Abstract
Human histocompatibility leukocyte antigen (HLA)-E is a nonclassical HLA class I molecule, the gene for which is transcribed in most tissues. It has recently been reported that this molecule binds peptides derived from the signal sequence of HLA class I proteins; however, no function for HLA-E has yet been described. We show that natural killer (NK) cells can recognize target cells expressing HLA-E molecules on the cell surface and this interaction results in inhibition of the lytic process. Furthermore, HLA-E recognition is mediated primarily through the CD94/NKG2-A heterodimer, as CD94-specific, but not killer cell inhibitory receptor (KIR)–specific mAbs block HLA-E–mediated protection of target cells. Cell surface HLA-E could be increased by incubation with synthetic peptides corresponding to residues 3–11 from the signal sequences of a number of HLA class I molecules; however, only peptides which contained a Met at position 2 were capable of conferring resistance to NK-mediated lysis, whereas those having Thr at position 2 had no effect. Interestingly, HLA class I molecules previously correlated with CD94/NKG2 recognition all have Met at residue 4 of the signal sequence (position 2 of the HLA-E binding peptide), whereas those which have been reported not to interact with CD94/NKG2 have Thr at this position. Thus, these data show a function for HLA-E and suggest an alternative explanation for the apparent broad reactivity of CD94/NKG2 with HLA class I molecules; that CD94/NKG2 interacts with HLA-E complexed with signal sequence peptides derived from “protective” HLA class I alleles rather than directly interacting with classical HLA class I proteins.
In humans, there are three classical class I MHC molecules (HLA-A, -B, and -C). These molecules consist of a peptide bound to a transmembrane glycoprotein subunit, the heavy chain, in association with a soluble light chain, β2-microglobulin (1). Nonclassical MHC class I molecules show homology to classical class I molecules but generally have limited polymorphism, low cell surface expression, and more restricted tissue distribution (2). The function of these nonclassical class I molecules remains unclear, but some of them may have more specialized antigen presentation activities, e.g., mouse H2-M3 presents N-formylated peptides (3). In humans, the nonclassical HLA-G molecule binds a wide range of peptides derived from cellular proteins and has been suggested to play an important role in the maintenance of maternal tolerance to the fetus by interacting with inhibitory receptors on NK cells (4). HLA-E is another nonclassical class I molecule and, like classical MHC class I loci, the HLA-E gene is highly transcribed in many tissues (5–7). Mouse cell lines transfected with HLA-E and human β2-microglobulin generally exhibit low levels of cell surface expression, which has been attributed to a lack of appropriate endogenous peptides (8). Recent in vitro studies have shown that HLA-E can bind peptides derived from the signal sequences of certain HLA class I molecules and that the primary peptide anchor residues are at positions 2 and 9 (9).
NK cells are one of the three lineages of lymphocytes and are thought to control viral infections and tumor development (10). They are capable of killing MHC class I negative target cells without prior sensitization. Furthermore, the expression of class I molecules on target cells renders them resistant to lysis by most NK cells (11). Inhibitory receptors expressed by human NK cells belong to either the immunoglobulin (killer cell inhibitory receptor; KIR) or the C-type lectin superfamily. The best characterized members of the KIR family contain two (p58 molecules: CD158) or three (p70 molecules: NKB1) immunoglobulin-like domains and different receptors of this family bind defined groups of classical HLA class I molecules (12, 13). The second family of receptors is composed of two subunits: CD94 paired with one member of the NKG2 family of proteins. The CD94–NKG2-A heterodimer transmits signals that lead to the inhibition of the lytic process. Numerous studies suggest that this heterodimer recognizes a broad panel of classical HLA class I molecules (14–19).
Recognition of nonclassical HLA class I molecules by NK cells has been recently described. Specifically, numerous reports suggest that HLA-G can confer protection from lysis to otherwise sensitive target cells, although the identity of the receptors involved in this interaction remains controversial (20–24). In this study we examined whether another nonclassical class I molecule, HLA-E, can be recognized by NK cells. Our results show that NK cells can interact with HLA-E complexed with specific peptides on target cells and that this recognition is mediated, at least partially, if not solely, by CD94–NKG2.
Materials and Methods
Reagents, Peptides, and mAbs.
Media and supplements were purchased from BioFluids (Rockville, MD) and FCS from Hyclone (Logan, UT). G418 and Hygromycin B were obtained from GIBCO BRL (Baltimore, MD). Recombinant IL-2 was a gift from Hoffman-LaRoche (Nutley, NJ). Peptides (Table 1) were synthesized and purified as previously described (25). The following mAbs were used: B9.12.1 (anti–HLA class I); GL183 (anti-CD158b); EB6 (anti-CD158a); HP-3B1 (anti-CD94; all from Immunotech, Westbrook, ME); 3G8 (anti-CD16 from PharMingen, San Diego, CA); Leu-19 (anti-CD56; from Becton-Dickinson, San Jose, CA); UCHT1 (anti-CD3; from DAKO, Carpinteria, CA); and fluorescein-labeled goat anti– mouse IgG F(ab′)2 (from the Jackson Immunoresearch Laboratories, West Grove, PA).
Table 1.
