining.
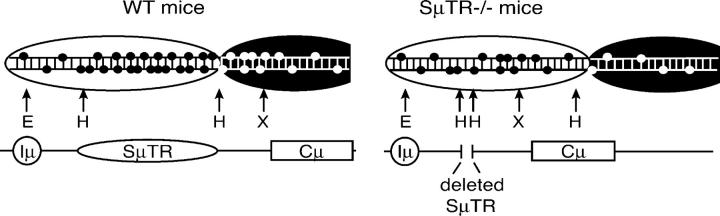
Antibody gene class switch recombination (CSR) occurs between switch (S) regions that are located upstream of each CH gene and contain tandem repeat sequences that are often G-rich on the nontranscribed DNA strand (1, 2). The unusual nature of S region tandem repeat (SμTR) sequences suggests that they play a role in CSR. However, mice lacking Sμ tandem repeats (SμTR−/−, previously designated as ΔSμ; reference 3) show only two- to threefold reductions in isotype switching, which indicates that sequences outside of the SμTR can target the switching mechanism (3). Larger deletions of Sμ sequences have greater defects in switching (4), and an extensive deletion of Sγ1 shows almost complete abrogation of γ1 switching (5), indicating that S region sequences are critical for CSR. The mechanisms that target CSR to S regions are unknown.
Activation-induced cytidine deaminase (AID) and proteins involved in nonhomologous end joining are critical for CSR activity. AID deamination of deoxycytidine residues appears to initiate events that result in S region DNA cleavages (6, 7), whereas nonhomologous end joining proteins may be involved in the religation of broken S region DNA ends (8–10). CSR also involves the uracil DNA glycosylase protein, possibly for the generation of abasic sites from AID-generated dU–dG DNA mismatches (11). DNA mismatch repair (MMR) proteins are also important for CSR (12, 13), but their roles are unclear. Studies of S sites located within a portion of Sμ showed that the absence of the Msh2 MMR protein caused CSR to focus on GAGCT/GGGGT sequences (12). Furthermore, the presence of the SμTR region is critical for class switching in Msh2-deficient mice, suggesting that Msh2 affects the sequences that are targeted or selected for CSR (14). Msh2 is also important for the low levels of CSR found in uracil DNA glycosylase–deficient mice, suggesting an Msh2 role in some DNA cleavages that initiate CSR (15).
We have analyzed the locations of S sites in two mutant mouse strains that lack either the SμTR region or the Msh2 protein to determine how these mutations affect CSR targeting. S site locations in wild-type and SμTR−/− mice show that CSR events occur within a 4–5-kb DNA region located downstream of the Iμ promoter and provide the first indication that CSR is targeted to a fixed-length “domain” that is linked to the location of a transcriptional promoter. Comparing S site locations in wild-type with Msh2− /− mice shows that the absence of Msh2 focuses CSR sites on the SμTR region. This finding indicates that Msh2 has a role in targeting or selecting specific sequences for CSR within the accessible DNA domain.
Results and Discussion
