Abstract
The three-dimensional structure of the complex between a T cell receptor (TCR) β chain (mouse Vβ8.2Jβ2.1Cβ1) and the superantigen (SAG) staphylococcal enterotoxin C3 (SEC3) has been recently determined to 3.5 Å resolution. To evaluate the actual contribution of individual SAG residues to stabilizing the β–SEC3 complex, as well as to investigate the relationship between the affinity of SAGs for TCR and MHC and their ability to activate T cells, we measured the binding of a set of SEC3 and staphylococcal enterotoxin B (SEB) mutants to soluble recombinant TCR β chain and to the human MHC class II molecule HLA-DR1. Affinities were determined by sedimentation equilibrium and/or surface plasmon detection, while mitogenic potency was assessed using T cells from rearrangement-deficient TCR transgenic mice. We show that there is a clear and simple relationship between the affinity of SAGs for the TCR and their biological activity: the tighter the binding of a particular mutant of SEC3 or SEB to the TCR β chain, the greater its ability to stimulate T cells. We also find that there is an interplay between TCR–SAG and SAG–MHC interactions in determining mitogenic potency, such that a small increase in the affinity of a SAG for MHC can overcome a large decrease in the SAG's affinity for the TCR. Finally, we observe that those SEC3 residues that make the greatest energetic contribution to stabilizing the β–SEC3 complex (“hot spot” residues) are strictly conserved among enterotoxins reactive with mouse Vβ8.2, thereby providing a basis for understanding why SAGs having other residues at these positions show different Vβ-binding specificities.
Superantigens (SAGs)1 are a class of disease-causing and immunostimulatory proteins of bacterial or viral origin. In addition to causing toxic shock syndrome and food poisoning (1, 2), SAGs have been implicated in a number of autoimmune disorders, including diabetes mellitus (3), multiple sclerosis (4), and rheumatoid arthritis (5), through the activation of T cells specific for self-antigens. SAGs are able to recognize particular elements on the Vβ domain of TCRs, largely irrespective of their peptide–MHC specificity, leading to stimulation of a disproportionally large fraction of the T cell population. The activated T cells release massive amounts of cytokines such as IL-2 and tumor necrosis factor, contributing to the symptoms caused by SAGs.
The structurally and immunologically best characterized group of SAGs are the Staphylococcus aureus enterotoxins, which are mainly associated with food poisoning and toxic shock syndrome (1, 2). The three-dimensional structure of the complex between staphylococcal enterotoxin C3 (SEC3) and the β chain (Vβ8.2Jβ2.1.Cβ1) of a mouse TCR (designated 14.3.d) specific for a peptide of influenza virus hemagglutinin (HA 110–120) in the context of I-Ed shows that CDR2 of the β chain, and to lesser extents CDR1 and the fourth hypervariable region (HV4), bind in a cleft between the small and large domains of the SAG (6). The structure of the TCR β–SEC3 complex agrees well with mutational and genetic studies implicating residues in Vβ CDR1, CDR2, and HV4 in SAG recognition (2, 7). In addition, mutagenesis of SAGs has revealed that the stimulatory activity of these molecules is affected when residues at the TCR binding site are altered (8). T cell stimulation by SAGs is generally thought to require simultaneous binding to MHC class II molecules on APCs and the Vβ element on T cells (9, 10). A model of the TCR–SAG– MHC complex constructed from the crystal structures of the TCR-β–SEC3 complex (6), of a TCR Vα domain (11), and of the complex between staphylococcal enterotoxin B (SEB) and an MHC class II molecule (12) suggests that the SAG acts like a wedge between the TCR and MHC molecules to displace the antigenic peptide away from the TCR combining site. In this way, the SAG circumvents the normal mechanism for T cell activation by recognition of specific peptide–MHC complexes (6).
