Abstract
T cells specific for nucleosomal autoepitopes are selectively expanded in lupus mice and these Th cells drive autoimmune B cells to produce pathogenic antinuclear antibodies. We transfected the TCR-α and -β chain genes of a representative, pathogenic autoantibody-inducing Th clone specific for the nucleosomal core histone peptide H471–94 into TCR-negative recipient cells. Although the autoimmune TCRs were originally derived from SNF1 (I-Ad/q) mice, the transfectants could recognize the nucleosomal autoepitope presented by APC-bearing I-A molecules of all haplotypes tested, as well as human DR molecules. Competition assays indicated that the autoepitopes bound to the MHC class II groove. Most remarkably, MHC-unrestricted recognition of the nucleosomal peptide epitope was conferred by the lupus TCR-α chain even when it paired with a TCR-β chain of irrelevant specificity. Several other disease-relevant Th clones and splenic T cells of lupus mice had similar properties. The TCR-α chains of these murine lupus Th clones shared related motifs and charged residues in their CDRs, and similar motifs were apparent even in TCR-α chains of human lupus Th clones. The lupus TCR-α chains probably contact the nucleosomal peptide complexed with MHC with relatively high affinity/avidity to sustain TCR signaling, because CD4 coreceptor was not required for promiscuous recognition. Indeed, pathogenic autoantibody-inducing, CD4-negative, TCR-αβ+ Th cells are expanded in systemic lupus erythematosus. These results have implications regarding thymic selection and peripheral expansion of nucleosome-specific T cells in lupus. They also suggest that universally tolerogenic epitopes could be designed for therapy of lupus patients with diverse HLA alleles. We propose to designate nucleosomes and other antigens bearing universal epitopes “Pantigens” (for promiscuous antigens).
In murine, as well as human systemic lupus erythematosus (SLE), the production of nephritogenic antinuclear autoantibodies by autoimmune B cells is driven by cognate interactions with specific autoimmune Th cells (1–7). The pathogenic autoantibody-inducing Th cells of lupus have been cloned from (SWR × NZB)F1 (SNF1)1 mice and also from patients with lupus nephritis. Representative Th clones from the SNF1 mice can precipitate glomerulonephritis upon transfer into preautoimmune animals, which establishes their relevance to disease (2). In SNF1 mice, the majority of these pathogenic Th clones are specific for nucleosomal peptides, which are processed and presented by the classical MHC II pathway (3, 6). Nucleosomes are routinely released from apoptotic cells and this event is not unique to lupus (8–10). However, the spontaneous expansion of nucleosome-specific T cells is a lupus-specific event that occurs very early in life (3, 6). These Th cells are essential for sustaining the pathogenic autoantibody-producing B cells of lupus (4). Without such T cell help, the potentially pathogenic B cells that arise even in normal subjects as an accompaniment of the immune response to common pathogens are destined to undergo apoptosis (11, 12). The presence of anionic residues in the junctional regions (CDR3) of the TCRs of these lupus Th cells suggested that they could be specific for peptides with cationic residues (2, 13, 14). Indeed, the Th clones of lupus were found to be specific for nucleosomal peptides containing multiple charged residues (3, 6).
To further investigate the structural basis for this autoimmune recognition event, we have cloned and expressed the TCR-α and -β chain genes of the prototypic pathogenic autoantibody-inducing Th clone, 3A, which accelerates lupus nephritis in SNF1 mice. The TCR of this representative pathogenic Th clone is specific for a peptide spanning residues 71–94 of the nucleosomal core histone H4. H471–94 is in contact with DNA in the native nucleosome particle, thus allowing this epitope to be protected during autoantigen processing (6). In this study, we report that recognition of nucleosomal autoepitopes is MHC-dependent, but unrestricted. Remarkably, the TCR-α chain of the pathogenic Th clone is critical for this promiscuous recognition and nucleosomal peptide specificity, and this recognition response is CD4 coreceptor–independent.
Materials and Methods
Mice.
BALB/c (H-2d), NZB (H-2d), NZW (H-2z), SWR (H-2q), C3H (H-2k), (BALB/c × SWR)F1 (H-2d/q), B6.C (H-2bm12), B10.M (H-2f), B10.S (H-2s), B10RIII (H-2r), and B10.PL (H-2u) mice were from The Jackson Laboratory (Bar Harbor, ME). SNF1 mice (H-2d/q) were bred at Northwestern University animal facility.
Antibodies.
Hybridomas producing mAbs against Thy-1.2 (TIB99), CD3 (145-2C11), CD4 (GK1.5), I-Ab/d/q (TIB120), and I-Ad (HB3/MKD6) were acquired from the American Type Culture Collection (ATCC, Rockville, MD). Anti–I-Aq (MKQ7) was from Phillipa Marrack (National Jewish Center for Immunology, Denver, CO). Anti–I-Ed (14-4-4S) was from Laurie Glimcher (Harvard University, MA) (3). Anti–HLA-DR (L243), anti-DP (B7/21), and anti-DQ (Sk10) were from Becton Dickinson (San Jose, CA). All antibodies were 10-times-concentrated by standard method of ammonium sulphate precipitation. Anti–IL-2, biotinylated anti–IL-2, biotinylated anti-Vβ4, biotinylated anti-Vβ8, FITC-conjugated anti-TCR-α/β, and purified anti-CD4 mAbs (RM4.5) were purchased from PharMingen (San Diego, CA).
Antigens.
Nucleosomes were prepared as previously described (3). Overlapping core histone peptides from nucleosomes corresponding to the regions found to bear pathogenic Th cell epitopes were synthesized by FMOC chemistry (Chiron Technologies, San Diego, CA; reference 6).
Cells.
The cloning of pathogenic autoantibody-inducing Th cells from SNF1 mice with lupus nephritis has been previously described (1, 2). Th clones were rested for 10 d before the assays. Fresh splenic CD4+ T cells were isolated from 4–5 mo-old SNF1 female mice by nylon wool column followed by the lysis of CD8+ cells and contaminating B cells with mAb and C′ (3, 6). Short-term T cell lines (polyclonal) were prepared by culturing the splenic CD4+ T cells with nucleosome-pulsed APCs plus rIL-2 (25 U/ml) for 5 d and then being rested for 7–10 d before the assays. The 4G4 clone (TCR-negative T cell hybridoma) was provided by Charles A. Janeway, Jr. (Yale University, New Haven, CT; reference 15). The EL-4 T cell lymphoma clone, EL-4.IL-2 (TIB39), was from ATCC. Splenic APCs were isolated from 4-wk old mice of different MHC backgrounds by depleting erythrocytes in Red Cell Lysis Buffer (Sigma Chemical Co., St. Louis, MO), followed by anti-Thy1.2 (TIB99) and rabbit complement treatment (3, 6). After irradiating (3,000 rad), the cells were pulsed with antigens by plating into 96-well plates (5 × 105 cells/ well) in 100 μl of serum-free HL-1 medium (Ventrex Laboratories, Portland, ME) at 37°C for 4–16 h, and then were washed for presentation. Two clones of EBV transformed B cell line from human subjects previously described (7) were also used as APCs.
Gene Constructs.
The TCR-α and -β shuttle vectors with or without the hen egg lysozyme (HEL)-specific 3A9α and 3A9β TCR constructs were provided by Mark M. Davis (Stanford University, CA; references 16, 17). The CD4 construct was obtained from Charles A. Janeway, Jr. (15). The functionally rearranged AV13S3J41 and BV4S1J2S6 region genes of the α and β chain of the TCR of nucleosome-specific lupus Th clone 3A were amplified by PCR from genomic DNA of 3A with oligonucleotide primers containing splice acceptor and donor sites. For the TCR-α gene, the forward primer, Vα13-5′-XhoI (AAA CTC GAG GGA AAG GGA TGA CAT CAC AGC), and the reverse primer, Jα41-3′-NotI (AAA ATT GCG GCC GCG ATA GCT CCT TCT GCA GGT TCA G), were designed to introduce a XhoI cloning site 57 bp 5′ of the ATG and a NotI site 74 bp 3′ of the splice donor sequence, respectively. This amplified fragment was inserted into the TCR-α shuttle vector at the XhoI and NotI sites. The TCR-α shuttle vector contains the Ecogpt gene which was used for selection. For the TCR-β gene, the forward primer, Vβ4-5′-ClaI (CGC TTG TAT CGA TTT CTA CAA TTA TCT TGA GGA T), and the reverse primer, Jβ2-3′-NotI (AAA ATT GCG GCC GCC GAG AGA TGT GAA TCT TAC CTA AA), were designed to introduce a ClaI cloning site 44 bp 5′ of the splice acceptor site and a NotI site 17 bp 3′ of the splice donor site, respectively. The genomic PCR-amplified fragment was then inserted into the β shuttle vector at the ClaI and NotI sites. Both constructs were sequenced to ensure fidelity during amplification, then excised by cleavage at the SalI site in the plasmid vector. The CD4 construct was linearized by cleavage at the ScaI site.
Transfection.
TCR-negative 4G4 cells were transfected with DNA constructs in four combinations: 3Aα/3Aβ; 3Aα/3A9β; 3A9α/3Aβ; and 3A9α/3A9β (10 μg/construct), along with a CD4 construct (5 μg) by electroporation using a GenePulser as suggested by the manufacturer (Bio-Rad, Hercules, CA). EL-4 cells were transfected with DNA constructs in six combinations: 3Aα; 3Aβ; 3Aα/3Aβ; 3A9α; 3A9β; and 3A9α/3A9β (2.5–5 μg/ construct) by cationic lipid–mediated DNA transfection using Lipofectin Reagent following the manufacter's protocol (GIBCO BRL, Gaithersburg, MD). After 48 h, HMX selection was started (Hypoxathine, 15 μg/ml; Mycophenolic Acid, 3 μg/ml; Xathine, 200 μg/ml). Resistant cells were then subcloned and screened for Vβ4 (for 3A) and Vβ8 (for 3A9) expression by flow cytometry, and Vα13 (for 3A) and Vα3 (for 3A9) by reverse transcription PCR. Positive clones were tested for antigen specificity.
