Abstract
Superantigens are defined as proteins that activate a large number of T cells through interaction with the Vβ region of the T cell antigen receptor (TCR). Here we demonstrate that the superantigen produced by Mycoplasma arthritidis (MAM), unlike six bacterial superantigens tested, interacts not only with the Vβ region but also with the CDR3 (third complementarity-determining region) of TCR-β. Although MAM shares typical features with other superantigens, direct interaction with CDR3-β is a feature of nominal peptide antigens situated in the antigen groove of major histocompatibility complex (MHC) molecules rather than superantigens. During peptide recognition, Vβ and Vα domains of the TCR form contacts with MHC and the complex is stabilized by CDR3–peptide interactions. Similarly, recognition of MAM is Vβ-dependent and is apparently stabilized by direct contacts with the CDR3-β region. Thus, MAM represents a new type of ligand for TCR, distinct from both conventional peptide antigens and other known superantigens.
In the first step of a specific immune response, T lymphocytes are activated by interaction between the TCR and peptide antigen bound to the MHC. Specific peptide recognition is achieved by the unique structure of TCRs, formed by somatic rearrangement of their composite gene segments (Vα, Jα, Vβ, Dβ, and Jβ). Most contacts with peptide are formed by very diverse CDR3 (third complementarity-determining region) regions that are encoded by the V-D-J junctional region (1, 2). During recombination, a second level of diversity is provided by the trimming and addition of nucleotides (N-additions)1 at the junctions (3). As a result of the huge diversity of TCRs created, one peptide antigen activates only a small number of T cells bearing specific receptors. However, several groups of microorganisms have developed a way to activate large numbers of T cells. They produce superantigens, potent stimulators of T cells, that form contacts with lateral surfaces of both the MHC and TCR. In superantigen recognition, the complexity of the TCR is ignored and interaction occurs between the Vβ region and the superantigen. In this way, a single superantigen can activate >10% of all T cells (4–6).
Superantigens can be produced by bacteria, viruses, and even plants (7–9). Their biological role remains unclear, except for several mouse mammary tumor virus (MMTV) superantigens that are essential during the viral life cycle. By activating T and B cells in neonatal mice, MMTV superantigens form a pool of dividing cells that amplify the viral load and allow viral persistance until maturation of the main target site, the mammary tissue. The endogenous form of MMTV superantigen, acquired by integration of the viral gene into the mouse genome, has the opposite role. Mice expressing endogenous superantigens are protected from viral infection because of deletion of the superantigen-reactive T cell Vβ subsets in early development (10, 11). Other superantigens also cause proliferation and expansion of T cells with a responding Vβ phenotype, often followed by deletion of the targeted subset. It has been hypothesized that nonspecific stimulation of large numbers of T cells by superantigens could include self-reactive T cells and lead to autoimmunity (5). A recent report suggests involvement of a human endogenous retrovirus (HERV-K)– encoded superantigen in induction of autoimmune diabetes (12). Unlike MMTV-encoded superantigens that are products of the 3′ LTR, this superantigen seems to be encoded by the env gene. It activates the Vβ7+ subset of T cells, which was previously found to be enriched in pancreatic infiltrates of diabetic patients (13).
The superantigen produced by Mycoplasma arthritides (MAM) does not have significant homology to the primary sequence of other known bacterial or viral superantigens (14). Nevertheless, MAM shares several features common to many superantigens, including requisite presentation by MHC class II to responding CD4 and CD8 T cells, lack of restriction of the presenting MHC class II alleles, lack of an antigen processing requirement, and the ability to stimulate T cells expressing specific Vβ genes (for review see reference 15). However, functional differences between MAM and other superantigens have been noted. MAM is a relatively weak mitogen for polyclonal human T cells, but it is an efficient stimulator of B cells and induces IgM production better than do staphylococcal superantigens (16). It has been suggested that the poor mitogenic activity of MAM reflects a lower frequency of MAM-reactive T cells among peripheral lymphocytes compared with other superantigens (16, 17).
Here we demonstrate that the MAM represents a new type of ligand for the TCR, intermediate between superantigens and conventional peptide antigens, because it forms contacts both within the Vβ region and the CDR3-β region of the TCR.
Materials and Methods
Human T Cell Clones.
CD4+ clones were obtained after stimulation of fresh PBL with MAM by limiting dilution in 96-well plates. Every 2 wk clones were stimulated by sodium periodate–treated non-T PBL as previously described (18) and maintained in RPMI 1640 with 10% FCS and 5% IL-2 (Pharmacia Biotech, Piscataway, NJ).
Proliferative Response.
2 × 104 T cells and 1.5 × 104 irradiated (15,000 rads) lymphoblastoid 8866P cells were incubated with 10-fold dilutions of superantigens (from 1 to 10−5 ng/ml) in triplicate microwells for 48 h, then labeled overnight with [3H]-thymidine and harvested. Reactivity was scored as cpm greater than twofold over background. Superantigens were purchased from Toxin Technology, (Sarasota, FL); MAM, purified to homogeneity, was provided by Dr. B.C. Cole (University of Utah School of Medicine, Salt Lake City, UT) and prepared according to reference 19.
TCR Sequencing.
cDNAs obtained from human T cell clones were amplified with primers specific for human Vβ families (18) and Vα families (20), cloned into a pCRII vector (Invitrogen Corp., Carlsbad, CA) and sequenced by the Sequenase 2 method (United States Biochemical, Cleveland, OH). Expression of Vβ 5.1, Vβ 8.1/2, Vβ12.2, and Vβ17.1 was confirmed by staining with anti-Vβ–specific monoclonal antibodies (21).
Transfection of TCR β Chains.
The retroviral cassette pTbeta was constructed by subcloning a SnabI-SalI fragment of mouse Cβ2 gene into the pBabe Puro vector (22). PCR products containing the leader sequence, Vβ region, N, Dβ, and Jβ were derived from T cell clones N17 and J17. Mutants were generated by overlapping extension PCR (23) cloned into pTbeta and transfected into the packaging cell line BOSC by the calcium phosphate method (24). TCR β chain–deficient T hybridoma cell lines DS 23.27 (mouse Vα2; reference 25) and YLβ- (mouse Vα3.1; reference 26) were infected with each virus, puromycin-selected, and sorted for Vβ17 expression using the C1 monoclonal antibody (21).
IL-2 Production Assay.
Activation of the TCR transfectants was tested according to reference 27. 105 cells in a 100 μl/well were mixed with an equal number of 8866P lymphoblastoid cells. 10-fold dilutions of each superantigen were tested in triplicates. After 24 h, supernatants were frozen at −20°C. Portions of these supernatants were thawed and incubated for 24–30 h with IL-2– dependent HT-2 cells at 4,000 cells/well. Viability of HT-2 cells was evaluated by the MTT (3-(dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide) assay and analyzed on an ELISA reader at 570 nm (28). The amount of IL-2 produced was determined in comparison with the standard curve established with recombinant IL-2 (BRMP units; Becton Dickinson).
