Abstract
Major histocompatibility complex class I–restricted cytotoxic T lymphocytes (CTLs) specific for epitopes within eight of the nine Epstein Barr Virus (EBV)-encoded latency-associated proteins have been recovered from EBV-infected human subjects by restimulation of lymphocytes in vitro. However, human class I–restricted CTL responses capable of recognizing EBNA-1 expressing cells were not detected in these studies. We have raised a murine CTL line that recognizes an epitope within EBNA-1 by immunizing mice with a vaccinia virus encoding a COOH-terminal EBNA-1 fragment. This novel CTL line was used to investigate whether the epitope (positions 509–517 in EBNA-1, presented through Kd) was presented to CTL by mouse cells expressing full-length EBNA-1 or a deletion mutant of EBNA-1, lacking the Glycine-Alanine (Gly-Ala)–rich region. Cells expressing full-length EBNA-1 are not lysed by the CTL line, whereas cells expressing the Gly-Ala deletion mutant are recognized. These results suggest that epitopes from full-length EBNA-1 are poorly presented, and that the Gly-Ala–rich region is responsible for this phenomenon. The inefficient presentation of EBNA-1–derived epitopes may explain the absence or rarity of EBNA-1–specific CTLs in vivo, a strategy that may allow EBV to maintain persistence within the immunocompetent host without being eliminated by CTLs.
Epstein-Barr virus (EBV) is a ubiquitous human herpesvirus that establishes latency in B cells (1, 2). In B cells immortalized by EBV infection in vitro (lymphoblastoid cell lines; LCLs) a restricted repertoire of nine “latency- associated” viral genes are expressed: six EBNAs (EBNAs 1–6), LMP-1, LMP-2a, and LMP-2b (1).
CTLs eliminate virus-infected cells, which they recognize by the presence of peptides derived from viral proteins in association with class I molecules (3). To analyze the specificity of the human CTL response against EBV latency-associated proteins, lymphocytes recovered from EBV-infected individuals were restimulated in vitro with autologous LCLs, which express the nine latency-associated proteins (4, 5). The target antigens of the restimulated CTL were then identified by expressing the EBV latency-associated proteins individually through recombinant vaccinia viruses. By this protocol, polyclonal CTL responses, or individual CTL clones, capable of lysing autologous cells expressing most EBV latency-associated antigens were identified. Surprisingly, no CTL response capable of lysing autologous cells expressing EBNA-1 were ever found (4–6). Immunization of several strains of mice with syngeneic tumour cell lines expressing EBNA-1 also failed to raise an EBNA-1–specific rejection response in vivo (7). Recently, MHC class II–restricted, CD-4 positive lymphocyte lines that recognize peptides within EBNA-1 have been identified; these lymphocytes were unable to lyse cells expressing full-length EBNA-1 (8).
Levitskaya et al. (9) created chimeric proteins containing regions of EBNA-1 inserted into another EBV latency-associated protein, EBNA-4, to investigate whether sequences within EBNA-1 affected the presentation of epitopes. Residues 39–498 of EBNA-1 were inserted, in frame, into the EBNA-4 protein. CTL specific for an EBNA-4 epitope, presented by HLA-A11, were unable to recognize cells expressing the EBNA-1–EBNA-4 chimera efficiently. Deletion of the Glycine-Alanine (Gly-Ala)–rich region (residues 93–325 in EBNA-1) from the chimera restored the presentation of the EBNA-4 epitope, thus implicating this region as a cis-acting inhibitor of antigen processing and presentation (9).
Here, we examine the processing and presentation of EBNA-1–derived epitopes using a novel murine class I–restricted CTL line, raised against a fragment of EBNA-1. Using the same CTL line, we also evaluate the role of the Gly-Ala repeat on the presentation of the EBNA-1–derived epitope via class I MHC molecules.
Materials and Methods
Construction of Recombinant Vaccinia for Expression of a 78 Amino Acid EBNA-1 Fragment.
The EBNA-1 COOH-terminal fragment, residues 505–583, was PCR amplified from the plasmid pJ130, described in reference 7. The primers were 5′CCCAAGCTTACCATGGCCGCTTGTTCGTATATGGACCTA3′ and 5′GAAGATCTCGAGTCAAAACAAGGTCCTTAATCGCATCCTTTA 3′ containing an NcoI site (5′) and a BglII site (3′ end, underlined); the ATG within the NcoI sequence supplied the start Methionine. The product was cloned into the vaccinia virus vector pSC113OR.2 to generate pSC11/C-EB1 and a recombinant vaccinia virus VVΔEB1 was generated as previously described (10).
Cell Lines.
The rabbit α-EBNA-1 polyclonal serum (11) was raised against a bacterially expressed EBNA-1 fragment (residues 7–37 and 420–617) and affinity purified against the recombinant protein. The 2C CTL line was a gift from Dr. Herman Eisen. The P-107 serum (12) is a human polyclonal serum from an EBV carrier, affinity purified against a Gly-Ala peptide.