Reverse Transcription PCR Analysis.
Total RNA was prepared from each transfectant using TRIzol Reagent (GIBCO BRL), converted to cDNA by reverse transcription using the GeneAmp RNA PCR Kit (Perkin-Elmer, Norwalk, CT), and amplified for 35 cycles (95°C for 45 s, 60°C for 45 s/cycle) with Taq polymerase. For 3A TCR-α detection, the forward primer 3ACDR3SEN (AGT GGG AAC ACG GGT TAC CA), located in the CDR3-Jα41 region of clone 3A TCR-α chain, and reverse primer Cα-EX1 (GAC CCA AAG CAG CTT TTG GA), located in the Cα exon 1, were used. For clone 3A9 TCR-α detection, the forward primer Vα3 (AAGTACTATTCCGGAGACCC) and reverse primer CαA (TGGCGTTGGTCTCTTTG AAG) were used (18).
Flow Cytometric Analysis.
106 cells were incubated on ice for 30 min with 20 μl of FITC-conjugated anti–TCR-α/β (1:40), FITC-conjugated anti-CD4 (1:40), biotinylated anti-Vβ4 (1:40), or biotinylated anti-Vβ8 (1:40) followed by staining with PE-conjugated streptavidin (GIBCO BRL) at 10 μg/ml. Stained cells were analyzed using a Becton Dickinson FACScan® flow cytometer. In all cases, dead cells were gated out using propidium iodide.
T Cell Stimulation and Inhibition Assay.
Transfectants (105), Th clones (105), fresh splenic CD4+ T cells (5 × 105), or short-term T cell lines (5 × 105) were cultured either with irradiated APCs (5 × 105) that had been prepulsed with various concentrations of antigens and washed, or with APC and anti-CD3 mAb. The cultures were done in triplicate in flat-bottomed 96-well plates with or without ionomycin (10–100 ng/ml). For antibody blocking, anti-CD4 mAb (purified RM4.5 or GK1.5) or mAb to MHC class II antigens were added to the cultures. After 24–36 h of incubation, IL-2 secretion was measured by harvesting 100 μl of culture supernatant from each well, which was added into a 96-well Nunc-Immuno Plate (MaxiSorbTM, Dynatech Laboratories, Chantilly, VA) that had been coated with anti–IL-2 (1 μg/ml), and blocked with 10% horse serum in PBS. After incubation at 4°C overnight, the plate was washed and revealed with biotinylated anti–IL-2 (1 μg/ml) and horseradish peroxidase–conjugated streptavidin (1:2000) (Sigma Chemical Co.), followed by the substrate TMB (Sigma Chemical Co.) at room temperature for ∼30 min, then stopped by using 1 N H2SO4 and read immediately at 450 nM. Standard curves for each assay were obtained with known quantities of mouse rIL-2.
Competitive Assay for MHC ClassII Binding.
Quantitative assays to determine the binding affinity of nucleosomal histone peptides to purified MHC class II molecules on the basis of competitive inhibition of binding of a known peptide ligand were performed as previously described (19, 20).
Results
Structural and Functional Properties of the Pathogenic Th Clones of Lupus.
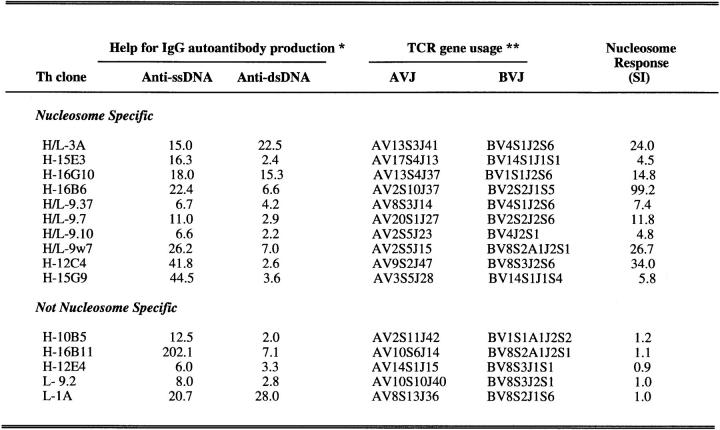
Approximately 15% of 268 T cell clones derived from T cells that are spontaneously activated in nephritic SNF1 mice have the ability to selectively augment the production of IgG anti-DNA and anti-nucleosome autoantibodies when cocultured with autologous B cells (1–3, 13). Similar Th cells are also found in patients with lupus nephritis (5, 7). Moreover, representative SNF1-derived clones (H/L-3A and L-1A) rapidly precipitate lupus nephritis upon transfer into young preautoimmune mice (2, 4). A compilation of such pathogenic Th clones is shown in Figure 1. The TCR gene usage of the clones previously published (2, 13) has been revised according to the most recent nomenclature (21). Each of the lupus Th clones expresses a single functional TCR-α and -β (2, 13). The nucleosome specificity of several additional hybridoma clones included in this panel was unmasked by using a suboptimal dose (50 ng/ml) of ionomycin that does not induce stimulation in the absence of antigen (Fig. 1). It is likely that the requirement for a suboptimal dose of ionomycin reflects a partially impaired antigen-specific signaling pathway in some of the Th hybridomas.
Figure 1.
Summary of pathogenic anti-DNA autoantibody-inducing Th clones derived from SNF1 mice with lupus nephritis (1–3, 13). L, cloned Th lines; H, Th hybridomas derived directly from splenic T cells; H/L, hybridomas derived from cloned Th lines by fusion with TCR-negative BW5147 thymoma, after initial functional characterization. These H/L hybridomas retained their nucleosome specificity and pathogenic autoantibody-inducing ability. A proportion of the Th clones that were previously found to be unresponsive to nucleosomes (3) were retested in this study in the presence of suboptimal doses of ionomycin that unmasked the nucleosome specificity of some Th clones (H-15E3 and H-16G10). *, help for IgG autoantibody production is expressed as fold increase in antibody production when Th clones were cocultured with syngeneic B cells as compared with control cultures containing B cells alone. **, the TCR-α and -β chain genes expressed by the Th clones were renamed according to WHO-IUIS nomenclature (21). Optimal results from >10 experiments are shown.
The TCR-α Chain of a Pathogenic Th Clone Confers Nucleosome Specificity.
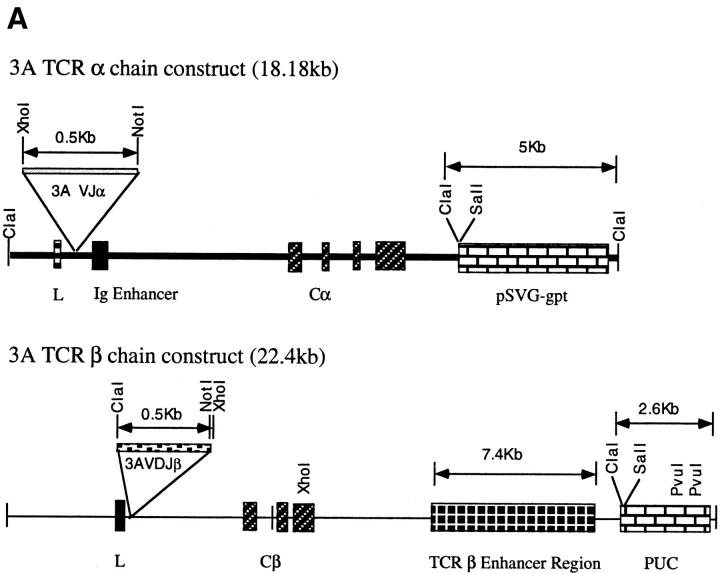
The functionally rearranged TCR-α and -β chain genes from the prototypic lupus Th clone 3A (H/L-3A) were inserted into TCR-α and -β shuttle vectors and transfected in different combinations into the TCR loss mutant 4G4, as well as the EL4 T-cell lymphoma lines. 3A is nucleosome specific, and its epitope has been mapped to the core histone H471–94 region. This peptide epitope can precipitate severe lupus nephritis in vivo by inducing similar Th cells, and T cells spontaneously primed to this epitope are prevalent in the spleens of very young SNF1 lupus-prone mice (6). Control transfections of different TCR-α and -β chain combinations were performed with HEL-specific 3A9 TCR-α and -β constructs (16, 17, 22). Figure 2 (A–C) shows stable transfectants expressing the respective TCRs. The 4G4 line was found to have surface TCR expression (as detected by anti-CD3 or anti–TCR-α/β mAb staining) when both α and β TCR constructs were used for transfections but not with either of them alone. The EL4 transfectants express the transfected TCR along with the EL4's own endogenous TCR, the specificity of which is not known. The sequence of the EL4 TCR-α is unknown, but we identified its TCR Vβ as Vβ 12 (BV12S1), and furthermore sequenced its junctional region (see below).
Figure 2.