Peptides Used in this Study
| Name | Sequence | Protein of origin and position in the intact protein | ||
|---|---|---|---|---|
| MAGE | EADPTGHSY | MAGE-1 161-169 | ||
| RT | ILKEPVHGV | HIV-1 RT 476-484 | ||
| B27 | VTAPRTLLL | HLA-B*1301, -B*1801, -B*2705 §, -B*3701, -B*4002, -B*4402, | ||
| -B*4701, -B*5401, -B*5501, and -B*5601 leader sequences 3–11‡ | ||||
| B15 | VTAPRTVLL | HLA-B*1501, -B*3501, -B*4101, -B*4501, -B*4601, -B*4901, | ||
| -B*5001, -B*5101, -B*5201, -B*5301, -B*5701, -B*5801, | ||||
| and -B*7801 leader sequences 3-11‡ | ||||
| B7 | VMAPRTVLL | HLA-B*0702 ‖, -B*0801, -B*1401, -B*1402, -B*3901, -B*3902, -B*4201, | ||
| and -B*4801 leader sequences 3–11‡ | ||||
| Cw3 | VMAPRTLIL | HLA-Cw*0302, -Cw*0304, -Cw*0401, -Cw*0801, -Cw*1201, -Cw*1401, | ||
| and -Cw*0602 leader sequences 3–11‡ |
This list is derived from the SWISS-PROT database using the PROWL search program (http://prowl.rockefeller.edu), but does not include all HLA class I molecules that possess these leader sequences.
Cell lines and NK Cells.
The human mutant lymphoblastoid cell lines 721.221 and 721.221 transfected with HLA-Cw*0304 (named here as 721.221-Cw3) have been previously described (26). The murine transporter associated with antigen processing (TAP)-2–deficient RMA-S cell line was obtained from Dr. Marika Pla (INSERM, Hopital St. Louis, Paris, France). Cells were cotransfected by electroporation with 15 μg of a BamHI-linearized 15-kb BamHI/SalI subclone of the human β2-microglobulin gene in pUC19 (27), together with 5 μg of a Sal I-linearized derivative of the Cos203 vector (28), in which the EBV sequences and Cos sites had been deleted by Eca digestion and that contained either no insert or a HindIII/BglII fragment of cosmid cd3.14 (7) encoding the HLA-E*01033 allele. Transfectants expressing human β2-microglobulin alone (referred to here as RMA-S) or human β2-microglobulin and HLA-E*01033 (RMA-S/HLA-E) were selected in complete RPMI supplemented with 0.4 mg/ml of Hygromycin B. NK leukocytosis 221707 cells were provided by Dr. J.P. Houchins (Department of Laboratory Medicine and Pathology, University of Minnesota, Minneapolis, MN; 29). NK clones from healthy donors were obtained and grown as previously described (18).
Immunostaining.
Target cells were incubated at 26°C or 37°C overnight in 5% CO2 in the absence or presence of saturating amounts of the indicated peptide (300 μM). Cells were then washed and analyzed for HLA class I expression by flow cytometry using the B9.12.1 mAb. All NK clones were CD3−, CD56+, CD16+, and CD94+.
Cytotoxicity Assays.
Target cells were prepared as described for immunostaining, except that for the last 90–120 min the cells were labeled with sodium [51Cr]chromate (DuPont, Charlotte, NC). The cytotoxic activity of NK cells was assayed in a 4-h 51Cr-release assay as previously described (18, 30).
Results and Discussion
The 721.221 cell line that does not express HLA-A, -B, -C, or -G has been shown to express the nonclassical class I molecule HLA-E (9). This ability to express HLA-E could explain the low but detectable level of HLA class I molecules on the surface of these cells when they are grown at 37°C (data not shown). In vitro studies suggest that HLA-E, like its murine homologue Qa-1b (31), binds signal sequence–derived peptides (9). In an attempt to increase cell surface expression of HLA-E, four synthetic peptides derived from the signal sequences of classical HLA class I molecules and two control peptides (see Table 1) were individually incubated with 721.221 cells. When the incubation was performed at 37°C, no significant change in the cell surface expression of HLA class I molecules was observed (data not shown). Since the incubation of a murine cell line transfected with HLA-E at 26°C increased its expression on the cell surface (8), 721.221 cells were incubated with these peptides at 26°C. Under these conditions the signal sequence–derived peptides induced a small, but reproducible increase in class I cell surface expression (Fig. 1 A). The melanoma-associated antigen (MAGE) and reverse transcriptase (RT) peptides did not enhance the class I expression with respect to cells incubated without peptides, in agreement with previous data indicating that these two peptides do not bind HLA-E (9). Thus, cell surface expression of HLA-E can be enhanced on 721.221 cells by incubation with appropriate peptides at 26°C.
Figure 1.
