Abstract
The orphan receptor CRF2-4 is a member of the class II cytokine receptor family (CRF2), which includes the interferon receptors, the interleukin (IL) 10 receptor, and tissue factor. CRFB4, the gene encoding CRF2-4, is located within a gene cluster on human chromosome 21 that comprises three interferon receptor subunits. To elucidate the role of CRF2-4, we disrupted the CRFB4 gene in mice by means of homologous recombination. Mice lacking CRF2-4 show no overt abnormalities, grow normally, and are fertile. CRF2-4 deficient cells are normally responsive to type I and type II interferons, but lack responsiveness to IL-10. By ∼12 wk of age, the majority of mutant mice raised in a conventional facility developed a chronic colitis and splenomegaly. Thus, CRFB4 mutant mice recapitulate the phenotype of IL-10–deficient mice. These findings suggest that CRF2-4 is essential for IL-10–mediated effects and is a subunit of the IL-10 receptor.
The class II cytokine receptor family (CRF2)1 has been defined on the basis of significant sequence and structural resemblance, notably evolutionary links to fibronectin type III within domains of the extracellular receptor portion of ∼100 amino acids (1). The family includes tissue factor (TF; 2, 3), a ligand-binding subunit of the IL-10 receptor (IL-10R1; 4, 5), two subunits of the IFN-γ receptor (IFN-γR1 and IFN-γR2; 6–8), two subunits of the IFN-α receptor (IFN-αR1 and IFNAR2; 9–11), and the orphan receptor CRF2-4 (12, 13). CRF2 members are characterized by the presence of a 200–amino acid extracellular domain composed of two distinct subdomains of 100 amino acids (1). These subdomains contain conserved cysteine, proline, and tryptophan residues that determine a characteristic folding pattern of seven beta strands similar to the constant domain of immunoglobulins (14). Family members also contain conserved intracellular domain regions that are involved in the interaction with downstream signaling molecules (15).
Full length CRFB4 (the gene that encodes CRF2-4) cDNA was cloned based on an expressed sequence tag corresponding to the D21S58 locus on human chromosome 21 that maps close to IFNAR1 (12, 16). The genes encoding IFN-αR2, CRF2-4, IFN-αR1, and IFN-γR2 form a cluster on human chromosome 21 (11). Human CRFB4 cDNA was also independently cloned through exon trapping and used as a probe to isolate a murine homologue that encodes a 349–amino acid polypeptide that is 69% identical to the 325-residue human counterpart (13). CRFB4 maps on chromosome 16 to a region of conserved synteny with human chromosome 21 (16). This clustering and the similarity to IFN receptor genes (17) suggested that CRF2-4 may be a component of IFN receptors, but coexpression of human CRF2-4 with human IFN-αR1 and/or human IFN-αR2 in a mouse L929 cell line did not affect the responsiveness of such cells to human IFNs (Gibbs, V.C., unpublished data).
Whereas functional type I and type II IFN receptors have been reconstituted from their known subunits (14, 18), there is evidence to suggest that other CRF2 members may need additional receptor subunits to transduce biological responses.
TF is a high affinity receptor for plasma factor VII/VIIa and is responsible for cellular initiation of the coagulation protease cascade (2). There is evidence to suggest that TF that has only a 20–amino acid cytoplasmic domain may participate in intracellular signaling through association with an unknown coreceptor. Thus, inactivation of the TF gene in the mouse resulted in early embryonic lethality, presumably due to anomalies in blood vessel formation (19, 20). There is some biochemical evidence to suggest that, in monocytes, TF may associate with a component of the IgE receptor (21), but TF-mediated signaling pathways remain elusive.
IL-10R1 is able to bind IL-10 and signal a biological response, but there is evidence to suggest that a coreceptor is involved. Intriguingly, human IL-10 proved more active on mouse Ba/F3 cells transfected with the murine IL-10R1 than on the same cells expressing the human IL-10R1 (5), suggesting that these murine cells may express a species-specific coreceptor. Characterization of an EBV-derived IL-10 homologous protein (22) provided further evidence for the requirement of additional receptor components for signaling. Notably, a soluble form of human IL-10R1 neutralized biologic responses only to human, but not to EBV-derived, IL-10 (5). Lastly, IL-10 treatment of T cells and monocytes triggers activation of the Jak1 and Tyk2 kinases (23, 24). The known receptor subunit coprecipitated only with Jak1, suggesting that Tyk2 may associate with a second receptor subunit (23, 24). Using hybrid IFN-γR2/CRF2-4 molecules, it has recently been shown that CRF2-4 associates with Tyk2 (25), and it was proposed that CRF2-4 may be a subunit of the IL-10R (26).
To clarify whether CRF2-4 may be a coreceptor of other CRF2 members, we generated mice with an inactivated CRFB4 gene.
Materials and Methods
Generation of CRFB4− /− Embryonic Stem Cells and Mice.
