Abstract
Microphthalmia (Mi) is a basic helix-loop-helix–leucine zipper (b-HLH-ZIP) transcription factor implicated in pigmentation, mast cells, and bone development. Two dominant-negative mi alleles (mi/mi and Mior/Mior) in mice cause osteopetrosis. In contrast, osteopetrosis has not been observed in a number of recessive mi alleles, suggesting the existence of Mi protein partners important in osteoclast function. An osteopetrotic rat of unknown genetic defect (mib) has been described whose skeletal sclerosis improves dramatically with age and that is associated with pigmentation defects reminiscent of mouse mi alleles. Here we report that this rat strain harbors a large genomic deletion encompassing the 3′ half of mi including most of the b-HLH-ZIP region. Osteoclasts from these animals lack Mi protein in contrast to wild-type rat, mouse, and human osteoclasts. Mi is not detectable in primary osteoblasts. In addition TFE3, a b-HLH-ZIP transcription factor related to Mi, was found to be expressed in osteoclasts, but not osteoblasts, and to coimmunoprecipitate with Mi. These results demonstrate the existence of members of a family of biochemically related transcription factors that may cooperate to play a central role in osteoclast function and possibly in age-related osteoclast homeostasis.
The osteoclast is a multinucleated giant cell derived from the monocyte/macrophage lineage which forms a ruffled membrane border and secretes a variety of specialized factors, including proteases, phosphatases, and protons which break down bone matrix, releasing mineral ions into the extracellular fluid (1, 2). Osteopetrosis is an abnormality of bone remodeling caused by decreased osteoclastic bone resorption producing skeletal sclerosis and reduced or absent marrow cavities. Several mouse models of osteopetrosis have led to new insights on bone remodeling and osteoclast function in development (3–11). Most, but not all, of these mouse mutants display cell autonomous defects in osteoclast development or function. The op/op (osteopetrotic) mouse contains an inactivating mutation in the M-CSF gene which results in a severe deficiency of macrophages and osteoclasts (10). The fos-null mouse develops osteopetrosis associated with a lack of osteoclasts and an increase in macrophage numbers (4, 5, 12). The src null mouse has multinucleated osteoclasts that do not form a ruffled border and that resorb bone matrix poorly (6, 9, 13). The mi/mi microphthalmia mutant mouse was recognized in the 1940's (14) as an allele that produces severe osteopetrosis associated with ocular and pigmentation defects (7, 8, 15, 16). mi/mi mice contain osteoclasts that harbor a variety of morphologic defects, including diminished formation of mature multinucleate osteoclasts, lack of ruffled membranes, and diminished bone resorption (17).
The gene encoding mouse microphthalmia was cloned in 1993 and found to encode a Myc-related basic helix-loop-helix–leucine zipper1 (b-HLH-ZIP) protein (18, 19). Biochemical studies (20) demonstrated a DNA binding specificity for consensus sequence CA(C/T)(G/A)TG (20) and its capacity to heterodimerize in vitro with three structurally related b-HLH-ZIP factors, TFEB (21), TFE3 (22, 23), and TFEC (24, 25), but not Myc/Max (26), USF, or other b-HLH-ZIP proteins (20). These three dimerization partners (TFEB, TFE3, and TFEC) are less well understood in terms of expression patterns, particularly in osteoclasts, although targeted disruption of TFE3 in lymphocytes impairs B cell activation (27). mi/mi mutant mice display defective eye development (related to pigment cell abnormalities), complete lack of skin melanocytes, deafness (related to absence of pigment cells in the inner ear), severe defects in mast cells, and osteopetrosis. Mutations in human microphthalmia have been detected in the autosomal dominant hereditary deafness and pigmentation condition Waardenburg Syndrome type 2A (28, 29).
Biochemical analysis of a series of mouse microphthalmia mutant alleles identified several dominant-negative mutations that produced corresponding dominant pigmentation defects in heterozygous mice (20, 30). Only two microphthalmia alleles (mi/mi and Mior/Mior) in the mouse (out of ∼15 described) display osteopetrosis, whereas all display defects in pigment cells (7, 16). Interestingly, the two osteopetrotic alleles both contain dominant negative mutations in the basic domain that disrupt DNA contact without perturbing dimerization (20, 31) and affect pigmentation even in heterozygotes. In b-HLH-ZIP proteins, the basic domain contacts DNA through an induced alpha helix while the HLH-ZIP mediates dimerization and positions the basic domain to lie optimally within the major groove of DNA (20, 26, 32–34). A variety of mutations for the pigmentation phenotype have been characterized. Mutations in the basic region which disrupt DNA binding of the protein confer pigmentation defects in a semidominant pattern of inheritance, whereas mutations in the helix-loop-helix or other essential parts of the gene result in the absence of pigmentation in a recessive pattern of inheritance. Osteopetrosis has not previously been recognized for any of the recessive microphthalmia (mi) mutant alleles. This implies that there may be another Mi family member expressed in osteoclasts that could substitute for Mi deficiency. However, basic region mutations in Mi could potentially disable the mutant as well as normal partner protein(s).
In 1989, a spontaneous osteopetrotic mutation in the rat was described in which homozygotes display osteopetrosis at birth, but this defect in bone resorption dramatically improves with age (35). Because of associated small eyes and white coat color, the allele was called microphthalmia blanc (mib), although the nature of the mutation is to date unknown. The mib mutation is autosomal recessive, showing expression in homozygotes (mib/mib) with absence of skin and fur pigmentation, small eyes, and defects of incisor tooth eruption (36). The mib locus has previously been suggested to be different from microphthalmia (mi), incisorless (ia), osteopetrotic (op), or hypodactyly (hd) loci based on phenotypic differences and rough linkage estimates (35). Cielinski and Marks (37, 38) have shown that the mib/mib mutant osteoclasts in newborn animals are decreased in number, do not form a ruffled border, and show decreased levels of mRNA for carbonic anhydrase II and tartrate-resistant ATPase. However, by the fourth postnatal week, the mib/mib mutant osteoclast number, resorptive function, and cell-specific gene expression have returned to normal, and the skeletal sclerosis is significantly reduced (37, 38).
In this paper, we demonstrate that Mi is abundantly expressed in wild-type mouse, rat, and human osteoclasts by immunohistochemistry and Western and Northern blot analyses. Immunohistochemical staining localizes Mi to osteoclast nuclei. Mouse calvarial osteoblasts in primary culture and the ST2 murine osteoblast cell line do not express Mi. Given the phenotypic similarities of the mi/mi mutant mouse and the mib/mib osteopetrotic rat, we have examined the microphthalmia locus in the mib rat and discovered a large deletion in the 3′ half of the mi gene starting in the basic region. The age-dependent remission of osteopetrosis in the mib mutant led us to search for Mi-related transcription factors in osteoclasts. We show here that TFE3 is abundantly expressed by osteoclasts and is found in coimmunoprecipitates with Mi, suggesting that a discrete transcription factor family may play a critical, age-dependent role in osteoclasts.
Materials and Methods
Animals.
Animals used in this study were the offspring of +/mib and mib/mib rats (originally derived from the stock of Dr. Rene Moutier, CNRS, Orleans, France [35] and generously donated by Dr. Sandy Marks, Jr., University of Massachusetts Medical School, Worcester, MA). The animals were bred and maintained at Harvard Medical School in accordance with Harvard guidelines. All animals were given powdered and solid rat food. Homozygous mutant animals (mib/mib) were easily identified by their white coat color and small eyes. +/mib and +/+ (wild-type) rats are indistinguishable phenotypically. +/+ rats are defined as animals that have delivered at least 20 phenotypically normal offspring when mated with mib/mib rats. Primary mouse osteoclast cultures were made using spleen and bone marrow from 6–12-wk-old C57 black mice.