The P815/EBNA-1 cells and the P815/ΔEBNA-1 cells were generated by transfecting P815 HTR (H2d) cells with a pJ130-EB1 and pJHISTdelEB1, respectively (7). The ΔEBNA-1 construct lacks the residues 93–325 (see Fig. 2 a; reference 9).
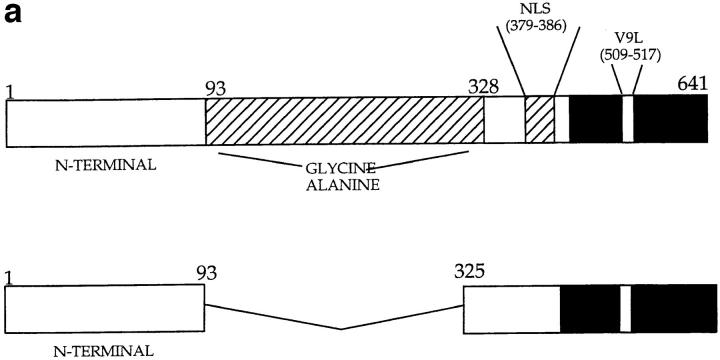
Figure 2.
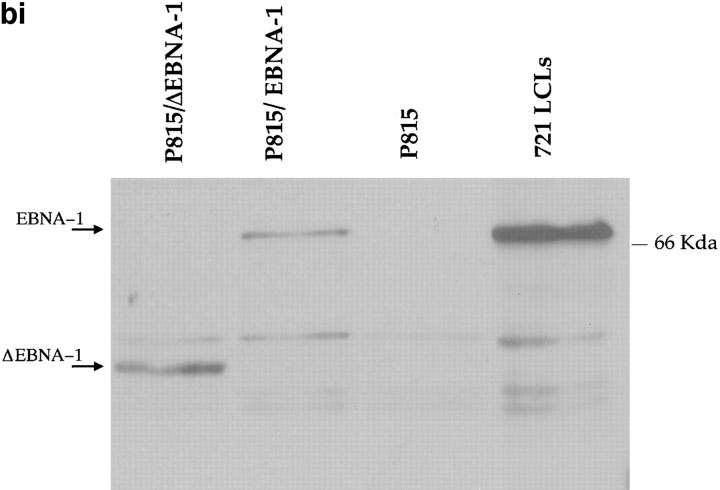
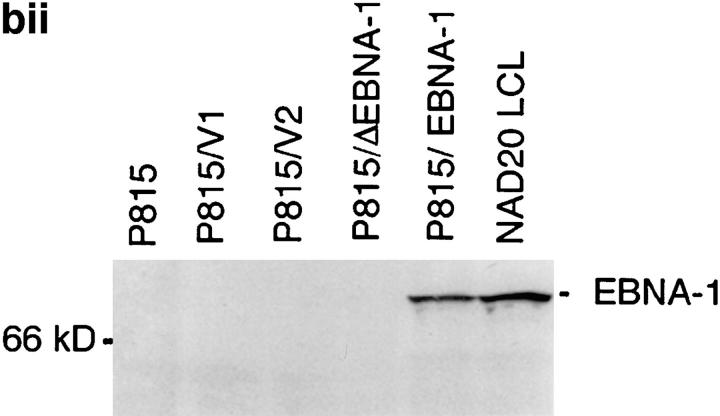
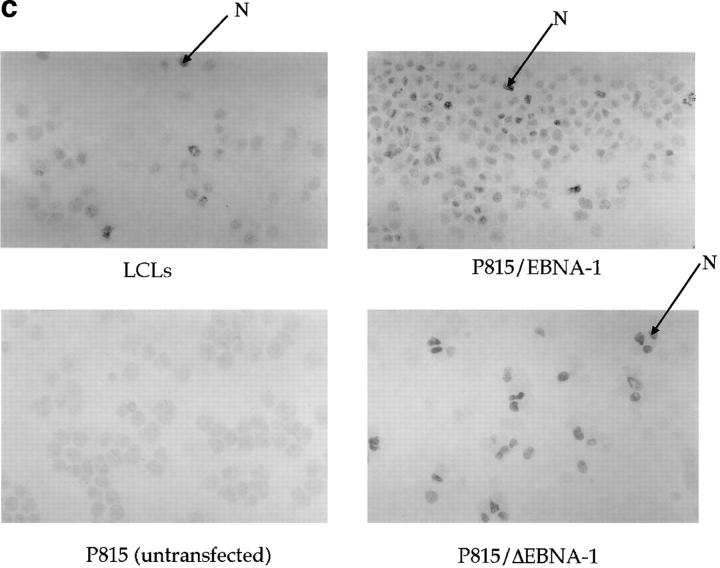
(a) Constructs used to transfect P815 cells. (Top) Full-length EBNA-1 of the B-95-8 sequence (641 amino acids total). Hatched region (93–328) shows the Gly-Ala–rich region. NLS, nuclear localisation sequence as described in reference 16. The position of the V9L epitope is 509–517 (indicated as V9L). (Bottom) ΔEBNA-1: residues 93–325 have been deleted, but the rest of EBNA-1 is unaffected. (b, i) Western Blot analysis of transfected cells using the Rbt-EBNA-1 antibody (11). Cells are as labeled. A 72-kD band is detected in P815/EBNA-1 cells; based on reactivity with the antiserum and the size of EBNA-1 detected in the 721 LCL line (lane 4), this is EBNA-1. A 43-kD band is detected in P815 cells transfected with EBNA-1 lacking the Gly-Ala repeat. (b, ii) Western Blot using the P-107 serum (12) that reacts against the Gly-Ala repeat only. Lanes 2 and 3 indicate P815 cells transfected with vectors alone (i.e., no EBNA-1). Lane 6, NAD20 cells are EBV-transformed LCLs. (c) Cell staining with Rbt-EBNA-1 serum (11) reactive against EBNA-1. Nuclear staining is indicated with N and arrow. Cells are as indicated in legend and text.