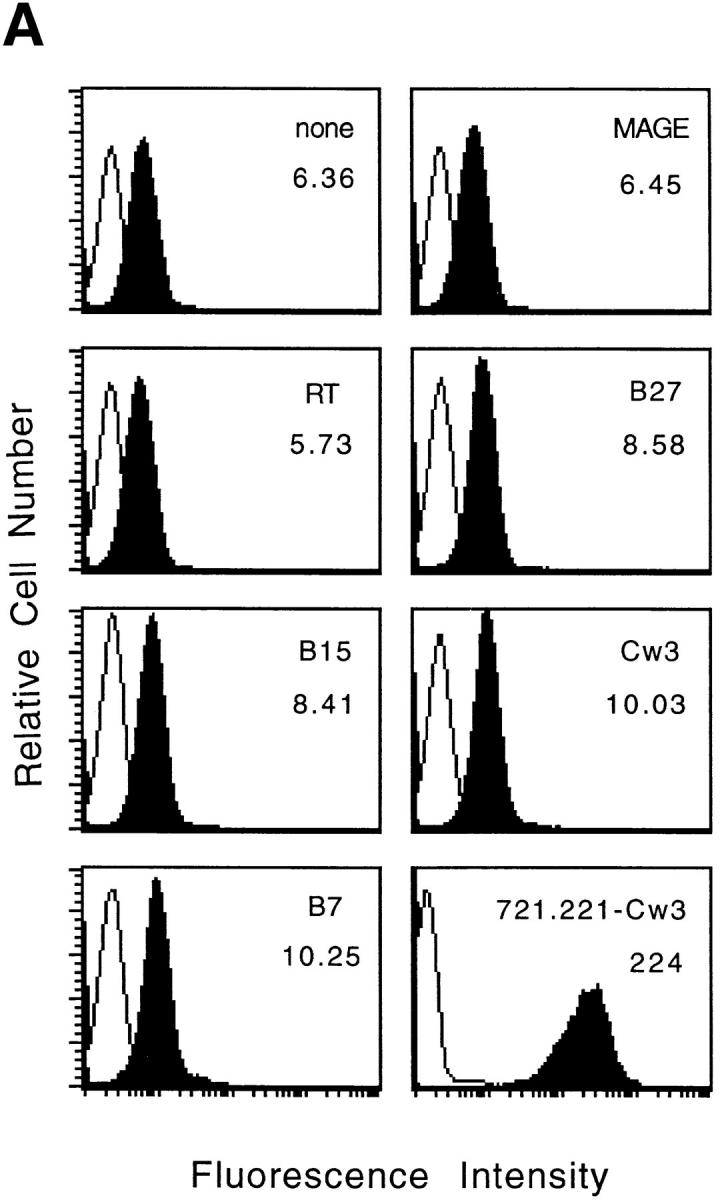
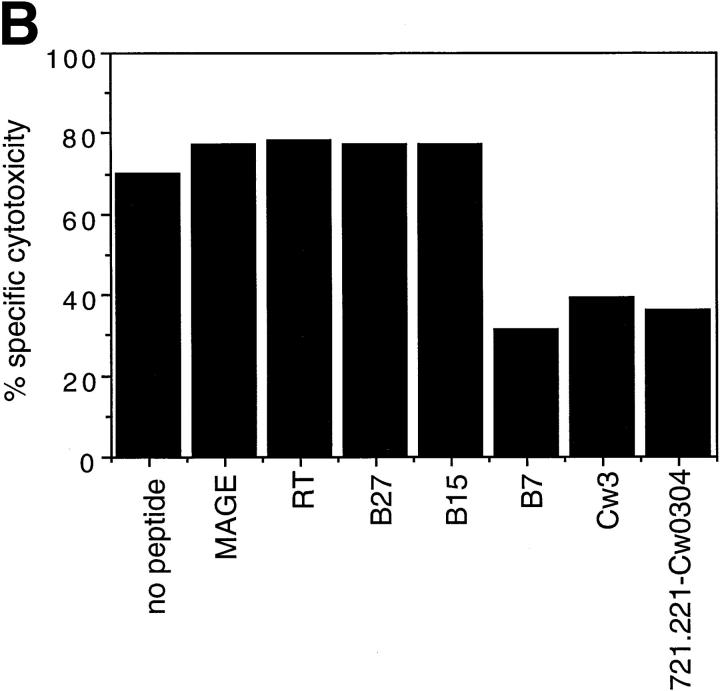
Cell surface expression of HLA-E molecules complexed with HLA class I signal-sequence derived peptides confers protection against NK cell– mediated lysis. (A) 721.221 target cells were incubated in the absence or presence of the indicated peptides at 26°C and analyzed for HLA class I expression. The empty histograms correspond to the secondary antibody (goat anti–mouse) alone and the filled histograms show the binding of the HLA class I–specific mAb B9.12.1. The histogram in the lower right hand corner shows the HLA class I expression on 721.221 cells transfected with HLA-Cw*0304. The mean fluorescence channel of B9.12.1 staining is shown inside the boxes. (B) 221707 NK cells were assayed for cytotoxicity against 721.221 target cells incubated with or without the indicated peptides at 26°C or 721.221-Cw3 cells. The E/T ratio was 15:1 and the percentage of specific lysis shown is the average of three experiments. The standard deviation was <10%.