Osteoclast Cultures.
mib/mib and wild-type rat bone marrow from 6-mo-old rats was harvested by flushing cold PBS through the femurs and tibiae with a 22-gauge needle. Rat osteoclast cultures (39) were made using 1.5 × 106 bone marrow cells/ml in MEM containing 10% fetal bovine serum and 10−8 M 1,25 dihydroxyvitamin D3 (1,25-(OH)2 D3; gift of Dr. Uskokovich, Hoffman-LaRoche, Nutley, NJ). Cultures were fed every 3 d by replacing one half of the spent medium with fresh medium. Mouse osteoclast cultures (40, 41) were made using primary mouse calvarial osteoblasts (4 × 104 cells/ml) obtained as previously described (42) and cocultured with spleen from 6–12-wk-old mice (6 × 105 cells/ml) in the above medium. Cultures were fed every 3 d by replacing one half of the old medium with fresh medium. Human osteoclasts cultures were made using 106 cells/ml of Ficoll-fractionated whole normal donor adult (ages 18–60 yr) bone marrow or 2 × 106 cells/ml of Ficoll-fractionated CD34-depleted normal human bone marrow. Bone marrow was cultured in MEM media with 20% horse serum (Hyclone Laboratories, Logan, Utah) and 10−8 M 1,25-(OH)2 D3) (43–45). Cultures were refed every week by changing one half of the media. After 14 d, cultures were refed with the above media supplemented with 10 ng/ml human recombinant M-CSF (R & D Systems, Inc., Minneapolis, MN).
Immunohistochemistry and Tartrate Resistant Acid Phosphatase Staining.
Mi antibodies (46) were generated against the NH2 terminus Taq-Sac fragment of human Mi expressed as a His-fusion and shown not to cross react with other b-HLH-ZIP factors by DNA mobility shift assay (data not shown). Cells were grown on glass chamber slides (Fisher Scientific, Pittsburgh, PA) and were fixed with 3% formaldehyde in PBS for 30 min and washed, followed by 10 min in 1% Triton X-100. After another PBS wash, slides were incubated with 3% H2O2 to remove endogenous peroxidase. The anti-Mi monoclonal antibody 1:10–1:40 or anti-TFE3 monoclonal antibody (PharMingen, San Diego, CA) 1:250–1:500 was added for 1 h. The Vecta-stain Elite kit (Vector Laboratories, Burlingame, CA) was then used for immunohistochemical staining according to the manufacturer's instructions. The diaminobenzidine reagent (Vector Laboratories) was applied for 2–4 min and the slides were analyzed under light microscopy. Tartrate resistant acid phosphatase (TRAP) staining was performed according to the manufacturer's instructions using the Sigma TRAP kit (Sigma Chemical Co., St. Louis, MO). TRAP is an osteoclast marker.
Immunoprecipitation and Western Blotting.
Immunoblots of cell cultures containing osteoclast-like cells were generally preceded by immunoprecipitation that concentrated the antigen and permitted efficient on-plate cell lysis in 1% Triton X-100 detergent with 150 mM NaCl, Tris, pH 7.6, 2 μg/ml aprotinin, 2 μg/ml leupeptin, 200 μg/ml trypsin inhibitor, 500 μg/ml antipain, 10 mM sodium fluoride, 1 mM sodium vanadate, 2 mM PMSF, 20 mM sodium pyrophosphate and 10 μg/ml pepstatin. The soluble fraction was incubated for 1–2 h on ice with appropriate antibodies and washed protein A agarose beads (GIBCO BRL, Gaithersburg, MD) were added. The mixture was rotated at 4°C for a minimum of 12 h. Beads were then washed three times with PBS, resuspended in 2% SDS/1% glycerol and boiled for 5 min, and eluted proteins were resolved on an 8% SDS–polyacrylamide gel (4% SDS–polyacrylamide stacking gel) run at 200 V for 6–8 h. Proteins were transferred to nitrocellulose with methanol–glycine electrotransfer (Bio-Rad Labs., Hercules, CA). The membrane was blocked in 5% milk for 1 h at room temperature or overnight at 4°C. After washing in TBST (10 mM Tris, pH 7.6, 150 mM NaCl, and 0.5% Tween 20), 1:40 dilution of the Mi antibody or 1: 500 dilution of TFE3 antibody or α tubulin 1:500 dilution (Sigma Chemical Co.) was added for 1 h at room temperature. After washing, goat anti–mouse horseradish peroxidase–conjugated antibody (Cappel, West Chester, PA) was added for 40 min. After washing, the enhanced chemiluminescence reaction was performed (Amersham Corp., Arlington Heights, IL).
PCR and Southern Analysis of Genomic DNA.
Genomic DNA was extracted from the tails of mib/mib, +/mib, and +/+ animals using the Puregene kit (Gentra Systems, Plymouth, MN) according to manufacturer's instructions. The genomic PCR reactions used 50 ng of purified genomic DNA under the following conditions: 94°C for 2 min, 57°C for 1 min, and 72°C for 2 min except for exon 9, where an annealing temperature of 60°C for 1 min was used for 30 cycles. For Southern analysis, 10 μg of genomic DNA was digested with each of the following restriction enzymes: Hinc II, Xba I, and Bam HI, using the manufacturer's instructions (New England Biolabs, Beverly, MA). The digested DNA was electrophoresed on a 1% agarose/Tris borate EDTA (TBE) gel. The gel was denatured with 0.25 M HCl followed by 0.5 M NaOH/1.5 M NaCl, and equilibrated with Tris, pH 8.0/1.5 M NaCl. The DNA was transferred to nylon membranes, UV cross-linked, and prehybridized for 30 min with Quik-Hyb (Stratagene, La Jolla, CA) at 65°C. 32P-dCTP radiolabeled full-length Mi cDNA was made using the Stratagene Prime-it random labeling kit. 106 cpm/ml of Quik-Hyb solution was used and hybridization was performed at 65°C for 2 h.
Reverse Transcriptase PCR and Northern Analysis.
Total cellular RNA from heart tissue and cultured cells was isolated using RNAzol (Tel-Test, Friendswood, TX) according to the manufacturer's instructions. A series of primers to mouse microphthalmia exons 5–9 were synthesized: 5′ exon 5 CCCGTCTCTGGAAACTTGATCG; 5′ exon 6 CGTGTATTTTCCCCACAGAGTC; 5′ exon 8 GACATGCGGTGGAACAAGGG; 5′ exon 9 GAGCTGGAGATGCAGGCTAG; 3′ exon 5 GTTGGGAAGGTTGGCTGGAC; 3′ exon 6 CTGCCTCTCTTTAGCCAATGC; 3′ exon 7 GGATCATTTGACTTGGGGATCAG; 3′ exon 8 CTGTACTCTGAGCAGCAGGTG; 3′ exon 9 GCTCTCCGGCATGGTGCCGAGG. cDNA was made using a reverse transcriptase (RT) (PCR kit) (GIBLO BRL, Gaithersburg, MD). 1–5 μg of total RNA was used for each CDNA reaction along with 30–100 pmol of 3′ primer and 200 U of SuperScript II TM reverse transcriptase and incubated for 50 min at 42°C. 2–4 μl of the 20 μl cDNA reaction mixture was used for PCR reactions including Taq polymerase (Fisher Scientific Co., Pittsburgh, PA), dNTP's (Pharmacia Biotech, Piscataway, NJ), and PCR buffer (Perkin-Elmer Corp., Norwalk, CT) and with incubations at 94°C for 2 min, 57°C for 1 min, and 72°C for 2 min for 30 cycles (annealing temperature for reactions containing 5′ exon 6 was 67°C; for 5′ exon 7 was 57°C; and for 5′ exon 8 and 5′ exon 9 was 62°C). 25–50 μl of the 100 μl reaction mixtures were resolved by electrophoresis in nondenaturing 8–10% polyacrylamide gels. Negative controls were performed with water in place of DNA. The RT-PCR product from 5′ exon 5 and 3′ exon 8 for wild-type rat was gel purified and sequenced. The sequence was confirmed from four independent PCR reactions.