Cells were transfected by electroporation (960 μF, 230 V). P815-EBNA-1 cells were grown in 1.5 μg/ml mycophenolic acid, 160 μg/ml xanthine, and 10 μg/ml hypoxanthine, whereas P815-ΔEBNA-1 cells were selected in histidine-free DMEM/ 10% FCS with 0.5 mM histidinol.
Establishment and Maintenance of CTL Cultures.
Splenocytes were recovered 14 d after priming BALB/c mice with 5 × 106 PFU of vaccinia virus, VVΔEB1 intravenously. Cells from a nonimmunized BALB/c spleen were coincubated with the V9L peptide (VYGGSKTSL, 1 μM for 1 h), washed extensively, and irradiated. These “stimulator” cells were cocultured with the “effector” spleen cells harvested from the immunized mouse, with 106 stimulators and 2.0 × 107 effectors in the absence of IL-2, in 10 ml. The cells were restimulated weekly by splitting the culture threefold and adding 2 × 106 fresh peptide-pulsed stimulators and IL-2 (10 U/ml; Cetus Corp., Emoryville, CA).
Chromium Release Assays for Cytotoxicity.
51Cr release cytotoxicity assays were performed as described (10). For peptide-pulsed cells, cells were incubated with peptide (10 μM unless otherwise indicated) for 30 min in RPMI 10% FCS, and then washed thrice. Target cells prepared thus were added at 10,000 cells in 100 μl/well to 96-well plates. Calculations for specific lysis were as previously described (10).
Expression of EBNA-1 and ΔEBNA-1(GA) by Western Blotting and Cell Staining.
Whole cell lysates were made by sonicating 5 × 107 cells suspended in 0.5 ml of PBS, at 4°C, for 2 min. Laemmli sample buffer was added to the lysate, boiled for 2 min, and then loaded on an SDS gel. Lysates were transferred onto nitrocellulose membranes (HiBOND C; Amersham Corp., Arlington Heights, IL) for a total of 1 AmpHour and blocked overnight with 10% milk in PBS. Blots were incubated with the sera indicated, at a dilution of 1:1,000 in 5% nonfat milk for 1 h, washed extensively with PBS/0.2% Tween, and visualized with a horseradish peroxidase–conjugated secondary antibodies.
Cytospin preparations of acetone-fixed cells were incubated with primary antibody for 1 h, washed in PBS, incubated with biotinylated secondary antibodies (Vector Labs., Burlingame, CA) for 1 h, and incubated then with avidin–biotinylated peroxide complex for 40 min followed by reaction with diaminobenzedine/hydrogen peroxide. Cells were stained with 2% methyl green.
Results
The CTL Line V9L Recognizes an Epitope in the COOH-terminal Region of EBNA-1.
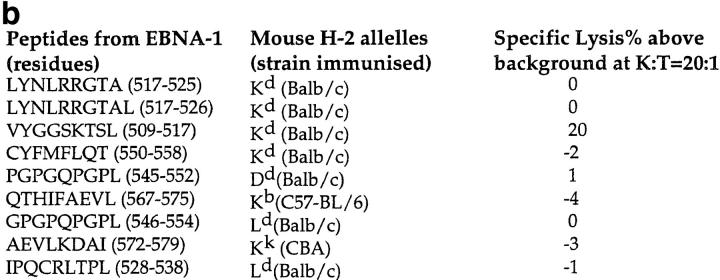
CTL against potential epitopes within EBNA-1 were raised using the protocol described in Fig. 1 a. “Candidate” epitopes, octa-, nona-, and decamers that matched the previously characterized consensus motifs for peptides eluted from common mouse MHC alleles (for Kb, Kd, Kk, Ld, and Dd) were identified (13). For instance, residues 509–517 (VYGGSKTSL), which contain a Tyrosine in position 2 and a Leucine in position 9, match the consensus motif for Kd-binding peptides (see Fig. 1 b).