Upstream S sites are not affected by SμTR deletion
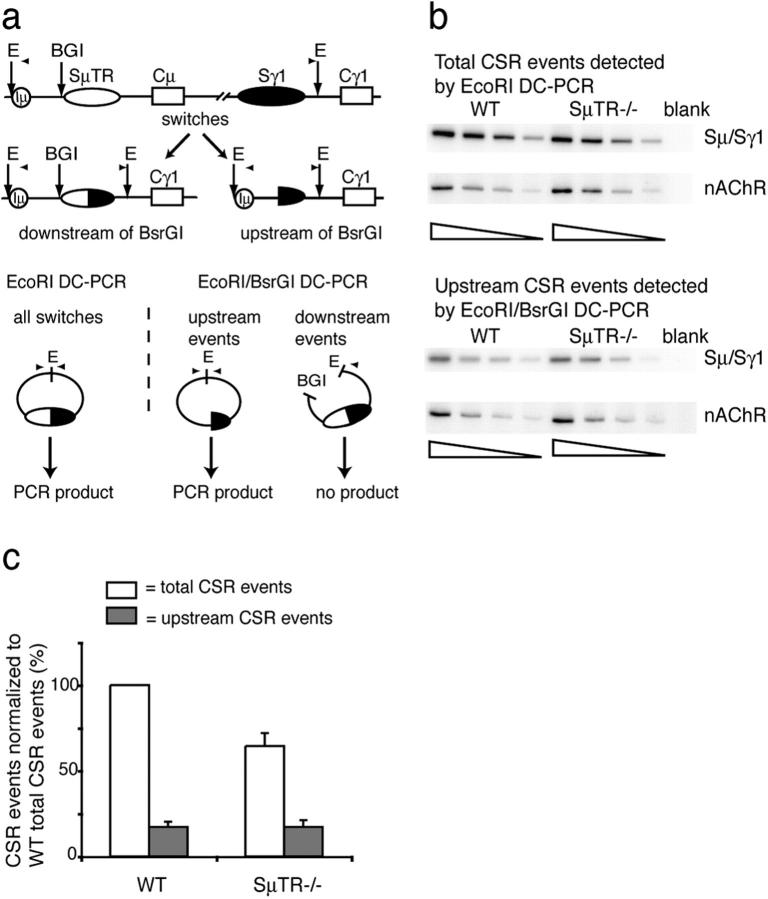
We used several digestion-circularization (DC)-PCR assays to analyze CSR sites in stimulated splenic B cells. To quantify Sμ/Sγ1 CSR sites located upstream of the SμTR region in wild-type and SμTR−/− mice, a BsrGI digestion was added to the standard EcoRI DC-PCR assay (16) before DNA circularization (Fig. 1 a). Only Sμ/Sγ1 junctions upstream of the Sμ BsrGI site will lack the BsrGI site and generate EcoRI fragments that can be self-ligated and PCR amplified. Comparing band intensities in EcoRI DC-PCR with those in EcoRI–BsrGI DC-PCR assays allows the percentages of junctions located upstream or downstream of the Sμ BsrGI site to be calculated.
Figure 1.
SμTR−/− and wild-type mice have similar numbers of upstream CSR events. (a) DC-PCR measures CSR events upstream of the SμTR region. A BsrGI site distinguished CSR junctions located upstream and downstream of the site. Only CSR sites in sequences upstream of BsrGI generate PCR signals. Total CSR events are measured by EcoRI DC-PCR (E, EcoRI; BGI, BsrGI). PCR primers are indicated by arrowheads. (b) Twofold dilutions of genomic DNAs were assayed by Sμ/Sγ1 EcoRI DC-PCR and EcoRI–BsrGI DC-PCR. Samples were normalized using nAChR DC-PCR reactions. (c) CSR events in wild-type or SμTR−/− mice as percentages of total wild-type CSR events (set to 100%). Mean values and SEM are from six independent experiments with two mice per group.
Genomic DNAs isolated from SμTR−/− or wild-type B cells were analyzed using the two assays (Fig. 1 b). Based on the EcoRI DC-PCR results, total levels of Sμ/Sγ1 CSR in SμTR−/− B cells were reduced about twofold relative to the wild type, which was consistent with previous findings (3, 14). However, analyses of Sμ/Sγ1 CSR upstream of the BsrGI site (Fig. 1 b) showed that these were the same in SμTR−/− and wild-type B cells (Fig. 1 c). The results indicate that, in this upstream region, CSR events are not influenced by the SμTR deletion in nearby downstream regions.
Switch recombinations in SμTR−/− mice shift to downstream JH-Cμ regions
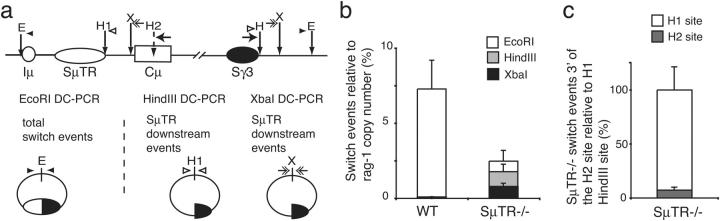
To further locate S sites within the JH-Cμ intron, downstream CSR events measured in HindIII and XbaI DC-PCR assays were compared with the total number of switch events measured by the EcoRI DC-PCR assay (Fig. 2, a and b). Because the three DC-PCRs can have different efficiencies, we used a control plasmid (pSμ/Sγ3) that contained Sμ and Sγ3 sequences together with the three enzyme sites to normalize the DC-PCR assays. The frequencies of CSR sites within different segments of the JH-Cμ region were determined by comparison with standard curves generated using pSμ/Sγ3.
Figure 2.