Results
MAM Reactivity Is Not Determined by TCR Vβ Alone.
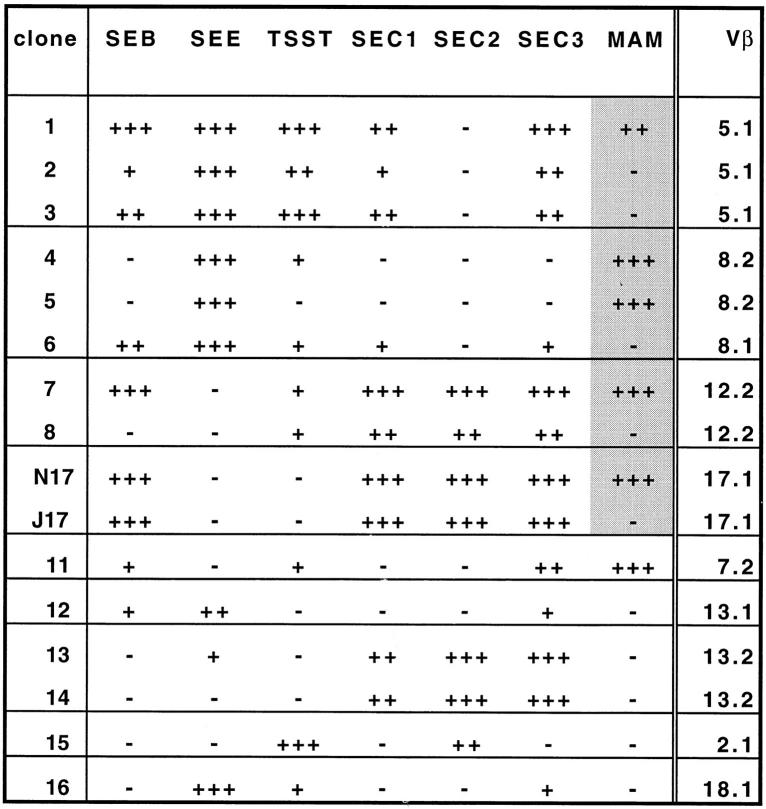
To characterize MAM-reactive TCRs, we tested a panel of 16 human T cell clones for proliferative responses to 7 different superantigens (Table 1, Fig. 1). Clonal T cells were incubated with APCs in the presence of gradual 10-fold dilutions of each superantigen, then pulsed with [3H]-thymidine and harvested. Among T cell clones using the same Vβ genes, similar patterns of reactivity were observed with most superantigens, whereas reactivity to MAM often differed (Fig. 1, shaded field). T cell clones expressing either Vβ5.1, Vβ8, Vβ12, or Vβ17 could respond to MAM, but within each of these Vβ subsets there were both reactive (MAM+) and nonreactive (MAM−) clones. Lack of reactivity to MAM was not dose dependent, as we were unable to find concentrations of MAM that would activate MAM− clones (Table 1). The MAM− cells were competent to respond to TCR-mediated signals from other superantigens (Fig. 1). In addition, the sequences of TCR Vβ regions from MAM+ and MAM− T cell clones were identical, ruling out the possibility that Vβ allelic variants resulted in differences in MAM reactivity.
Table 1.
Proliferative Response of Two Vβ17+ Clones to Several Superantigens
| Concentration | ||||||||
|---|---|---|---|---|---|---|---|---|
| 10−1 | 10−2 | 10−3 | 10−4 | |||||
| ng/ml | ||||||||
| Clone N17 (Vβ17.1)* | ||||||||
| SEB | 28,261 | 19,957 | 11,042 | 6,378 | ||||
| SEE | 1,480 | 1,128 | 1,189 | 1,189 | ||||
| TSST | 2,110 | 1,921 | 1,731 | 1,110 | ||||
| MAM | 13,043 | 16,165 | 10,888 | 3,891 | ||||
| SEC1 | 13,870 | 19,182 | 13,218 | 9,241 | ||||
| SEC2 | 16,884 | 16,204 | 12,381 | 11,922 | ||||
| SEC3 | 16,048 | 18,197 | 13,877 | 8,039 | ||||
| Clone J17 (Vβ 17.1)‡ | ||||||||
| SEB | 23,107 | 20,380 | 19,807 | 2,687 | ||||
| SEE | 555 | 424 | 493 | 420 | ||||
| TSST | 578 | 611 | 576 | 432 | ||||
| MAM | 496 | 455 | 496 | 480 | ||||
| SEC1 | 15,534 | 17,898 | 15,191 | 6,453 | ||||
| SEC2 | 12,958 | 15,322 | 12,419 | 4,035 | ||||
| SEC3 | 15,957 | 20,429 | 14,608 | 2,038 | ||||
Background: 1,239 CPM
Background: 394 CPM Numbers show mean CPM of triplicates. Each T cell clone was tested at least four times. Data on 16 clones are summarized in Figure 1.
Figure 1.
Proliferative response of human T cell clones to bacterial superantigens. The shaded boxes represent examples of clones differing in MAM reactivity despite shared Vβ usage (Vβ5.1, 8, 12.2 and 17.1). Vβ13.1 and 13.2 differ primarily in the HVR4 region, a component of the lateral TCR surface that interacts with superantigens (24, 37). We confirm that TCRs expressing the two closely related Vβ13 gene segments have few superantigen reactivities in common. Key to symbols: −, no reactivity with 1 ng/ml or any concentration; +, reactivity with 10−1 ng/ml or greater; ++, reactivity with 10−2 ng/ml or greater; +++, reactivity with 10−3 ng/ml or greater. See Table 1 for example of raw data.