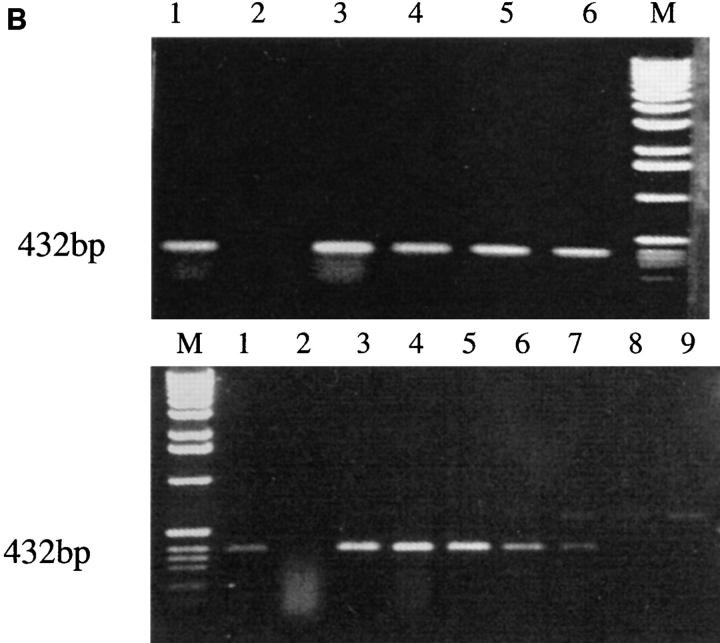
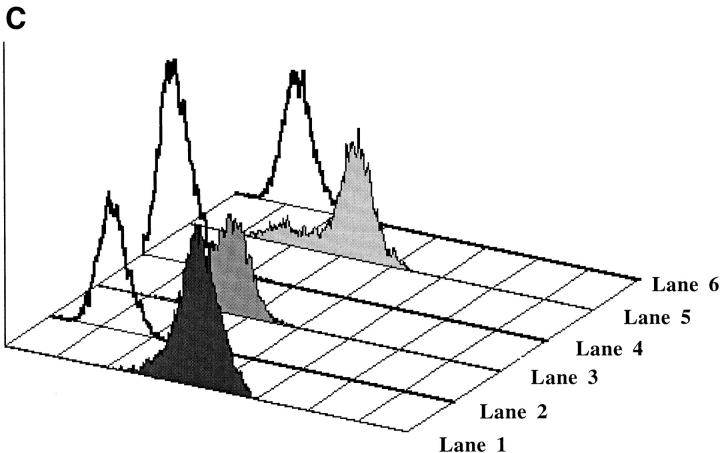
(A) 3A TCR-α and -β chain gene constructs. Functionally rearranged segments of TCR genes AV13S3J41 and BV4S1DJ2S6, expressed by the prototypic pathogenic autoantibody-inducing Th clone 3A (H/L-3A), were amplified from genomic DNA and inserted into the TCR expression shuttle vectors (17). (B, top) Stable transfection of the 3Aα chain gene in 4G4 cells. Transcription of TCR-α chain gene is shown. Total RNA was prepared from each transfectant, converted to cDNA by reverse transcription, amplified by Taq polymerase using 3ACDR3SEN forward and Cα-EX1 reverse primers, then separated on a 1% agarose. Lane 1, H/L-3A (positive control, the TCR donor lupus Th clone 3A); lane 2, 4G4 (TCR-negative recipient); lanes 3 and 5, examples of two separate clones of 4G4 transfected with 3Aα/3Aβ genes; lanes 4 and 6, two clones of 3Aα/3A9β-transfected 4G4. Integration of 3Aα construct into genomic DNA of respective transfectants was detected by using the primers Vα13-5′-XhoI and Jα41-3′-NotI to amplify the specific AV13S3J41 fragment by PCR (data not shown). (Bottom) Stable transfection of 3Aα chain gene in EL-4 cells. Transcription of TCR-α chain gene was detected by reverse transcription PCR as above. The method was specific because EL-4's own endogenous TCR-α was not amplified. Lane 1, H/L-3A; lane 2, EL-4 (recipient); lanes 3–5, three separate clones of 3Aα gene-transfected EL-4; lanes 6 and 7: two examples of clones with 3Aα/3Aβ genes in EL-4; lanes 8 and 9, two clones of 3Aβ gene-transfected EL-4. Integration of 3Aα construct into genomic DNA of the transfectants was detected as above (data not shown). Stable transfection of the HEL-specific T cell–derived 3A9α chain gene was detected by reverse transcription PCR using primers Vα3 and CαA (data not shown). (C) Expression of transfected 3Aβ chain (Vβ4) and 3A9β chain (Vβ8) in 4G4 and EL-4 cells, analyzed by flow cytometry after staining with anti-Vβ4-biotin or anti-Vβ8-biotin followed by PE-conjugated streptavidin. Lane 1, 3Aα/3Aβ in 4G4; lane 2, 4G4; lane 3, 3Aβ in EL-4; lane 4, EL-4; lane 5, 3Aα/3A9β in 4G4; lane 6, 4G4. PCR of genomic DNA showed stable integration of the transfected TCR-β genes (data not shown). The mean fluorescence intensities of Vβ4 or Vβ8 staining of the transfectants were similar to the donor Th clone 3A or 3A9, respectively (data not shown).
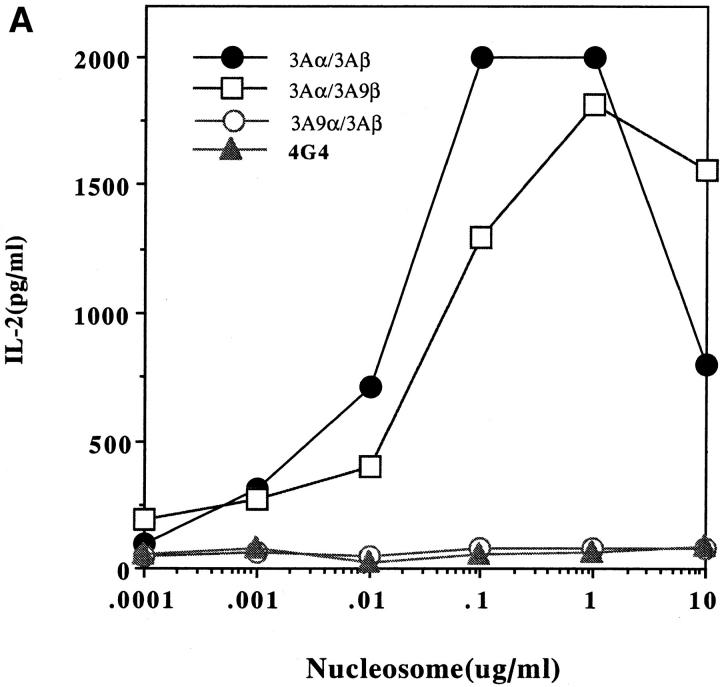
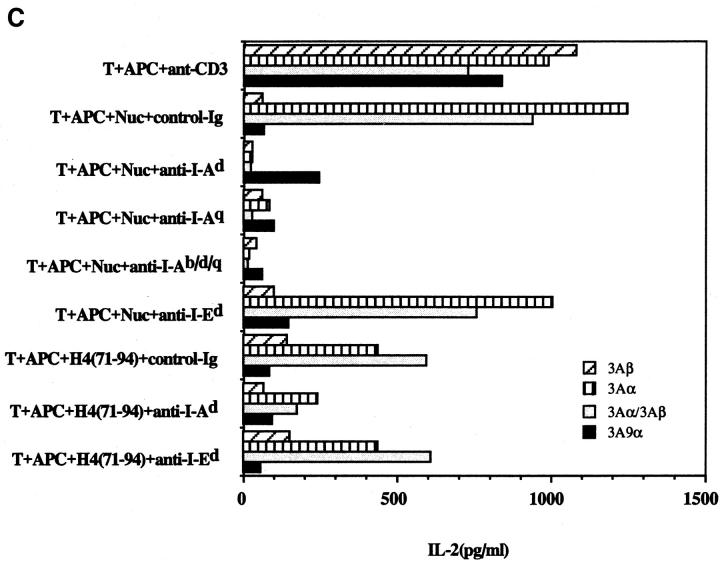
Figure 3 shows that the transfectants bearing the TCR of Th clone 3A have the same antigenic specificity and MHC-dependence as the donor pathogenic Th clone from SNF1 lupus-prone mice. The nucleosomal epitope is presented by I-Ad or I-Aq molecules of the SNF1 (H-2d/q) APCs but not I-Ed. Moreover, the response of the transfected cells to the nucleosomal epitope was as robust as that to anti-CD3 antibody stimulation. Surprisingly, the TCR-α chain of Th clone 3A in combination with either the TCR-β of HEL-specific clone 3A9 or the EL4 TCR-β chain (unknown specificity) was sufficient to confer the MHC-dependent nucleosome specificity (Fig. 3). In contrast, the TCR-β chain of the Th clone 3A in combination with the TCR-α of EL4 or the HEL-specific clone 3A9 could not reconstitute nucleosome recognition (Fig. 3). Moreover, unlike the nucleosome-specific TCR-α of lupus clone 3A, the TCR-α chain of 3A9 required pairing with 3A9 TCR-β to reconstitute HEL specificity that was restricted to I-Ak (reference 16 and data not shown).
Figure 3.
IL-2 production by 4G4 (A) and EL-4 (B) transfectants in response to the nucleosome presented by SNF1 APCs. Concentrations between 0.1 and 1 μg/ml were selected for subsequent assays except where indicated. MHC class II is required for nucleosomal antigen presentation to the EL-4 (C) and 4G4 (D) transfectants. Transfectants were cultured with irradiated SNF1 (H-2d/q) APCs that had been prepulsed with nucleosomes (Nuc, 1 μg/ ml) or the H4 (71–94) peptide (1 μM) in the presence of anti–I-Ad, anti–I-Aq, anti–I-Ab/d/q, anti–I-Ed, or control Ig. Culture supernatants were collected and tested for the presence of IL-2 by ELISA. Data are representative of more than five experiments and the data shown are means of assays done in triplicate. SD <10%.