Although NK recognition of the nonclassical HLA-G molecules has recently been a focus of attention, NK recognition of other nonclassical class I molecules has not been reported. Previous data have shown that 721.221 cells are susceptible to NK lysis (14–16), indicating that the small amount of HLA-E expressed on the surface of these cells is not sufficient to inhibit NK-mediated lysis or that the HLA-E is not complexed with the appropriate peptide(s). To determine whether HLA-E could serve as a ligand for NK receptors, 721.221 cells were cultured with the peptides described above before their use as target cells in cytotoxicity assays with 221707 NK cells. When incubated at 37°C, 721.221 cells were susceptible to NK–mediated lysis in both the presence and absence of peptides (data not shown). However, when target cells were incubated with peptide at 26°C, NK-mediated lysis was dramatically inhibited by the B7 and Cw3 peptides. This decrease in lysis was comparable to that obtained by transfection of 721.221 cells with HLA-Cw*0304 (Fig. 1 B). Incubation with the other four peptides had no effect on lysis. These data suggest that HLA-E in association with specific peptides can be recognized by NK cells, even when the levels of class I expression are relatively low (compare the class I expression on 721.221 cells in the presence of B7 and Cw3 peptides with that on 721.221-Cw3 cells in Fig. 1 A).
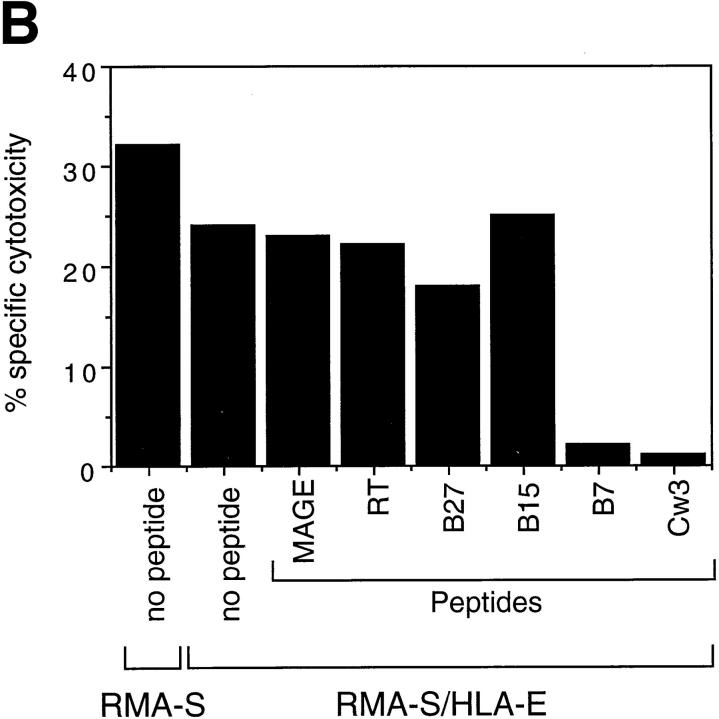
To eliminate the possibility that other HLA-like molecules (e.g., HLA-F) expressed by 721.221 cells are involved in NK cell recognition, we made use of murine RMA-S cells transfected with HLA-E. Incubation with appropriate peptides increased the levels of class I expression on these cells both at 26°C (data not shown) and at 37°C (Fig. 2 A). Although all four class I signal sequence–derived peptides induced HLA-E expression in comparison to the two control peptides, only the B7 and Cw3 peptides induced resistance to NK-mediated lysis (Fig. 2 B). These results clearly show that HLA-E molecules can provide protection from NK-mediated lysis. The observation that only the B7 and Cw3 peptides protected target cells from lysis, suggests a degree of peptide specificity. The biochemical basis for the observed “peptide specificity” remains to be determined.