Mi expression was examined using Northern analysis with 20 μg of total RNA loaded on a formaldehyde gel, transferred to nylon membranes, and probed with radiolabeled full length mi DNA for 2 h at 65°C in Quik-hyb solution. For loading control, a radiolabeled 500-bp Xba I/Hind III fragment of the human glyceraldehyde 3′ phosphate dehydrogenase was used.
Cell Lines.
NIH3T3 (mouse fibroblasts), L cells (murine connective tissue line), BHK (hamster kidney cell line), and B16 (murine melanoma) were grown in DMEM with 10% fetal bovine serum. ST2 (Riken Cell Bank, Tsukuba Science City, Japan) was grown in RPMI 1640 and 10% FBS. The mast cell line C57 (gift of Dr. Stephen Galli, Harvard Medical School, Cambridge, MA) was grown in DMEM with 10% fetal bovine serum and 2 × 10−5 M 2-β mercaptoethanol. The human melanoma cell line, 501MEL (gift of Dr. Ruth Haliban, Yale Medical School, New Haven, CT) was grown in F10 media with 10% fetal bovine serum.
Results
Expression of Mi in Cultured Osteoclasts.
Osteoclasts were prepared by two standard techniques. Mouse calvarial osteoblasts and spleen cells were cocultured in the presence of 1,25-(OH)2D3 to generate osteoclast-like cells. This technique produces osteoclasts that express calcitonin receptors and TRAP, and make resorption pits in bone (2, 42). Human osteoclast-like cells (HOCs) were grown in the presence of 1,25-(OH)2D3 and M-CSF. HOCs cultured in this manner have also been shown to express calcitonin receptors and TRAP, and make resorption pits in bone (43, 44). Immunohistochemical staining was performed on day 12 for mouse osteoclast-like cells and day 21 for HOCs using two separate monoclonal antibodies prepared against a novel peptide from Mi (see Materials and Methods). Fig. 1 shows nuclear staining by the anti-Mi antibody in the multinucleated osteoclast-like cells. The surrounding calvarial osteoblasts showed no Mi staining. The human melanoma cell line (501 MEL) served as a positive control (46). L-cells, NIH3T3, and BHK showed no Mi staining (data not shown).
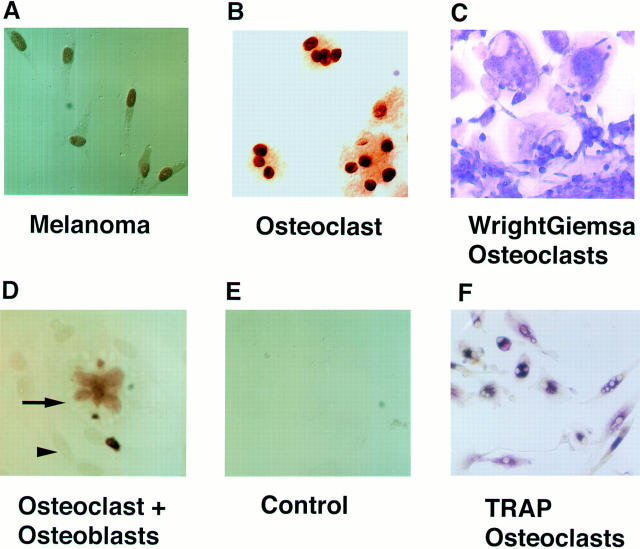
Figure 1.
Immunohistochemical staining for Mi in osteoclasts. A, B, D, and E show nuclear localization of Mi; there is no counterstaining. (A) 501 Mel human melanoma cell line. (B) Multinucleate human osteoclast-like cells after 21 d in culture. (D) Murine osteoclast-like cocultures after 12 d in culture; arrowhead points to nonstaining calvarial-derived osteoblast-like cells and arrow points to multinucleated osteoclast. (E) Murine osteoclast-like cocultures (as in D) with second step antibody alone showing little background staining. (C) Human osteoclast cultures (day 21) stained with Wright-Giemsa. (F) Human osteoclast cultures (day 21) stained with TRAP stain.
Protein extracts were made from various cell types including ST2, L cells, NIH3T3, as well as HOCs. There are two known forms of Mi differing by 66 amino acids at the NH2 terminus that likely initiate from different promoters and are spliced to the same downstream exon (31). The shorter forms are expressed in melanocytes (31) and run as two bands at 54 and 60 kD. These melanocyte isoforms have been shown by two-dimensional tryptic mapping to differ in c-Kit–induced phosphorylation (46). The longer nonmelanocyte Mi form runs as a cluster of species at ∼70 kD in osteoclasts (Fig. 2 A) and in B16 melanoma cells (but not other melanoma cell lines [46] as well as mast cells and heart (data not shown). The presence of multiple closely migrating bands suggests that Mi may undergo modifications in osteoclasts, a possibility which would be consistent with its kit-induced phosphorylation in melanoma cells (46). 3T3, ST2, and L cells do not show any Mi-specific bands (Fig. 2 A). It should be noted that although immunohistochemistry appears to identify Mi (and TFE3, see below) only in osteoclast-like cells, these cultures do represent mixed populations and we cannot exclude the possibility of a contribution in gel signal from nonosteoclasts. All of these studies were carried out in mixed primary bone marrow and spleen cultures because of the lack of availability of purified osteoclast-derived cell lines (2). Previously, Mi expression has been demonstrated at the RNA level in melanocytes, heart, and mast cells (18, 31). Northern analysis from cultured primary osteoclast-like cells reveals Mi mRNA at 5.6 Kb (Fig. 2 B). Mast cells exhibit the same size species (Fig. 2 B), in agreement with previous studies (18, 31).
Figure 2.
Mi immunoprecipitation and Northern blotting. (A) Immunoprecipitation and Western blotting shows that the human long-term marrow osteoclast cultures have an ∼70 kD form of Mi (nonmelanocyte form). B16 murine melanoma has this ∼70 kD species in addition to the more predominant melanocyte specific forms at 54 kD and 60 kD. NIH3T3, L-cells, and ST2-like cells do not express detectable Mi. (B) Northern blot analysis shows that the Mi mRNA transcripts in osteoclast cultures comigrates with Mi mRNA from mast cells with a size of ∼5.6 kb. Glyceraldehyde 3′ phosphate dehydrogenase probe confirms levels of RNA loaded in each lane.
Mapping the Mutation in the mib Rat.
To examine the microphthalmia gene in the mib/mib rat, a series of PCR primers was used to amplify both genomic DNA and RNA from mib/mib and genetically matched wild-type rats. Exon mapping of genomic DNA was performed using 5′ and 3′ primers at the ends of each exon from exon 5 through 9 except for exon 7, which is short and very AT-rich, thus rendering PCR mapping of this exon technically difficult. Bands of the expected sizes were identified for exons 5 and 6 in both the mib/mib and wild-type DNA samples (Fig. 3 B). However, exons 8 and 9 were only identified in wild-type samples, and not in mib/mib.
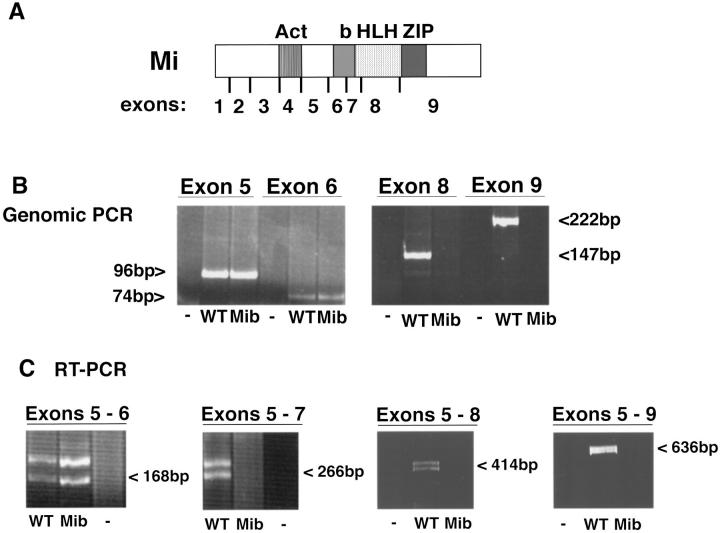
Figure 3.