To investigate the relationship between the affinity of SAGs for TCR and MHC and their ability to activate T cells, we have measured the binding of a set of SEC3 and SEB mutants to soluble recombinant 14.3.d β chain and to a human MHC class II molecule, HLA-DR1, loaded with influenza virus hemagglutinin peptide 306–318 (HA 306– 318). These mutants were generated by alanine-scanning mutagenesis of all SEC3 residues in contact to the TCR β chain in the β-SEC3 crystal structure, or by mutating selected TCR-contacting residues of SEB (which is structurally similar to SEC3 but binds the TCR more weakly) to those of SEC3. We show that there is a direct correlation between affinity and mitogenic potency, with SAGs that have the highest affinity for the TCR β chain being the most biologically active. We also show that a relatively small increase in the affinity of the SAG–MHC interaction is able to compensate a large decrease in SAG–TCR affinity. Finally, a comparison of the so-called “functional epitope” of SEC3 (those residues that contribute most to TCR binding) with the “structural epitope” (all SEC3 residues contacting the β chain in the crystal structure) enables us to explain the ability of different SAGs to recognize the same Vβ elements.
Materials and Methods
Reagents.
All chemicals were of analytical grade. Restriction endonucleases and DNA modifying enzymes were purchased from New England Biolabs, Inc. (Beverly, MA). Oligonucleotides were obtained from Integrated DNA Technologies (Coralville, IA). Radiolabeled [35S]dATP was from Amersham Corp. (Arlington Heights, IL).
Production of Recombinant TCR β Chain.
Unglycosylated 14.3.d VβCβ chain was obtained by elimination of four out of five potential N-linked glycosylation sites through site-directed mutagenesis and introduction of a termination codon at the end of the first C region exon of the β chain (13). Asparagines at positions 24, 74, and 121 were mutated to glutamine, while the glycosylation site at position 236 was eliminated by mutation of Ser238 to valine. The 14.3.d β chain was produced in J558L cells and affinity purified using the anti–mouse Cβ monoclonal antibody H57-597 (13, 14). The unmutated N-linked glycosylation site at position Cβ 186 was used in <10% of the chains, as judged by SDS-PAGE. The recombinant β chain was further purified by gel filtration on a Superdex 75 fast protein liquid chromatography column (Pharmacia Biotech AB, Uppsala, Sweden) before sedimentation equilibrium or BIAcore experiments to eliminate aggregated material that could interfere with affinity measurements (15).
Mutagenesis and Production of SEC3 and SEB Mutants.
The gene for SEB, seb, was obtained from C. Jones and S. Khan (University of Pittsburgh School of Medicine, Pittsburgh, PA; reference 16). Cloning and sequencing of the sec gene encoding SEC3 from Staphylococcal aureus strain FRI913 was reported previously by Hovde et al. (17). All SEB mutants and certain SEC3 mutants were produced by site-directed mutagenesis using a Muta-Gene M13 In Vitro Mutagenesis Kit (Bio-Rad, Richmond, CA). The corresponding genes, on a BamHI/HindIII fragment in the case of SEC3 and on a KpnI/HindIII fragment in the case of SEB, were subcloned into M13mp18. Mutagenic oligonucleotides were designed to replace wild-type codons with ones for alanine in the SEC3 gene or with codons for threonine, tyrosine, or valine in the SEB gene. The mutations were confirmed by DNA sequencing using a Sequenase Version 2.0 Kit (USB, Cleveland, OH). Mutated SEC3 genes were cloned into the expression vector pMIN164 (17) and mutated SEB genes into pUC18. The enterotoxins were expressed in BL21(DE3) Escherichia coli cells, and the periplasmic fraction, in which the SAGs are mostly contained, was obtained by osmotic shock as previously described (18). After dialysis against 20 mM Tris-HCl, pH 7.5, the periplasmic extract was applied to a RedA-Dye column (Amicon, Beverly, MA) as previously described (19). Bound proteins were eluted using a linear 0–500 mM NaCl gradient. The SAG-containing fractions were pooled and concentrated. Alternatively, SEC3 mutants were produced by subcloning the gene for SEC3 on a BamHI/HindIII fragment into pALTER. Oligonucleotide site-directed mutagenesis was accomplished using the Altered Sites In Vitro Mutagenesis Systems Kit (Promega, Madison, WI). Mutations were confirmed by sequencing as above. The mutated genes were cloned into pMIN164 (17) for expression in E. coli DH5α or S. aureus RN 9220. The SAGs were initially purified from stationary phase cultures by ethanol precipitation and preparative thin layer isoelectric focusing was as previously described (1). All SEC3 and SEB mutants were further purified on a MonoQ ion exchange column (Pharmacia Biotech AB) before binding and activation studies.