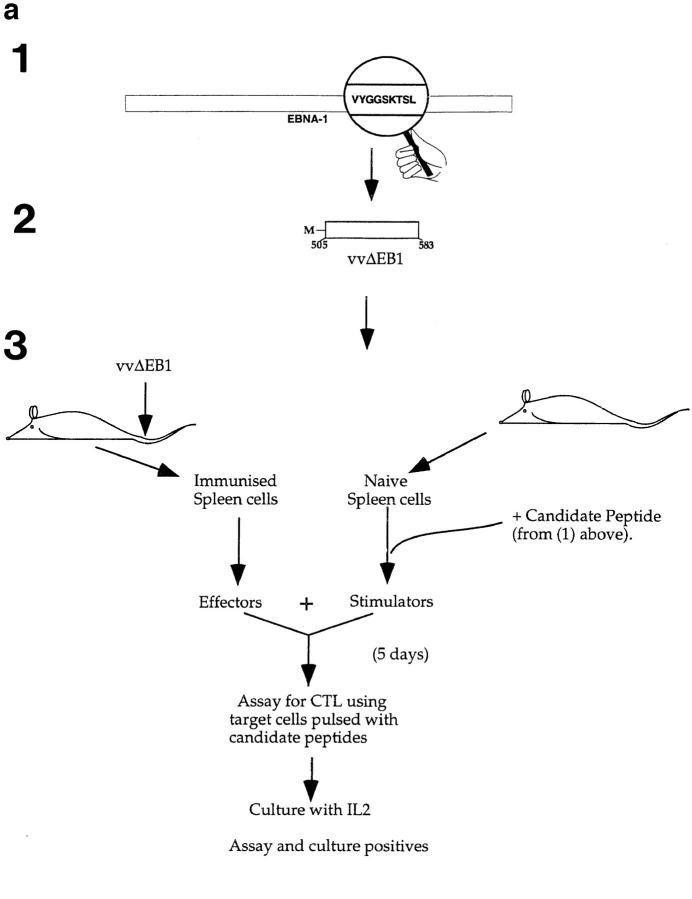
Figure 1.
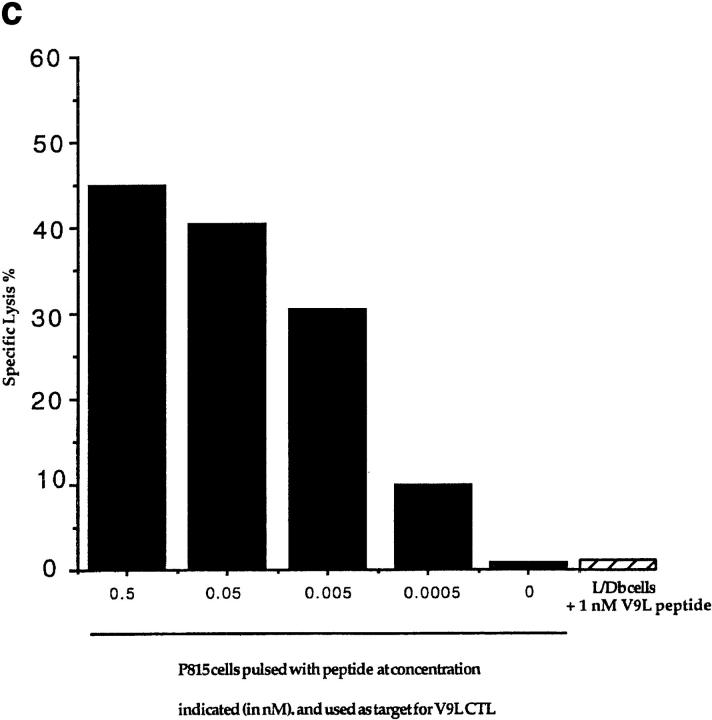
(a) Scheme for rescue of peptide-specific CTLs by coculturing immunized splenocytes with naive splenocytes pulsed with candidate peptides (see text). (1) Identification of candidate epitopes. (2) Construction of vaccinia virus expressing the EBNA-1 (505–583) fragment (called VVΔEB-1) containing the candidate epitopes. (3) Immunization of mice and recovery of CTLs. (b) Mouse strains initially immunized with VVΔEB-1 and the peptides used to restimulate them in vitro. (Column 1) Candidate epitopes derived from the COOH-terminal of EBNA-1 using consensus motifs described in reference 13. (Column 2) Class I molecule for which the candidate peptide in column 1 carries the consensus motif (and strains immunized with VVΔEB-1). For example, VYGGSKTSL contains a Kd motif (i.e., Y at position 2, and L at position 9, underlined). Splenocytes from BALB/c mice (H2-d) immunized with VVΔEB-1 were restimulated with splenocytes pulsed with the V9L peptide, as described in a. Cocultures restimulated on peptides (as in column 1) were used as effectors in a Cr–release assay using peptide-pulsed cells as targets (see below for the target cells). (Column 3) Specific lysis above background in 51Cr release assays 5 d after in vitro restimulation with peptide-pulsed cells, was obtained for each of the effector cultures. Target cells used: P815 for all H2-d, L929 for H2-k, and EL-4 for H2-b. (c) V9L CTLs are peptide specific and MHC restricted. P815 cells were pulsed with the peptide dose indicated (in nM) and used as targets in Cr–release assay. K/T ratio was 20:1. Unpulsed cells (0 nM) are not lysed. Hatched bar shows L/Db cells that were pulsed with V9L peptide; these cells do not express Kd and were not lysed. The x-axis represents P815 cells pulsed with peptide at concentration indicated (in nM) and used as target for V9L CTLs.