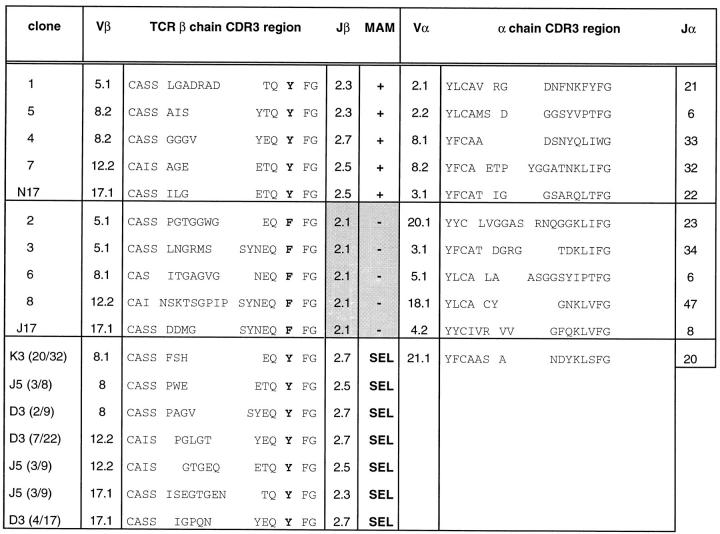
Strikingly, all T cell clones that did not respond to MAM used the TCR Jβ2.1 gene segment, whereas MAM-reactive clones used other Jβ2 segments (Fig. 2). Alignment of the CDR3 regions showed that a tyrosine (Y), present within the conserved motif Q(Y/F)FG, correlated with responsiveness to MAM. Nonreactive clones had phenylalanine (F) at this position. The same correlation was observed in polyclonal cell lines obtained after repeated stimulation of fresh T cells with MAM. Dominant clones within these cultures (Fig. 2; SEL) represent T cells expanded in vitro after stimulation with MAM. All Vβ17 MAM+ and SEL clones have isoleucine at position 94 in addition to Y within the Q(Y/F)FG motif in their CDR3-β region.
Figure 2.
TCR α and β chains from human T cell clones with different MAM reactivity. The sequences represent MAM-reactive clones (+), MAM nonreactive clones (−), and dominant clonal sequences (SEL) obtained from three polyclonal cell lines (K3, J5, D3) that were repeatedly stimulated with MAM, respectively, 3×, 5×, or 3× consecutively at 2-wk invervals. The ratio shown after the name of the cell line indicates the number of identical CDR3-β sequences over the total number of sequences obtained for the indicated Vβ subset. Vα and Vβ nomenclature is according to reference 63, and Jα is according to EMBL/Genbank/ DDBJ accession No. M94081.
Staining data indicated that in the polyclonal cell lines only the Vβ17+ subset was expanded in response to MAM, to ∼50% of all T cells, in agreement with previous data (21). The minor MAM-responsive phenotypes (Vβ5, Vβ8, and Vβ12) were not expanded in these polyclonal cell lines. However, within these minor Vβ subsets, dominant clones were also found after three to five stimulations with MAM. We hypothesize that these clones represented relatively infrequent cells within the Vβ5, Vβ8, or Vβ12 subsets, which may depend on the other TCR elements for MAM reactivity. Consistent with this view, Vα2 and Vα8 were frequently observed in clones of minor Vβ phenotypes (Fig. 2). Thus, reactivity may depend on Vα usage with these minor Vβ phenotypes, as previously described for low affinity TCR Vβ–superantigen interactions (29–31).
However, the major MAM-responsive Vβ17 phenotype is not α chain–dependent. To prove this point, we transfected β chains derived from clones N17 and J17 into two mouse T hybridoma cell lines, DS 23.27 and YLβ-, that are β chain–deficient (Table 2). Surface expression of the endogenous mouse TCR-α chain, either Vα2 or Vα3.1, respectively, is rescued by TCR complex formation with the transfected β chain. β chains N17 and J17 retained their differential MAM reactivity regardless of which α chain they were paired with (Table 2).
Table 2.
TCR α chain Does Not Contribute to the Recognition of MAM by Vβ17+ TCRs
| N17 | J17 | Recipient cell line | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 10 ng/ml | 1 ng/ml | 0.1 ng/ml | 10 ng/ml | 1 ng/ml | 0.1 ng/ml | |||||||||
| SEC2 | 605 | 465 | 216 | 823 | 784 | 167 | DS 23.27 | |||||||
| MAM | 559 | 604 | 452 | 0 | 0 | 0 | DS 23.27 | |||||||
| SEC2 | 550 | 414 | 67 | 590 | 502 | 126 | YLβ- | |||||||
| MAM | 362 | 334 | 43 | 0 | 0 | 0 | YLβ- | |||||||
Numbers indicate IL-2 U/ml produced after stimulation of transfectants with superantigens. Two TCR β chains derived from clones N17 (MAM+) and J17 (MAM−) were expressed in mouse T hybridomas deficient for TCR β chain (DS 23.27, with endogenous mouse Vα2; or YLβ-, with endogenous mouse Vα3.1). Resulting transfectants expressed functional TCRs formed by coupling of their endogenous TCR α chains with N17 or J17 β chains. TCR β chains N17 and J17 remained MAM+ and MAM−, respectively, when paired with the same TCR α chain (either DS or YL).
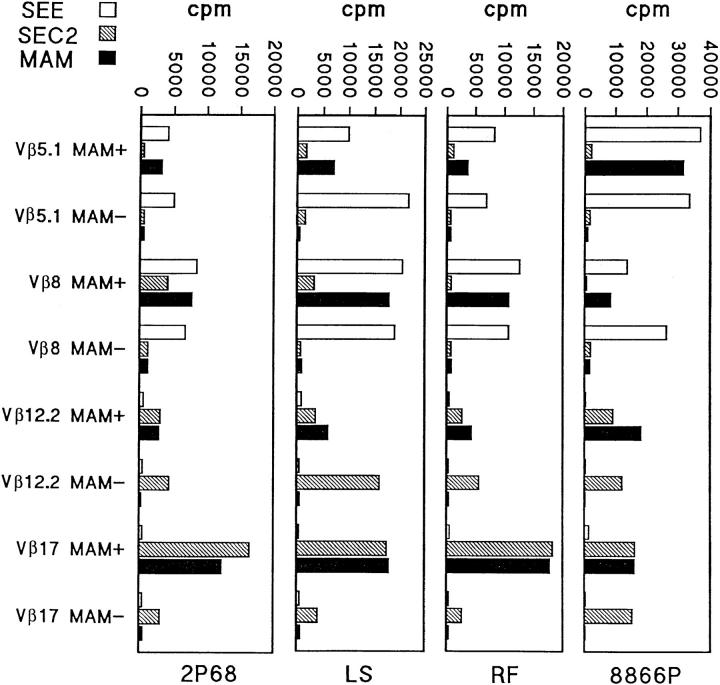
Presentation of superantigens to T cell clones can be sensitive to minor differences among MHC class II alleles (32) and the nature of the peptide in the MHC class II groove (33). Therefore, we tested the reactivity of MAM+ and MAM− clones with three superantigens presented by lymphoblastoid cell lines expressing different MHC class II alleles (Fig. 3). In each of four groups (Vβ17, Vβ8, Vβ12, and Vβ5), MAM+ and MAM− clones retained their specific reactivity regardless of the MHC class II alleles expressed on APCs. The magnitude of the proliferative responses differed from one APC type to another, probably due to different levels of expression of MHC class II or accessory molecules, but the pattern of reactivity remained the same. MAM+ clones were always MAM-reactive and MAM− clones did not respond to MAM presented by other APCs.