Production of Recombinant Peptide–HLA-DR1 Complex.
The pACDR1 virus encoding both soluble DRα and DRβ1*0101 chains was provided to R.-P. Sékaly by Dr. L. Stern (Massachusetts Institute of Technology, Boston, MA). The virus was amplified and cloned by plaque assay using procedures described by Summers and Smith (20). Spodoptera frugiperda (Sf21) cells (obtained from Dr. C. Richardson, Ontario Cancer Institute, Ontario, Canada) were grown in μ-carrier spinner flasks (Bellco Technology, Vineland, NJ) in Grace's medium (GIBCO BRL, Gaithersburg, MD) supplemented with 10% fetal calf serum, 1.66% mass/vol lactalbumin hydrolysate, 1.66% mass/vol Yeastolate, and 1% pluronic acid (all from GIBCO BRL). Sf21 cells (3–5 × 105/ml) were infected with a stock of recombinant baculovirus encoding DRα/DRβ1*0101 at a multiplicity of infection of 5–10 PFU/cell and grown at 26°C for 12 h. The infected cells were collected and returned to culture in an equal volume of Grace's medium containing 1% pluronic acid for 72 h after infection. The supernatant was collected and filtered, and protease inhibitors (0.1 μM pepstatin A, 0.5 mM phenylmethylsulfonylfluoride, 2.5 mM EDTA) and 0.02% mass/vol NaN3 were added. The DR αβ dimer was purified by affinity chromatography according to a slight modification of a protocol used by Stern and Wiley (21). In brief, Sf21 supernatants were passed over a Sepharose column coupled with L243 anti-DR monoclonal antibody. After loading, the column was extensively washed with PBS. The MHC protein was eluted using 50 mM 3-[cyclohexylamino]-1-propanesulfonic acid, pH 11.5, and was immediately neutralized in 20% (vol/vol) 0.5 M Tris-HCl, pH 7.0. The empty αβ DR1 dimer was loaded with the antigenic HA 306–318 peptide as previously described (21). A solution of 1.0 μM HLA-DR1 was incubated in PBS containing protease inhibitors (0.1 mM aminoethylbenzenesulfonylfluoride, 4 μg/ml E-64, and 1 mM EDTA; all from Boehringer Mannheim, Indianapolis, IN) and 0.02% NaN3 in the presence of 10 μM HA 306–318 peptide (Tana Laboratories, Houston, TX). The mixture was incubated for 3 d at 37°C and HA306-318/HLA-DR1 complex was concentrated and purified on a Sephadex G75 (Pharmacia Biotech AB) gel filtration column.
Sedimentation Equilibrium.
Equilibrium measurements of the interactions between the TCR-β chain and the various SAG mutants, as well as between HLA-DR1 and SAGs, were carried out with a Beckman XL-A Optima analytical ultracentrifuge using a four-hole, An 55 rotor. The samples were prepared by dialysis against 50 mM Tris-HCl, pH 7.5, at concentrations of the 1:1 complexes ranging from 4 to 10 μM. The experiments were performed at 25°C at rotor speeds between 20,000 and 24,000 rpm. The molar extinction coefficients used were: 43.2 mM−1cm−1 for the TCR-β chain; 34.2 mM−1cm−1 for SEC3 and SEC3 mutants; 39.6 mM−1cm−1 for SEB and SEB mutants; and 50.0 mM−1cm−1 for HLA-DR1. The concentration distributions of the samples at sedimentation equilibrium were acquired by an average of 25 measurements of absorbance at each radial position, with a nominal spacing of 0.001 cm between each radial position. The data obtained were analyzed as described by Malchiodi et al. (15). Errors on the dissociation constants (K ds) were <25% of parameter values.
BIAcore Analysis.