Exon mapping of Mi in the mib/mib rat. (A) Schematic of the microphthalmia coding region. Act, transactivation domain; b, basic region; HLH, helix-loop-helix; ZIP, leucine zipper. 5′ and 3′ primers were constructed at the boundaries of exons 5–9. (B) Genomic PCR: PCR reactions using primers from the 5′ and 3′ ends of exons 5, 6, 8, and 9 on genomic DNA from mib/mib (Mib) and wild-type (WT) rats are shown. Control (-) lacked template DNA. Exon 5 PCR product is 96 bp, exon 6 is 74 bp, exon 8 is 222 bp and exon 9 is 147 bp. Exon 7 is too small and AT rich to map by this method. (C) RT-PCR: a series of 3′ primers downstream from exon 5 were used to map the deletion in the mib/mib rat. In all cases the 5′ primer is at the 5′ end of exon 5. Exons 5 and 6 (5′ exon 5 to 3′ exon 6) encodes a 168-bp doublet seen in both mib/mib RNA (Mib) and wild-type (WT) RNA. Exons 5–7 (5′ exon 5 to 3′ exon 9) is 266 bp in length. Exons 5–8 (5′ exon 5 to 3′ exon 8) is 414 bp and exons 5–9 (5′ exon 5 to 3′ exon 9) is 636 bp. The doublet arises from the previously described 18-bp alternative splice at the exon 5 and 6 boundary (31) and was confirmed by sequencing (data not shown). Absence of amplifiable sequences in exons 8 and 9 by genomic PCR and in 7–9 by RT-PCR suggest the presence of a large deletion within Mi.
RT-PCR was performed on heart RNA from wild-type and mib/mib rats using a 5′ primer in exon 5 and a series of 3′ primers spanning exon 6 through exon 9 (Fig. 3 C). A doublet is seen throughout this series of RT-PCR products due to an 18-bp alternative splice at the beginning of exon 6 (31), which was confirmed by sequencing (data not shown). Using 5′ exon 5 and 3′ exon 6 primers, Mi-specific bands of the expected size were identified in both the mib/mib and wild-type RNA samples. 5′ exon 5 and 3′ exon 7 primers gave the appropriate products only in the wild-type RNA sample with no corresponding band in the mib/mib samples. Similarly for 3′ exons 8 and 9, only the wild-type sample showed bands of the expected sizes. Data from genomic exon mapping and RT-PCR thus indicate that there is a deletion in Mi of all or part of exon 7, and in exons 8 and 9 in the mib/mib rat.
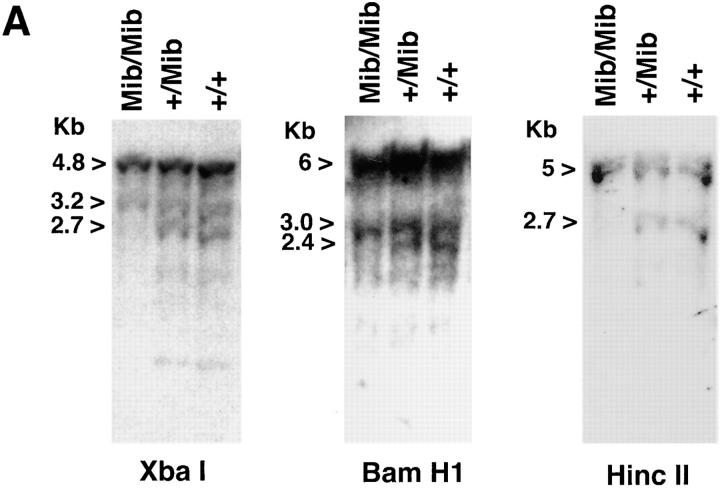
Mi Southern blot analysis of mib/mib, +/mib, and +/+ genomic DNA was performed using BamHI, HincII, and XbaI (Fig. 4 A). At least one distinct band was absent in the mib/mib lanes for each of these enzymes, consistent with the PCR data indicating genomic deletion of a region within mi. For example, with XbaI, the 2.7-kb band is absent in the mib/mib lane. The lack of apparent new bands in mib is consistent with lack of a reciprocal translocation (also suggested by absence of signal in 3′ exons by genomic exon mapping). These data indicate that the microphthalmia gene has undergone a large genomic deletion in the mib/ mib osteopetrotic rat.
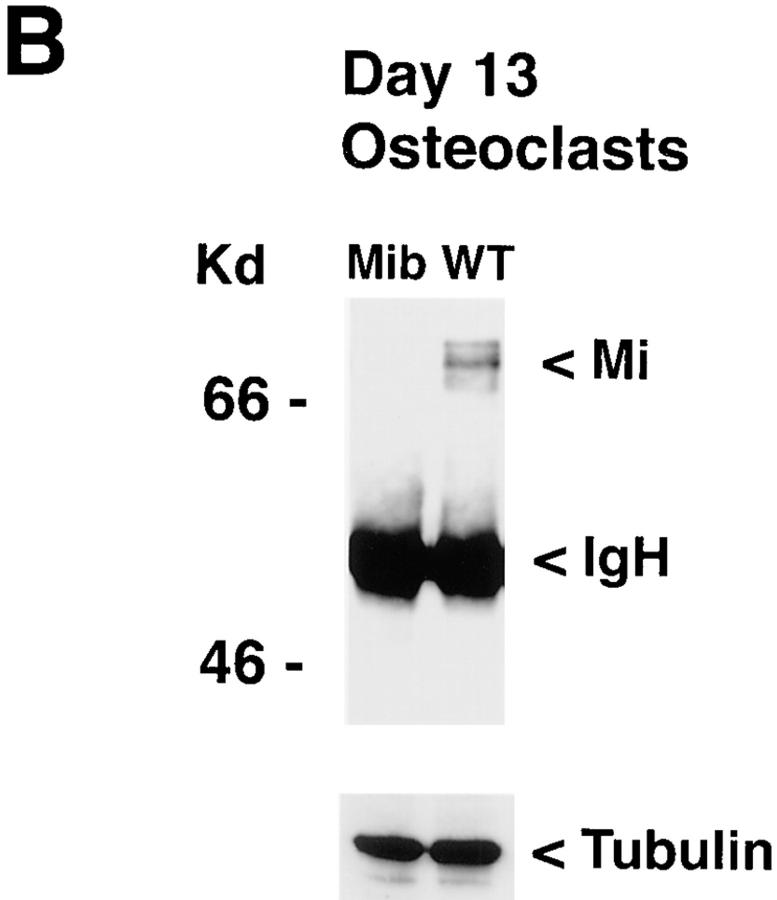
Figure 4.
Partial deletion of mi in the mib/mib rat and mutant osteoclasts. (A) Southern Blot: genomic DNA from mib/mib, +/mib, and +/+ rats were digested with XbaI, BamH1, and HincII, and probed with radiolabeled full-length mi mouse cDNA. The pattern is noteworthy for absent bands in the mib/ mib samples compared with heterozygote or wild-type samples. (B) Protein extracts were made from day 13 cultured mib/mib osteoclasts and wild-type rat osteoclasts (WT) and immunoprecipitated with Mi antibody. Western blotting with the Mi antibody after SDS-PAGE is shown. Immunoglobulin heavy chain (IgH) runs at 50 kD and Mi osteoclast form runs at ∼70 kD. Western blotting with anti–rat α tubulin antibody demonstrates protein loading. No Mi is detected in mib/ mib osteoclasts.