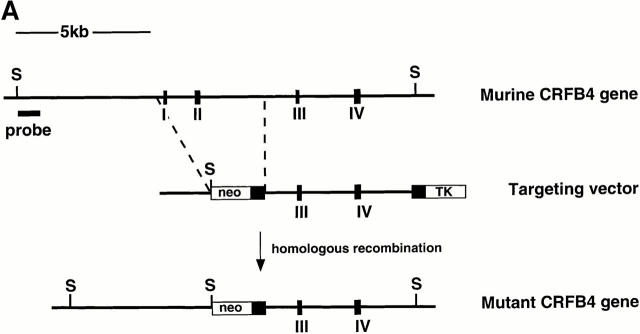
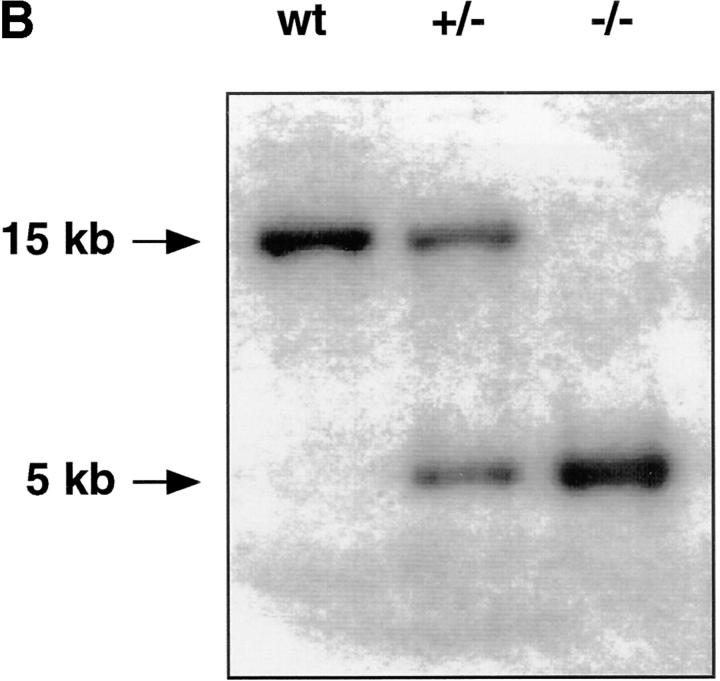
A 32P-labeled oligonucleotide derived from exon II of murine CRFB4 was used to screen an isogenic 129Sv/Ev embryonic stem (ES) cell library (27). A clone containing an 18-kb gene fragment encoding exons I–V of CRFB4 was identified and subcloned into a Bluescript vector as four SacI fragments. To generate the CRFB4 targeting vector, a 5.0-kb ClaI fragment containing exons III and IV was placed into the ClaI site of the pTK.neo.ums vector (28) such that its 3′ end was adjacent to the polyoma enhancer promoter-driven herpes simplex virus thymidine kinase gene. The 5′ end of the targeting construct was generated by inserting a 2.0-kb EcoRI fragment containing sequences upstream of exon I into the blunted NotI site of the pTK.neo.ums (see Fig. 1). ES-D3 mouse ES cells were cultured in high glucose DME with 10% FCS and leukemia inhibitory factor (LIF) on mouse embryonic feeder cells. ES cells were electroporated with 10 μg of SstI linearized CRFB4 targeting vector per 107 cells and grown under double selection in 400 μg/ml G-418 and 1 μM gancyclovir (Roche Labs., Nutley, NJ). Cells were analyzed for homologous recombination by Southern blot analysis as previously described (29). Genomic DNA was digested with SpeI and the disrupted allele was detected by hybridization with a 32P-labeled 1.3-kb XbaI fragment derived from genomic sequences upstream of the targeting vector. 3 positive clones were identified by the presence of a 5-kb SpeI fragment in addition to the 15-kb SpeI band typical of the CRFB4 wild-type allele. To generate ES cells with a homozygous mutation of the CRFB4 gene, ES cells carrying the CRFB4+/− mutation were cultured in high concentrations of G-418 as described (30). CRFB4+/− ES cells were injected into C57Bl/6 blastocysts and resulting male chimeras mated to C57Bl/6 females. CRFB4+/− offspring were intercrossed to generate homozygous mutant mice. Mutant mice were kept in conventional animal facilities.
Figure 1.
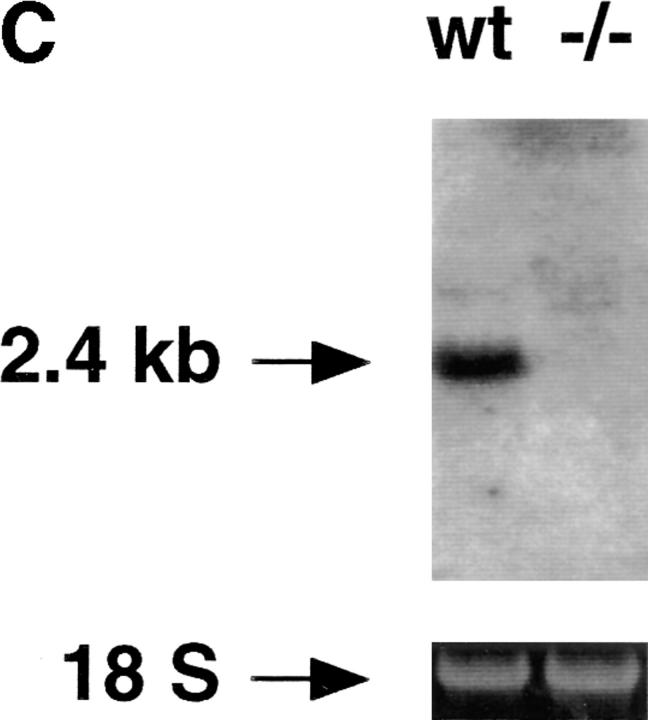
(A) The structure of the murine gene was predicted from the published human gene (17). Exons (black boxes) are numbered I–IV. The targeting vector was designed to replace exons I and II with the neomycin resistance gene. Homologous insertion of the targeting vector resulted in the introduction of an additional SpeI site (S). (B) Southern blot analysis of SpeI-digested ES cell DNA. Probe: 1.3-kb XbaI fragment derived from genomic sequences upstream of the targeting vector. (C) Northern blot of total RNA derived from wild-type or CRFB4−/− ES cells probed with a 1 kb XbaI-HindIII random labeled fragment derived from murine CRFB4 cDNA.
Macrophage Cell Culture and IL-10 Bioassay.
Biological activity of IL-10 was determined by its ability to inhibit LPS-stimulated TNF-α production in macrophages isolated from bone marrow as previously described (31, 32). In brief, bone marrow– derived macrophages were obtained by culturing 107 bone marrow cells in 50 ml of high glucose DME containing 20% inactivated horse serum, 4 mM l-glutamine, 1 mM sodium pyruvate, 50 U/ml of penicillin, and 50 μg/ml of streptomycin, and supplemented with 30% (v/v) L929-conditioned medium in bacterial culture 10 mm Petri dishes for 7 d. Adherent cells were reseeded in 96-well plates at a density of 5 × 104 cells/well. IL-10 was added at various concentrations and cells were incubated for 1 h at 37°C before the addition of 50 ng LPS. Incubation was continued for 24 h at 37°C, and culture supernatants were assayed for TNF-α activity as previously described (33).
Activation of IFN-inducible Genes and Northern Blot Analysis.
Wild-type and CRFB4−/− ES cell lines were allowed to differentiate by growth on gelatin in the absence of LIF for 10 d. Differentiated ES cells were treated for 4 h with 1 μg/ml of murine IFN-γ or 1,000 U/ml of recombinant human IFN-α2/α1, which is cross-reactive on mouse cells (34). Northern analysis was performed on 10 μg total RNA loaded per lane. Probes were as follows: a 1.9-kb EcoRI fragment encoding murine IFN regulatory factor 1 (35); a 1.2-kb PCR fragment containing nucleotide 51– 1240 of murine PKR (double stranded RNA-dependent protein kinase) (36), derived by reverse transcriptase PCR from RNA of IFN-treated L929 cells; a 0.9-kb fragment containing nucleotide 71–949 of murine 2,5-A oligoadenylate synthetase (37), derived by reverse transcriptase PCR from RNA of IFN-treated L929 cells; a 1 kb XbaI-HindIII fragment derived from plasmid pRK.muCRFB4 containing the open reading frame of CRF2-4; and a 0.5-kb XhoII fragment of the GAPDH (glutaraldehyde 3-phosphate dehydrogenase) gene (38).