Figure 2.
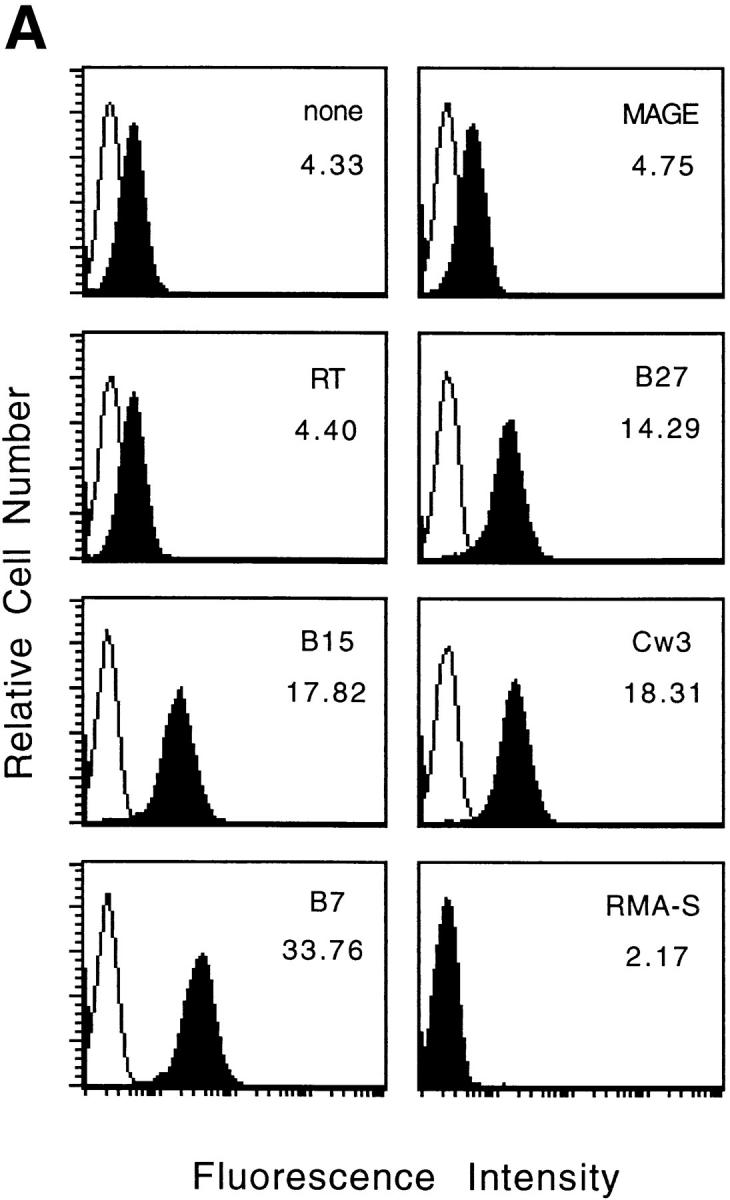
Expression of HLA-E by RMA-S cells with appropriate peptides confers resistance to NK-mediated lysis. (A) RMA-S/HLA-E cells were incubated in the absence or presence of the indicated peptide at 37°C and analyzed for cell surface HLA class I expression. The empty histograms represent secondary Ab controls (goat anti–mouse) and the filled histograms show the binding of the B9.12.1 mAb. The histogram in the lower right hand corner shows staining of RMA-S cells transfected with human β2-microglobulin alone. The mean fluorescence channel of B9.12.1 staining is shown inside the boxes. (B) 221707 NK cells were assayed for cytotoxicity against RMA-S and RMA-S/ HLA-E cells incubated in the presence or absence of the indicated peptides at 37°C. The E/T ratio was 40:1.
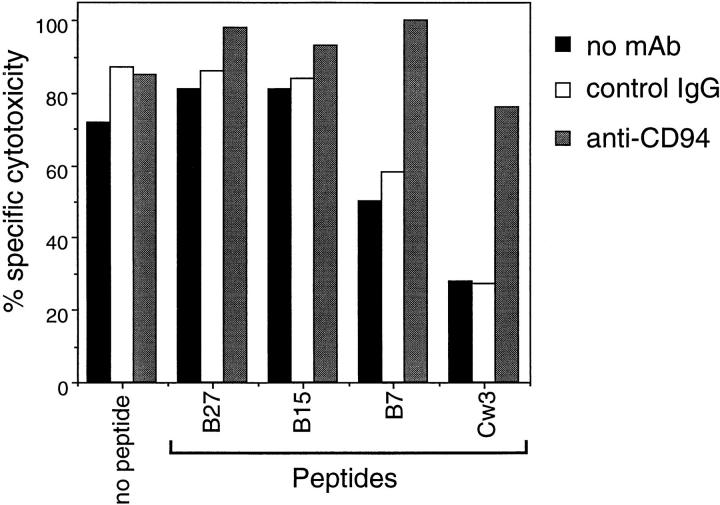
To identify the inhibitory receptor involved in HLA-E– mediated protection from NK cell lysis, we made use of anti-CD94 and anti-CD158 specific mAbs and a panel of NK clones. The 221707 NK cells used in the above experiments are CD158a− and CD158b−, but express the inhibitory CD94–NKG2-A heterodimer as determined by immunoblot and redirected antibody-dependent cellular cytotoxicity assay (data not shown). CD94–NKG2-A has been implicated in the recognition of a broad panel of HLA class I molecules including the nonclassical class I molecule HLA-G (14–17, 22–24). Thus, we examined whether anti-CD94 mAb could block the protection conferred on target cells by peptide-induced stabilization of HLA-E. The presence of anti-CD94 mAb did not affect the levels of lysis obtained in the presence of both B27 and B15 peptides. In contrast, the addition of anti-CD94 mAb blocked the inhibition of lysis conferred by the B7 and Cw3 peptides, suggesting that CD94–NKG2-A is the receptor involved in HLA-E recognition (Fig. 3).
Figure 3.
HLA-E mediated protection is blocked by anti-CD94 mAb. 721.221 target cells were incubated with or without the indicated peptides at 26°C and then used in a cytotoxicity assay with 221707 NK cells in the presence of anti-CD94 or control antibodies at a final concentration of 10 μg/ml. The E/T ratio was 15:1.