Although candidate epitopes were interspersed throughout EBNA-1, the largest number of such epitopes for common mouse alleles was found between residues 505 and 583. The length of the immunogen was minimized to this 78–amino acid region, to avoid any potential cis-acting inhibitory sequences. All the candidate peptides were synthesized chemically (see Fig. 1 b for the candidate peptides).
The EBNA-1 fragment was PCR amplified, with a start Methionine and a stop codon added (Fig. 1 a), and a vaccinia virus (VVΔEB1) encoding this fragment was created (see Materials and Methods). By DNA sequencing and PCR from the virus, we confirmed that the recombinant virus generated carried the appropriate fragment. Three strains of mice were immunized with a vaccinia virus encoding the EBNA-1 fragment, corresponding to residues 505–583 of EBNA-1 (of the prototypic B-95-8 sequence) (see Fig. 1 b for mouse genotypes and strains). To recover CTLs specific for EBNA-1–derived epitopes, splenocytes derived from the immunized mice (effectors) were cocultured with stimulators, autologous splenocytes pulsed with candidate peptide epitopes (as shown in Fig. 1 a) to restimulate effector cells that were specific for the candidate peptide.
To determine whether CTLs had been generated upon secondary restimulation, a 51Cr–release assay was used after 5 d of coculture of effectors with stimulators. 51Cr-labeled target cells expressing the relevant class I MHC molecule (Fig. 1 b) were pulsed with 100 nM of the candidate peptide and the effector cultures tested for the ability to lyse these peptide-pulsed targets. The level of lysis was compared against control targets, in which no peptide was used.
For eight of the nine candidates, no cytotoxicity above background on peptide-pulsed targets was observed (Fig. 1 b, column 3). The ninth candidate, the peptide VYGGSKTSL (V9L), containing a putative Kd-binding motif, showed lysis above background (Fig. 1 b, column 3).
All lines of CTLs were then grown for another 7 d on peptide-pulsed stimulators with IL-2 (100 U/ml) and tested again for cytotoxicity. Again, only the V9L line showed lysis above background. This CTL line grew continuously in culture with weekly restimulation with peptide-pulsed spleen cells supplemented with IL-2. In total, three BALB/c mice immunized at separate times with VVΔEB1 yielded a V9L-specific CTL response.
The V9L CTL line was unable to lyse Kd-expressing cells unless the V9L peptide was added exogeneously (i.e., it was peptide specific) and was unable to lyse cells that lacked the Kd MHC molecule even in the presence of peptide (i.e., MHC class I restricted) (Fig. 1 c). The dose of peptide required for the recognition of target cells by the V9L CTL line was determined. P815 cells were pulsed with varying doses of peptide (as indicated in Fig. 1 c), washed extensively to remove peptide, and used as targets in a 51Cr–release assay. The V9L CTL line was capable of recognizing cells pulsed with low doses of peptides (up to 0.005 nM).
Expression of EBNA-1 in Murine Cells.
To determine whether the V9L epitope was presented from murine cells expressing full-length EBNA-1, or EBNA-1 lacking the Gly-Ala region, Kd-expressing murine cells expressing full-length and Gly-Ala–deleted EBNA-1 were generated.
P815 (H2-d, mouse mammary mastocytoma cells) were transfected with an expression vector containing full-length EBNA-1, previously described in reference 7. P815 cells were also transfected with a vector containing an EBNA-1 construct, with the Glycine-Alanine rich region deleted, called ΔEBNA-1, in which residues 93–325 have been deleted (Fig. 2 a and reference 9).
The expression of EBNA-1 and the Gly-Ala–deleted EBNA-1 (ΔEBNA-1) in transfected murine cells were determined by Western blots and immunocytochemistry (Fig. 2, b and c). A rabbit (Rbt) polyclonal serum (11), Rbt-α-EBNA-1, raised against a fragment of EBNA-1 and an affinity-purified human serum, P-107, with reactivity against the Gly-Ala repeat only (12) were used in two separate Western Blots (Fig. 2, b i and ii). With the Rbt-α-EBNA-1 serum, a 72-kD band was detected in P815/ EBNA-1 cells, which comigrated with a 72-kD band detected in the EBV-transformed human 721 LCL line, used as a positive control. The 72-kD band was not present in P815 untransfected cells. In P815 cells transfected with EBNA-1 lacking the Gly-Ala repeat (P815/ΔEBNA-1), the size of the mutant protein is expected to be between 40 and 43 kD. Indeed, a band of appropriate size (43 kD) was detected (Fig. 2 b, i). With the P-107 serum, which had been affinity purified to retain reactivity against the Gly-Ala repeat alone, a 72-kD band was detected in P815/ EBNA-1 cells, but no 43-kD band was detected in cells transfected with the Gly-Ala deleted construct.