Flow Cytometry.
Single-cell suspensions from thymocytes, spleen cells, and peritoneal cells were isolated from 15–21-wk-old mice, resuspended in PBS and incubated with the appropriate mAbs, for 1 h at 4°C. In most experiments, splenocytes and peritoneal cells were preincubated with purified rat IgG anti–mouse CD16/CD32 mAb (Fc Block; PharMingen, San Diego, CA) that blocked the nonspecific adherence of subsequent antibodies to murine Fc receptors. The following mAbs were purchased from PharMingen, except where designated, and were used in flow cytometric analysis: biotin-conjugated anti-CD90.2 (Thy-1.2; clone 53-2.1); FITC-conjugated anti-CD4 (clone YTS 177.9; Serotec Ltd., Kidlington, Oxford, UK); PE-conjugated anti-CD8a (Ly-2; clone 53-6.7); biotin-conjugated anti-CD19 (clone 1D3); FITC-conjugated anti-CD45R/B220 (clone RA3-6B2); biotin-conjugated anti-IgM (clone R6-60.2); FITC-conjugated anti-IgD (clone 11-26c.2a); FITC-conjugated anti-CD11b (Mac 1; clone M1/70; Serotec Ltd.); PE-conjugated anti-CD5 (Ly-1; clone 53-7.3); PE-conjugated anti-CD24 (heat stable antigen; clone M1/69); anti-H-2Kb (clone E121.46; Seikagaku Corporation, Rockville, MD); and PE-conjugated anti-I-a (clone MRC OX-3; Serotec Ltd.). Biotinylated mAbs were detected in a second staining step with streptavidin-PE or streptavidin-FITC. Unconjugated mAbs were detected with PE- or FITC-labeled goat anti–mouse IgG antisera (Caltag Antibodies, South San Francisco, CA). Cells were analyzed with a FACScan® cytometer and Cellquest software (Becton Dickinson, San Jose, CA). Dead cells were excluded by propidium iodide staining. Cells present in the lymphocyte or monocyte gate as defined by forward light scatter were analyzed. In stimulation experiments, thymocytes or splenocytes were isolated and cultured for 20 h in microtiter plates in the presence or absence of various concentrations of IFN-α, IFN-γ, IL-4, IL-10, or LPS. Cells were harvested and washed once with PBS and then incubated with the indicated antibodies and analyzed by flow cytometry.
Histopathology.
Mice were killed by asphyxiation and complete necropsy was performed. Terminal body weight, absolute organ weights, and organ to body weight ratios were determined. Blood samples collected in EDTA were analyzed on a Technicon H1E Analyzer (Bayer Diagnostics Division, Tarrytown, NY) according to the manufacturer's instructions. Tissues were fixed with buffered 1% glutaraldehyde, 4% paraformaldehyde, then embedded in plastic and 2-μm-thick sections were stained with Giemsa.
Results
Generation of CRF2-4–deficient ES Cells and Mice.
The CRFB4 gene was inactivated by transfecting ES cells with a replacement vector containing 7 kb of the murine gene (Fig. 1 A). Homologous integration resulted in the replacement with the neomycin resistance gene of exons I and II of the CRFB4 gene, which encode the first 39 amino acid residues of the protein, including the signal peptide. The structure of the mutated CRFB4 gene in targeted ES cells and in the progeny of germline chimeras was identified by Southern blot analysis (Fig. 1 B). Homologous recombination occurred with a frequency of 1 in 180 G-418 and Gancyclovir-resistant ES cell colonies. Homozygous mutant clones were obtained in high G-418 concentrations as previously described (30). Northern blot analysis confirmed the disruption of the CRFB4 gene as evidenced by the absence of the 2.4-kb CRF2-4 mRNA (Fig. 1 C). Homozygous CRFB4 mutant mice were obtained from three independent heterozygous ES cell clones with a Mendelian frequency, proved fertile, and had no apparent phenotypic anomalies by 12 mo of age. Gross anatomical examination revealed no phenotypic differences except for macroscopic signs of colitis and splenomegaly in a majority of mutant mice. CRFB4−/− mice housed in a conventional facility were fertile but had poor reproductive success, presumably due to disease development (see below).
CRFB4− /− Cells Respond Normally to IFN-α and IFN-γ.
To address the possibility that CRF2-4 may have a role in interferon signaling, the biological responses of CRFB4−/− ES cells to IFN-α and IFN-γ were examined. As ES cells constitutively express some IFN-dependent genes, such as IRF-1 (39), CRFB4−/− and control ES cells were cultured for 10 d in the absence of LIF on gelatin-coated plates to induce fibroblastoid differentiation and were then either left untreated or incubated with IFN-α or IFN-γ. Total RNA was isolated and subjected to Northern blot analysis using probes derived from several IFN-inducible genes as indicated (Fig. 2, A–C). No significant differences were observed in the responses of CRFB4−/− versus wild-type cells. Next, we examined the ability of IFN-α and IFN-γ to upregulate expression of MHC class I antigen on thymocytes. Cells from either normal or mutant mice had comparable basal MHC class I antigen levels and responded to a similar extent to either IFN-α or IFN-γ (data not shown).
Figure 2.