SμTR−/− mice have increased levels of CSR events targeted to downstream sequences. (a) DC-PCR measuring CSR junction sites downstream of the SμTR region. HindIII and XbaI sites downstream of the SμTR region were used in DC-PCR assays to measure CSR junctions in sequences downstream of each site. Total CSR sites were determined by EcoRI DC-PCR (E, EcoRI; H1 and H2, HindIII; X, XbaI). Primer pairs used for the different DC-PCR assays are indicated by small arrows of different types. (b) Wild-type and SμTR−/− CSR events measured by EcoRI, HindIII, and XbaI real-time DC-PCR. Copy numbers for CSR events were derived from pSμ/Sγ3 standard curves. Rag-1 PCR normalized CSR events relative to Rag-1 copy number. Mean values and SEM are from seven independent experiments with four mice per group. (c) Percentage of SμTR−/− switch events downstream of the H2 relative to events downstream of H1 (set to 100%). Mean values and SEM are from three independent experiments per group.
We first assessed total Sμ/Sγ3 recombinations by EcoRI DC-PCR assays (Fig. 2 a) and used DC-PCR quantitation of the Rag-1 gene to normalize the content of genomic DNA from different samples. The total frequency of Sμ/Sγ3 switch events in wild-type B cells was estimated to be ∼7.3% (Fig. 2 b). This frequency from real-time DC-PCR matches the levels of in vitro γ3 switching as assessed by flow cytometry (14). Sμ/Sγ3 CSR in SμTR−/− mice was reduced about threefold relative to the wild type (Fig. 2 b). Of the total Sμ/Sγ3 junctions in SμTR−/− mice, ∼67% were located downstream of the HindIII site located at the 3′ end of the SμTR region (Fig. 2 a). This high frequency of junctions downstream of H1 in SμTR−/− mice is in striking contrast to the negligible number of Sμ/Sγ3 sites located within the same sequence in wild-type mice (<1% of all switch events; Fig. 2 b).
Of the Sμ/Sγ3 recombinations in SμTR−/− mice located downstream of the H1 site, about half occurred downstream of the XbaI site located near the 5′ end of the Cμ exons (Fig. 2 b). Comparisons of EcoRI DC-PCR with XbaI DC-PCR assays indicated that ∼30% of Sμ/Sγ3 recombinations in SμTR−/− mice occurred at sequences downstream of the XbaI site.
For wild-type and SμTR−/− mice, we compared the locations of CSR sites determined by DC-PCR with the locations of CSR events in hybridoma panels (3). For wild-type mice, 23 hybridomas show five (22%) CSR sites upstream of the SμTR, 17 (78%) sites within the SμTR, and no sites downstream of the SμTR. DC-PCR analyses of wild-type CSR events show 17% upstream, 82% SμTR, and 1% downstream. For CSR sites in SμTR−/− mice, 10 hybridomas show three (30%) upstream of the H1 site and seven (70%) downstream of H1, whereas DC-PCR shows 33% upstream and 67% downstream. Thus, although the hybridoma panels are small and will miss CSR events that result in nonfunctional Ig proteins, the comparisons show a good correlation between the distributions of CSR sites as determined by the two methods.
The CSR site frequency between the HindIII and XbaI sites might be slightly underestimated because of the H2 HindIII site in the Cμ segment (Fig. 2 a). For SμTR−/− mice, CSR events occurring downstream of H2 were estimated using the HindIII-circularized B cell DNA samples. Among SμTR−/− mice with CSR sites located downstream of H1, only ∼7% were located downstream of H2 (Fig. 2 c), assuming that both HindIII DC-PCR assays have equal efficiencies. An examination of the threshold cycles of the two HindIII PCR assays indicated that the efficiencies differ by less than a factor of two (not depicted). Thus, although the majority of switch recombinations in SμTR−/− mice are skewed toward downstream JH-Cμ intron sequences, there are few junction sites downstream of the Cμ exons.
Lack of the MMR Msh2 protein shifts the targeting of CSR to focus on the SμTR region
The role of Msh2 in the CSR process is unclear. Previous analyses of SμTR−/−:Msh2− /− mice have shown that the SμTR region is critical for isotype switching when Msh2 is lacking (14). These studies suggested that Msh2 might be more important for switching in the sequences flanking the SμTR region than within the SμTR itself. To directly test this model, S site locations were determined in Msh2− /− mice that retain the SμTR region.