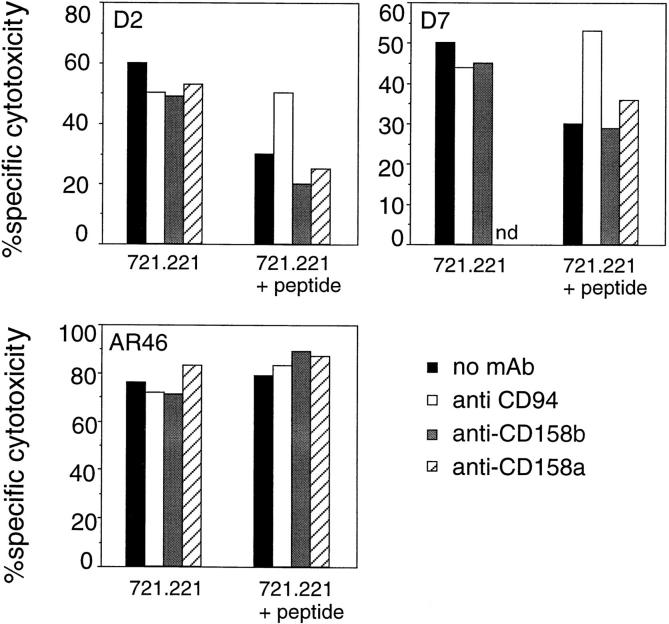
To determine if members of the KIR family were involved in HLA-E recognition, three NK clones were studied: D2 (CD94+, CD158a−, CD158b−); D7 (CD94+, CD158a+, CD158b+); and AR46 (CD94+, CD158a+, CD158b+). NK clone D2 has the same phenotype as the 221707 NK cells used above. Consequently, the addition of anti-CD94 mAb restored NK-mediated lysis when HLA-E was complexed with a protective peptide. Similar data were obtained with the CD158a+, CD158b+ NK clone D7; once again the presence of anti-CD94 mAb restored lysis. In contrast, the addition of anti-CD158a and anti-CD158b mAb had no effect on lysis, suggesting that CD158a and CD158b receptors did not interact with HLA-E. Finally, data obtained from NK clone AR46 confirmed that the CD158a and CD158b receptors were not involved in HLA-E recognition (Fig. 4). The AR46 NK clone kills target cells incubated with the Cw3 peptide despite the presence of the inhibitory CD158a and CD158b molecules. No CD94–NKG2 mediated protection was observed in AR46 cells, as they express a CD94–NKG2 receptor that activates NK killing as determined by rADCC (data not shown). The ligation of CD94 on NK cells has been previously shown to provide either activation or inhibition of NK cell lysis, depending on the NK clone. These divergent responses depend on the particular NKG2 protein associated with CD94; although NKG2-A transmits negative signals leading to the inhibition of the cytolytic activity, signaling through NKG2-C is thought to activate NK cells (19, 23, 32, 33).
Figure 4.
HLA-E is not recognized by CD158a and CD158b KIR receptors. 721.221 target cells were incubated at 26°C with or without the Cw3 peptide and then used in a cytotoxicity assay with the indicated NK clones (D2, D7, and AR46) in the presence of anti-CD94, anti-CD158a, or anti-CD158b mAb at a final concentration of 10 μg/ml. The E/T ratio was 5:1. nd, not determined.
The CD94–NKG2-A inhibitory NK receptor has been previously implicated in the recognition of a broad panel of HLA class I molecules, but little is known about the structural basis for this interaction. Previous data has shown that the low levels of cell surface expression of HLA-E was limited, at least in part, by the lack of appropriate peptides (8), and that a potential source of such peptides are the signal sequences of certain class I molecules (9). It is worth noting that the molecules encoded by transfected HLA class I genes that apparently interact with CD94–NKG2-A all possess Met at position 4 of the signal sequence, whereas those alleles that do not confer resistance all encode Thr at the same position (14–16). This suggests an alternative explanation for the apparently broad reactivity of CD94–NKG2-A with HLA class I molecules, namely that CD94–NKG2-A interacts directly with HLA-E complexed with peptides derived from the signal sequence of HLA class I molecules. Overexpression of transfected classical HLA class I alleles in the HLA-A–, -B–, and -C–negative, but HLA-E–positive 721.221 cells likely provides sufficient leader sequence– derived peptides to increase the levels of cell surface HLA-E to protect the otherwise sensitive targets. Moreover, the use of anti-HLA class I mAb to reverse the protection induced by classical HLA class I molecules may in fact be due to antibody masking of HLA-E rather than of the transfected HLA class I allele.
These data demonstrate a novel mechanism by which NK cells can monitor self-HLA class I expression through the recognition of HLA class I–derived peptides in the context of HLA-E. This may be important in preventing autoreactivity of NK cells, in particular by NK cells that lack KIR specific for self HLA class I molecules.
Footnotes
We would like to thank Dr. J.P. Houchins for the 221707 NK cells and Hoffman-LaRoche for rIL-2. We also thank Drs. E. Fernandez, J. Ochoa, J. Shuman, and F. Zappacosta for advice and comments on the manuscript.
Address correspondence to John E. Coligan, Laboratory of Molecular Structure, National Institute of Allergy and Infectious Diseases, National Institute of Health, 12441 Parklawn Dr., Rockville, MD 20852. Phone: 301-496-8247; Fax: 301-402-0284.