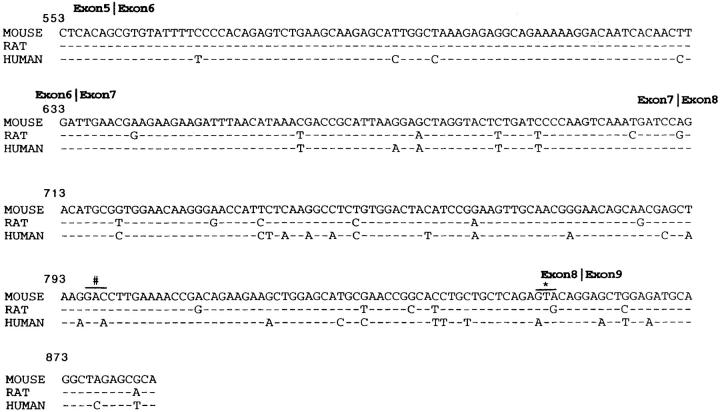
The sequence of rat microphthalmia for exons 5–9 was determined from multiple independent PCR products and is shown in Fig. 5. Mi is highly conserved between human and mouse as well as in the rat in the b-HLH-ZIP region. Fig. 5 shows the sequence of the rat as compared to mouse and human mi. Over the 331 bp examined, there are 20 single base differences between mouse and rat, none of which alter the coding sequence. Of the 20 positions where mouse and rat differ, the human sequence matched mouse in 8, matched rat in 8, and was different from both in 4.
Figure 5.
Rat mi sequence for exons 6–9. The sequence is highly conserved between rat, human, and mouse with only minor nucleotide changes. The only mutations that alter the amino acid sequence are depicted by (#) aspartic acid (mouse and rat) to glutamic acid (human) and (*) valine (mouse and rat) to isoleucine (human).
Evaluation of mib/mib Mutant Osteoclasts.
mib/mib and wild-type rat bone marrow from 6-mo-old rats were harvested and cultured in the presence of 1,25-(OH)2 D3 to produce osteoclast-like cells. TRAP stains were performed at day 13 and revealed equivalent numbers of multinucleated TRAP positive cells per high powered field in both cultures (data not shown) consistent with previous reports that mib/mib osteoclast number and function return to normal after 4 wk of age (38). However, immunoblots for Mi revealed no detectable Mi protein in the mutant osteoclasts compared to wild-type (Fig. 4 B). Although the antibody epitope is not deleted, no truncated forms of Mi were detected, suggesting instability of the truncated RNA or protein (or both).
Expression of TFE3 in Cultured Osteoclasts.
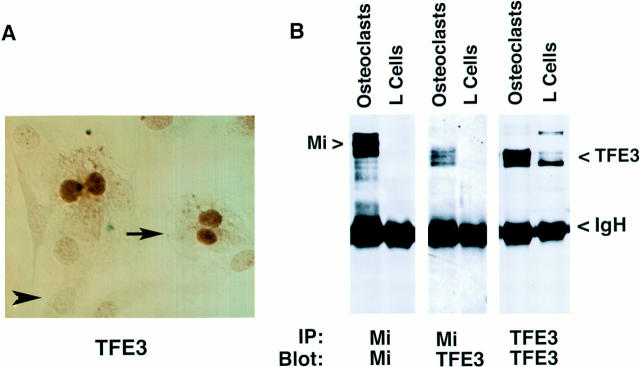
Improvement of the osteoclast defect in the mib/mib rat and the recessive inheritance pattern of osteopetrosis in dominant negative mouse mutants (20) suggest that an unidentified partner protein may exist for Mi in osteoclasts. We have previously demonstrated the ability of several related b-HLH-ZIP factors to heterodimerize in vitro with Mi (20). Immunohistochemistry and immunoblot analyses were carried out to examine mouse osteoclasts for the presence of TFE3. Immunostaining demonstrated strong staining for TFE3 in the nuclei of osteoclasts (Fig. 6 A). In contrast, osteoblasts do not express detectable TFE3. Mi runs at ∼70 kD in osteoclasts and TFE3 migrates at 59 kD in osteoclasts. L-cells (from which murine TFE3 was cloned) express TFE3 but not detectable Mi. In addition, anti-TFE3 blotting of Mi immunoprecipitates from osteoclasts revealed significant TFE3 protein (Fig. 6 B). Approximately 10% of the input TFE3 was detected in Mi immunoprecipitates indicating significant Mi TFE3 heterodimerization (Fig. 6 B). As a specificity control, no TFE3 was seen in anti-Mi immunoprecipitates of L cells (Fig. 6 B) or in isotype-matched (to anti-Mi) control immunoprecipitates of osteoclasts (data not shown). Other experiments have demonstrated the reciprocal coimmunoprecipitation of Mi by anti-TFE3 (data not shown). These findings suggest that both TFE3 and Mi proteins are present in osteoclast-like cells and may heterodimerize in vivo, although this analysis cannot discriminate heterodimers that form after extract preparation.
Figure 6.
TFE3 expression and coimmunoprecipitation with Mi in osteoclasts. (A) TFE3 immunohistochemical staining of murine cocultured osteoclast-like cells. Arrow points to staining of multinucleated osteoclast-like cells with TFE3 antibody. Arrowhead points to nonstaining murine calvaria– derived osteoblast-like cells. (B) Extracts from murine osteoclast-like cells or L cells were subjected to immunoprecipitation (IP) with monoclonal antibodies against Mi or TFE3, resolved by SDS-PAGE, and immunoblotted with antibodies to Mi or TFE3 as indicated. Anti-Mi coimmunoprecipitates TFE3 from osteoclasts (but not from L cells, which lack Mi, but contain TFE3). IP using a monoclonal antibody isotype matched to anti-Mi fails to precipitate TFE3 from osteoclasts as well (data not shown).
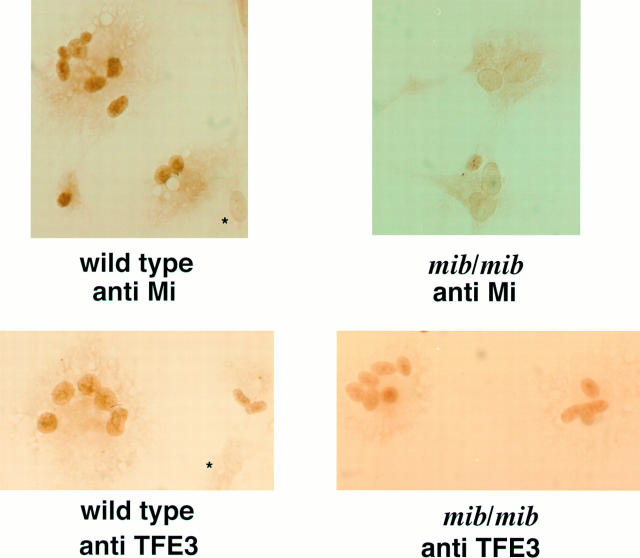
Immunohistochemical staining for TFE3 in wild-type and mib/mib cultured rat osteoclasts is shown in Fig. 7. TFE3 is present in both wild-type and mib/mib osteoclast-like cells in a nuclear pattern. Mi staining was also performed and is seen in wild-type rat osteoclast-like cells but not in mib/mib. Second step antibody staining alone shows little background staining (data not shown). The asterisk denotes the nonstaining stromal cells in these bone marrow cultures. TFE3 is thus detectable in both mouse and rat osteoclast-like cells including mib/mib, consistent with a potential ability to interact with or partially compensate for Mi deficiency.
Figure 7.
Immunohistochemical staining for Mi and TFE3 in wild-type and mib/mib cultured rat osteoclasts from day 12 bone marrow cultures. Mi localizes to the nucleus in wild-type multinucleated osteoclast-like cells, but there is no Mi nuclear staining in the mib/mib osteoclast-like cells. TFE3 nuclear staining can be seen in both wild-type and mib/mib osteoclast-like cells. Nonstaining stromal cells can be seen in the wild-type representative fields for Mi and TFE3 (denoted by *).