Figure 3.
Reactivity to MAM is not MHC-restricted. Four lymphoblastoid cell lines, 2P68 (HLA DR4), LS (HLA DR5,2), RF (HLA DR2,6), and 8866P (HLA DR15), were used for presentation of MAM, SEC2, and SEE to MAM+ and MAM− T cell clones. In each group of T cell clones (Vβ17, Vβ8, Vβ12, and Vβ5) specific reactivity with the tested superantigens was not dependent on the HLA alleles expressed. MAM+ clones remained reactive regardless of the APCs used and MAM− clones remained nonreactive.
TCR-β Chain CDR3 Mutants.
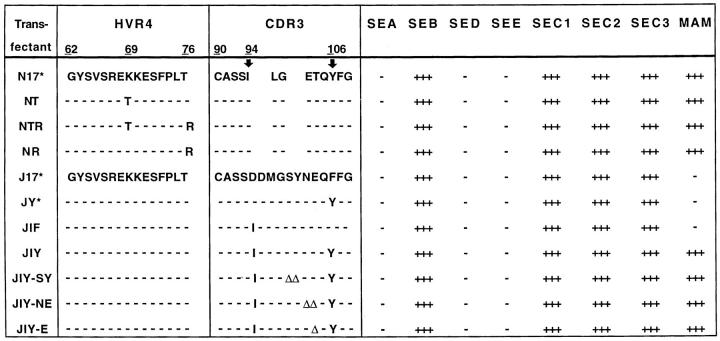
Sequence alignment of TCR-β chains implicated a single residue (Y/F) of the CDR3 region in determining MAM reactivity (Fig. 2). To test whether Y103 of clone N17 (MAM+) and the equivalent residue, F106, of clone J17 (MAM−) determine MAM reactivity, we attempted to reconstitute reactivity of J17 by mutagenesis in the CDR3-β region (Fig. 4). Wild-type and mutant TCRs were expressed in DS23.27 cells and tested for IL-2 production in response to several superantigens. A single amino acid substitution (JY, F106Y) at position 106 was insufficient to restore reactivity to MAM. A second candidate amino acid was isoleucine in position 94. This position is encoded by the Vβ–Db junctional region and in all Vβ17+ clones reactive with MAM, it was represented by isoleucine (Fig. 2). Therefore, we introduced a second substitution at position D94 of J17. Strikingly, the combination of D94I and F106Y (transfectant JIY) resulted in full reconstitution of MAM reactivity (Fig. 4) while the single change, D94I, was insufficient (JIF).
Figure 4.
Mapping of the TCR residues required for reactivity with MAM. Mutations based on alignment between N17 and J17 map the positions involved in interaction with MAM (I and Y). Deletions in the middle of the CDR3 loop (Δ) did not affect reactivity, as well as several mutations in the HVR4 region. Constructs marked with a * were transfected into both DS23.27 (mouse Vα2) and YLβ- (mouse Vα3) cells, and had identical reactivity regardless of the α chain used. 10-fold dilutions of superantigens were tested in IL-2 assays as described in reference 26, (see also Table 2). Reactivity was scored as the minimal concentration of the tested superantigens required for an IL-2 response and the key to the symbols is: +++, 0.1 ng/ml, ++, 1 ng/ml, and +, 10 ng/ml.
To demonstrate that other regions of CDR3 were not involved, we tested three deletion mutants including either one or two residues between I94 and Y106 (JIY-SY, JIY-NE, JIY-E). These mutants would be expected to differ in their peptide/MHC reactivity, because recognition of conventional peptide antigen is extremely sensitive to changes in CDR3 (34, 35). All three deletion mutants retained MAM reactivity (Fig. 4). Together with the data on MAM presentation by different MHC class II alleles (Fig. 3) these experiments exclude a role for MHC II–bound peptides in these MAM–TCR interactions. A direct contact between TCR CDR3-β and MAM is the most likely explanation for dependence on residues I94 and Y106. Based on recent crystallographic data, these two amino acids are located at the base of the CDR3 loop lying in close proximity to one another (36). The side chain of Y106 extends towards the exposed surface of the TCR (Y107 in reference 2). It appears that residues I94 and Y106 are accessible for direct interaction with MAM. These interactions are specific for MAM since none of the tested mutations affected reactivity with staphylococcal superantigens (Fig. 4).
Discussion
CDR3-dependent Superantigen.
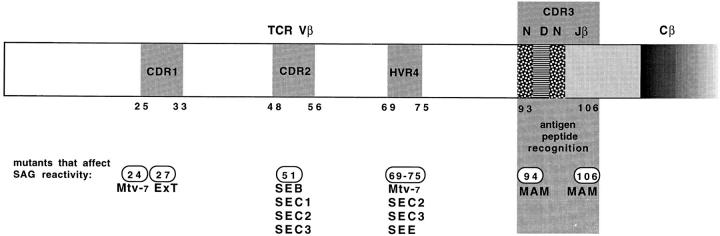
In this study we show that the CDR3 region of TCR-β chain mediates specific recognition of the MAM superantigen. Complementarity-determining regions (CDR loops) (2, 37) represent hypervariable loops of the TCR that face the MHC during antigen recognition. On the β chain, both CDR1 and CDR2 as well as the HVR4 loops can be involved in interactions with superantigens. TCR-β point mutations that affect specific interaction with a particular superantigen are shown in Fig. 5. The cocrystals of TCR β chain with staphylococcal enterotoxins SEC2 and SEC3 revealed that these staphylococcal superantigens interact with CDR1, CDR2, FR3, and HVR4 but not with the CDR3 loop (38). Most contacts (47%) are formed between the superantigen and the CDR2 loop. Since Y106 of CDR3 (2) and exposed CDR2 residues are located at opposing faces of the TCR β chain (1), our data suggest that recognition of MAM and staphylococcal superantigens is substantially different. That CDR2 regions are not involved in MAM recognition is supported by the observation that the primary sequence of the homologous human Vβ17 and mouse Vβ6 chains, both MAM-reactive, differs in the CDR2 region.
Figure 5.
Linear model of TCR-β indicating residues implicated as superantigen (SAG) interaction sites by mutation studies. Typically, superantigens interact with solvent-exposed residues of CDR1, CDR2, and HVR4 encoded by Vβ genes (24, 64–67). The interaction of MAM with TCR-β is clearly distinct, as it interacts both with the Vβ region because of its Vβ specificity (the exact site is not known), and with two residues located at the base of the CDR3-β loop.