Nucleosome Presentation and Recognition by Lupus T Cells are Promiscuous.
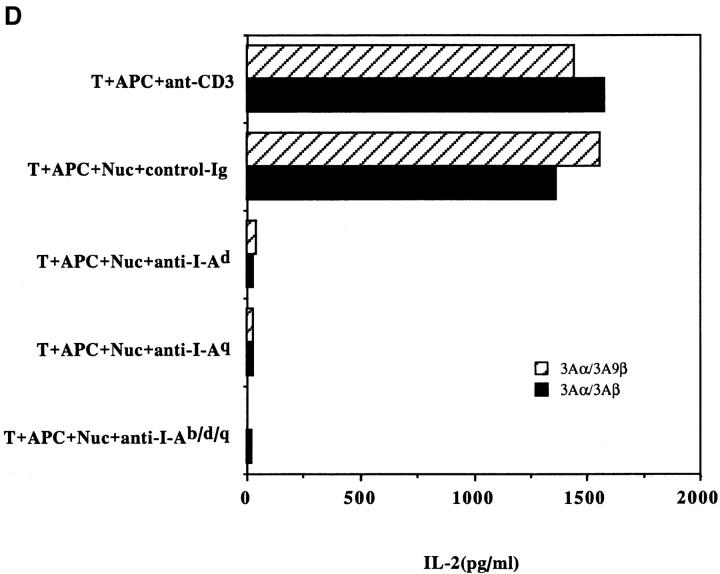
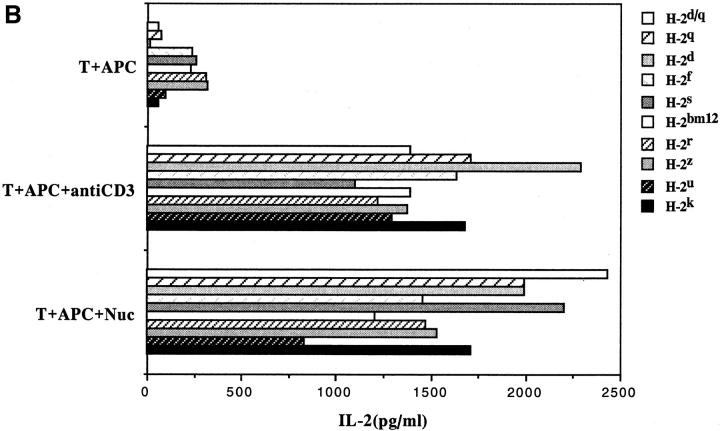
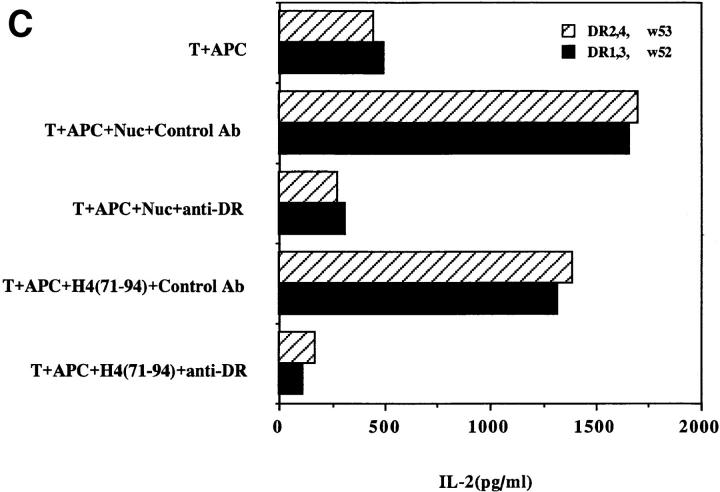
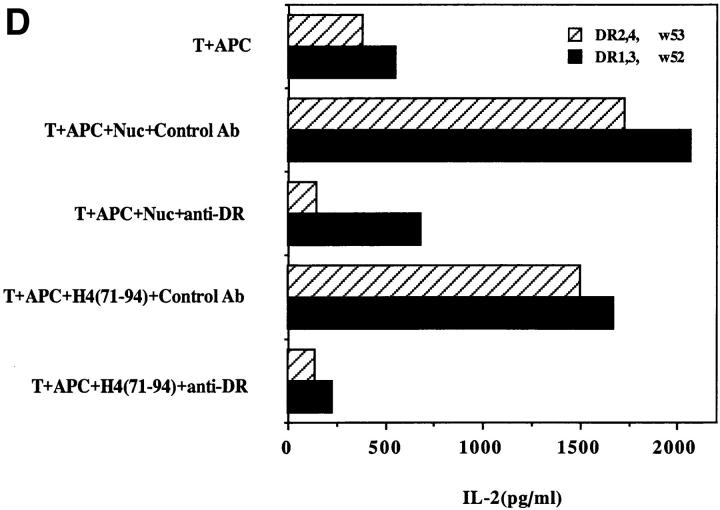
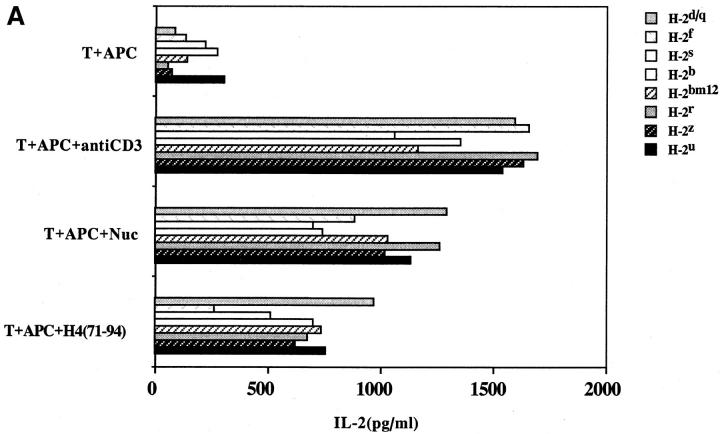
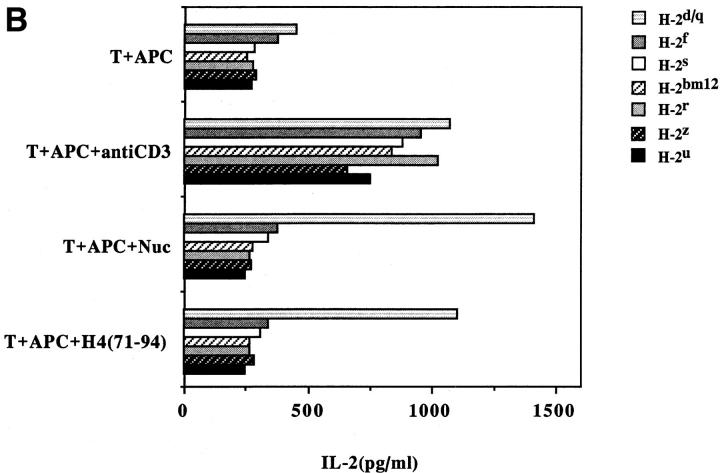
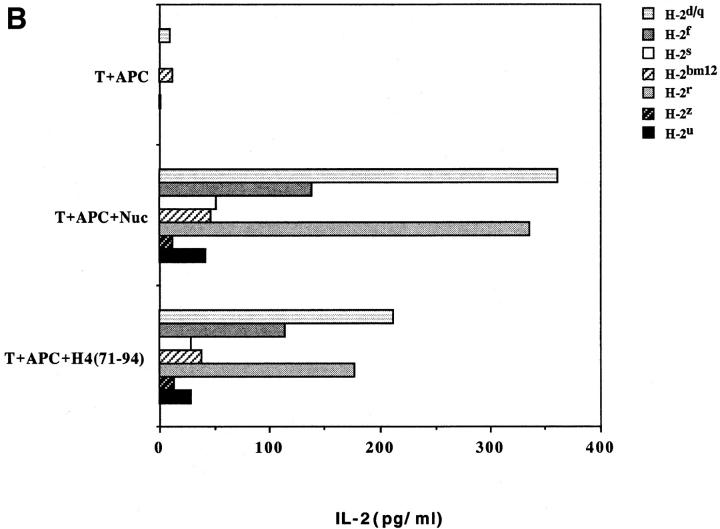
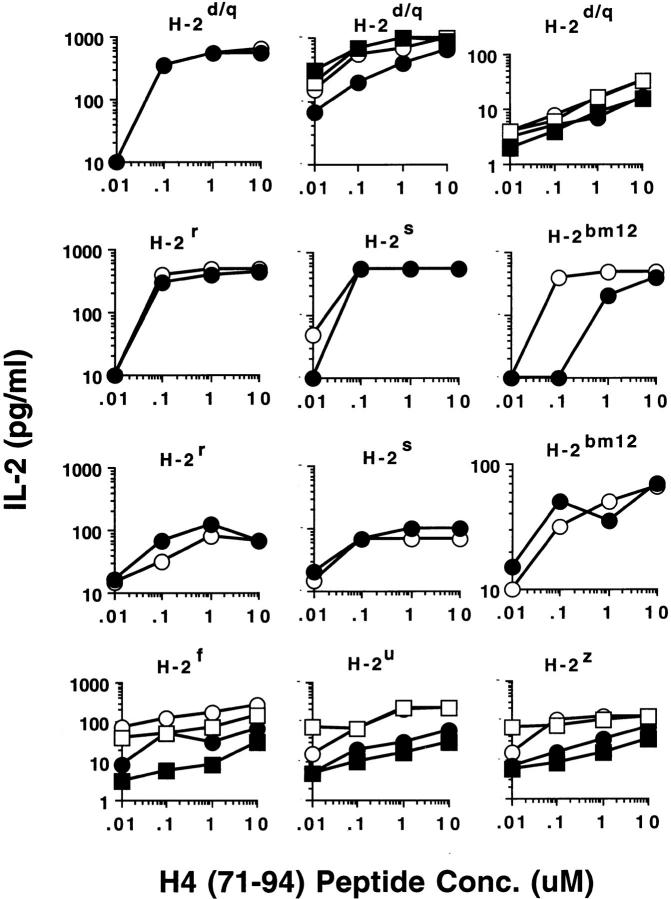
Another surprising finding was that the nucleosomal epitopes could be presented to the transfectants bearing colne 3A TCR by almost all MHC II haplotype APCs tested, including human DR molecules (Fig. 4). Again, the promiscuous recognition was dependent upon the TCR-α chain of the pathogenic lupus Th clone (Fig. 4, A and C), and pairing with its TCR-β chain did not restrict its promiscuity (Fig. 4, B and D). Moreover, the nucleosome-specific TCR did not mount an alloreactive response against the different APCs. The promiscuous recognition of nucleosomal antigen was also a property of three other lupus Th clones (H-16G10, H-15E3, and H-16B6) in this SNF1-derived panel (Fig. 1), that shared specificity for the H471–94 peptide; however, two Th clones (H/L-9w7 and H-12C4) in the group were restricted to syngeneic MHC (Fig. 5 and data not shown). Interestingly, the promiscuous Th clone H-16G10 expresses the same TCR Vα family as clone 3A, but the others use members from various other TCR families (Fig. 1).