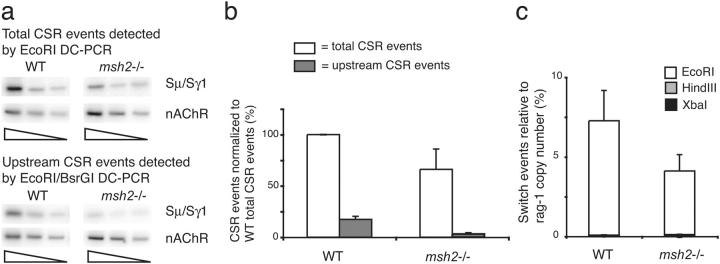
CSR sites throughout the JH-Cμ region were analyzed by DC-PCR in Msh2− /− mice. CSR sites upstream of the SμTR in Msh2− /− mice were decreased about sixfold relative to the wild type, whereas switching within the SμTR was only reduced by a factor of 1.4 (Fig. 3, a and b). The fraction of CSR events located upstream in Msh2− /− mice was significantly different from the wild type (two-sided P < 0.0001 by χ2 test; Fig. 3 b). Very few CSR events occurred downstream of the SμTR in both Msh2− /− and wild-type mice (Fig. 3 C), and we cannot assess whether any specific reductions occur. Within the regions that exhibited the majority of CSR activity, however, our results show a shift of CSR site focus into the SμTR region in Msh2− /− mice (Fig. 4) and support the model that Msh2 is more important for CSR in non-SμTR sequences and less important for CSR within the SμTR.
Figure 3.
CSR within sequences upstream of the SμTR are reduced in Msh2 −/− mice. (a) Twofold dilutions of genomic DNAs were assayed by Sμ/Sγ1 EcoRI DC-PCR and EcoRI–BsrGI DC-PCR. Samples were normalized using nAChR DC-PCR. (b) CSR sites in wild-type or Msh2 −/− mice measured relative to total wild-type CSR (set to 100%). Mean values and SEM of at least four independent experiments are presented. (c) CSR events in Msh2 −/− mice measured by EcoRI, HindIII, and XbaI real-time DC-PCR and compared with wild-type mice. Mean values and SEM are from six independent experiments with two Msh2 −/− mice and seven independent experiments with four wild-type mice.
Figure 4.
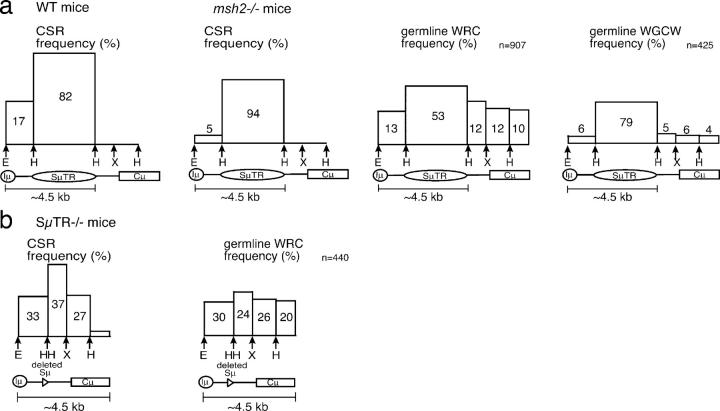
CSR junction sites and AID target sites within different segments of the JH-Cμ intron. Histograms of CSR site, WRC motif, and WGCW motif frequencies are shown for different segments of the JH-Cμ region. (a) Comparisons of wild-type with Msh2 −/− mice that both have wild-type Sμ regions. (b) Comparisons of SμTR−/− mice with a shortened Sμ region. DNA segments are drawn in proportion on the x axis. Areas of boxes in the histogram are proportional to frequencies in each segment, resulting in box heights that indicate relative recombinational frequencies per nucleotide. Boxes for mutant mice are drawn to scale relative to wild-type mice. Switching reductions in SμTR−/− varied slightly in different DC-PCR assays and were averaged. Junction site distributions in each segment were assumed to be uniform. Wild-type CSR frequencies are set to 100%; other CSR frequencies are normalized to this total. Total WRC and WGCW motif frequencies were set to 100% for each mouse strain. The SμTR is only partially sequenced (available from GenBank/EMBL/DDBJ under accession no. AC073553). Densities of motifs are assumed conserved throughout the SμTR. The total numbers of each motif in the Iμ–Cμ region are indicated in the top right of each graph.