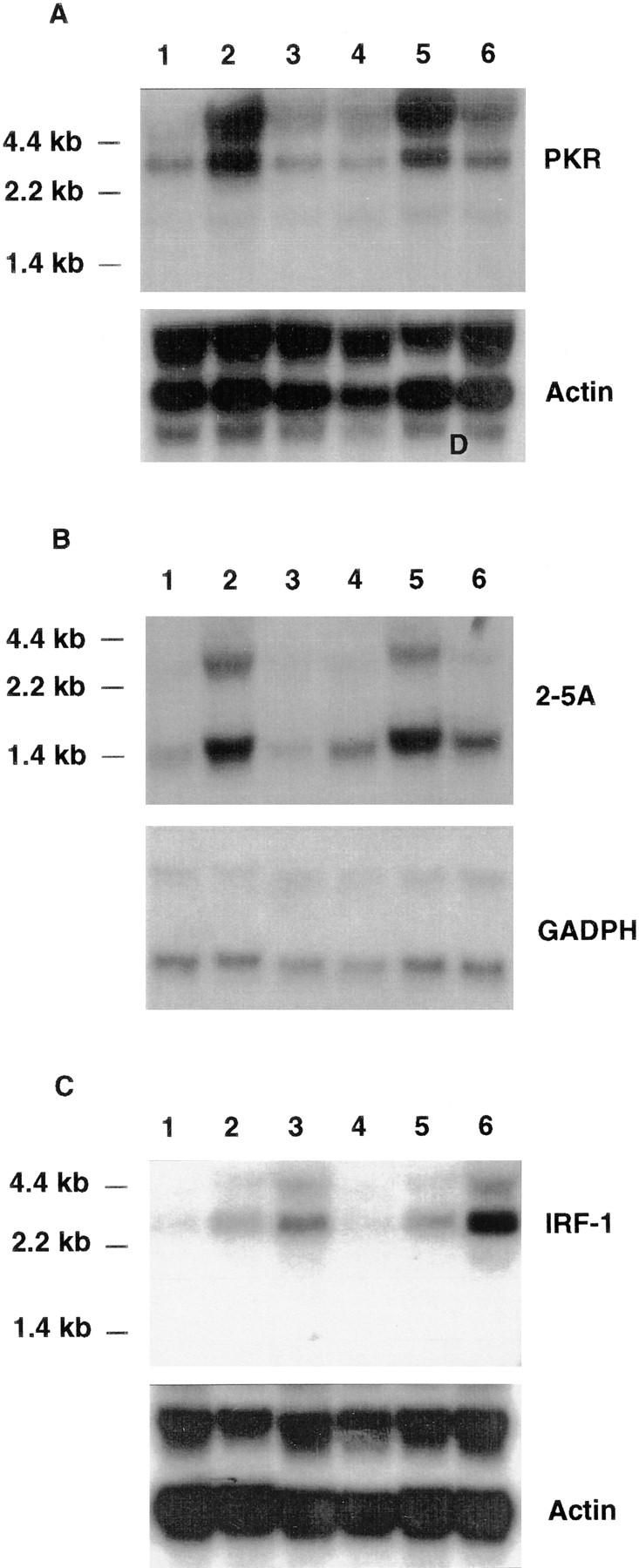
Northern blot analysis of IFN-inducible genes. Wild-type (lanes 1–3) or CRFB4−/− ES cells (lanes 4–6) were cultured under differentiating conditions and treated for 4 h at 37°C with 1,000 U/ml IFN-α2/α1 (lanes 2 and 5), 1 μg/ml muIFNγ (lanes 3 and 6; Genentech, Inc., South San Francisco, CA) or left untreated (lanes 1 and 4). Northern blot analysis was carried out with 10 μg of total RNA per lane and blots were hybridized as indicated with the probes described in Materials and Methods.
Macrophages and Lymphocytes from CRFB4−/− Mice are Unresponsive to IL-10.
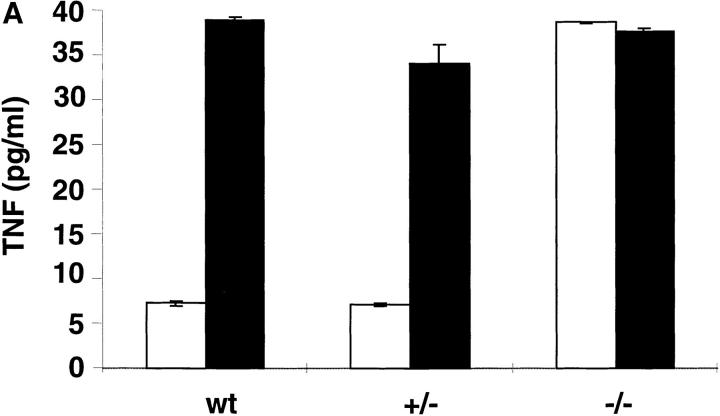
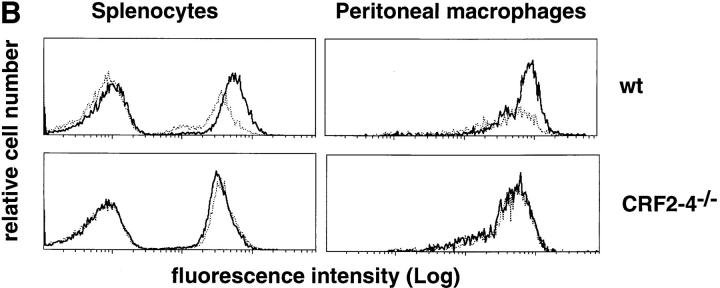
To assess whether the IL-10 system was affected by the lack of CRF2-4, IL-10 responsiveness was assayed on bone marrow–derived macrophages as previously described (31, 32). The ability of IL-10 to inhibit LPS-stimulated production of TNF-α (31) was lost in bone marrow–derived macrophages from CRFB4−/− mice (Fig. 3 A). Likewise, the ability of IL-10 to induce FcγIII/II receptor expression on lymphocytes and monocytes (40) was abolished in cells derived from CRFB4−/− mice (Fig. 3 B).
Figure 3.
Macrophages and splenocytes derived from CRFB4−/− mice are unresponsive to IL-10. (A) TNF production in LPS-activated macrophages. Bone marrow macrophages from individual wild-type (wt), heterozygous (+/−), or homozygous mutant mice (−/−) were pretreated with 100 ng of IL-10 (white bars) or left untreated (black bars) and subsequently stimulated with 50 ng/ml of LPS before determination of TNF secretion. Indicated values are means ± SD of duplicate wells from two independent experiments. (B) 105 splenocytes or 5 × 104 peritoneal monocytes from wild-type or CRF2-4–deficient mice were treated with 10 ng of IL-10 (bold solid line) or left untreated (dotted line) for 20 h at 37°C. Cell surface expression of FcγIII/II receptor was determined by flow cytometry with anti-FcγIII/II mAb as described in Materials and Methods. The first peak of cells in the splenocyte population represents cells that do not express FcγIII/II receptors.
Cytofluorometry.