Mutational and structural studies have shown that the CDR3 loops of the α and β chains are central in peptide recognition (1, 2, 34, 35, 39, 40). CDR3 regions are the most diverse parts of the TCR produced by extensive processing of end joints during antigen receptor rearrangement (3). Junctional diversity is increased by N-additions contributed by the lymphoid-specific enzyme terminal deoxytransferase (TdT; references 41–43). Although an N-region– depleted TCR repertoire from TdT-knockout mice was shown to be as efficient as normal TCR repertoire (44), the N-region-depleted TCRs were more promiscuous in peptide recognition. These TCRs recognized a significantly larger number of different peptides than did the wild-type TCRs when tested for reactivity against a library of random peptides (45).
The structural basis for the role of N-encoded TCR residues was revealed in a recently solved structure of the TCR–MHC interface (1). In this crystal, three interactions are formed between residue p5 of the peptide and the TCR. Two of these are contacts of p5 with N-encoded residues of the Vα–Dα and Vβ–Dβ junctional regions.
The specific interaction of MAM with the TCR appears to imitate the natural process of peptide/MHC recognition. During peptide recognition, CDR3 regions of α and β chains form contacts with MHC. The complex is stabilized by CDR3–peptide interactions (46, 47). Recognition of MAM is clearly Vβ-dependent, as is the case with other superantigens, but it appears to be stabilized by additional contacts with the CDR3-β region.
In all MAM-reactive Vβ17+ clones that we have sequenced, position 94 was represented by isoleucine. I94 is adjacent to the conserved end of Vβ region CASS and corresponds to position 2 of the CDR3 region (48, 49). In random CDR3 sequences, position 2 is rarely represented by isoleucine (49) but in Vβ17+ sequences I94 is more common, occurring in ∼20% of the sequences (50). This residue is often assumed to be part of the N-region, but in the case of isoleucine it may be encoded by the 3′ end of the germline Vβ17 sequence, codon ATA of the genomic sequence (51). However, in ∼80% of the cases this codon is removed during Vβ–Dβ recombination and replaced by N-additions.
The second CDR3-β position critical for interaction with MAM is tyrosine within the Q(Y/F)FG motif of Jβ2 (T cell clones using Jβ1 are rare and they were not analyzed in this study). At this position Y is encoded by Jβ2.3, Jβ2.4, Jβ2.5, and Jβ2.7. F, which does not allow MAM reactivity (Fig. 4), is encoded by Jβ2.1 and Jβ2.2. The Jβ2.1 segment, which was used by the MAM− T cell clones studied here (Fig. 2), is used by ∼40% of all T cells from peripheral blood (52).
The major subset of T cells targeted by MAM is the Vβ17 subset (21). Yet not all Vβ17+ T cells respond to MAM, and the response is limited to cells expressing two appropriate residues at the base of the CDR3-β loop. This suggests that T cell activation by MAM is dependent on TCR junctional diversity, thus limiting the number of potential MAM-reactive T cell clones. The frequency of T cells responding to this superantigen could be significantly lower than to other superantigens (for example SEB, SEC1, SEC2, and SEC3). Among the minor MAM-responsive TCR phenotypes (Vβ5, Vβ8, and Vβ12), the frequency is even lower than within the Vβ17 subset, probably because Vα usage is an additional requirement as suggested by the data in Fig. 2. As previously shown, MAM-responsive T cells expressing Vβ5, Vβ8, and Vβ12 can only be detected by testing individual clones and these Vβ phenotypes do not expand in polyclonal cultures exposed to MAM (18). However, TCRs of the minor phenotype also appear to use the Jβ-encoded tyrosine for MAM recognition (Fig. 2).
It is possible that Jβ usage can affect low affinity interactions of TCR with other superantigens. Incomplete deletion of Vβ8+ or Vβ6+ cells in mice carrying Mls-1a (Mtv-7) revealed that TCRs that escaped deletion used distinct Jβ regions (53, 54). In this experimental system, Jβ1.2 seemed to protect Vβ6+ TCR from interaction with a retroviral superantigen, Mls-1a. However, these studies did not find any conserved motifs in the CDR3 region that could be implicated in unresponsiveness to Mls-1a (54). Staphylococcal superantigens seem to be CDR3-independent (Fig. 4 and reference 55). However, in the case of SEB and Urtica dioica superantigens, it was suggested that the Jβ segment, but not the CDR3 region, can affect superantigen binding by influencing the quaternary structure of the TCR-β chain (55).
The major MAM-responsive Vβ phenotype is strikingly dependent on discrete residues of CDR3. This fact clearly distinguishes MAM from other superantigens. Thus, recognition of MAM is dependent on junctional diversity of the TCR β chain.
Autoimmunity.
Mycoplasma arthritides is a microorganism that causes a disease in rodents that is remarkably similar to human rheumatoid arthritis (RA). It is also the only mycoplasma that produces MAM. It should be emphasized that Mycoplasma arthritides is not a human pathogen. Surprisingly, preferentially expanded T cell clonotypes found in human RA often belong to the MAM-reactive TCR phenotype. Dominant clones from RA patients, characterized in three independent studies, were found to use Vβ17 followed by isoleucine at position 94 (50, 56, 57). In one of the studies, these T cells were shown to be autoreactive against a lymphoblastoid cell line expressing HLA-DR4 (50). Similar autoreactivity was reported in a TCR transgenic mouse model of RA (58). Inflammatory sinovitis in mice was triggered by the transgenic TCR recognition of host MHC class II (Ag7). This transgenic TCR-β chain used a mVβ6 followed by isoleucine (CASSI) (D. Mathis, personal communication). Murine Vβ6 is the closest homologue of human Vβ17, and mouse Vβ6+ T cells are also MAM-reactive (15). Thus, both murine and human TCRs associated with RA appear to express a conserved isoleucine at the COOH-terminal end of the Vβ region. The role of the conserved isoleucine in the CDR3-β region of arthritogenic TCRs remains to be determined. Is it necessary for interaction with MHC class II or a putative joint-specific peptide? Is it possible that tolerized self-reactive clones can be reactivated by MAM in mice infected by Mycoplasma arthritides?
There is a strong genetic link between certain DR alleles (class II MHC) and RA in humans (59, 60). Position 71 of the particular DR4 or DR1 β chains, a lysine residue, is associated with susceptibility for RA (61). This residue determines the nature of the peptides in the class II groove by interacting with P4/P5 of the peptide (for review see reference 62). Conserved CDR3-β motifs that include isoleucine 94 have been proposed as evidence for antigen-driven immune response in RA (50). Whether superantigens like MAM have a role in RA pathogenesis is still an open question.