Patterns of S sites in wild-type and SμTR−/− mice suggest a 4–5-kb switching domain
SμTR−/− mice exhibit shifts of S site locations into downstream JH-Cμ intron sequences that show almost no CSR in wild-type mice. These shifts indicate that, in wild-type mice, there is a mechanism that restricts switching in these downstream regions because the same sequences can clearly support numerous switch events when the SμTR region is deleted. This mechanism does not appear to reflect competition between different sequences for the recruitment of the CSR machinery because the sequences upstream of the SμTR, which can clearly support CSR even in wild-type mice, show no increases in CSR when the SμTR is absent. Based on S site locations in wild-type mice, it appears that a 4–5-kb-long region downstream of the Iμ promoter is accessible for CSR in stimulated B cells. The pattern of S sites in SμTR−/− B cells suggests that a similarly sized region is also accessible for CSR, but, because of the SμTR deletion, this region now extends into Cμ sequences (Fig. 5).
Figure 5.
Model depicting the targeting domain for CSR in the JH-Cμ region for wild-type and SμTR−/− mice. The JH-Cμ intron region is shown with small circles cartooning WRC motifs (potential AID cleavage sites) in the JH-Cμ region. Open ovals represent domains accessible to CSR cleavage; regions not accessible to CSR are indicated by shaded ovals.
The mechanism that limits the CSR domain is not known. The Iμ promoter appears to be the upstream boundary of most CSR events (4, 17) and may localize the upstream limit of the CSR domain (Fig. 5). The CSR domain might be explained by a transcriptional-tracking model similar to models for the targeting of somatic hypermutation (18) in which switch cleavage factors are carried with RNA polymerase during the transcription process, but dissociate after a distance. AID interactions with RNA polymerase (19) provide some support for such a transcription-based model.
The CSR-accessible DNA domain could also involve a chromatin structure that regulates the switching machinery, similar to the ability of chromatin structure to regulate V(D)J recombination (20, 21). Studies of B cells undergoing somatic hypermutation indicate that mutating V(D)J segments are found in chromatin where the histones H3 and H4 are hyperacetylated relative to the C-region of the same gene (22). For switching, however, relationships between CSR and S region H3/H4 hyperacetylation are complex, suggesting that other modifications or protein factors might be needed for CSR targeting (23). In pro–B cells, dimethylation of histone 3 at lysine 4 in the JH-Cμ region peaks around Cμ with a rapid drop off within 2 kb of this central peak (21). Thus, the dimethylation of histone 3 at lysine 4 modification rises rapidly in the region just downstream of the SμTR that forms the 3′ boundary of the switching domain and could be involved in limiting CSR accessibility.
In wild-type mice, CSR sites correlate with the distribution of WRC motifs (Fig. 4 a) that are known sites for AID deamination on single-stranded DNAs (24, 25). In SμTR−/− mice, however, CSR sites do not correlate with WRC motifs (Fig. 4 b). The HindIII–XbaI focus of SμTR−/− CSR sites could reflect the high concentration of nontranscribed G residues in this region (3). Such G-rich sequences promote RNA–DNA R-loops in S regions and might provide single-stranded DNA targets for AID activity (5, 26).
Mice with a larger Sμ-region deletion show more drastic CSR reductions and have Sμ sites mostly constrained to the Iμ exon region (4). Sequence elements important for the 4–5-kb CSR domain proposed in Fig. 5 may be disrupted by the larger Sμ deletion. Identifying sequences that establish the CSR domain might distinguish transcriptional and chromatin structure targeting models and lead to the identification of targeting factors. S regions clearly contain sequences important for CSR because replacing S region sequences with a random DNA sequence does not provide for CSR (27).
Msh2 may be important for processing “frayed” switch DNA cleavages
Our results show that S sites in Msh2− /−mice are strongly focused into the SμTR region, indicating that CSR events within SμTR sequences are much less dependent on Msh2 than are events within non-SμTR sequences. The Msh2 activity that leads to this sequence-specific effect is not clear. Msh2 is known to recognize base pair mismatches, and recent studies suggest that Msh2 recognition of G–U might lead to S region DNA breaks (15). However, Msh2 G–U recognition appears to play a minor role in switch cleavage (15), and there is no apparent reason for this G–U recognition activity to affect switching outside of SμTR sequences to a greater extent than within the SμTR.