To address the possibility that inactivation of the CRFB4 gene may affect other immune functions, thymocytes, splenocytes, and peritoneal cells from unchallenged 13–21-wk-old mutant and control mice were subjected to cytofluorimetric analysis. At these time points the T cell compartment in the thymus and spleen was similar in mutant and wild-type mice. Therefore, CRF2-4 is not essential for the generation of CD4+ and CD8+ T cells (data not shown). CRFB4−/− mice consistently had increased numbers of splenocytes, due to an expansion of CD11b, Mac1-positive polymorphonuclear, monocytic cells. IgM/IgD-expressing B cells and CD5+ (Ly-1) B cells were similar in mutant and control mice. There were slightly fewer MHC class II positive B cells in CRFB4−/− mice than in control mice, but IL-4 treatment of splenocytes increased expression of MHC class II antigens in all CD19+ cells to a similar extent in both the wild-type and CRFB4−/− cells. Serum IgG, IgM, and IgA, was not significantly different in wild-type and mutant mice (data not shown). Peritoneal cells in CRFB4−/− mice were also composed of a higher percentage of CD11b+ cells than in wild-type littermates.
CRFB4− /− Mice Develop Splenomegaly and Colitis.
Histopathological examination was performed on two groups of mice. The brain, heart, kidney, thymus, pancreas, and reproductive organs from CRF−/− mice that had been kept in a conventional animal facility were grossly and histologically normal. In contrast, marked alterations were present in the large intestine, spleen, and the hematopoietic system. There was thickening and narrowing of the large intestine in CRFB4−/− mice relative to wild-type littermate controls, and there was a fourfold increase in the spleen/body weight ratio. The splenomegaly was correlated with a dramatic increase in extramedullary hematopoiesis in the splenic red pulp involving erythroid, myeloid, and megakaryocytic lineages. There was mesenteric lymphadenopathy with an increase in plasma cells in the medullary cords and increased granulocytopoiesis in the sternal bone marrow. These changes were reflected in the peripheral hematology, which showed a slightly decreased hemoglobin concentration and a leukocytosis composed primarily of neutrophils and monocytes. Platelet counts were normal. The microscopic findings in the large intestine consisted of moderate to severe diffuse chronic colitis characterized by a mixed inflammatory cell mucosal inflammation (Fig. 4). The colonic mucosa was diffusely thickened by the inflammatory cell infiltrate, and in severe cases also by hyperplasia of the crypt epithelium. There was no involvement of the small intestine. Immunohistochemistry on the large intestine demonstrated that the inflammatory cell infiltrate was composed predominately of macrophages (histiocytes), neutrophils, and T lymphocytes. The histopathological changes of chronic colitis and splenomegaly in CRFB4−/− mice were variable. In an examination of 4 15–20-wk-old CRFB4−/− mice raised in the colony in Switzerland, only two of the mice demonstrated this phenotype. Among 9 13–32-wk-old CRFB4−/− mice examined from the colony in the United States, 7 animals had splenomegaly and colitis. It is unclear whether these differences represent solely environmental differences or whether they reflect genetic heterogeneity (129Sv/Ev × C57Bl/6 background).
Figure 4.
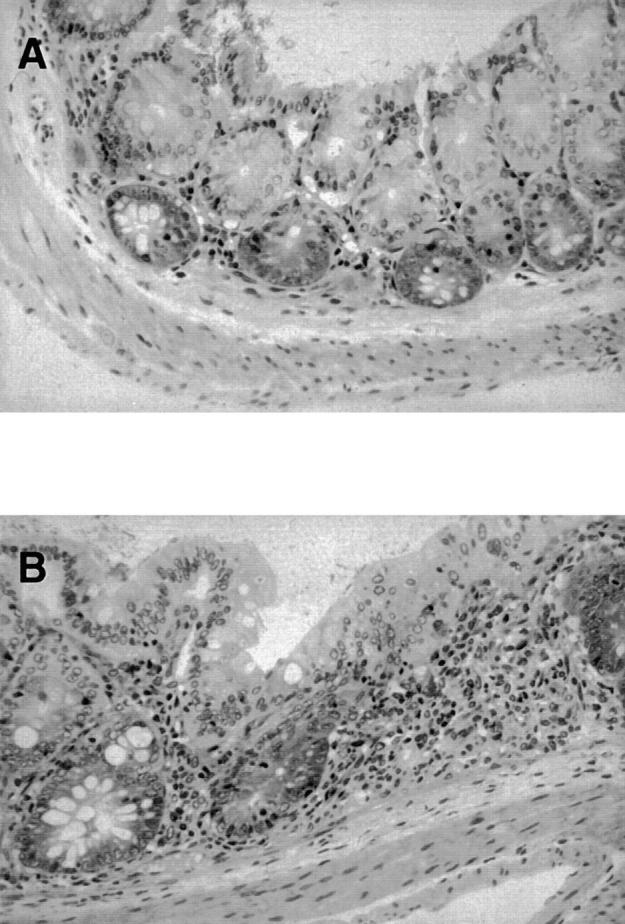
Histopathology of the large bowel from CRFB4−/− mice and control wild-type littermates. In contrast to controls showing a normal histology (A), the colon of mutant mice contains foci of colitis with mucosal inflammatory cell infiltrates composed of neutrophils, monocytes, macrophages, and lymphocytes (B). Original magnification: ×450.
Discussion
We report the phenotypic analysis of mice lacking the orphan receptor CRF2-4. This study was initiated by the observation that the CRFB4 gene is located within a gene cluster comprising the genes encoding three known subunits of IFN receptors. Therefore, before the second subunits of the IFN-γ (7, 8) and the type I IFN receptor (10, 11) were known, CRF2-4 was considered a candidate subunit of IFN receptors. The discovery of these IFN receptor components allowed it to reconstitute functional IFN receptors, and the role of CRF2-4 remained elusive.
The detailed elucidation of the events that link the ligation of the IFN-γR to the Jak–STAT signaling pathway (18) provides a paradigm for other cytokine receptors. The IFN-γR consists of two subunits that presumably form a heterotetramer upon receptor ligation. The ligand-binding subunit (IFN-γR1) confers signal specificity through binding of the signal transducer Stat1and associates with the tyrosine kinase Jak1. The second obligatory subunit (IFN-γR2) recruits Jak-2 into the ligated complex (18). The cytoplasmic domain of R2 can be replaced with the cytoplasmic domain of CRF2-4 (25) and still form a functional receptor, confirming that the Jak family kinases are redundant in terms of signaling specificity, and indicating that CRF2-4, like the IFN-γR1, may function as a coreceptor.