The binding of the TCR-β chain to immobilized enterotoxins was monitored with a BIAcore instrument (Pharmacia Biosensor, Piscataway, NJ). The SAG mutants were coupled to the dextran matrix of CM5 sensor chips using the Amine Coupling Kit (Pharmacia Biosensor). All SEC3 or SEB mutants, whose concentration were ∼100 μg/ml, were dialyzed against 10 mM sodium acetate, pH 5.3, before coupling. The activation and immobilization periods ranged from 5 to 7 min, which yielded between 2,000 and 3,500 resonance units (RU) of coupled SAG. The 14.3.d β chain was dialyzed against Hepes-buffered saline (HBS) containing 150 mM NaCl, 10 mM Na-Hepes, pH 7.5, 3.4 mM EDTA, and 0.005% Surfactant P20 (Pharmacia Biosensor). Twofold dilutions were made in the same buffer. The measurements were carried out at 25°C at a flow rate of 5 μl/min over a period of 6 min. Dissociation was done with HBS and the chip surface was regenerated by a pulse of 10 mM HCl. The data were analyzed using the BIAevaluation 2.1 software package (Pharmacia Biosensor); K ds were determined under equilibrium binding conditions using Scatchard plots as previously described in (15, 22).
T Cell Proliferation Assay.
4 × 104 lymph node T cells from RAG-2−/− TCR transgenic mice expressing the 14.3.d TCR (23) were cultured together with 2 × 105 irradiated (10,000 rad) BALB/c spleen cells as APCs in the presence of varying amounts of different SAGs in 200 μl of IMDM supplemented with 10% FCS as duplicates in flat-bottomed 96-well plates. After 48 or 96 h of incubation at 37°C, 1 mCi/well of [3H]thymidine was added for the next 12 h. Incorporation of radioactivity was then measured using a Betaplate 1250 system (Wallacy, Turku, Finland).
Results
Mutation of SEC3 Residues N23, Y90, and Q210 Has the Most Effect on Binding to the TCR β Chain Whereas Mutations in the 93-110 Disulfide Loop Have the Least.
The functional contribution of individual SEC3 residues to stabilization of the complex with the 14.3.d β chain was determined by alanine-scanning mutagenesis of all SEC3 residues in contact to the β chain in the crystal structure (6). Alanine was chosen because it eliminates the side chain without altering the main-chain conformation and does not impose any extreme steric or electrostatic effects (24). All affinity measurements were performed using an engineered, unglycosylated version of the 14.3.d β chain because of its stability and monodisperse behavior in solution. In addition, earlier studies had shown that this mutant β chain binds to a variety of SAGs with the same K d as the associated αβ TCR heterodimer, or as the glycosylated β chain alone (15).
Binding affinities were determined by sedimentation equilibrium, a technique with which even weak interactions can be analyzed in solution. Before measuring complex formation between SAGs and the TCR-β chain, the behavior of the individual proteins was assessed by separate runs. All species behaved well under the conditions used, with no tendency to aggregate. The calculated molecular weights for the β chain and the enterotoxins deviated <8% from the expected values based on amino acid composition. To assure the accuracy of our results, we measured β chain–SAG interactions at two different rotor speeds and/ or protein concentrations. The affinity values obtained under these different conditions differed by 30% at most. The K ds for the interaction of the 14.3.d β chain with a panel of SEC3 alanine mutants are listed in Table 1, and a representative sedimentation profile is shown in Fig. 1. As can be seen, the affinities of the various SEC3 mutants for the β chain vary between 3 and 150 μM, with all mutants binding similarly to, or weaker than, the wild-type SAG. Mutation to alanine of three residues (G102, K103, and G106) within the flexible 93–110 disulfide loop of SEC3 had no or little effect on binding to the β chain. In contrast, SEC3 mutants N23A, Y90A, and Q210A bind the β chain at least 50–100-fold less tightly than does the wild-type. At the protein concentrations used (4–10 μM), the K ds of all three mutants could be only estimated as ∼150 μM, but the actual values may be significantly greater. Trials using higher protein concentrations (up to 50 μM) in order to favor complex formation were unsuccessful due to nonspecific aggregation of the proteins. Mutagenesis of other SEC3 residues in contact with β chain (T20, Y26, N60, V91, and F176) resulted in smaller decreases in affinity, with K ds ranging from 25 to 120 μM.