The shifts of CSR sites in Msh2− /− mice are consistent with a model that we have previously proposed in which Msh2 processes DNA “flaps” found predominantly on cleavage ends generated from sequences outside of the SμTR (14). This model suggests a postcleavage end-processing function for Msh2 that converts staggered single-stranded breaks to the double-stranded breaks required for CSR, in addition to its role in initiating some switch cleavages.
We analyzed the JH-Cμ sequence to determine whether motifs associated with CSR might account for the S site distribution in Msh2− /− mice. WRC motifs are preferred sites for AID (24, 25), and the WGCW motif is a site where AID activity on opposing strands could provide a double-stranded cleavage with a single base overhang (24). WRC motifs within the JH-Cμ intron correlate well with distributions of CSR sites in the 4–5-kb CSR domain for wild-type mice (Fig. 4 a). For Msh2− /− mice, on the other hand, CSR sites correlate better with WGCW motifs (Fig. 4 a). CSR downstream of the SμTR is rare in both wild-type and Msh2− /− mice, so that Msh2 effects are not discernable. However, SμTR−/− mice show numerous S sites in this downstream region and previous analyses have shown that SμTR−/−:Msh2− /− mice show large decreases in all CSR events (14). Thus, Msh2 is also important when CSR occurs downstream of the SμTR, consistent with the paucity of WGCW sites within these sequences (Fig. 4 a). CSR sites in SμTR−/−:Msh2− /− mice also show a significant focus on WGCW motifs (two-tailed P = 0.0043 by Fisher's exact test) relative to CSR sites in SμTR−/− mice (14), emphasizing the correlation between WGCW motifs and CSR sites when Msh2 is not present. Furthermore, the number of WGCW motifs in the wild-type Sμ region is about half the number of WRC motifs in the same region (Fig. 4 a). Perhaps the twofold reduction in CSR observed between wild-type and Msh2− /− mice correlates roughly with the fraction of cleavage sites that cannot complete CSR because of frayed ends.
The involvement of Msh2 in processing S region breaks would suggest that Msh2 alters switch targeting by affecting the selection of switch breaks that can undergo CSR. Thus, Msh2 has an important role to broaden the types of DNA sequences that can successfully participate in CSR. Analyses of SμTR−/− mice crossed with mice deficient in other MMR proteins, such as ExoI, Mlh1, and Pms2, should indicate whether these proteins are involved in the same processes of the CSR mechanism that are affected by Msh2.
Materials and Methods
Mice and cell culture
SμTR−/− and Msh2 −/− mice were used in these studies. Animal experiments were approved by Tufts University Institutional Animal Care and Use Committee. Splenic B cells were cultured with 50 μg/ml LPS (Sigma-Aldrich) with or without 0.3 ng/ml anti–δ-dextran, or with LPS and IL-4 (800 U/ml) as described previously (14).
DC-PCR
DC-PCR was performed as previously described (3, 14). Circularized DNAs were diluted twofold, and PCR was amplified as follows. For switch events located upstream of the SμTR, DNAs were digested with BsrGI and EcoRI before circularization. Sμ/Sγ1 DC-PCR products were amplified and phosphorimager quantification was normalized using the nicotinic acetylcholine receptor (nAChR) gene (16). BsrGI digestion was monitored by PCR of the BsrGI site using primers Sμ_out (14) and BsrGI_bk (5′-TCATTACTGTGGCTGGAGAG-3′). Switching upstream was quantitated by comparing EcoRI–BsrGI DC-PCR with EcoRI DC-PCR products. For switch events located downstream of the SμTR, genomic DNAs were digested with EcoRI, HindIII, or XbaI, and circularized for real-time PCR using fluorescent SYBR green dye (Applied Biosystems). The pSμ/Sγ3 control plasmid provided standard curves for each real-time DC-PCR. Rag-1 copy numbers were measured by real-time PCR using Rag-1 plasmid standards (Rag-1 plasmid provided by E. Marchlik, Tufts University, Boston, MA). Gel electrophoresis of PCR products and melting temperature analyses were used to confirm the specificity of real-time PCR amplifications. Total Sμ/Sγ3 junctions were measured in an EcoRI DC-PCR assay using published primer pairs DC-μ.2 and DC-γ3.2 (28).