Figure 4.
Promiscuous recognition of nucleosomal antigen presented by APCs of various MHC haplotypes by 4G4 transfected with 3Aα/3A9β (A) or 3Aα/3Aβ (B) TCRs. In Figs. 4–6, the concentration of nucleosomes used to prepulse the APCs was 1 μg/ml and that of H4 peptide was 1 μM. The APC haplotypes are indicated, H-2d/q here designates (BALB/c × SWR)F1 APCs. Nucleosomal antigen presented by xenogeneic (human) MHC class II molecules is also recognized by 4G4 transfected with 3Aα/3A9β (C) or 3Aα/3Aβ (D) TCRs. Transfectants were cultured with irradiated human HLA-DR2,4, w53 or DR1,3, w52 haplotype bearing EBV-B cell lines that had been prepulsed with antigen in the presence of anti–HLA-DR or control antibody (anti-DP, anti-DQ). Data are representative of more than five experiments. Mean IL-2 production of triplicate assays are shown; SD <10%.
Figure 5.
Promiscuous recognition of nucleosomal antigen by other lupus Th clones from SNF1 mice: clones H-16G10 (A), H-15E3, and H-16B6 (data not shown) are promiscuous. However, recognition by Th clones H/L-9W7 (B) and H-12C4 (data not shown) are restricted to syngeneic MHC (H-2d/q indicates SNF1 APCs). Data from representative experiments are presented as in previous figures.
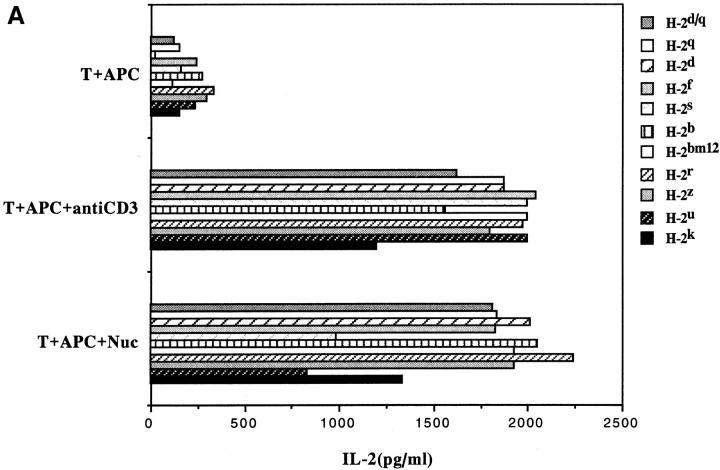
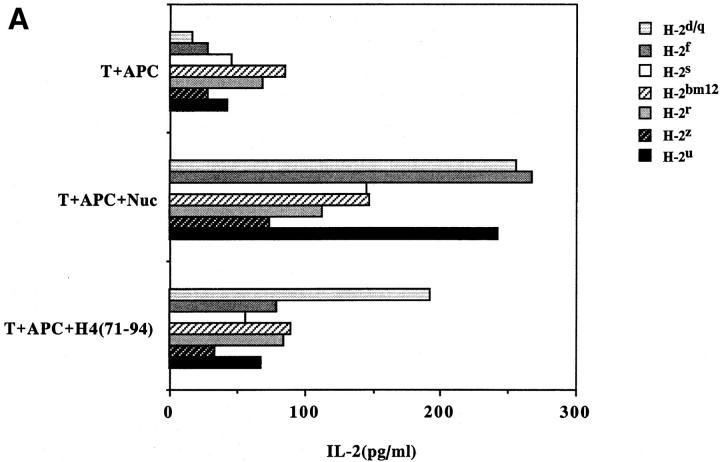
The universal presentation and promiscuous recognition of nucleosome epitopes were not just limited to the transfectants or the pathogenic Th clones. Freshly obtained CD4+ T cells or short-term T cell lines from SNF1 lupus mice also had this property (Fig. 6). In lupus-prone mice, unlike normal strains, T cells are spontaneously primed in vivo against nucleosomal epitopes (3, 6). Therefore, the response of such nucleosome-primed T cells could be detected early, such as in a 24 h, IL-2 production assay, before alloreactive T cells could obscure the autoantigen-specific signal by mounting a primary mixed lymphocyte reaction. Promiscuous recognition of the nucleosomal H471–94 peptide became detectable after just one round of nucleosome stimulation of the splenic CD4+ T cell, which probably increased the frequency of such Th cells in the bulk population (Fig. 6, A versus B).
Figure 6.
Promiscuous recognition of nucleosomes by splenic CD4+ T cells of SNF1 mice. Freshly isolated splenic CD4+ T cells (A) or shortterm CD4+ T cell line from SNF1 mice (B) were cultured with irradiated APCs of various MHC haplotypes that had been prepulsed with antigen. Culture supernatants were assayed after 24 h for IL-2. For making short-term lines, splenic CD4+ T cells were stimulated once with nucleosome ex vivo in the presence of IL-2 and then rested for 10 d. Mean of triplicate assays are shown (SD <10%). Data are representative of three experiments.
Nucleosomal Peptide Epitopes Bind to Diverse MHC II Molecules.
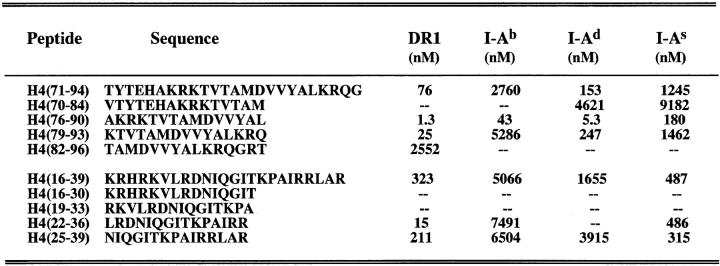
In competition assays with known conventional peptide antigens that bind to the class II groove, H471–94 and a series of truncated peptides spanning this region bound several murine I-A molecules and also human DR1 (Fig. 7). Markedly higher affinity for all the class II alleles tested was noted for the peptide spanning residues 76–90 of H4, located nearer to the COOH terminus of the parent peptide H471–94. The TCRs of 3A and other lupus Th clones in the panel (Fig. 1), are mainly specific for H471–94. However, despite having a single pair of functional TCR-α/β, each of them also recognized H416–39, thus revealing a degenerate or cross-reactive specificity (6, 13) analogous to other TCR–antigen systems (23, 24). H416–39 and its truncations also bound to the different class II alleles, particularly the peptides spanning position 22–39 in this region. Thus, this region of nucleosomal core histone may also be promiscuously recognized. However, another epitope for other pathogenic Th cells of lupus, which was previously localized in histone H2B10–33 (6), did not show promiscuous binding (data not shown), suggesting that such a binding pattern is not found in every lupus-associated autoepitopes.
Figure 7.
Binding of histone H4-derived truncated peptides to mouse and human MHC class II molecules in competition asssays. The data is presented as 50% inhibitory concentration in nanomoles (nM). Peptide binding was ranked as follows: good binders (binding affinity <100 nM); intermediate binders (binding affinity = 100–1,000 nM); low binders (binding affinity = 1,000–10,000 nM); and negative binders (binding affinity >10,000 nM).
Although the truncated H476–90 bound to MHC with the highest affinity, it did not stimulate the 3A clone TCR as strongly as did the parental H471–94 peptide (data not shown). And, unlike the case of the H471–94 epitope (Fig. 6 B), reactivity to H476–90 could not be detected in freshly obtained CD4+ T cells or short-term T cell lines from SNF1 mice (data not shown).