Table 1.
Dissociation Constants for the Binding of Staphylococcal Enterotoxin Mutants to the TCR 14.3.d β Chain and Stimulatory Activity of these Mutants
| SAG mutant | K d analytical centrifuge | K d BIAcore | ΔΔG | Stimulatory capacity | ||||
|---|---|---|---|---|---|---|---|---|
| × 10−6 M | × 10−6 M | kcal/mol | ng/ml | |||||
| SEC3 wild-type | 4.5 | 3.0 | 200 | |||||
| SEC3 T20A | 42 | 49 | 1.4 | 4,000 | ||||
| SEC3 N23A | ⩾150* | NB | >2.5 | >30,000§ | ||||
| SEC3 Y26A | 86 | 60 | 1.7 | 3,000 | ||||
| SEC3 N60A | 25 | 48 | 1.3 | 2,000 | ||||
| SEC3 Y90A | ⩾150* | 240 | >2.5 | 9,000 | ||||
| SEC3 V91A | 117 | 130 | 2.1 | 6,000 | ||||
| SEC3 G102A | 4.6 | ND | 0.1 | 400 | ||||
| SEC3 K103A | 5.6 | 9.4 | 0.4 | 1,000 | ||||
| SEC3 G106A | 3.4 | ND | 0.1 | 400 | ||||
| SEC3 F176A | 92 | 110 | 1.9 | 30,000 | ||||
| SEC3 Q210A | ⩾150* | NB | >2.5 | >30,000§ | ||||
| SEB wild-type | 120 | 140‡ | 20 | |||||
| SEB L20T | 100 | 120 | 30 | |||||
| SEB V26Y | 26 | 12 | 5 | |||||
| SEB Y91V | 140 | 160 | 100 | |||||
| SEB L20T, V26Y, Y91V | ⩾150 | NB | 7,000 |
Affinity measurements by sedimentation equilibrium and BIAcore, as well as the biological activity assays, were performed as described in Materials and Methods. Differences in free energy changes are calculated as the differences between ΔGs of mutant and wild-type SAGs (ΔΔG = ΔGmutant − ΔGwild-type). The values of the individual ΔGs are calculated from the average K ds obtained from sedimentation equilibrium and BIAcore measurements according to the equation ΔG = −RT ln (1/K d), where R is the universal gas constant and T is the absolute temperature in Kelvin. For the three weakest binding mutants, N23A, Y90A, and Q210, a ΔΔG value of >2.5 kcal/mol (which corresponds to a 70-fold decrease in affinity) is estimated, as these mutants bind at least 50–100-fold weaker than wild-type SEC3. The stimulatory capacity of different SAGs was estimated from the dose (ng/ml) required to induce proliferation 100-fold above background. Background cpm were typically less than 100. NB, no detectable binding observed in the BIAcore measurements at TCR-β chain concentrations up to 128 μM; ND, not determined.
150 mM should be considered as a lower limit for these mutants.
Value for SEB wild-type was taken from the earlier work by Malchiodi et al. (15).
For these SEC3 mutants, SAG concentrations of 30,000 ng/ml were not sufficient to induce proliferation 100-fold above background.
Figure 1.