Acknowledgments
We are grateful to N. Bhardwaj for her clones of human T cells, B.C. Cole for purified MAM protein, J.P. Morgenstern for the pBabe vector, W.S. Pear for the BOSC cell line, and D. Kostyu for HLA typing. We also thank G. Kelsoe, A. Chervonsky, and S. Santagata for discussion of the manuscript.
This work was supported by an Aaron Diamond Foundation fellowship to A.S. Hodtsev and by National Institutes of Health grants to D.N. Posnett (AI-31140 and AI-33322) and to Y. Choi (CA-59751).
Footnotes
Abbreviations used in this paper: MAM, Mycoplasma arthritidis mitogen; MMTV, mouse mammary tumor virus; SE, staphylococcal enterotoxin.
References
- 1.Garboczi DN, Ghosh P, Ultz U, Fan QR, Biddison WE, Wiley DC. Structure of the complex between human T-cell receptor, viral peptide and HLA-A2. Nature. 1996;384:134–141. doi: 10.1038/384134a0. [DOI] [PubMed] [Google Scholar]
- 2.Garcia KC, Degano M, Stanfield RL, Brunmark A, Jackson MR, Peterson PA, Teyton L, Wilson IA. An αβ T cell receptor structure at 2.5 A and its orientation in the TCR–MHC complex. Science. 1996;274:209–219. doi: 10.1126/science.274.5285.209. [DOI] [PubMed] [Google Scholar]
- 3.Schatz DG, Oettinger MA, Schlissel MS. V(D)J recombination: molecular biology and regulation. Annu Rev Immunol. 1992;10:359–383. doi: 10.1146/annurev.iy.10.040192.002043. [DOI] [PubMed] [Google Scholar]
- 4.Janeway CA, Yagi J, Conrad PC, Katz ME, Jones B, Vroegop S, Buxser S. T-cell responses to Mls and to bacterial proteins that mimic its behavior. Immunol Rev. 1989;107:61–88. doi: 10.1111/j.1600-065x.1989.tb00003.x. [DOI] [PubMed] [Google Scholar]
- 5.Marrack P, Kappler J. The staphylococcal enterotoxins and their relatives. Science. 1990;248:705–711. doi: 10.1126/science.2185544. [DOI] [PubMed] [Google Scholar]
- 6.Acha-Orbea H, MacDonald HR. Superantigens of mouse mammary tumor virus. Annu Rev Immunol. 1995;13:459–486. doi: 10.1146/annurev.iy.13.040195.002331. [DOI] [PubMed] [Google Scholar]
- 7.Choi Y, Kappler JW, Marrack P. A superantigen encoded in the open reading frame of the 3′ long terminal repeat of mouse mammary tumour virus. Nature. 1991;350:203–207. doi: 10.1038/350203a0. [DOI] [PubMed] [Google Scholar]
- 8.Lafon M, Lafage M, Martinez-Arends A, Ramirez R, Vuillier F, Charron D, Lotteau V, Scott-Algara D. Evidence in humans of a viral superantigen. Nature. 1992;358:507–510. doi: 10.1038/358507a0. [DOI] [PubMed] [Google Scholar]
- 9.Galelli A, Delcourt M, Wagner MC, Peumans W, Truffa-Bachi P. Selective expansion followed by profound deletion of mature Vβ 8.3+ cells in vivo after exposure to the superantigenic lectin Urtica dioicaagglutinin. J Immunol. 1995;54:2600–2611. [PubMed] [Google Scholar]
- 10.Golovkina T, Chervonsky AV, Dudley JP, Ross SR. Transgenic mouse mammary tumor virus superantigen expression prevents viral infection. Cell. 1992;69:637–645. doi: 10.1016/0092-8674(92)90227-4. [DOI] [PubMed] [Google Scholar]
- 11.Held W, Waanders GA, Shakhov AN, Scarpellino L, Acha-Orbea H, MacDonald HR. Superantigen-induced immune stimulation amplifies mouse mammary tumor virus infection and allows virus transmission. Cell. 1993;74:529–540. doi: 10.1016/0092-8674(93)80054-i. [DOI] [PubMed] [Google Scholar]
- 12.Conrad B, Weidmann E, Trucco G, Rudert WA, Behboo R, Ricordi C, Rodriques-Rilo H, Finegold D, Trucco M. Evidence for superantigen involvement in insulin-dependent diabetes mellitus aetiology. Nature. 1994;371:351–355. doi: 10.1038/371351a0. [DOI] [PubMed] [Google Scholar]
- 13.Conrad B, Weissmahr RN, Boni J, Arcari R, Schupbach J, Mach B. A human endogenous retroviral superantigen as candidate autoimmune gene in type I diabetes. Cell. 1997;90:303–313. doi: 10.1016/s0092-8674(00)80338-4. [DOI] [PubMed] [Google Scholar]
- 14.Cole BC, Knudtson KL, Oliphant A, Sawitzke AD, Pole A, Manohar M, Benson LS, Ahmed E, Atkin CL. The sequence of the Mycoplasma arthritidissuperantigen, MAM: identification of functional domains and comparison with microbial superantigens and plant lectin mitogens. J Exp Med. 1996;183:1105–1110. doi: 10.1084/jem.183.3.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cole B, Atkin C. The Mycoplasma arthritidisT-cell mitogen, MAM: a model superantigen. Immunol Today. 1991;12:271–276. doi: 10.1016/0167-5699(91)90125-D. [DOI] [PubMed] [Google Scholar]
- 16.Crow MK, Zagon G, Chu Z, Ravina B, Tumang JR, Cole BC, Friedman SM. Human B cell differentiation induced by microbial superantigens: unselected peripheral blood lymphocytes secrete polyclonal immunoglobulin in response to Mycoplasma arthritidesmitogen. Autoimmunity. 1992;14:23–32. doi: 10.3109/08916939309077353. [DOI] [PubMed] [Google Scholar]
- 17.Matthes N, Schrezenmeier H, Hemfeld J, Fleischer S, Malissen B, Kirchner H, Fleischer B. Clonal analysis of human cell activation by the Mycoplasma arthritidismitogen (MAS) Eur J Immunol. 1988;18:1733–1737. doi: 10.1002/eji.1830181112. [DOI] [PubMed] [Google Scholar]
- 18.Bhardwaj N, Hodtsev AS, Nisanian A, Kabak S, Friedman SM, Cole BC, Posnett DN. Human T-cell responses to Mycoplasma arthritidis-derived superantigen. Infect Immun. 1994;62:135–144. doi: 10.1128/iai.62.1.135-144.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Atkin CL, Wei S, Cole BC. The Mycoplasma arthritidissuperantigen MAM: purification and identification of an active peptide. Infect Immun. 1994;62:5367–5375. doi: 10.1128/iai.62.12.5367-5375.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Genevee C, Diu A, Nierat J, Caignard A, Dietrich PY, Ferradini L, Roman-Roman S, Triebel F, Hercend T. An experimentally validated panel of subfamily-specific oligonucleotide primers (Vα1-w29/Vβ1-w24) for the study of human T cell receptor variable V gene segment usage by polymerase chain reaction. Eur J Immunol. 1992;22:1261–1269. doi: 10.1002/eji.1830220522. [DOI] [PubMed] [Google Scholar]