Discussion
Mice with certain mutations in mi develop osteopetrosis that, based on Mi's abundant expression in osteoclasts but not stromal cells, likely reflects a cell autonomous defect. This is also consistent with hematopoietic transplantation studies that corrected the mi/mi osteopetrotic phenotype (8). The mib rat develops osteopetrosis with dramatic age-dependent changes. mib/mib osteoclasts resorb bone poorly and do not have a ruffled border at birth. However, after birth, osteoclasts begin to regain normal morphology and osteoclastic activity, and the osteopetrosis largely resolves, with the exception of a persistent long-term failure of incisor eruption (36). In this report we demonstrate that mib/ mib rats harbor a large deletion in the 3′ half of the mi gene that eliminates most of the b-HLH-ZIP region. The deletion probably starts within the intron separating exons 6 and 7 and truncates the gene in the region coding for the basic domain. The age-resolving osteopetrotic phenotype prompted us to search for a transcription factor partner for Mi that might in principal rescue mib/mib osteoclasts. TFE3, a b-HLH-ZIP transcription factor (21, 22), was found within the nuclei of mouse and rat osteoclasts (including mib/mib), but not osteoblasts. Moreover TFE3 was also found to coimmunoprecipitate with Mi from wild-type cells. These data indicate that a distinct family of related b-HLH-ZIP transcription factors is expressed in osteoclasts and suggests that these factors may collectively play a role in the function of osteoclasts in bone resorption.
Interestingly, the majority of mouse strains harboring mi mutations have not been recognized to develop osteopetrosis. Only mi/mi and Mior/Mior strains display osteopetrosis, and these two notably contain basic region defects (31) that function as dominant negative proteins in vitro (20). The only other known basic domain mutant of mi (Miwh/Miwh) differs in that the presence of an alternatively spliced 6-amino acid insert NH2-terminal to the basic domain rescues its heterodimeric DNA binding capacity to wild-type levels (20), which may explain why Miwh/Miwh animals do not develop osteopetrosis. These observations suggest the functional relevance of Mi heterodimers involving other family member(s) in vivo. It was anticipated that the mutation in mib rats might also be dominant negative, by crippling DNA binding independently of dimerization. However, the large genomic deletion removes most of the basic domain and all of the dimerization region (HLH-ZIP) precluding the formation of species analogous to the mouse osteopetrotic (basic region mutations) mi/mi and Mior/Mior alleles. Athough the resulting protein should be profoundly truncated and could in principal be dominant negative through expression of a remaining domain, such a species is not readily detected within mib osteoclasts by Western blotting or immunohistochemistry, and it is likely that the protein or its mRNA undergo accelerated degradation. An alternative hypothesis is that TFE3 or other heterodimeric protein partners are expressed later in osteoclast development than Mi, resulting in delayed osteoclast maturation and function. Mi could also regulate the transcription of TFE3 and other family members and this could explain the delay in osteoclast restoration of function. The dominant-negative mouse alleles display dominant phenotypes with respect to pigmentation (white belly spots in heterozygotes). Unfortunately ventral midline pigmentation cannot be assessed in mib heterozygotes because the background rat strain is black hooded (i.e., wild-type animals have a completely white belly).
There are several potential explanations for the presence of osteopetrosis with a mi allele (mib) that is unlikely to act as a dominant negative mutation. The osteopetrosis in mib is less severe than that seen in the mouse mi alleles. It may thus represent a more subtle osteoclast defect that was heretofore unrecognized in mice with recessive mi mutations. The improvement with age may, in particular, prevent recognition of the phenotype in older animals. The osteoclast defect may occur during gestation and already begin improving by the neonatal period. In fact, careful characterization of the osteopetrosis in Mio r/Mio r has also revealed evidence of phenotypic changes with age (16). Interestingly, in this case the osteopetrotic indices first worsen and then improve somewhat over 3–8 mo. The age-dependent remission may be the result of either diminished requirement for Mi protein or enhanced rescue by another factor(s). Additional studies that compare temporal expression of Mi, TFE3, and other b-HLH-ZIP factors may shed more light on the age-specific osteoclast phenotype.
A direct comparison of the mib mutation with the other osteopetrotic mutations in Mi thus reveals a correlation in the relative mildness of mib skeletal defects with the inability of this deletion to dominantly affect partner proteins. It is not currently known whether expression of Mimi or Mior is maintained within the mutant osteoclasts, but it is reasonable to expect that these small single residue mutations are likely to produce stable gene products, as has been demonstrated in heart tissue of the mutant mice (31). In contrast, the large deletion in mib results in no detectable gene product (mRNA or protein), presumably due to premature degradation or precursor gene products. Although it is important to consider the contribution of deleted domains COOH-terminal to the HLH-ZIP region in mib, these are as yet poorly characterized although the proline-rich COOH-terminal region is a plausible transactivation motif.
Future studies will be needed to determine whether expression levels of TFE3 (as well as Mi and other b-HLH-ZIP factors) change during skeletal development and aging. Although it is plausible that the age-effect and lack of severe osteopetrosis in mouse recessive mi alleles is due to compensation by other b-HLH-ZIP factors, this possibility is by no means proven by the current studies. Genetic manipulations, such as analysis of knockout mice, may be more decisive. It also remains to be seen whether these age-dependent changes reflect simple compensation in a pathologic setting or a change in transcription factor usage that occurs during normal maturation/aging. Since osteoclast function clearly changes with age and other extracellular signals, the identification of factors that reflect these changes may provide mechanistic clues to the problems of osteopetrosis and osteoporosis.
Our findings place microphthalmia among a handful of genes that may be important to osteoclast function. Mi may be of particular interest because of recent evidence that it is a signal responsive factor that is phosphorylated and becomes more transcriptionally active upon c-Kit stimulation in melanocytes (46). Similar signaling pathways may exist in osteoclasts and could regulate both Mi and its partners. The presence of multiple migrating species of Mi and TFE3 in osteoclasts is consistent with this possibility, which is currently being examined. It will be of interest to determine whether Src, M-CSF, and/or Fos modulate the activity of Mi because mice defective in these factors also harbor osteoclast defects of varying magnitudes. Tyrosinase and related pigment enzyme genes are important targets of Mi in melanocytes (47–49). Mouse mast cell protease 6 (MMCP-6) and c-Kit are likely to be important targets of Mi in mast cells (46, 50–52). Mi and/or other b-HLH-ZIP family members probably regulate genes related to osteoclast resorptive function because of the poor bone resorption of mutant mi/mi osteoclasts. Transcriptional targets for Mi and TFE3 in osteoclasts have yet to be identified, but their elucidation may shed important light on osteoclast biology.
Footnotes
The authors thank Glenn Belinski and Edward Voelkel for care and breeding of the mib rats and calvaria preparation; Dr. Todd Golub, Dr. Michael Tomasson, and Dr. Cornelius Schmaltz for thoughtful discussions; Dr. Richard Sidman and Dr. Bela Kosaris for advice on immunohistochemical staining; Dr. Colin Sieff, Dr. Michael Pulsipher, Dr. Miyumi Tanaka, and Dr. Vivienne Rebel for help with human bone marrow cultures; Dr. Stephen Galli for the C57 mast cell line; and Dr. Ruth Haliban for 501 Mel cells.
This research was supported partly by National Institutes of Health grant DK-10206 (A.H. Tashjian, Jr.). D.E. Fisher is a Pew Foundation scholar and a James S. McDonnell Fellow. C.L. Hershey has a predoctoral fellowship from National Science Foundation.
Address correspondence to David E. Fisher, Dana Farber Cancer Institute, 44 Binney Street, Boston, MA 02115. Phone: 617-632-4916; Fax: 617-632-2085; E-mail: david_fisher@dfci.harvard.edu
Abbreviations used in this paper: 1,25-(OH)2D3, 1,25 dihydroxyvitamin D3; b-HLH-ZIP, basic helix-loop-helix-leucine zipper; Mi, microphthalmia; RT, reverse transcriptase; TF, transcription factor; TRAP, tartrate-resistant acid phosphatase.