Clearly, CRF2-4 is not an obligatory component of the IFN system. Wild-type ES cells cultured under conditions that induce differentiation are responsive to both type I and type II IFNs. ES cells that lack CRF2-4 showed normal responses to both type I and type II IFNs (Fig. 2). Likewise, splenocytes from wild-type and mutant mice showed comparable type I and type II IFN-mediated induction of MHC class I and II antigens, respectively (data not shown). The absence of any overt phenotype in mutant mice was in contrast to the severe phenotype observed in mice that lack TF (19, 20). Thus, the phenotypic analysis of mice lacking CRF2-4 focused on the IL-10 receptor system as the last known member of the CRF2 (4).
Some recent findings pointed to a potential role of CRF2-4 as a component of the IL-10R; CRF2-4 associates with Tyk-2 (25), and aside from the type I IFN receptor (41) the IL-10R is the only CRF2 member that activates Tyk2 (23, 24). Most recently, IL-10R1 was shown to associate with CRF2-4 to form a functional receptor (26). Our study unequivocally identifies CRF2-4 as an obligatory element of the IL-10 system. Bone marrow–derived macrophages isolated from mice lacking CRF2-4 were unresponsive to IL-10, which normally inhibits LPS-stimulated production of TNF in those cells (Fig. 3 A). Likewise, splenocytes and peritoneal cells derived from mutant mice were unable to upregulate FcγIII/II receptors in response to IL-10 (Fig. 3 B). Lastly, mice lacking CRF2-4 developed an inflammatory bowel disease reminiscent of the one seen in mice lacking IL-10 (42, 43; Fig. 4).
IL-10 is produced by activated Th2 cells, Ly-1 B cells, monocytes, macrophages, thymocytes, and keratinocytes. It has pleiotropic effects on a number of different cell types but in general is notable for its suppressive effects upon cellular activation. For example, one action of IL-10 is to suppress the effector functions of macrophages and Th1 cells by blocking their ability to produce cytokines such as IFN-γ, TNF-α, and IL-2 (40). IL-10–deficient mice raised in a conventional facility were growth retarded, anemic, and suffered from chronic enterocolitis (42). Mutants kept under specific pathogen-free conditions had attenuated phenotypes with a bowel disease limited to the proximal colon, but still had an iron deficiency anemia, probably reflective of a degree of malabsorption and of chronic blood loss through the diseased bowel. The chronic intestinal inflammation seen in IL-10–deficient mice was interpreted as a disorder in the normal control mechanisms that ordinarily downregulate responses to mucosal antigens.
Approximately 60% of CRFB4−/− mice kept under conventional conditions developed moderate to severe colitis without small intestinal involvement, and none developed severe anemia. There was a slightly lower hemoglobin concentration in the CRFB4−/− mice but without any corresponding changes in other red cell values (data not shown). IL-10–deficient mice did not develop splenomegaly and had hypoplastic red pulp in the spleen, whereas CRFB4−/− mice typically developed splenomegaly with a hyperproliferative splenic red pulp. Assessing the significance of these phenotypic differences between IL-10– and CRF2-4–deficient mice would require a side by side histopathologic examination of the two mutant strains under matched genetic and environmental conditions.
Functional IL-10 is a homodimer with a tertiary structure similar to that of IFN-γ (44). Binding of IL-10 to cells that express the IL-10R leads to activation of the Jak1 and Tyk2 kinases and to tyrosine phosphorylation of Stat3, which associates with a paired tyrosine domain within the cytoplasmic receptor region (45). The findings presented here showing that CRF2-4 is involved in IL-10 signaling, in conjunction with the report that CRF2-4 can associate with Tyk2 (25), and with a chimeric IL-10R1 (26), suggest that a functional IL-10R complex is composed of the IL-10R1 and CRF2-4, in which IL-10R1 associates with Jak1 and recruits Stat3, and CRF2-4 associates with Tyk2. Thus, the function of CRF2-4 would be similar to the function of the IFN-γR2 chain, whose only role is to recruit the Jak2 kinase into the ligand-assembled receptor complex (46).
Our study raises a number of questions. Since CRF2-4 is expressed in most human tissues except the brain (data not shown), whereas IL-10R1 expression is restricted mainly to human hemopoietic cells and cell lines (5), CRF2-4 may be shared by other receptor systems, in analogy to other cytokine systems (47, 48). Inversely, whether IL-10R1 and CRF2-4 alone are sufficient to mediate IL-10 signaling remains to be clarified. However, we believe the data presented here justify that CRF2-4 be designated IL-10R2.
Footnotes
We thank Kelly Hagler, Laurie Leong, Lorena Cabote, Julio Ramirez, Fatima da Conceicao-Silva for histopathologic and hematologic examinations, and Dr. R.D. Schreiber for advice and suggestions.
Part of this work was supported by National Institutes of Health grant R29 DK-48748 (to V.C. Gibbs).
Address correspondence to Michel Aguet, Swiss Institute for Experimental Cancer Research (ISREC), 155, Ch. des Boveresses, 1066 Epalinges s/Lausanne, Switzerland. Phone: 41-21-692-5918; Fax: 41-21-652-6933; E-mail: michel.aguet@isrec.unil.ch
Abbreviations used in this paper: CRF2, class II cytokine receptor family; ES, embryonic stem; LIF, leukemia inhibitory factor; TF, tissue factor.