- 21.Friedman SM, Crow MK, Tumang JR, Tumang M, Xu Y, Hodtsev AS, Cole BC, Posnett DN. Characterisation of human T cells reactive with the Mycoplasma arthritidis-derived superantigen (MAM): generation of a monoclonal antibody against Vβ17, the T cell receptor gene product expressed by a large fraction of MAM-reactive human T cells. J Exp Med. 1991;174:891–900. doi: 10.1084/jem.174.4.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morgenstern JP, Land H. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 24.Pear WS, Nolan GP, Scott ML, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi Y, Herman A, DiGusto D, Wade T, Marrack P, Kappler J. Residues of the variable region of the T-cell-receptor β-chain that interact with S. aureustoxin superantigens. Nature. 1990;346:471–473. doi: 10.1038/346471a0. [DOI] [PubMed] [Google Scholar]
- 26.Saito H, Kranz DM, Takagaki Y, Hayday AC, Eisen H, Tonegawa S. A third rearranged and expressed gene in a clone of cytotoxic T lymphocytes. Nature. 1984;312:36–40. doi: 10.1038/312036a0. [DOI] [PubMed] [Google Scholar]
- 27.Kubo RT, Born W, Kappler JW, Pigeon M. Characterization of a monoclonal antibody which detects all murine αβ T cell receptors. J Immunol. 1989;142:2736–2742. [Google Scholar]
- 28.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 29.Vacchio MS, Kanagawa O, Tomonari K, Hodes RJ. Influence of T cell receptor Vα expression on Mlsasuperantigen-specific T cell responses. J Exp Med. 1992;175:1405–1408. doi: 10.1084/jem.175.5.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Waanders GA, Lussow AR, Macdonald HR. Skewed T cell receptor Vα repertoire among superantigen reactive murine T cells. Int Immunol. 1993;5:55–60. doi: 10.1093/intimm/5.1.55. [DOI] [PubMed] [Google Scholar]
- 31.Woodland DL, Blackman MA. How do T-cell receptors, MHC molecules and superantigens get together? . Immunol Today. 1993;14:208–212. doi: 10.1016/0167-5699(93)90164-G. [DOI] [PubMed] [Google Scholar]
- 32.Herman A, Croteau G, Sekaly RP, Kappler J, Marrack P. HLA-DR alleles differ in their ability to present staphylococcal enterotoxins to T cells. J Exp Med. 1990;172:709–718. doi: 10.1084/jem.172.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thibodeau J, Cloutier I, Lavoie PM, Labrecque N, Mourad W, Jardetzky T, Sekaly RP. Subsets of HLA-DR1 molecules defined by SEB and TSST-1 binding. Science. 1994;266:1874–1878. doi: 10.1126/science.7997881. [DOI] [PubMed] [Google Scholar]
- 34.Engel I, Hedrick SM. Site-directed mutations in the VDJ junctional region of a T cell receptor β chain cause changes in antigenic peptide recognition. Cell. 1988;54:473–484. doi: 10.1016/0092-8674(88)90068-2. [DOI] [PubMed] [Google Scholar]
- 35.Jorgensen JL, Esser U, Fazekas de St B, Groth, Reay PA, Davis MM. Mapping T-cell receptor–peptide contacts by variant peptide immunization of single-chain transgenics. Nature. 1992;355:224–230. doi: 10.1038/355224a0. [DOI] [PubMed] [Google Scholar]
- 36.Bentley GA, Boulot G, Karjalainen K, Mariuzza RA. Crystal structure of the β chain of a T cell antigen receptor. Science. 1995;267:1984–1987. doi: 10.1126/science.7701320. [DOI] [PubMed] [Google Scholar]
- 37.Chothia C, Boswell DR, Lesk AM. The outline structure of the T cell αβ receptor. EMBO (Eur Mol Biol Organ) J. 1988;12:3745–3755. doi: 10.1002/j.1460-2075.1988.tb03258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fields BA, Malchiodi EL, Li H, Ysern X, Stauffacher CV, Schlievert PM, Karjalainen K, Mariuzza RA. Crystal structure of a T-cell receptor β-chain complexed with a superantigen. Nature. 1996;384:188–192. doi: 10.1038/384188a0. [DOI] [PubMed] [Google Scholar]
- 39.Danska JS, Livingstone AM, Paragas V, Ishihara T, Fathman CG. The presumptive CDR3 regions of both T cell receptor α and β chains determine T cell specificity for myoglobin peptides. J Exp Med. 1990;172:27–33. doi: 10.1084/jem.172.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hsu BL, Donermeyer DL, Allen PM. TCR recognition of the Hb(64–76)/I-Ekdeterminant. Single conservative amino acid changes in the complementarity-determining region 3 dramatically alter antigen fine specificity. J Immunol. 1996;157:2291–2298. [PubMed] [Google Scholar]
- 41.Bogue M, Candeias S, Benoist C, Mathis D. A special repertoire of a α:β T cells in neonatal mice. EMBO (Eur Mol Biol Organ) J. 1991;10:3647–3654. doi: 10.1002/j.1460-2075.1991.tb04931.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gilfillan S, Dietrich A, Lemeur M, Benoist C, Mathis D. Mice lacking TdT: mature animals with an immature lymphocyte repertoire. Science. 1993;261:1175–1178. doi: 10.1126/science.8356452. [DOI] [PubMed] [Google Scholar]
- 43.Komori T, Okado A, Stewart V, Alt F. Lack of N regions in antigen receptor variable region genes of TdT-deficient lymphocytes. Science. 1993;261:1171–1175. doi: 10.1126/science.8356451. [DOI] [PubMed] [Google Scholar]
- 44.Gilfillan S, Bachmann M, Trembleau S, Adorini L, Kalinke U, Zinkernagel R, Benoist C, Mathis D. Efficient immune responses in mice lacking N-region diversity. Eur J Immunol. 1995;25:3115–3122. doi: 10.1002/eji.1830251119. [DOI] [PubMed] [Google Scholar]
- 45.Gavin MA, Bevan MJ. Increased peptide promiscuity provides a rationale for the lack of N regions in the neonatal T cell repertoire. Immunity. 1995;3:793–800. doi: 10.1016/1074-7613(95)90068-3. [DOI] [PubMed] [Google Scholar]