References
- 1.Chambers TJ. The pathobiology of the osteoclast. J Clin Pathol. 1985;38:241–252. doi: 10.1136/jcp.38.3.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Suda T, Takahashi N, Martin T., Jr Modulation of osteoclast differentiation. Endocr Rev. 1992;13:66–80. doi: 10.1210/edrv-13-1-66. [DOI] [PubMed] [Google Scholar]
- 3.Popoff S, Marks S. The heterogeneity of the osteopetroses reflects the diversity of cellular influences during skeletal development. Bone. 1995;17:437–445. doi: 10.1016/8756-3282(95)00347-4. [DOI] [PubMed] [Google Scholar]
- 4.Grigoriadis AE, Wang Z, Cecchini M, Hofstetter W, Felix R, Fleisch H, Wagner E. c-Fos: a key regulator of osteoclast-macrophage lineage determination and bone remodeling. Science. 1994;266:443–448. doi: 10.1126/science.7939685. [DOI] [PubMed] [Google Scholar]
- 5.Grigoriadis AE, Wang ZQ, Wagner EF. Fos and bone cell development: lessons from a nuclear oncogene. Trends Genet. 1995;11:436–441. doi: 10.1016/s0168-9525(00)89142-8. [DOI] [PubMed] [Google Scholar]
- 6.Soriano P, Montgomery C, Geske R, Bradley A. Targeted disruption of the s-src proto-oncogene leads to osteopetrosis in mice. Cell. 1991;64:693–702. doi: 10.1016/0092-8674(91)90499-o. [DOI] [PubMed] [Google Scholar]
- 7.Murphy H. The osteopetrotic syndrome in the microphthalmic mutant mouse. Calcif Tissue Res. 1973;13:19–26. doi: 10.1007/BF02015392. [DOI] [PubMed] [Google Scholar]
- 8.Walker D. Bone resorption restored in osteopetrotic mice by transplants of normal bone marrow and spleen cells. Science. 1975;190:784–785. doi: 10.1126/science.1105786. [DOI] [PubMed] [Google Scholar]
- 9.Boyce B, Yoneda T, Lowe C, Soriano P, Mundy G. Requirement of pp60c-src expression for osteoclasts to form ruffled borders and resorb bone in mice. J Clin Invest. 1992;90:1622–1627. doi: 10.1172/JCI116032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoshida H, Hayashi S, Kunisada T, Ogawa M, Nishikawa S, Okamura H, Sudo T, Shultz LD, Nishikawa S. The murine mutation osteopetrosis is in the coding region of the macrophage colony stimulating factor gene. Nature. 1990;345:442–444. doi: 10.1038/345442a0. [DOI] [PubMed] [Google Scholar]
- 11.Wisner-Lynch L, Shalhoub V, Marks SC., Jr Administration of colony stimulating factor–1 to toothless osteopetrotic rats normalizes osteoblast, but not osteoclast, gene expression. Bone. 1995;16:611–618. doi: 10.1016/8756-3282(95)00114-s. [DOI] [PubMed] [Google Scholar]
- 12.Mitchell R, Henning-Chubb C, Huberman E, Verma I. c-fos expresssion is neither sufficient nor obligatory for differentiation of monomyelocytes to macrophages. Cell. 1986;45:497–504. doi: 10.1016/0092-8674(86)90281-3. [DOI] [PubMed] [Google Scholar]
- 13.Tanaka S, Amling M, Neff L, Peyman A, Uhlmann E, Levy JB, Baron R. c-Cbl is downstream of c-Src in a signalling pathway necessary for bone resorption. Nature. 1996;383:528–531. doi: 10.1038/383528a0. [DOI] [PubMed] [Google Scholar]
- 14.Hertwig P. Neue Mutationen und Kopplungsgruppen bei der Hausmaus. Z Indukt Abstammungs-Vererbungsl. 1942;80:220–246. [Google Scholar]
- 15.West, J., G. Fisher, J. Loutit, M. Marshall, N. Nisbet, and H. Perry. 1985. A new allele of microphthalmia induced in the mouse: microphthalmia-defective iris (midi). Genet. Res., Camb. 46:309–324. [DOI] [PubMed]
- 16.Nii A, Steingrimsson E, Copeland N, Jenkins N, Ward J. Mild osteopetrosis in the microphthalmia-oak ridge mouse. Am J Pathol. 1995;147:1871–1882. [PMC free article] [PubMed] [Google Scholar]
- 17.Thesingh C, Scherft J. Fusion disability of embryonic osteoclast precursor cells and macrophages in the microphthalmic osteopetrotic mouse. Bone. 1985;6:43–52. doi: 10.1016/8756-3282(85)90406-5. [DOI] [PubMed] [Google Scholar]
- 18.Hodgkinson CA, Moore KJ, Nakayama A, Steingrimsson E, Copeland NG, Jenkins NA, Arnheiter H. Mutations at the mouse microphthalmia locus are associated with defects in a gene encoding a novel basic-helix-loop-helix-zipper protein. Cell. 1993;74:395–404. doi: 10.1016/0092-8674(93)90429-t. [DOI] [PubMed] [Google Scholar]
- 19.Hughes JJ, Lingrel JB, Krakowsky JM, Anderson KP. A helix-loop-helix transcription factor–like gene is located at the milocus. J Biol Chem. 1993;268:20687–20690. [PubMed] [Google Scholar]
- 20.Hemesath TJ, Steingrimsson E, McGill G, Hansen MJ, Vaught J, Hodgkinson CA, Arnheiter H, Copeland NG, Jenkins NA, Fisher DE. Microphthalmia, a critical factor in melanocyte development, defines a discrete transcription factor family. Genes Dev. 1994;8:2770–2780. doi: 10.1101/gad.8.22.2770. [DOI] [PubMed] [Google Scholar]
- 21.Carr CS, Sharp PA. A helix-loop-helix protein related to the immunoglobulin E box-binding proteins. Mol Cell Biol. 1990;10:4384–4388. doi: 10.1128/mcb.10.8.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beckman H, Su LK, Kadesch T. TFE3: a helix-loop-helix protein that activates transcription through the immunoglobulin enhancer uE3 motif. Genes Dev. 1990;4:167–179. doi: 10.1101/gad.4.2.167. [DOI] [PubMed] [Google Scholar]
- 23.Roman C, Matera A, Cooper C, Artandi S, Blain S, Ward D, Calame L. mTFE3, an X-linked transcriptional activator containing basic helix-loop-helix and zipper domains, utilizes the zipper to stabilize both DNA binding and multimerization. Mol Cell Biol. 1992;12:817–827. doi: 10.1128/mcb.12.2.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao GQ, Zhao Q, Zhou X, Mattei MG, De Crombrugghe B. TFEC, a basic helix-loop-helix protein, forms heterodimers with TFE3 and inhibits TFE3-dependent transcription activation. Mol Cell Biol. 1993;13:4505–4512. doi: 10.1128/mcb.13.8.4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yasumoto K, Shibahara S. Molecular cloning of cDNA encoding a human TFEC isoform, a newly identified transcriptional regulator. Biochim Biophys Acta. 1997;1353:23–31. doi: 10.1016/s0167-4781(97)00034-1. [DOI] [PubMed] [Google Scholar]
- 26.Blackwood EM, Eisenman RN. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science. 1991;251:1211–1217. doi: 10.1126/science.2006410. [DOI] [PubMed] [Google Scholar]
- 27.Merrell K, Wells S, Henderson A, Gorman J, Alt F, Stall A, Calame K. The absence of the transcription activator TFE3 impairs activation of B cells in vivo. Mol Cell Biol. 1997;17:3335–3344. doi: 10.1128/mcb.17.6.3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tassabehji M, Newton VE, Read AP. Waardenburg syndrome type 2 caused by mutations in the human microphthalmia (MITF) gene. Nat Genet. 1994;8:251–255. doi: 10.1038/ng1194-251. [DOI] [PubMed] [Google Scholar]