The intracellular location of EBNA-1 in transfected cells was determined by immunohistochemistry, using the Rbt-EBNA-1 antibody. Cell staining demonstrated that both EBNA-1 and the Gly-Ala–deleted EBNA-1 was located primarily in the nucleus (Fig. 2 c).
The V9L Epitope Is Not Presented to CTLs by Cells Expressing Full-length EBNA-1, whereas Cells Expressing ΔEBNA-1 Are Lysed.
51Cr–release assays were performed to determine whether the V9L epitope was being presented by murine cells expressing either the full-length EBNA-1, or EBNA-1 lacking the Gly-Ala–rich region. As targets, the transfected P815 cell lines expressing full-length EBNA-1 or ΔEBNA-1 were used. The presence of the MHC restriction element of the V9L CTLs, Kd, in the target cells, had been previously confirmed by using these cells as targets for other Kd-restricted CTLs (not shown).
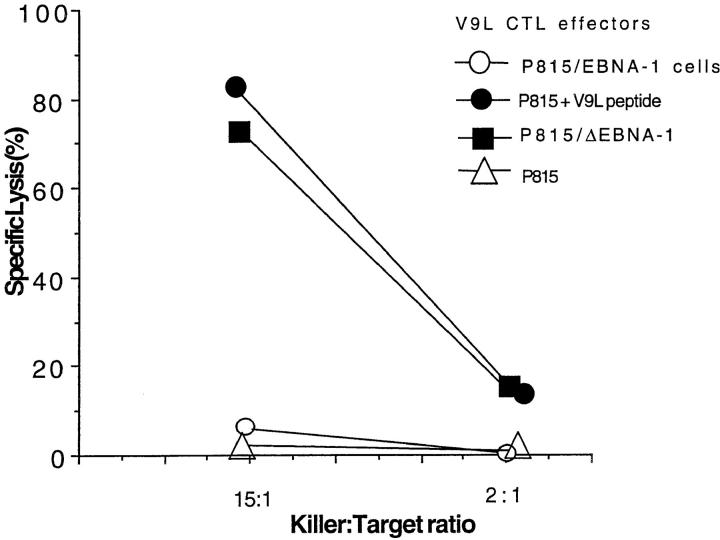
The results of a representative 51Cr–release assay using the murine cells appears in Fig. 3. Untransfected P815 cells, used as negative control, were not lysed by V9L CTLs. P815 cells pulsed with the V9L peptide were efficiently lysed. In three separate instances, the P815/EBNA-1 cells were not lysed above background. In contrast, P815/ ΔEBNA-1 cells, which express EBNA-1 lacking the Gly-Ala–rich region, were lysed efficiently.
Figure 3.
V9L CTLs used as effectors in 51Cr–release assays. Targets: P815/EBNA-1 (open circles), P815/ΔEBNA-1 (filled squares), P815 with no peptide (open triangles), and a P815 + V9L peptide pulsed at 10 μm (filled circles).
To determine whether EBNA-1 expression altered the ability of cells to present endogenous antigens, P815/ EBNA-1 cells were used as targets in 51Cr–release assays with the 2C CTLs line as effectors. The 2C line recognizes an endogenous mitochondrial protein presented in association with Ld molecules (14). In a 51Cr–release assay, both P815 and P815/EBNA-1 cells showed no impairment in lysis by 2C CTLs, whereas L/Db cells, which do not express Ld molecules, were not lysed (not shown). Thus, cells expressing EBNA-1 or the Gly-Ala–deleted EBNA-1 appear to be no different in the processing and presentation of epitopes derived from other proteins, and the inhibition of antigen presentation of the V9L epitope occurs strictly in cis.
Discussion
Polyclonal CTLs or CTL clones derived from EBV carriers by restimulation in vitro with autologous LCLs, are capable of lysing autologous cells expressing most EBV latency-associated antigens (4–6), but not cells expressing EBNA-1. Since several peptides from EBNA-1 are capable of binding human HLA alleles in vitro (15), the nonrecognition of EBNA-1–expressing cells was unlikely to be due to the absence of antigenic epitopes within this protein. Using EBNA-1–EBNA-4 chimeric molecules, Levitskaya et al. demonstrated that the Gly-Ala–rich region within EBNA-1 inhibited the generation of class I–restricted epitopes (9).
We have generated a novel, murine CTL line against an epitope within EBNA-1 itself, to study the processing and presentation of this protein without placing exogenous epitopes within EBNA-1 or creating chimeric molecules. This CTL line, called V9L, was recovered by immunizing mice with a vaccinia virus encoding a short EBNA-1 fragment, followed by restimulation in vitro using candidate epitopes, a method described in Fig. 1 a. The shortest COOH-terminal stretch of EBNA-1 where the largest number of putative epitopes could be identified was expressed through vaccinia and this virus was used to immunize mice.