References
- 1.Bazan JF. Structural design and molecular evolution of a cytokine receptor superfamily. Proc Natl Acad Sci USA. 1990;87:6934–6938. doi: 10.1073/pnas.87.18.6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morrissey JH, Fakhrai H, Edgington TS. Molecular cloning of the cDNA for tissue factor, the cellular receptor for the initiation of the coagulation protease cascade. Cell. 1987;50:129–135. doi: 10.1016/0092-8674(87)90669-6. [DOI] [PubMed] [Google Scholar]
- 3.Harlos K, Martin D, O'Brien DP, Jones EY, Stuart DI, Polikarpov I, Miller A, Tuddenham E, Boys C. Crystal structure of the extracellular region of human tissue factor. Nature. 1994;370:662–666. doi: 10.1038/370662a0. [DOI] [PubMed] [Google Scholar]
- 4.Ho AS, Liu Y, Khan TA, Hsu DH, Bazan JF, Moore KW. A receptor for interleukin 10 is related to interferon receptors. Proc Natl Acad Sci USA. 1993;90:11267–11271. doi: 10.1073/pnas.90.23.11267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu Y, Wei SH, Ho AS, de Waal R, Malefyt, Moore KW. Expression cloning and characterization of a human IL-10 receptor. J Immunol. 1994;152:1821–1829. [PubMed] [Google Scholar]
- 6.Aguet M, Dembic Z, Merlin G. Molecular cloning and expression of the human interferon-gamma receptor. Cell. 1988;55:273–280. doi: 10.1016/0092-8674(88)90050-5. [DOI] [PubMed] [Google Scholar]
- 7.Hemmi S, Böhni R, Stark G, DiMarco F, Aguet M. A novel member of the interferon receptor family complements functionality of the murine interferon gamma receptor in human cells. Cell. 1994;76:803–810. doi: 10.1016/0092-8674(94)90355-7. [DOI] [PubMed] [Google Scholar]
- 8.Soh J, Donnelly RJ, Kotenko S, Mariano TM, Cook JR, Wang N, Emanuel S, Schwartz B, Miki T, Pestka S. Identification and sequence of an accessory factor required for activation of the human interferon gamma receptor. Cell. 1994;76:793–802. doi: 10.1016/0092-8674(94)90354-9. [DOI] [PubMed] [Google Scholar]
- 9.Uzé G, Lutfalla G, Gresser I. Genetic transfer of a functional human interferon alpha receptor into mouse cells: cloning and expression of its cDNA. Cell. 1990;60:225–234. doi: 10.1016/0092-8674(90)90738-z. [DOI] [PubMed] [Google Scholar]
- 10.Novick D, Cohen B, Rubinstein M. The human interferon alpha/beta receptor: characterization and molecular cloning. Cell. 1994;77:391–400. doi: 10.1016/0092-8674(94)90154-6. [DOI] [PubMed] [Google Scholar]
- 11.Lutfalla G, Holland SJ, Cinato E, Monneron D, Reboul J, Rogers NC, Smith JM, Stark GR, Gardiner K, Mogensen KE, et al. Mutant U5A cells are complemented by an interferon-alpha beta receptor subunit generated by alternative processing of a new member of a cytokine receptor gene cluster. EMBO (Eur Mol Biol Organ) J. 1995;14:5100–5108. doi: 10.1002/j.1460-2075.1995.tb00192.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lutfalla G, Gardiner K, Uzé G. A new member of the cytokine receptor gene family maps on chromosome 21 at less than 35 kb from IFNAR. Genomics. 1993;16:366–373. doi: 10.1006/geno.1993.1199. [DOI] [PubMed] [Google Scholar]
- 13.Gibbs VC, Pennica D. Crf2-4: isolation of cDna clones encoding the human and mouse proteins. Gene. 1997;186:97–101. doi: 10.1016/s0378-1119(96)00690-7. [DOI] [PubMed] [Google Scholar]
- 14.Uzé G, Lutfalla G, Mogensen KE. Alpha and beta interferons and their receptor and their friends and relations. J Interferon Cytokine Res. 1995;15:3–26. doi: 10.1089/jir.1995.15.3. [DOI] [PubMed] [Google Scholar]
- 15.Mullersman JE, Pfeffer LM. A novel cytoplasmic homology domain in interferon receptors. Trends Biochem Sci. 1995;20:55–56. doi: 10.1016/s0968-0004(00)88956-2. [DOI] [PubMed] [Google Scholar]
- 16.Cheng S, Lutfalla G, Uzé G, Chumakov IM, Gardiner K. GART, SON, IFNAR, and CRF2-4 genes cluster on human chromosome 21 and mouse chromosome 16. Mamm Genome. 1993;4:338–342. doi: 10.1007/BF00357094. [DOI] [PubMed] [Google Scholar]
- 17.Lutfalla G, McInnis MG, Antonarakis SE, Uze G. Structure of the human CRFB4 gene: comparison with its IFNAR neighbor. J Mol Evol. 1995;41:338–344. doi: 10.1007/BF00186545. [DOI] [PubMed] [Google Scholar]
- 18.Bach EA, Aguet M, Schreiber RD. The IFN-gamma receptor: a paradigm for cytokine receptor signaling. Annu Rev Immunol. 1997;15:563–591. doi: 10.1146/annurev.immunol.15.1.563. [DOI] [PubMed] [Google Scholar]
- 19.Carmeliet P, Mackman N, Moons L, Luther T, Gressens P, Van Vlaenderen I, Demunck H, Kasper M, Breier G, Evrard P, et al. Role of tissue factor in embryonic blood vessel development. Nature. 1996;383:73–75. doi: 10.1038/383073a0. [DOI] [PubMed] [Google Scholar]
- 20.Toomey JR, Kratzer KE, Lasky NM, Stanton JJ, Broze GJ., Jr Targeted disruption of the murine tissue factor gene results in embryonic lethality. Blood. 1996;88:1583–1587. [PubMed] [Google Scholar]
- 21.Masuda M, Nakamura S, Murakami T, Komiyama Y, Takahashi H. Association of tissue factor with a gamma chain homodimer of the IgE receptor type I In cultured human monocytes. Eur J Immunol. 1996;26:2529–2532. doi: 10.1002/eji.1830261037. [DOI] [PubMed] [Google Scholar]
- 22.Vieira P, de Waal R, Malefyt, Dang MN, Johnson KE, Kastelein R, Fiorentino DF, deVries JE, Roncarolo MG, Mosmann TR, Moore KW. Isolation and expression of human cytokine synthesis inhibitory factor cDNA clones: homology to Epstein-Barr virus open reading frame BCRFI. Proc Natl Acad Sci USA. 1991;88:1172–1176. doi: 10.1073/pnas.88.4.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ho AS, Wei SH, Mui AL, Miyajima A, Moore KW. Functional regions of the mouse interleukin-10 receptor cytoplasmic domain. Mol Cell Biol. 1995;15:5043–5053. doi: 10.1128/mcb.15.9.5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Finbloom DS, Winestock KD. IL-10 induces the tyrosine phosphorylation of Tyk2 and Jak1 and the differential assembly of STAT1 alpha and STAT3 complexes in human T cells and monocytes. J Immunol. 1995;155:1079–1090. [PubMed] [Google Scholar]