- 29.Hughes AE, Newton VE, Liu XZ, Read AP. A gene for Waardenburg syndrome type 2 maps close to the human homologue of the microphthalmia gene at chromosome 3p12-p14.1. Nat Genet. 1994;7:509–512. doi: 10.1038/ng0894-509. [DOI] [PubMed] [Google Scholar]
- 30.Moore KJ. Insight into the microphthalmiagene. Trends Genet. 1995;11:442–448. doi: 10.1016/s0168-9525(00)89143-x. [DOI] [PubMed] [Google Scholar]
- 31.Steingrimsson E, Moore KJ, Lamoreux ML, Ferre-D'Amare AR, Burley SK, Sanders-Zimring DC, Skow LC, Hodgkinson CA, Arnheiter H, Copeland NG, Jenkins NA. Molecular basis of mouse microphthalamia(mi) mutations helps explain their developmental and phenotypic consequences. Nat Genet. 1994;8:256–263. doi: 10.1038/ng1194-256. [DOI] [PubMed] [Google Scholar]
- 32.Fisher DE, Carr CS, Parent LA, Sharp PA. TFEB has DNA-binding and oligomerization properties of a unique helix-loop-helix/leucine-zipper family. Genes Dev. 1991;5:2342–2352. doi: 10.1101/gad.5.12a.2342. [DOI] [PubMed] [Google Scholar]
- 33.Beckman H, Kadesch T. The leucine zipper of TFE3 dictates helix-loop-helix dimerization specificity. Genes Dev. 1991;5:1057–1066. doi: 10.1101/gad.5.6.1057. [DOI] [PubMed] [Google Scholar]
- 34.Fisher DE, Parent LA, Sharp PA. High affinity DNA-binding Myc analogs: recognition by an α helix. Cell. 1993;72:467–476. doi: 10.1016/0092-8674(93)90122-7. [DOI] [PubMed] [Google Scholar]
- 35.Moutier R, Ostrowski K, Lamendin H. Microphthalmia: a new recessive mutation in the Norway rat. J Hered. 1989;90:76–78. doi: 10.1093/oxfordjournals.jhered.a110798. [DOI] [PubMed] [Google Scholar]
- 36.Cielinski M, Iizuka T, Marks SC., Jr Dental abnormalities in the osteopetrotic rat mutation microphthalmia blanc. Arch Oral Biol. 1994;39:985–990. doi: 10.1016/0003-9969(94)90083-3. [DOI] [PubMed] [Google Scholar]
- 37.Cielinski M, Marks SC., Jr Neonatal reductions in osteoclast number and function account for the transient nature of osteopetrosis in the rat mutation microphthalmia blanc (mib) . Bone. 1994;15:707–715. doi: 10.1016/8756-3282(94)90321-2. [DOI] [PubMed] [Google Scholar]
- 38.Cielinski MJ, Marks SC., Jr Bone metabolism in the osteopetrotic rat mutation microphthalmia blanc. Bone. 1995;165:567–574. doi: 10.1016/8756-3282(95)00080-w. [DOI] [PubMed] [Google Scholar]
- 39.Takahashi N, Yamana H, Yoshiki S, Roodman D, Mundy G, Jones S, Boyde A, Suda T. Osteoclast-like cell formation and its regulation by osteotropic hormones in mouse bone marrow cultures. Endocrinology. 1988;122:1373–1382. doi: 10.1210/endo-122-4-1373. [DOI] [PubMed] [Google Scholar]
- 40.Shiina Y, Yamaguchi A, Yamana H, Abe E, Yoshiki S, Suda T. Comparison of the mechanisms of bone resorption induced by 1,25-dihydroxyvitamin D3 and lipopolysaccharides. Calcif Tissue Int. 1986;39:28–34. doi: 10.1007/BF02555737. [DOI] [PubMed] [Google Scholar]
- 41.Takahashi N, Akatsu T, Sasaki T, Nicholson G, Moseley J, Martin J, Suda T. Induction of calcitonin receptors by 1 alpha 25-dihydroxyvitamin D3 in osteoclast- like multinucleated cells formed from mouse bone marrow cells. Endocrinology. 1988;123:1504–1510. doi: 10.1210/endo-123-3-1504. [DOI] [PubMed] [Google Scholar]
- 42.Takahashi N, Akatsu T, Udagawa N, Sasaki T, Yamaguchi A, Moseley J, Martin J, Suda T. Osteoblastic cells are involved in osteoclast formation. Endocrinology. 1988;123:2600–2602. doi: 10.1210/endo-123-5-2600. [DOI] [PubMed] [Google Scholar]
- 43.Sarma U, Flanagan A. Macrophage colony-stimulating factor induces substantial osteoclast generation and bone resorption in human bone marrow cultures. Blood. 1996;88:2531–2540. [PubMed] [Google Scholar]
- 44.MacDonald B, Takahashi N, McManus L, Holahan J, Mundy G, Roodman G. Formation of multinucleated cells that respond to osteotropic hormones in long term human bone marrow cultures. Endocrinology. 1987;120:2326–2333. doi: 10.1210/endo-120-6-2326. [DOI] [PubMed] [Google Scholar]
- 45.Tanaka S, Takahashi N, Udagawa N, Tamura T, Akatsu T, Stanley R, Kurokawa T, Suda T. Macrophage colony-stimulating factor is indispensable for both proliferation and differentiation of osteoclast progenitors. J Clin Invest. 1993;91:257–263. doi: 10.1172/JCI116179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hemesath T, Price ER, Takemota C, Badalian T, Fisher D. MAPK links transcription factor microphthalmia to c-Kit signaling in melanocytes. Nature. 1998;391:298–301. doi: 10.1038/34681. [DOI] [PubMed] [Google Scholar]
- 47.Lowings P, Yavuzer U, Goding CR. Positive and negative elements regulate a melanocyte-specific promoter. Mol Cell Biol. 1992;56:3653–3662. doi: 10.1128/mcb.12.8.3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yasumoto K, Yokoyama K, Shibata K, Tomita Y, Shibahara S. Microphthalmia-associated transcription factor as a regulator for melanocyte-specific transcription of the human tyrosinase gene. Mol Cell Biol. 1994;14:8058–8070. doi: 10.1128/mcb.14.12.8058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yasumoto K, Mahalingam H, Suzuki H, Yoshizawa M, Yokoyama K, Shibahara M. Transcriptional activation of the melanocyte-specific genes by the human homolog of the mouse microphthalmia protein. J Biochem. 1995;118:874–881. doi: 10.1093/jb/118.5.874. [DOI] [PubMed] [Google Scholar]
- 50.Morii E, Tsujimura T, Jippo T, Hashimoto K, Takebayashi K, Tsujino K, Nomura S, Yamamoto M, Kitamura Y. Regulation of mouse mast cell protease 6 gene expression by transcription factor encoded by the milocus. Blood. 1996;88:2488–2494. [PubMed] [Google Scholar]
- 51.Ebi Y, Kanakura Y, Jippo-Kanemoto T, Tsujimura T, Furitsu T, Ikeda H, Adachi S, Kasugai T, Nomura S, Kanayama Y, Yamatodani A, et al. Low c-kit expression of cultured mast cells of mi/mi genotype may be involved in their defective responses to fibroblasts that express the ligand for c-kit. . Blood. 1992;80:1454–1462. [PubMed] [Google Scholar]
- 52.Tsujimura T, Morii E, Nozaki M, Hashimoto K, Moriyama Y, Takebayashi K, Kondo T, Kanakura Y, Kitamura Y. Involvement of transcription factor encoded by the mi locus in the expression of c-kitreceptor tyrosine kinase in cultured mast cells of mice. Blood. 1996;88:1225–1233. [PubMed] [Google Scholar]