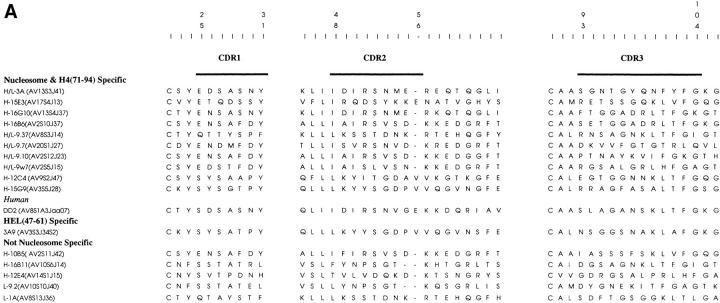
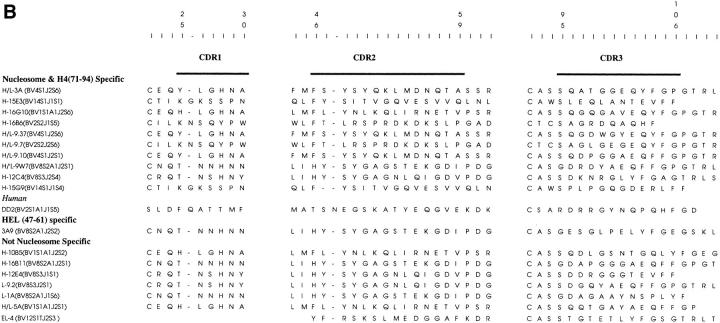
Shared Motif in the Nucleosome-specific TCR-α Chains.
A comparison of the sequences of the various TCRs analyzed here revealed that the sequence motif of IRSNMERE (or conservative variations of it) was frequently present in the CDR2 region (position 50–57) of the nucleosome-specific TCR Vα chains (Fig. 8 A). Furthermore, charged residues in the flanking region (position 57–62) of the CDR2 were also prevalent. These features were not found in the HEL-specific TCR-α chain.
Figure 8.
Amino acid sequences of CDRs of TCR-α (A) and -β (B), chains relevant to this study, compiled from references 2, 7, and 13, and aligned (21). All sequences are from mouse T cells except for DD2, which is a human lupus Th clone (7). Updated TCR-α chain sequence of HEL-specific clone 3A9 (52) was provided by Paul Allen (Washington University, St. Louis, MO). The EL-4 TCR-β chain (B) was sequenced in this study (available from EMBL/GenBank/DDBJ under accession number BankIt 13531 AF020206).
It should be noted that TCR-α of H-10B5, another pathogenic autoantibody-inducing Th clone in the panel that did not respond to nucleosomal antigens, shared similar motifs with the nucleosome-specific TCR-α. The antigen specificity of this lupus clone is not known but it may possibly be specific for other chromosomal proteins with charged residues similar to the nucleosomal histones (7).
Interestingly, the TCR Vβ chains of EL-4 or HEL-specific 3A9 that could pair with 3A TCR-α for its promiscuous recognition of nucleosomal H4 epitope contained several negatively charged residues in their CDR3, just like the TCR Vβ chains of other nucleosome-specific Th clones (Fig. 8 B).
CD4 Coreceptor Is Not Required for Promiscuous Recognition of Nucleosomes.
The anti-CD4 mAbs, GK1.5 and RM4.5, which are known to inhibit antigen-specific responses (25), could not block the recognition of nucleosomal antigens by 4G4 and EL-4 transfectants expressing 3A TCR-α chain (Fig. 9 and data not shown). Moreover, the promiscuous recognition of nucleosome antigens by these transfectants (Figs. 3 and 4) could not be blocked by the anti-CD4 mAb even at saturating concentrations (data not shown). The transfectants were surface CD4+ by flow cytometry (data not shown). In addition, 4G4 transfectants that expressed the 3A TCR-α chain (with 3A9 TCR-β), but had lost surface expression of CD4, also responded to nucleosomes and H471–94 peptide in a promiscuous but MHC class II–dependent manner, and their responses were equivalent to those shown in Figures 3 and 4 (data not shown). Furthermore, we tested the pathogenic autoantibody-inducing CD4+ Th clones, H/L- 3A, H-15E3, and H-16G10, and found that they were also relatively resistant to anti-CD4 blocking in their ability to promiscuously recognize the nucleosomal H471–94 peptide (Fig. 9 and data not shown). Moreover, the addition of anti-CD4 mAb did not result in a significant shift in the concentrations of peptide required to stimulate the Th clones in most cases. Finally, independently derived, short-term CD4+ T cell lines freshly obtained from different SNF1 animals also contained a substantial population of T cells that were CD4-independent in recognition of the nucleosomal epitope. Anti-CD4 blocking could only partially inhibit (from 50 to 70%) promiscuous recognition of nucleosomal H471–94 peptide by the CD4+ T cell lines at all the different concentrations of antigen tested (Fig. 9).
Figure 9.
Effect of the purified anti-CD4 mAb, RM4.5, on the responses of lupus T cells to nucleosomal peptide H471–94. Solid symbols represent anti-CD4 mAb containing cultures and the corresponding cultures without anti-CD4 are designated by open symbols. The MHC haplotypes of different APCs used to present the peptide are shown at the top of each panel; H-2d/q represents syngeneic SNF1 APCs. Background values of IL-2 production in corresponding cultures (T cells + APC) without antigen have been subtracted from each data point. (Top row) Left panel shows 4G4 transfected with 3Aα/3a9β TCRs, middle shows Th clones H/L-3A (circles) and H-15E3 (squares), and right panel shows two short-term, CD4+ T cell lines from SNF1 spleen (squares and circles). Responses of pathogenic autoantibody-inducing Th clones H/L-3A and H-15E3 to nucleosomal peptide presented by different allogeneic APC are shown in second and third rows, respectively, and that of the short-term T cell lines are shown in the bottom row. The anti-CD4 mAb, RM4.5, was used at 1 μg/ml, which is the saturating concentration (25), but similar results were obtained at a concentration of 10 μg/ml. Another anti-CD4 mAb (GK1.5), concentrated 10 and 5 times from culture supernatants by ammonium sulphate precipitations, gave similar results (data not shown). Mean of triplicate assays are shown above. SD <10%.
Discussion
These studies show that certain nucleosomal peptides recognized by spontaneously arising, autoimmune Th cells that cause lupus nephritis are degenrate MHC binders, and the corresponding Th cells that recognize these peptides are MHC unrestricted although they were educated in the I-Ad/q thymus of SNF1 mice. The presentation of the nucleosomal H4 peptides (71–94 region) was clearly MHC dependent as shown by anti–class II antibody blocking. Either mAb to I-Ad or I-Aq could almost completely block nucleosome presentation by SNF1 (I-Ad/q) APCs, because the antibodies have cross-reactive specificity due to the highly homologous structure of I-Ad and I-Aq molecules (Fig. 3, reference 26, and data not shown). Presentation by class I (classical or nonclassical) is ruled out because transfectants derived from the mouse T cell lymphoma EL-4, which lacks class II but is capable of presenting antigen with class I molecules (27), did not produce IL-2 by themselves when exposed to the nucleosomal peptide (Fig. 3). Moreover, the nucleosomal peptides most likely bound the diverse set of class II molecules tested through engagement of the classical binding groove of MHC, because they efficiently competed with well-characterized peptides known to bind to the grooves. Furthermore, previous work has established that nucleosomes and their peptide epitopes do not behave as superantigens (3, 6, 13). Truncation of the H471–94 peptide increased its affinity for the class II alleles, and indeed its core region (amino acid position 78–89) does have a sequence motif (RKTVTAMDVVYA) that is homologous to other universal epitopes (20). However, truncation of the 24-mer H4 peptide to 15-mer decreased T cell recognition, suggesting that residues outside the MHC-binding core region can either directly or indirectly affect TCR engagement of MHC–peptide complexes (28, 29). Another degenerate MHC-binding and promiscuous T cell recognition epitope was mapped to residues 16–39 of nucleosomal histone H4. However, another T cell epitope in histone H2B (6) did not share those properties. These results suggest that nucleosomal autoantigens are frequently but not always associated with degenerate MHC binding and promiscuous recognition by lupus T cells.
Our results demonstrate that promiscuous recognition of nucleosomal epitopes by single α/β pairs of autoimmune TCRs is a property conferred by the TCR-α chain. Indeed, lupus susceptibility is not restricted to any particular MHC haplotype. Different MHC alleles are found in different lupus strains, and the same ones are also present in normal strains. Interestingly, degenerate MHC binding of certain myelin basic protein peptides and their recognition by T cell lines from multiple sclerosis patients have also been described. However, weakly cross-reactive binding to only DR molecules was reported, and TCRs responsible for the degenerate recognition were not analyzed (30).
TCR promiscuity was thought to occur in the context of DR and its murine homologue, the I-E molecules, assuming that the TCRs were recognizing the peptide epitopes along with the nonpolymorphic α chain in these MHC molecules (31, 32). However, this is not the case with the nucleosomal peptide-specific TCRs described here, or the universal peptide epitopes described in other systems (20, 33, 34), where the epitopes either bind or are recognized in the context of both human DR and murine I-A molecules, the latter being polymorphic in both α and β chains. In general, promiscuous or cross-reactive recognition by a TCR is thought to be the basis for positive selection in the thymus and alloreactivity, i.e., TCRs can somehow “learn” from one MHC allele to interact with others (35–37). However, it is notable that although the lupus TCRs in this study recognized the histone peptide in the context of the diferent MHC alleles, they were not reactive to APCs bearing those alleles in the absence of the histone peptide. Indeed, promiscuous recognition of nucleosomal epitope was not only a feature of the pathogenic Th clones, but was also detectable in freshly obtained splenic T cell populations from the SNF1 mice. In lupus mice, nucleosome-primed T cells arise spontaneously and are prevalent in vivo from an early age (3, 6).
The CDR2 regions of these Th clones appear to have a related sequence motif and multiple negative residues, which in combination with other sequences of their TCR-α and -β chains could confer the property of promiscuous recognition of nucleosomal antigen. TCR-α chain CDR2 has been implicated in other systems for MHC-antigen recognition (15, 38–41). It is noteworthy that a pathogenic autoantibody-inducing Th clone of human lupus that also recognizes the nucleosomal core histone H4 expresses TCR Vα8 (7), which is homologous to the mouse Vα13 gene family (42) expressed by the murine lupus Th clones 3A and 16G10. More remarkably, in contrast with an expression rate of 9% among unselected T cells, the Vα8 genes are expressed by 18 out of 42 (i.e., 43%, but ranging up to 100% in some patients) pathogenic autoantibody-inducing Th clones from lupus patients, and they also recognize chromosomal proteins (7). Notably, the CDR1 and 2 regions of this human TCR Vα8 gene family share sequence motifs with the murine Vα13 family (42).