- 25.Kotenko SV, Izotova LS, Pollack BP, Muthukumaran G, Paukku K, Silvennoinen O, Ihle JN, Pestka S. Other kinases can substitute for Jak2 in signal transduction by interferon-gamma. J Biol Chem. 1996;271:17174–17182. doi: 10.1074/jbc.271.29.17174. [DOI] [PubMed] [Google Scholar]
- 26.Kotenko SV, Krause CD, Izotova LS, Pollack BP, Pestka S. Identification and functional characterization of a second chain of the interleukin-10 receptor complex. EMBO (Eur Mol Biol Organ) J. 1997;16:5894–5903. doi: 10.1093/emboj/16.19.5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Müller U, Steinhoff U, Reis LF, Hemmi S, Pavlovic J, Zinkernagel RM, Aguet M. Functional role of type I and type II interferons in antiviral defense. Science. 1994;264:1918–1921. doi: 10.1126/science.8009221. [DOI] [PubMed] [Google Scholar]
- 28.Reis LFL, Ruffner H, Stark G, Aguet M, Weissmann C. Mice devoid of interferon regulatory factor 1 (IRF-1) show normal expression of type I interferon genes. EMBO (Eur Mol Biol Organ) J. 1994;13:4798–4806. doi: 10.1002/j.1460-2075.1994.tb06805.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hasty P, Rivera PJ, Chang C, Bradley A. Target frequency and integration pattern for insertion and replacement vectors in embryonic stem cells. Mol Cell Biol. 1991;11:4509–4517. doi: 10.1128/mcb.11.9.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mortensen RM, Conner DA, Chao S, Geisterfer-Lowrance AA, Seidman JG. Production of homozygous mutant ES cells with a single targeting construct. Mol Cell Biol. 1992;12:2391–2395. doi: 10.1128/mcb.12.5.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fiorentino DF, Zlotnik A, Mosmann TR, Howard M, O'Garra A. IL-10 inhibits cytokine production by activated macrophages. J Immunol. 1991;147:3815–3822. [PubMed] [Google Scholar]
- 32.Weber-Nordt RM, Meraz MA, Schreiber RD. Lipopolysaccharide-dependent induction of IL-10 receptor expression on murine fibroblasts. J Immunol. 1994;153:3734–3744. [PubMed] [Google Scholar]
- 33.Sheehan KC, Ruddle NH, Schreiber RD. Generation and characterization of hamster monoclonal antibodies that neutralize murine tumor necrosis factors. J Immunol. 1989;142:3884–3893. [PubMed] [Google Scholar]
- 34.Weber H, Valenzuela D, Lujber G, Gubler M, Weissmann C. Single amino acid changes that render human IFN-alpha 2 biologically active on mouse cells. EMBO (Eur Mol Biol Organ) J. 1987;6:591–598. doi: 10.1002/j.1460-2075.1987.tb04795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miyamoto M, Fujita T, Kimura Y, Maruyama M, Harada H, Sudo Y, Miyata T, Taniguchi T. Regulated expression of a gene encoding a nuclear factor, IRF-1, that specifically binds to IFN-beta gene regulatory elements. Cell. 1988;54:903–913. doi: 10.1016/s0092-8674(88)91307-4. [DOI] [PubMed] [Google Scholar]
- 36.Feng GS, Chong K, Kumar A, Williams BR. Identification of double-stranded RNA-binding domains in the interferon-induced double-stranded RNA-activated p68 kinase. Proc Natl Acad Sci USA. 1992;89:5447–5451. doi: 10.1073/pnas.89.12.5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ichii Y, Fukunaga R, Shiojiri S, Sokawa Y. Mouse 2-5A synthetase cDNA: nucleotide sequence and comparison to human 2-5A synthetase. Nucleic Acids Res. 1986;14:10117. doi: 10.1093/nar/14.24.10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fort, P., L. Marty, M. Piechaczyk, S. el Sabrouty, C. Dani, P. Jeanteur, and J.M. Blanchard. 1985. Various rat adult tissues express only one major mRNA species from the glyceraldehyde-3-phosphate-dehydrogenase multigenic family. Nucleic Acids Res. 13:1431–1442. [DOI] [PMC free article] [PubMed]
- 39.Ruffner H, Reis L, Naf D, Weissmann C. Induction of type-I interferon genes and interferon-inducible genes in embryonal stem cells devoid of interferon regulatory factor-1. Proc Natl Acad Sci USA. 1993;90:11503–11507. doi: 10.1073/pnas.90.24.11503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moore KW, O'Garra A, de Waal R, Malefyt, Vieira P, Mosmann TR. Interleukin-10. Annu Rev Immunol. 1993;11:165–190. doi: 10.1146/annurev.iy.11.040193.001121. [DOI] [PubMed] [Google Scholar]
- 41.Velazquez L, Fellous M, Stark GR, Pellegrini S. A protein tyrosine kinase in the interferon-alpha/beta signaling pathway. Cell. 1992;70:313–322. doi: 10.1016/0092-8674(92)90105-l. [DOI] [PubMed] [Google Scholar]
- 42.Kühn R, Lohler J, Rennick D, Rajewsky K, Muller W. Interleukin-10–deficient mice develop chronic enterocolitis. Cell. 1993;75:263–274. doi: 10.1016/0092-8674(93)80068-p. [DOI] [PubMed] [Google Scholar]
- 43.Davidson NJ, Leach MW, Fort MM, Thompson-Snipes L, Kühn R, Müller W, Berg DJ, Rennick DM. T helper cell 1–type CD4+T cells, but not B cells, mediate colitis in interleukin 10–deficient mice. J Exp Med. 1996;184:241–251. doi: 10.1084/jem.184.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walter MR, Nagabhushan TL. Crystal structure of interleukin 10 reveals an interferon gamma–like fold. Biochemistry. 1995;34:12118–12125. doi: 10.1021/bi00038a004. [DOI] [PubMed] [Google Scholar]
- 45.Weber-Nordt RM, Riley JK, Greenlund AC, Moore KW, Darnell JE, Schreiber BD. Stat-3 recruitment by two distinct ligand-induced, tyrosine-phosphorylated docking sites in the interleukin-10 receptor intracellular domain. J Biol Chem. 1996;271:27954–27961. doi: 10.1074/jbc.271.44.27954. [DOI] [PubMed] [Google Scholar]
- 46.Bach EA, Tanner JW, Marsters S, Ashkenazi A, Aguet M, Shaw AS, Schreiber RD. Ligand-induced assembly and activation of the gamma interferon receptor in intact cells. Mol Cell Biol. 1996;16:3214–3221. doi: 10.1128/mcb.16.6.3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kishimoto T, Taga T, Akira S. Cytokine signal transduction. Cell. 1994;76:253–262. doi: 10.1016/0092-8674(94)90333-6. [DOI] [PubMed] [Google Scholar]
- 48.Leonard WJ. The defective gene in X-linked severe combined immunodeficiency encodes a shared interleukin receptor subunit: implications for cytokine pleiotropy and redundancy. Curr Opin Immunol. 1994;6:631–635. doi: 10.1016/0952-7915(94)90152-x. [DOI] [PubMed] [Google Scholar]