Switch events downstream of the H1 site (Fig. 2) were measured by HindIII DC-PCR using primers H3-IM (5′-CCTACACCAGATCATCCAGTACAGCT-3′) and H3-G3F (5′-GATAGGACAGATGGAGCAGTTACAGA-3′). XbaI DC-PCR measured switch events downstream of the XbaI site located 5′ of the Cμ exon (Fig. 2 a) using primer pairs XBA-IM (5′-GAACGTGACTTTGGAAGCCTTCA-3′) and X1-G3F (5′-GCTGAGAGGAGACCTGGGTTG-3′). HindIII DC-PCR measured S junctions downstream of the H2 site (Fig. 2 c) using primers CMH3-R (5′-GTGGAGGGACCTGCAGTCAAG-3′) and H3-G3F.
Statistics
χ2 tests were used to assess the significance of Msh2 −/− upstream CSR differences. Fisher's exact test was used to assess the significance of differences in CSR at WGCW motifs between SμTR−/−:Msh2 −/− and SμTR−/− mice.
Acknowledgments
We thank Erica Marchlik for the Rag-1 plasmid and Dr. Naomi Rosenberg for discussions and critical readings of the manuscript.
I.M. Min, J. Stavnezer, and E. Selsing were supported by grants from the National Institutes of Health (CA65441, AI23283, and AI24465, respectively).
The authors have no conflicting financial interests.
References
- 1.Dunnick, W., G.Z. Hertz, L. Scappino, and C. Gritzmacher. 1993. DNA sequences at immunoglobulin switch region recombination sites. Nucleic Acids Res. 21:365–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kataoka, T., T. Miyata, and T. Honjo. 1981. Repetitive sequences in class-switch recombination regions of immunoglobulin heavy chain genes. Cell. 23:357–368. [DOI] [PubMed] [Google Scholar]
- 3.Luby, T.M., C.E. Schrader, J. Stavnezer, and E. Selsing. 2001. The μ switch region tandem repeats are important, but not required, for antibody class switch recombination. J. Exp. Med. 193:159–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khamlichi, A.A., F. Glaudet, Z. Oruc, V. Denis, M. Le Bert, and M. Cogne. 2004. Immunoglobulin class-switch recombination in mice devoid of any S mu tandem repeat. Blood. 103:3828–3836. [DOI] [PubMed] [Google Scholar]
- 5.Shinkura, R., M. Tian, M. Smith, K. Chua, Y. Fujiwara, and F.W. Alt. 2003. The influence of transcriptional orientation on endogenous switch region function. Nat. Immunol. 4:435–441. [DOI] [PubMed] [Google Scholar]
- 6.Muramatsu, M., K. Kinoshita, S. Fagarasan, S. Yamada, Y. Shinkai, and T. Honjo. 2000. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 102:553–563. [DOI] [PubMed] [Google Scholar]
- 7.Petersen-Mahrt, S.K., R.S. Harris, and M.S. Neuberger. 2002. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature. 418:99–103. [DOI] [PubMed] [Google Scholar]
- 8.Rolink, A., F. Melchers, and J. Andersson. 1996. The SCID but not the RAG-2 gene product is required for S mu-S epsilon heavy chain class switching. Immunity. 5:319–330. [DOI] [PubMed] [Google Scholar]
- 9.Casellas, R., A. Nussenzweig, R. Wuerffel, R. Pelanda, A. Reichlin, H. Suh, X.F. Qin, E. Besmer, A. Kenter, K. Rajewsky, and M.C. Nussenzweig. 1998. Ku80 is required for immunoglobulin isotype switching. EMBO J. 17:2404–2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manis, J.P., D. Dudley, L. Kaylor, and F.W. Alt. 2002. IgH class switch recombination to IgG1 in DNA-PKcs-deficient B cells. Immunity. 16:607–617. [DOI] [PubMed] [Google Scholar]
- 11.Rada, C., G.T. Williams, H. Nilsen, D.E. Barnes, T. Lindahl, and M.S. Neuberger. 2002. Immunoglobulin isotype switching is inhibited and somatic hypermutation perturbed in UNG-deficient mice. Curr. Biol. 12:1748–1755. [DOI] [PubMed] [Google Scholar]