Cells expressing full-length EBNA-1 were not lysed by the V9L CTL (Fig. 3), whereas cells expressing the Gly-Ala–deleted EBNA-1 (ΔEBNA-1) were effectively lysed. The presentation of EBNA-1–derived epitopes has recently been examined using class I–restricted human CTL clones that recognize peptides from EBNA-1. Like the murine CTL described here, these CTLs were unable to lyse human cells expressing the full-length protein.
To investigate the means by which EBNA-1 is able to resist antigen processing and presentation, the intracellular locations of EBNA-1 and Gly-Ala–deleted EBNA-1 (ΔEBNA-1) was examined by staining transfected P815 cells with the Rbt-EBNA-1 antiserum. P815/EBNA-1 cells and P815/ ΔEBNA-1 cells were both stained prominently in the nucleus (Fig. 2 c). Previously, the nuclear localization sequence in EBNA-1 had been mapped to residues 379–387, outside the region deleted in ΔEBNA-1 (17). Since EBNA-1 and EBNA-1 are both found in the nucleus, it is unlikely that the deletion of the Gly-Ala repeat exposes the protein to more efficient cytosolic proteolysis (18) and thus restores the presentation of the V9L epitope.
Interestingly, the Gly-Ala–rich region is not required for the replication function of EBNA-1, since vectors carrying the EBV-OriP can be maintained episomally in cells expressing the Gly-Ala–deleted EBNA-1. We speculate that this region may have evolved due to the selective advantage it confers on latently infected cells that are known to express EBNA-1 (but not EBNAs 2–6) by protecting them from CTL-mediated rejection.
Since only EBNA-1 is required for the maintenance of the EBV episome in latently infected cells (19), the ability of the Gly-Ala repeat to inhibit the presentation of epitopes derived from this protein may be related to the ability of EBV to maintain a persistent infection of B cells, in the face of a host immune response (2, 9). A subpopulation of latently infected B cells expressing only EBNA-1 (or EBNA-1 in conjunction with LMP-2) have been found in vivo (20). If epitopes derived from EBNA-1 are not presented efficiently, latently infected B cells, in which EBNA-1 is expressed, may be partially protected from elimination by host CTLs. This may allow the reservoir of latently infected cells to seed other cellular compartments, such as epithelial cells, where EBV can replicate and be transmitted. The fact that epitopes from EBNA-1 are not presented effectively to CTLs may also explain why EBV-positive Burkitt's lymphoma cells, which appear to express only EBNA-1 in vivo (21), can survive in immunocompetent hosts, although several other features in these cells may contribute to their lack of immunogenicity for CTLs (20, 22).
Acknowledgments
The authors wish to thank Dr. M.G. Masucci, Dr. B. Sugden, Dr. M.G. Kurilla, and Dr. A.B. Rickinson for valuable reagents and discussion.
Footnotes
S. Mukherjee was supported by the Rhodes Trust. P. Trivedi was supported by a grant from the Istituto Superiore di Sanita, Italy.
References
- 1.Kieff, E. 1996. Epstein Barr virus and its replication. In Virology. B.N. Fields, D.M. Knipe, P. Howley, R. Chanock, J. Melnick, T. Monath, B. Rozman, S.E. Straus, editors. Lippincott-Raven Press, New York. 2243–2396.
- 2.Klein G. Viral latency and transformation: the strategy of Epstein Barr virus. Cell. 1989;58:5–8. doi: 10.1016/0092-8674(89)90394-2. [DOI] [PubMed] [Google Scholar]
- 3.Townsend A, Bodmer H. Antigen recognition by class I–restricted T lymphocytes. Annu Rev Immunol. 1989;7:601–624. doi: 10.1146/annurev.iy.07.040189.003125. [DOI] [PubMed] [Google Scholar]
- 4.Khanna R, Burrows SR, Kurilla MG, Jacob CA, Misko IS, Sculley TB, Kieff E, Moss DJ. Localisation of Epstein-Barr virus–specific cytotoxic T cell epitopes using recombinant vaccinia: implications for vaccine development. J Exp Med. 1992;176:169–176. doi: 10.1084/jem.176.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murray RJ, Kurilla MG, Brooks JM, Thomas WA, Rowe M, Kieff E, Rickinson AB. Identification of target antigens for the human cytotoxic T cell response to Epstein-Barr virus (EBV): implications for immune control of EBV-positive malignanacies. J Exp Med. 1992;176:157–168. doi: 10.1084/jem.176.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Masucci M, Ernberg I. Epstein Barr virus: adaptation to a life within the immune system. Trends Microbiol. 1994;2:125–130. doi: 10.1016/0966-842x(94)90599-1. [DOI] [PubMed] [Google Scholar]