References
- 1.Madden DR. The three-dimensional structure of peptide-MHC complex. Annu Rev Immunol. 1995;13:587–622. doi: 10.1146/annurev.iy.13.040195.003103. [DOI] [PubMed] [Google Scholar]
- 2.Shawar SM, Vyas JV, Rodgers JR, Rich RR. Antigen presentation by major histocompatibility complex class I-bmolecules. Annu Rev Immunol. 1994;12:839–880. doi: 10.1146/annurev.iy.12.040194.004203. [DOI] [PubMed] [Google Scholar]
- 3.Shawar SM, Cook RG, Rodgers JR, Rich RR. Specialized functions of MHC class I molecules. I. An N-formyl peptide receptor is required for construction of the class I antigen Mta. J Exp Med. 1990;171:897–912. doi: 10.1084/jem.171.3.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee N, Malacko AR, Ishitani A, Chen MC, Bajorath J, Marquardt H, Geraghty DE. The membrane-bound and soluble forms of HLA-G bind identical sets of peptides but differ with respect to TAP association. Immunity. 1995;3:591–600. doi: 10.1016/1074-7613(95)90130-2. [DOI] [PubMed] [Google Scholar]
- 5.Koller BH, Geraghty DE, Shimizu Y, DeMars R, Orr HT. HLA-E: a novel HLA class I gene expressed in resting T-lymphocytes. J Immunol. 1988;141:897–904. [PubMed] [Google Scholar]
- 6.Wei X, Orr HT. Differential expression of HLA-E, HLA-F, and HLA-G transcripts in human tissue. Hum Immunol. 1990;29:131–142. doi: 10.1016/0198-8859(90)90076-2. [DOI] [PubMed] [Google Scholar]
- 7.Ulbrecht M, Honka T, Person S, Johnson JP, Weiss EH. The HLA-E gene encodes two diferentially regulated transcripts and a cell surface protein. J Immunol. 1992;149:2945–2953. [PubMed] [Google Scholar]
- 8.Ulbrecht M, Kellermann J, Johnson JP, Weiss EH. Impaired intracellular transport and cell surface expression of nonpolymorphic HLA-E: evidence for inefficient peptide binding. J Exp Med. 1992;176:1083–1090. doi: 10.1084/jem.176.4.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braud V, Jones EY, McMichael A. The human major histocompatibility complex class Ib molecule HLA-E binds signal sequence–derived peptides with primary anchor residues at positions 2 and 9. Eur J Immunol. 1997;27:1164–1169. doi: 10.1002/eji.1830270517. [DOI] [PubMed] [Google Scholar]
- 10.Trinchieri G. Biology of natural killer cells. Adv Immunol. 1989;47:187–376. doi: 10.1016/S0065-2776(08)60664-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ljunggren H-G, Karre K. In search of the “missing self”: MHC molecules and NK cell recognition. Immunol Today. 1990;11:237–244. doi: 10.1016/0167-5699(90)90097-s. [DOI] [PubMed] [Google Scholar]
- 12.Long EO, Burshtyn DN, Clark WP, Peruzzi M, Rajagopalan S, Rojo S, Wagtmann N, Winter CC. Killer cell inhibitory receptors: diversity, specificity and function. Immunol Rev. 1997;155:135–144. doi: 10.1111/j.1600-065x.1997.tb00946.x. [DOI] [PubMed] [Google Scholar]
- 13.Moretta A, Biassoni R, Bottino C, Pende D, Vitale M, Poggi A, Mingari MC, Moretta L. Major histocompatibility complex class I–specific receptors on human natural killer and T lymphocytes. Immunol Rev. 1997;155:105–117. doi: 10.1111/j.1600-065x.1997.tb00943.x. [DOI] [PubMed] [Google Scholar]
- 14.Phillips JH, Chang C, Mattson J, Gumperz JE, Parham P, Lanier LL. CD94 and a novel associated protein (94AP) form a NK cell receptor involved in the recognition of HLA-A, HLA-B, and HLA-C allotypes. Immunity. 1996;5:163–172. doi: 10.1016/s1074-7613(00)80492-6. [DOI] [PubMed] [Google Scholar]
- 15.Lazetic S, Chang C, Houchins JP, Lanier LL, Phillips JH. Human natural killer cell receptors involved in MHC class I recognition are disulfide-linked heterodimers of CD94 and NKG2 subunits. J Immunol. 1996;157:4741–4745. [PubMed] [Google Scholar]
- 16.Sivori S, Vitali M, Bottino C, Marcenaro E, Sanseverino L, Parolini S, Moretta L, Moretta A. CD94 functions as a natural killer cell inhibitory receptor for different HLA class I alleles: identification of the inhibitory form of CD94 by the use of novel monoclonal antibodies. Eur J Immunol. 1996;26:2487–2492. doi: 10.1002/eji.1830261032. [DOI] [PubMed] [Google Scholar]
- 17.Carretero M, Cantoni C, Bellon T, Bottino C, Biassoni R, Rodriguez A, Perez-Villar JJ, Moretta L, Moretta A, Lopez-Botet M. The CD94 and NKG2-A C-type lectins covalently assemble to form a NK cell inhibitory receptor for HLA class I molecules. Eur J Immunol. 1997;27:563–567. doi: 10.1002/eji.1830270230. [DOI] [PubMed] [Google Scholar]