Crystallographic data has recently illustrated how recognition of MHC–peptide complexes by monogamous TCR molecules (43, 44) is largely dependent on a single TCR pocket engaging a prominent side chain of the peptide antigen. It is thus possible to speculate that nucleosomal peptide antigens that can bind multiple MHC molecules (degenerate binders) and carry multiple positive charges can engage certain lupus TCR-α chains that carry multiple binding pockets and reciprocally charged residues, generating an interaction of such strength as to overcome the usual apparent requirement for MHC restriction (promiscuous T cell recognititon). This hypothesis is further supported by the lack of the requirement of CD4 coreceptor for signaling the promiscuous recognition response by the lupus TCR, even at the lowest stimulatory concentrations of the autoantigen. These results cannot be attributed to possible expression of TCR at high density in the transfectants, because other nucleosome-specific lupus Th clones and freshly prepared CD4+ T cell lines from the spleens of SNF1 mice also shared this property, whereas the HEL-specific TCR (3A9) transfectants were not promiscuous. Indeed, CD4-negative, TCR-α/β+ T cells that help in the production of pathogenic antinuclear autoantibodies are markedly expanded in both human and murine SLE (1, 2, 5, 7). The beneficial effect of chronic anti-CD4 antibody administration to lupus mice in vivo (45) is apparently contradictory to the above observations. However, the therapeutic effect of anti-CD4 in vivo could be due to a combination of factors and not exclusively due to an inhibition of CD4+ T cells. The CD4 molecule is also expressed on the surface of hematopoietic stem cell precursors, which could affect the generation of autoantigen-presenting APCs and autoimmune B cells, not just T cells (46). Significantly, the CD4 molecule is also expressed on APCs such as dendritic cells, monocytes, and macrophages (47). Thus, anti-CD4 therapy could interfere with autoantigen presentation and other effector and inflammatory components of lupus. Moreover, chronic anti-CD4 antibody therapy generates downregulatory T cells in vivo (48). Indeed, for anti-CD4 therapy to be effective it has to be administered repeatedly and for the long term to lupus-prone mice, beginning early in their lives (45), suggesting that something more than just inhibition of CD4+ T helper cells is responsible for the therapeutic effect.
Crystallographic and modeling data also indicate that different TCR–peptide–MHC complexes will have distinct structural orientations and contact points (14, 23, 41, 43, 44, 49–51). Therefore, analysis of the promiscuous lupus TCRs in contact with the H4 peptide complexed with different MHC molecules could reveal some unique features of this trimolecular interaction.
Since the lupus TCR-α chains could confer nucleosomal peptide specificity as well as promiscuity in combination with TCR-β chains of unrelated specificities, the nucleosome-specific T cell repertoire is potentially vast. How it is kept in check in normal mice and why they are selected in the lupus thymus are pressing questions that will be addressed in the transgenic mice we are developing with these TCR constructs. These studies also open up the possibility of designing “universally tolerogenic” epitopes for the therapy of patients with lupus, despite the diversity of their HLA alleles.
Acknowledgments
We thank Wendy Smith-Begolka for typing the V family member expressed by EL-4. We also thank Dr. Vijay Kuchroo for helpful suggestions.
This work was supported by grants from the National Institutes of Health (RO1 AI-41985 and AR-39157) and the Arthritis Foundation to S.K. Datta and from the Arthritis National Research Foundation to A. Kaliyaperumal.
Footnotes
Dr. Syamal K. Datta, Arthritis Division, Ward 3-315, Northwestern University Medical School, 303 East Chicago Ave., Chicago, IL 60611. Phone: 312-503-0535; Fax: 312-503-0994; E-mail: skd257@nwu.edu
Abbreviations used in this paper: HEL, hen egg lysozyme; SNF1, (SWR × NZB)F1.
References
- 1.Sainis K, Datta SK. CD4+ T-cell lines with selective patterns of autoreactivity as well as CD4−/CD8−T helper cell lines augment the production of idiotypes shared by pathogenic anti-DNA autoantibodies in the NZB × SWR model of lupus nephritis. J Immunol. 1988;140:2215–2224. [PubMed] [Google Scholar]
- 2.Adams S, Leblanc P, Datta SK. Junctional region sequences of T-cell receptor β chain genes expressed by pathogenic anti-DNA autoantibody-inducing T helper cells from lupus mice: possible selection by cationic autoantigens. Proc Natl Acad Sci USA. 1991;88:11271–11275. doi: 10.1073/pnas.88.24.11271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mohan C, Adams S, Stanik V, Datta SK. Nucleosome: a major immunogen for the pathogenic autoantibody-inducing T cells of lupus. J Exp Med. 1993;177:1367–1381. doi: 10.1084/jem.177.5.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mohan C, Shi Y, Laman JD, Datta SK. Interaction between CD40 and its ligand gp39 in the development of murine lupus nephritis. J Immunol. 1995;154:1470–1480. [PubMed] [Google Scholar]
- 5.Rajagopalan S, Zordan T, Tsokos GC, Datta SK. Pathogenic anti-DNA autoantibody-inducing T helper cell lines from patients with active lupus nephritis: isolation of CD4−/CD8−T helper cell lines that express the γ/δ T-cell receptor. Proc Natl Acad Sci USA. 1990;87:7020–7024. doi: 10.1073/pnas.87.18.7020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaliyaperumal A, Mohan C, Wu W, Datta SK. Nucleosomal peptide epitopes for nephritis-inducing T helper cells of murine lupus. J Exp Med. 1996;183:2459–2469. doi: 10.1084/jem.183.6.2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Desai-Mehta A, Mao C, Rajagopalan S, Robinson T, Datta SK. Structure and specificity of T-cell receptors expressed by pathogenic anti-DNA autoantibody-inducing T cells in human lupus. J Clin Invest. 1995;95:531–541. doi: 10.1172/JCI117695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casciola-Rosen LA, Anhalt G, Rosen A. Autoantigens targeted in systemic lupus erythematosus are clustered in two populations of surface structures on apoptotic keratinocytes. J Exp Med. 1994;179:1317–1330. doi: 10.1084/jem.179.4.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lorenz H-M, Grunke M, Hieronymus T, Herrmann M, Kuhnel A, Manger B, Kalden JR. In vitro apoptosis and expression of apoptosis-related molecules in lymphocytes from patients with systemic lupus erythematosus and other autoimmune diseases. Arthritis Rheum. 1997;40:306–317. doi: 10.1002/art.1780400216. [DOI] [PubMed] [Google Scholar]
- 10.Yu D, Rumore PM, Liu Q, Steinman CR. Soluble oligonucleosomal complexes in synovial fluid from inflamed joints. Arthritis Rheum. 1997;40:648–654. doi: 10.1002/art.1780400409. [DOI] [PubMed] [Google Scholar]
- 11.Ray SK, Putterman C, Diamond B. Pathogenic autoantibodies are routinely generated during the response to foreign antigen: a paradigm for autoimmune disease. Proc Natl Acad Sci USA. 1996;93:2019–2024. doi: 10.1073/pnas.93.5.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Datta SK, Stollar BD, Schwartz RS. Normal mice express idiotypes related to autoantibody idiotypes of lupus mice. Proc Natl Acad Sci USA. 1983;80:2723–2727. doi: 10.1073/pnas.80.9.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mao C, Osman GE, Adams S, Datta SK. T cell receptor α-chain repertoire of pathogenic autoantibody-inducing T cells in lupus mice. J Immunol. 1994;152:1462–1470. [PubMed] [Google Scholar]
- 14.Jorgensen JL, Esser U, Reay PA, Fazekas de St B, Groth, Davis MM. Mapping T cell receptor/peptide contacts by variant peptide immunization of single-chain transgenics. Nature. 1992;355:224–230. doi: 10.1038/355224a0. [DOI] [PubMed] [Google Scholar]
- 15.Hong S-C, Chelouche A, Lin R-H, Shaywitz D, Braunstein NS, Glimcher L, Janeway CA., Jr An MHC interaction site maps to the amino-terminal half of the T cell receptor alpha chain variable domain. Cell. 1992;69:999–1009. doi: 10.1016/0092-8674(92)90618-m. [DOI] [PubMed] [Google Scholar]
- 16.Patten PA, Rock EP, Sonoda T, Fazekas de St B, Groth, Jorgensen JL, Davis MM. Transfer of putative complementarity-determining region loops of T cell receptor V domains confers toxin reactivity but not peptide/MHC specificity. J Immunol. 1993;150:2281–2294. [PubMed] [Google Scholar]