- 7.Trivedi P, Masucci MG, Winberg G, Klein G. The Epstein-Barr-virus–encoded membrane protein LMP but not the nuclear antigen EBNA-1 induces rejection of transfected murine mammary carcinoma cells. Int J Cancer. 1991;48:794–800. doi: 10.1002/ijc.2910480527. [DOI] [PubMed] [Google Scholar]
- 8.Khanna R, Burrows SR, Steigerwald-Mullen PM, Thomson SA, Kurilla MG, Moss KDJ. Isolation of cytotoxic T lymphocytes from healthy seropositive individuals specific for peptide epitopes from Epstein-Barr virus nuclear antigen 1: implications for viral persistence and tumor surveillance. Virology. 1995;214:633–637. doi: 10.1006/viro.1995.0076. [DOI] [PubMed] [Google Scholar]
- 9.Levitskaya J, Coram M, Levitsky V, Imreh S, Steigerwald-Mullen PM, Klein G, Kurilla MG, Masucci MG. Inhibition of antigen presentation by the internal repeat region of the Epstein Barr virus nuclear antigen-1. Nature. 1995;375:685–688. doi: 10.1038/375685a0. [DOI] [PubMed] [Google Scholar]
- 10.Townsend A, Bastin J, Gould K, Brownlee G, Andrew M, Coupar B, Boyle D, Chan S, Smith G. Defective presentation to class I–restricted cytotoxic T lymphocytes in vaccinia-infected cells is overcome by enhanced degradation of antigen. J Exp Med. 1988;168:1211–1224. doi: 10.1084/jem.168.4.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sternas L, Middleton T, Sugden B. The average number of molecules of Epstein-Barr nuclear antigen 1 per cell does not correlate with the average number of Epstein-Barr virus (EBV) DNA molecules per cell among different clones of EBV-immortalized cells. J Virol. 1990;64:2407–2410. doi: 10.1128/jvi.64.5.2407-2410.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dillner J, Sternas L, Kallin B, Alexander H, Ehlin-Henriksson B, Jornvall H, Klein G, Lerner R. Antibodies against a synthetic peptide identify the Epstein-Barr virus–determined nuclear antigen. Proc Natl Acad Sci USA. 1984;81:4652–4656. doi: 10.1073/pnas.81.15.4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rammensee HKF, Rotzschke O. Peptides naturally presented by MHC class I molecules. Annu Rev Immunol. 1993;11:213–244. doi: 10.1146/annurev.iy.11.040193.001241. [DOI] [PubMed] [Google Scholar]
- 14.Sha WC, Nelson CA, Newberry RD, Kranz DM, Russell JH, Loh DY. Selective expression of an antigen receptor on CD8-bearing T lymphocytes in transgenic mice. Nature. 1988;335:271–274. doi: 10.1038/335271a0. [DOI] [PubMed] [Google Scholar]
- 15.Stuber G, Dillner J, Modrow S, Wolf H, Szekely L, Klein G, Klein E. HLA-A0201 and HLA B7 binding petides in EBV-encoded EBNA-1, EBNA 2 and BZLF-1 detected in the MHC class I stabilization assay. Int Immunol. 1995;7:653–663. doi: 10.1093/intimm/7.4.653. [DOI] [PubMed] [Google Scholar]
- 16.Blake N, Lee S, Redchenko I, Thomas W, Steven N, Leese A, Steigerwald-Mullen P, Kurilla MG, Frappier L, Rickinson A. Human CD8+T cell responses to EBV EBNA1: HLA class I presentation of the (GLY-ALA) containing protein requires exogenous processing. Immunity. 1997;7:791–802. doi: 10.1016/s1074-7613(00)80397-0. [DOI] [PubMed] [Google Scholar]
- 17.Ambinder RM, Mullen M, Chang Y, Hayward G, Hayward SD. Functional domains of Epstein Barr virus nuclear antigen EBNA-1. J Virol. 1991;65:1466–1478. doi: 10.1128/jvi.65.3.1466-1478.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cerundolo V, Benham A, Braud V, Mukherjee S, Gould K, Macino B, Neefjes J, Townsend A. The proteasome-specific inhibitor lactacystin blocks presentation of cytotoxic T lymphocyte epitopes in human and murine cells. Eur J Immunol. 1997;27:336–341. doi: 10.1002/eji.1830270148. [DOI] [PubMed] [Google Scholar]
- 19.Yates JL, Warren N, Sugden B. Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. Nature. 1985;313:812–815. doi: 10.1038/313812a0. [DOI] [PubMed] [Google Scholar]
- 20.Chen F, Zou JZ, di Renzo L, Winberg G, Hu LF, Klein E, Klein G, Ernberg I. A subpopulation of normal B cells latently infected with Epstein Barr virus resembles Burkitt lymphoma cells in expressing EBNA-1 but not EBNA-2 or LMP-1. J Virol. 1995;69:3752–3758. doi: 10.1128/jvi.69.6.3752-3758.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rooney CM, Rowe M, Wallace LE, Rickinson AB. Epstein-Barr virus positive Burkitt's lymphomas not recognised by virus specific T-cell surveillance. Nature. 1985;317:629–631. doi: 10.1038/317629a0. [DOI] [PubMed] [Google Scholar]
- 22.Frisan T, Zhang QJ, Levitskaya J, Coram M, Kurilla MG, Masucci MG. Defective presentation of MHC class I–restricted cytotoxic T cell epitopes in Burkitt's lymphoma cell. Int J Cancer. 1996;68:251–258. doi: 10.1002/(SICI)1097-0215(19961009)68:2<251::AID-IJC19>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]