- 46.Alam SM, Travers PJ, Wung JL, Nasholds W, Redpath S, Jameson SC, Gascoigne NR. T-cell-receptor affinity and thymocyte positive selection. Nature. 1996;381:616–620. doi: 10.1038/381616a0. [DOI] [PubMed] [Google Scholar]
- 47.Reich Z, Boniface JJ, Lyons DS, Borochov N, Wachtel EJ, Davis MM. Ligand-specific oligomerization of T-cell receptor molecules. Nature. 1997;387:617–620. doi: 10.1038/42500. [DOI] [PubMed] [Google Scholar]
- 48.Candéias S, Waltzinger C, Benoist C, Mathis D. The Vβ17+ T cell repertoire: skewed Jβ usage after thymic selection; dissimilar CDR3s in CD4+ versus CD8+cells. J Exp Med. 1991;174:989–1000. doi: 10.1084/jem.174.5.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moss PAH, Bell JI. Sequence analysis of the human αβ T-cell receptor CDR3 region. Immunogenetics. 1995;42:10–18. doi: 10.1007/BF00164982. [DOI] [PubMed] [Google Scholar]
- 50.Li Y, Sun GR, Tumang J, Crow MK, Friedman SM. CDR3 sequence motifs shared by oligoclonal rheumatoid arthritis synovial T cells. Evidence for an antigen-driven response. J Clin Invest. 1994;94:2525–2531. doi: 10.1172/JCI117624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li Y, Sun G-R, Zheng Q, Yoo DH, Bhardwaj N, Posnett DN, Crow MK, Friedman SM. Allelic variants of human TCR BV17S1 defined by restriction fragment length polymorphism, single strand conformation polymorphism and amplification refractory mutation system analyses. Hum Immunol. 1996;49:8585–8595. doi: 10.1016/0198-8859(96)00062-6. [DOI] [PubMed] [Google Scholar]
- 52.Grunewald J, Jeddi-Tehrani M, Pisa E, Janson CH, Anderson R, Wigzell H. Analysis of Jβ gene segment usage by CD4+ and CD8+human peripheral blood T lymphocytes. Int Immunol. 1992;4:643–650. doi: 10.1093/intimm/4.6.643. [DOI] [PubMed] [Google Scholar]
- 53.Woodland DL, Smith HP, Surman S, Le P, Wen R, Blackman MA. Major histocompatibility complex–specific recognition of Mls-1 is mediated by multiple elements of the T cell receptor. J Exp Med. 1993;177:433–442. doi: 10.1084/jem.177.2.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chies JAB, Marodon G, Joret AM, Regnault A, Lembezat MP, Rocha B, Freitas AA. Persistence of Vβ6+T cells in Mls-1a mice. J Immunol. 1995;155:4171–4178. [PubMed] [Google Scholar]
- 55.Musette P, Galelli A, Truffa-Bachi P, Peumans W, Kourilsky P, Gachelin G. The Jβ segment of the T cell receptor contributes to the Vβ-specific T cell expansion caused by staphylococcal enterotoxin B and Urtica dioicasuperantigens. Eur J Immunol. 1996;26:618–622. doi: 10.1002/eji.1830260317. [DOI] [PubMed] [Google Scholar]
- 56.Howell MD, Diveley JP, Lundeen KA, Esty A, Winters ST, Carlo DJ, Brostoff SW. Limited T-cell receptor β-chain heterogeneity among interleukin 2 receptor–positive synovial T cells suggests a role for superantigen in rheumatoid arthritis. Proc Natl Acad Sci USA. 1991;88:10921–10925. doi: 10.1073/pnas.88.23.10921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lim A, Toubert A, Pannetier C, Dougados M, Charron D, Kourilsky P, Even J. Spread of clonal T-cell expansions in rheumatoid arthritis patients. Hum Immunol. 1996;48:77–83. doi: 10.1016/0198-8859(96)00089-4. [DOI] [PubMed] [Google Scholar]
- 58.Kouskoff V, Korganow A-S, Duchatelle V, Degott C, Benoist C, Mathis D. Organ-specific disease provoked by systemic autoimmunity. Cell. 1996;87:811–822. doi: 10.1016/s0092-8674(00)81989-3. [DOI] [PubMed] [Google Scholar]
- 59.Gregersen PK, Silver J, Winchester RJ. The shared epitope hypothesis: an approach to understanding the molecular genetics of susceptibility to rheumatoid arthritis. Arthritis Rheum. 1987;30:1205–1213. doi: 10.1002/art.1780301102. [DOI] [PubMed] [Google Scholar]
- 60.Nepom GT, Erlich H. MHC class-II molecules and autoimmunity. Annu Rev Immunol. 1991;9:493–525. doi: 10.1146/annurev.iy.09.040191.002425. [DOI] [PubMed] [Google Scholar]
- 61.Weyand CM, McCarthy TG, Goronzy JJ. Correlation between disease phenotype and genetic heterogeneity in rheumatoid arthritis. J Clin Invest. 1995;95:2120–2126. doi: 10.1172/JCI117900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wucherpfennig KW, Strominger JL. Selective binding of self peptides to disease-associated major histocompatibility complex (MHC) molecules: a mechanism for MHC-linked susceptibility to human autoimmune diseases. J Exp Med. 1995;181:1597–601. doi: 10.1084/jem.181.5.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Arden B, Clark SP, Kabelitz D, Mak TW. Human T-cell receptor variable gene segment families. Immunogenetics. 1995;42:455–500. doi: 10.1007/BF00172176. [DOI] [PubMed] [Google Scholar]
- 64.Pullen AM, Wade T, Marrack P, Kappler JW. Identification of the region of T cell receptor β chain that interacts with the self-superantigen Mls-1a. Cell. 1990;61:1365–1374. doi: 10.1016/0092-8674(90)90700-o. [DOI] [PubMed] [Google Scholar]
- 65.Bellio M, Lone YC, Calle-Martin O, Malissen B, Abastado JP, Kourilsky P. The Vβ complementarity determining region 1 of a major histocompatibility complex (MHC) class I–restricted T cell receptor is involved in the recognition of peptide/MHC I and superantigen/MHC II complex. J Exp Med. 1994;179:1087–1097. doi: 10.1084/jem.179.4.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liao L, Marinescu A, Molano A, Ciurli C, Secaly R-P, Fraser JD, Popowicz A, Posnett DN. TCR binding differs for a bacterial superantigen (SEE) and a viral superantigen (Mtv-9) J Exp Med. 1996;184:1–12. doi: 10.1084/jem.184.4.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hong S-C, Waterbury G, Janeway CA. Different superantigens interact with distinct sites in the Vβ domain of a single T cell receptor. J Exp Med. 1996;183:1437–1446. doi: 10.1084/jem.183.4.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]