- 12.Ehrenstein, M.R., and M.S. Neuberger. 1999. Deficiency in Msh2 affects the efficiency and local sequence specificity of immunoglobulin class-switch recombination: parallels with somatic hypermutation. EMBO J. 18:3484–3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schrader, C., W. Edelmann, R. Kucherlapati, and J. Stavnezer. 1999. Reduced isotype switching in splenic B cells from mice deficient in mismatch repair enzymes. J. Exp. Med. 190:323–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Min, I.M., C.E. Schrader, J. Vardo, T.M. Luby, N. D'Avirro, J. Stavnezer, and E. Selsing. 2003. The Smu tandem repeat region is critical for Ig isotype switching in the absence of Msh2. Immunity. 19:515–524. [DOI] [PubMed] [Google Scholar]
- 15.Rada, C., J.M. Di Noia, and M.S. Neuberger. 2004. Mismatch recognition and uracil excision provide complementary paths to both Ig switching and the A/T-focused phase of somatic mutation. Mol. Cell. 16:163–171. [DOI] [PubMed] [Google Scholar]
- 16.Chu, C.C., W.E. Paul, and E.E. Max. 1992. Quantitation of immunoglobulin μ-γ1 heavy chain switch region recombination by a digestion-circularization polymerase chain reaction method. Proc. Natl. Acad. Sci. USA. 89:6978–6982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee, C., S. Kondo, and T. Honjo. 1998. Frequent but biased class switch recombination in the Sμ flanking regions. Curr. Biol. 8:227–230. [DOI] [PubMed] [Google Scholar]
- 18.Storb, U., A. Peters, E. Klotz, N. Kim, H.M. Shen, K. Kage, B. Rogerson, and T.E. Martin. 1998. Somatic hypermutation of immunoglobulin genes is linked to transcription. Curr. Top. Microbiol. Immunol. 229:11–19. [DOI] [PubMed] [Google Scholar]
- 19.Nambu, Y., M. Sugai, H. Gonda, C.G. Lee, T. Katakai, Y. Agata, Y. Yokota, and A. Shimizu. 2003. Transcription-coupled events associating with immunoglobulin switch region chromatin. Science. 302:2137–2140. [DOI] [PubMed] [Google Scholar]
- 20.Su, I.H., A. Basavaraj, A.N. Krutchinsky, O. Hobert, A. Ullrich, B.T. Chait, and A. Tarakhovsky. 2003. Ezh2 controls B cell development through histone H3 methylation and Igh rearrangement. Nat. Immunol. 4:124–131. [DOI] [PubMed] [Google Scholar]
- 21.Morshead, K.B., D.N. Ciccone, S.D. Taverna, C.D. Allis, and M.A. Oettinger. 2003. Antigen receptor loci poised for V(D)J rearrangement are broadly associated with BRG1 and flanked by peaks of histone H3 dimethylated at lysine 4. Proc. Natl. Acad. Sci. USA. 100:11577–11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woo, C.J., A. Martin, and M.D. Scharff. 2003. Induction of somatic hypermutation is associated with modifications in immunoglobulin variable region chromatin. Immunity. 19:479–489. [DOI] [PubMed] [Google Scholar]
- 23.Li, Z., Z. Luo, and M.D. Scharff. 2004. Differential regulation of histone acetylation and generation of mutations in switch regions is associated with Ig class switching. Proc. Natl. Acad. Sci. USA. 101:15428–15433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu, K., F.T. Huang, and M.R. Lieber. 2004. DNA substrate length and surrounding sequence affect the activation-induced deaminase activity at cytidine. J. Biol. Chem. 279:6496–6500. [DOI] [PubMed] [Google Scholar]
- 25.Pham, P., R. Bransteitter, J. Petruska, and M.F. Goodman. 2003. Processive AID-catalysed cytosine deamination on single-stranded DNA simulates somatic hypermutation. Nature. 424:103–107. [DOI] [PubMed] [Google Scholar]
- 26.Yu, K., F. Chedin, C.L. Hsieh, T.E. Wilson, and M.R. Lieber. 2003. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat. Immunol. 4:442–451. [DOI] [PubMed] [Google Scholar]
- 27.Zarrin, A.A., F.W. Alt, J. Chaudhuri, N. Stokes, D. Kaushal, L. Du Pasquier, and M. Tian. 2004. An evolutionarily conserved target motif for immunoglobulin class-switch recombination. Nat. Immunol. 5:1275–1281. [DOI] [PubMed] [Google Scholar]
- 28.Wuerffel, R.A., L. Ma, and A.L. Kenter. 2001. NF-kappa B p50-dependent in vivo footprints at Ig S gamma 3 DNA are correlated with mu-->gamma 3 switch recombination. J. Immunol. 166:4552–4559. [DOI] [PMC free article] [PubMed] [Google Scholar]