- 18.Brooks AG, Posch PE, Scorzelli CJ, Borrego F, Coligan JE. NKG2A complexed with CD94 defines a novel inhibitory natural killer cell receptor. J Exp Med. 1997;185:795–800. doi: 10.1084/jem.185.4.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Houchins JP, Lanier LL, Niemi EC, Phillips JH, Ryan JC. Natural killer cell cytolytic activity is inhibited by NKG2-A and activated by NKG2-C. J Immunol. 1997;158:3603–3609. [PubMed] [Google Scholar]
- 20.Pazmany L, Mandelboim O, Vales-Gomez M, Davis DM, Reyburn HT, Strominger JL. Protection from natural killer cell–mediated lysis by HLA-G expression on target cells. Science. 1996;274:792–795. doi: 10.1126/science.274.5288.792. [DOI] [PubMed] [Google Scholar]
- 21.Munz C, Holmes N, King A, Loke YW, Colonna M, Schild H, Rammensee H-G. Human histocompatibility leukocyte antigen (HLA)-G molecules inhibit NKAT3 expressing natural killer cells. J Exp Med. 1997;185:385–391. doi: 10.1084/jem.185.3.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soderstrom K, Corliss B, Lanier LL, Phillips JH. CD94/NKG2 is the predominant inhibitory receptor involved in recognition of HLA-G by decidual and peripheral blood NK cells. J Immunol. 1997;159:1072–1075. [PubMed] [Google Scholar]
- 23.Pende D, Sivori S, Accame L, Pareti L, Falco M, Geraghty D, Le Bouteiller P, Moretta L, Moretta A. HLA-G recognition by human natural killer cells. Involvement of CD94 both as inhibitory and as activating receptor complex. Eur J Immunol. 1997;27:1875–1880. doi: 10.1002/eji.1830270809. [DOI] [PubMed] [Google Scholar]
- 24.Perez-Villar JJ, Melero I, Navarro F, Carretero M, Bellon T, Llano M, Colonna M, Geraghty DE, Lopez-Botet M. The CD94/NKG2-A inhibitory receptor complex is involved in natural killer cell–mediated recognition of cells expressing HLA-G1. J Immunol. 1997;158:5736–5743. [PubMed] [Google Scholar]
- 25.Parker KC, Dibrino M, Hull L, Coligan JE. The β2-microglobulin dissociation rate is an accurate measure of the stability of MHC class I heterotrimers and depends on which peptide is bound. J Immunol. 1992;149:1896–1904. [PubMed] [Google Scholar]
- 26.Wagtmann N, Rajagopalan S, Winter CC, Peruzzi M, Long EO. Killer cell inhibitory receptors specific for HLA-C and HLA-B identified by direct binding and by functional transfer. Immunity. 1995;3:801–809. doi: 10.1016/1074-7613(95)90069-1. [DOI] [PubMed] [Google Scholar]
- 27.Perarnau BM, Guillet AC, Hakem R, Barad M, Lemonnier FA. Human β2-microglobulin specifically enhances cell-surface expression of HLA class I molecules in transfected murine cells. J Immunol. 1988;141:1383–1389. [PubMed] [Google Scholar]
- 28.Kioussis D, Wilson F, Daniels C, Leveton C, Taverne J, Playfair JH. Expression and rescuing of a cloned human tumour necrosis factor gene using an EBV-based shuttle cosmid vector. EMBO (Eur Mol Biol Organ) J. 1987;6:355–361. doi: 10.1002/j.1460-2075.1987.tb04762.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Houchins JP, Yabe T, McSherry C, Miyokawa N, Bach FH. Isolation and characterization of NK cell or NK/T cell-specific cDNA clones. J Mol Cell Immunol. 1990;4:295–306. [PubMed] [Google Scholar]
- 30.Zappacosta F, Borrego F, Brooks AG, Parker KC, Coligan JE. Peptides isolated from HLA-Cw*0304 confer different degrees of protection from natural killer cell– mediated lysis. Proc Natl Acad Sci USA. 1997;94:6313–6318. doi: 10.1073/pnas.94.12.6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aldrich CJ, DeCloux A, Woods AS, Cotter RJ, Soloski MJ, Forman J. Identification of a Tap-dependent leader peptide recognized by alloreactive T cells specific for class Ib antigen. Cell. 1994;79:649–658. doi: 10.1016/0092-8674(94)90550-9. [DOI] [PubMed] [Google Scholar]
- 32.Perez-Villar JJ, Melero I, Rodriguez A, Carretero M, Aramburu J, Sivori S, Orengo AM, Moretta A, Lopez-Botet M. Functional ambivalence of the Kp43 (CD94) NK cell–associated surface antigen. J Immunol. 1995;154:5779–5788. [PubMed] [Google Scholar]
- 33.Brumbaugh KM, Perez-Villar JJ, Dick CJ, Schoon RA, Lopez-Botet M, Leibson PJ. Clonotypic differences in signalling from CD94 (Kp43) on NK cells lead to divergent cellular responses. J Immunol. 1996;157:2804–2812. [PubMed] [Google Scholar]
- 34.Moretta A, Vitale M, Sivori S, Bottino C, Morelli L, Augugliaro R, Barbaresi M, Pende D, Ciccone E, Lopez-Botet M, Moretta L. Human natural killer cell receptors for HLA class I molecules: evidence that Kp43 (CD94) molecule functions as receptor for HLA-B alleles. J Exp Med. 1994;180:545–555. doi: 10.1084/jem.180.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]