- 17.Ho WY, Cooke MP, Goodnow CC, Davis MM. Resting and anergic B cells are defective in CD28-dependent costimulation of naive CD4+T cells. J Exp Med. 1994;179:1539–1549. doi: 10.1084/jem.179.5.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Casanova JL, Romero P, Widmann C, Kourilsky P, Maryanski JL. T cell receptor genes in a series of class I major histocompatibility complex–restricted cytotoxic T lymphocyte clones specific for a plasmodium berghei nonapeptide: implications for T cell allelic exclusion and antigen-specific repertoire. J Exp Med. 1991;174:1371–1383. doi: 10.1084/jem.174.6.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sette A, Buus S, Colon S, Miles C, Grey HM. Structural analysis of peptides capable of binding to more than one Ia antigen. J Immunol. 1989;142:35–40. [PubMed] [Google Scholar]
- 20.Alexander J, Sidney J, Southwood S, Rupert J, Oseroff C, Maewal A, Snoke K, Serra HM, Kubo RT, Sette A, Grey HM. Development of high potency universal DR-restricted helper epitopes by modification of high- affinity DR-blocking peptides. Immunity. 1994;1:751–761. doi: 10.1016/s1074-7613(94)80017-0. [DOI] [PubMed] [Google Scholar]
- 21.Arden B, Clark SP, Kabelitz D, Mak TW. Mouse T-cell receptor variable gene segment families. Immunogenetics. 1995;42:501–530. doi: 10.1007/BF00172177. [DOI] [PubMed] [Google Scholar]
- 22.Allen P, McKean DJ, Beck BN, Sheffield J, Glimcher LH. Direct evidence that a class II molecule and a simple globular protein generate multiple determinants. J Exp Med. 1985;162:1264–1274. doi: 10.1084/jem.162.4.1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nanda NK, Arzoo KK, Geysen HM, Sette A, Sercarz EE. Recognition of multiple peptide cores by a single T cell receptor. J Exp Med. 1995;182:531–539. doi: 10.1084/jem.182.2.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Evavold BD, Sloan-Lancaster J, Wilson KJ, Rothbard JB, Allen PM. Specific T cell recognition of minimally homologous peptides: evidence for multiple endogenous ligands. Immunity. 1995;2:655–663. doi: 10.1016/1074-7613(95)90010-1. [DOI] [PubMed] [Google Scholar]
- 25.Madrenas J, Chau LA, Smith J, Bluestone JA, Germain RN. The efficacy of CD4 requirement to ligand-engaged TCR controls the agonist/partial agonist properties of peptide–MHC molecule ligands. J Exp Med. 1997;185:219–229. doi: 10.1084/jem.185.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lundsberg AS, McDevitt HO. Evolution of major histocompatibility complex class II allelic diversity: direct descent in mice and humans. Proc Natl Acad Sci USA. 1992;89:6545–6549. doi: 10.1073/pnas.89.14.6545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaliyaperumal A, Falchetto R, Cox A, Dick R, II, Shabanowitz J, Chien YH, Matis L, Bluestone JA. Functional expression and recognition of nonclassical MHC class I T10b is not peptide-dependent. J Immunol. 1995;155:2379–2386. [PubMed] [Google Scholar]
- 28.Vignali DAA, Strominger JL. Amino acid residues that flank core peptide epitopes and the extracellular domains of CD4 modulate differential signaling through the T cell receptor. J Exp Med. 1994;179:1945–1956. doi: 10.1084/jem.179.6.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moudgil KD, Grewal IS, Jensen PE, Sercarz EE. Unresponsiveness to a self-peptide of mouse lysozyme owing to hindrance of T cell receptor–major histocompatibility complex/peptide interaction caused by flanking epitopic residues. J Exp Med. 1996;183:535–546. doi: 10.1084/jem.183.2.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Valli A, Sette A, Kappos L, Oseroff C, Sidney J, Miescher G, Hochberger M, Albert ED, Adorini L. Binding of myelin basic protein peptides to human HLA class II molecules and their recognition by T cells of multiple sclerosis patients. J Clin Invest. 1993;91:616–628. doi: 10.1172/JCI116242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Panina-Bordignon P, Tan A, Termijtelen A, Demotz S, Corradin G, Lanzavecchia A. Universally immunogenic T cell epitopes: promiscuous binding to human MHC class II and promiscuous recognition by T cells. Eur J Immunol. 1989;19:2237–2242. doi: 10.1002/eji.1830191209. [DOI] [PubMed] [Google Scholar]
- 32.Blank U, Boitel B, Mege D, Ermonval M, Acuto O. Analysis of tetanus toxin peptide/DR recognition by human T cell receptors reconstituted into a murine T cell hybridoma. Eur J Immunol. 1993;23:3057–3065. doi: 10.1002/eji.1830231203. [DOI] [PubMed] [Google Scholar]
- 33.Vordermeier HM, Harris DP, Moreno C, Ivanyi J. Promiscuous T cell recognition of an H-2 IA-presented mycobacterial epitope. Eur J Immunol. 1994;24:2061–2067. doi: 10.1002/eji.1830240919. [DOI] [PubMed] [Google Scholar]
- 34.Sinigaglia F, Guttinger M, Kilgus J, Doran DM, Matile H, Etlinger H, Trzediak A, Gillessen D, Pink JRL. A malaria T-cell epitope recognized in association with most mouse and human class II molecules. Nature. 1988;336:778–780. doi: 10.1038/336778a0. [DOI] [PubMed] [Google Scholar]
- 35.Ignatowicz L, Kappler J, Marrack P. The repertoire of T cells shaped by a single MHC/peptide ligand. Cell. 1996;84:521–529. doi: 10.1016/s0092-8674(00)81028-4. [DOI] [PubMed] [Google Scholar]
- 36.Nakano N, Rooke R, Benoist C, Mathis D. Positive selection of T cells induced by viral delivery of neopeptides to the thymus. Science. 1997;275:678–683. doi: 10.1126/science.275.5300.678. [DOI] [PubMed] [Google Scholar]
- 37.Zerrahn J, Held W, Raulet DH. The MHC reactivity of the T cell repertoire prior to positive and negative selection. Cell. 1997;88:627–636. doi: 10.1016/s0092-8674(00)81905-4. [DOI] [PubMed] [Google Scholar]
- 38.Brawley JV, Concannon P. Modulation of promiscuous T cell receptor recognition by mutagenesis of CDR2 residues. J Exp Med. 1996;183:2043–2051. doi: 10.1084/jem.183.5.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sim BC, Zerva L, Greene MI, Gascoigne NRJ. Control of MHC restriction by TCR Vα CDR1 and CDR2. Science. 1996;273:963–966. doi: 10.1126/science.273.5277.963. [DOI] [PubMed] [Google Scholar]
- 40.Kasibhatla S, Nalefski EA, Rao A. Simultaneous involvement of all six predicted antigen binding loops of the T cell receptor in recognition of the MHC/antigenic peptide complex. J Immunol. 1993;151:3140–3151. [PubMed] [Google Scholar]
- 41.Sant'Angelo DB, Waterbury G, Preston-Hulbert P, Medzhitov R, Hong SC, Janeway CA., Jr The specificty and orientation of a TCR to its peptide-MHC class II ligands. Immunity. 1996;4:367–376. doi: 10.1016/s1074-7613(00)80250-2. [DOI] [PubMed] [Google Scholar]
- 42.Clark SP, Arden B, Kabelitz D, Mak TW. Comparison of human and mouse T-cell receptor variable gene segment families. Immunogenetics. 1995;42:531–540. doi: 10.1007/BF00172178. [DOI] [PubMed] [Google Scholar]
- 43.Garboczi DN, Ghosh P, Utz U, Fan QR, Biddison WE, Wiley D C. Structure of the complex between human T-cell receptor, viral peptide and HLA-A2. Nature. 1996;384:134–141. doi: 10.1038/384134a0. [DOI] [PubMed] [Google Scholar]
- 44.Garcia KC, Degano M, Stanfield RL, Brunmark A, Jackson MR, Peterson PA, Teyton L, Wilson IA. An alpha-beta T cell receptor structure at 2.5 angstrom and its orientation in the TCR–MHC complex. Science. 1996;274:209–219. [Google Scholar]
- 45.Carteron NL, Schimenti CL, Wofsy D. Treatment of murine lupus with (Fab′)2 fragment of monoclonal antibody to L3T4. Suppression of autoimmunity does not depend on T helper cell depletion. J Immunol. 1989;142:1470–1475. [PubMed] [Google Scholar]
- 46.Fredrickson GG, Basch RS. L3T4 antigen expression by hemopoietic precursor cells. J Exp Med. 1989;169:1473–1478. doi: 10.1084/jem.169.4.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Steinman RM. The dendritic cell system and its role in immunogenicity. Annu Rev Immunol. 1991;9:271–296. doi: 10.1146/annurev.iy.09.040191.001415. [DOI] [PubMed] [Google Scholar]
- 48.Scully R, Qin S, Cobbold S, Waldmann H. Mechanisms in CD4 antibody-mediated transplantation tolerance: kinetics of induction, antigen dependency and role of regulatory T cells. Eur J Immunol. 1994;24:2383–2392. doi: 10.1002/eji.1830241019. [DOI] [PubMed] [Google Scholar]
- 49.Sun R, Shepherd SE, Geier SS, Thomson CT, Sheil JM, Nathenson SG. Evidence that the antigen receptors of cytotoxic T lymphocytes interact with a common recognition pattern on the H-2Kb molecule. Immunity. 1995;3:573–582. doi: 10.1016/1074-7613(95)90128-0. [DOI] [PubMed] [Google Scholar]
- 50.Brock R, Wiesmuller KH, Jung G, Walden P. Molecular basis for recognition of two structurally different MHC–peptide complexes by a single T cell receptor. Proc Natl Acad Sci USA. 1996;93:13108–13113. doi: 10.1073/pnas.93.23.13108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ehrich EW, Devaux B, Rock EP, Jorgensen JL, Davis MM, Chien Y-h. T cell receptor interaction with peptide/Major histocompatibilily complex (MHC) and superantigen/MHC ligands is dominated by antigen. J Exp Med. 1993;178:713–723. doi: 10.1084/jem.178.2.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Johnson NA, Carland F, Allen PM, Glimcher LH. T cell receptor gene segment usage in a panel of hen-egg lysozyme–specific, I-Akrestricted T helper hybridomas. J Immunol. 1989;142:3298–3304. [PubMed] [Google Scholar]