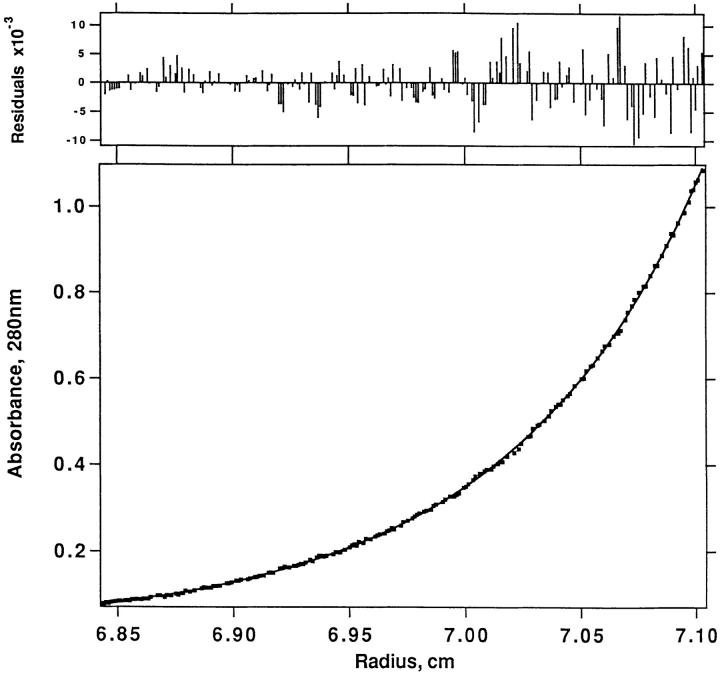
Sedimentation equilibrium profile of an equimolar mixture of the 14.3.d TCR-β chain with SEC3 F176A. Sedimentation was performed at 22,000 rpm in 50 mM Tris-HCl, pH 7.5, at 25°C and a starting concentration of 7.5 μM. (Bottom) Absorbance at 280 nm versus distance from the rotation center in centimeters. (Top) The residuals (A280-nm, theoretical − A280-nm, observed) for the equilibrium between the two components, yielding a K d of 92 μM, are small and random. Similar results were obtained for the other SEC3 and SEB mutants.
The Affinity of SEB for the TCR-β Chain Can Be Increased by Grafting Residues from the TCR Binding Site of SEC3.
We also attempted to obtain mutants of SEC3 and SEB with higher affinity for the 14.3.d β chain in order to see whether such tighter binding mutants would also show increased ability to stimulate T cells. In the case of the weakly binding SEB (K d = 120 μM by sedimentation equilibrium; Table 1) our approach was to graft residues from the TCR-binding site of SEC3 (K d = 4.5 μM), which differ between the two SAGs. As SEB and SEC3 have very similar three-dimensional structures (25, 26) and bind the 14.3.d β chain in essentially the same orientation (reference 6, Li and Mariuzza, unpublished results), engineering such chimeric SEB molecules was expected to be straightforward. Excluding the flexible 93–110 disulfide loop of SEC3 (which, as demonstrated above, does not functionally contribute to binding the β chain), the TCR binding sites of SEC3 and SEB differ only at positions 20, 26, and 91. We therefore replaced SEB residues L20, V26, and Y91 with SEC3 residues T20, Y26, and V91. We constructed three single mutants (SEB L20T, V26Y, and Y91V) in which these residues were individually substituted, as well as a triple mutant in which all three SEC3 residues were grafted onto the TCR binding site of SEB. As can be seen in Table 1, only SEB V26Y showed improved affinity: its K d (as determined by sedimentation equilibrium) was 26 μM, approximately five times lower than that of wild-type SEB, but still six times higher than that of SEC3. Surprisingly, mutations L20T and Y91V did not significantly alter the affinity of SEB for the β chain, whereas no binding whatsoever could be detected in the case of the the triple mutant.
We also attempted to design mutants of SEC3 with enhanced affinity for the 14.3.d β chain on the basis of the crystal structure of the β–SEC3 complex. For instance, SEC3 residues Y26 and Y90 were replaced by tryptophan and V91 by isoleucine with the expectation that the larger side chains of these residues would make additional van der Waals contacts with the β chain and contribute to hydrophobic stabilization of the complex. In addition, SEC3 residue G19 was mutated to lysine and F176 to glutamic acid, since the crystal structure suggested that charged amino acids at these positions could form salt bridges with residues of opposite charge on the TCR β chain. However, these mutants either bound the β chain with the same K d as wild-type SEC3 (Y26W or V91I), or even displayed up to 10-fold weaker affinities (G19K, Y90W, and F176E; data not shown).
The Dissociation Constants from Sedimentation Equilibrium Are Confirmed by Surface Plasmon Resonance (BIAcore) Measurements.
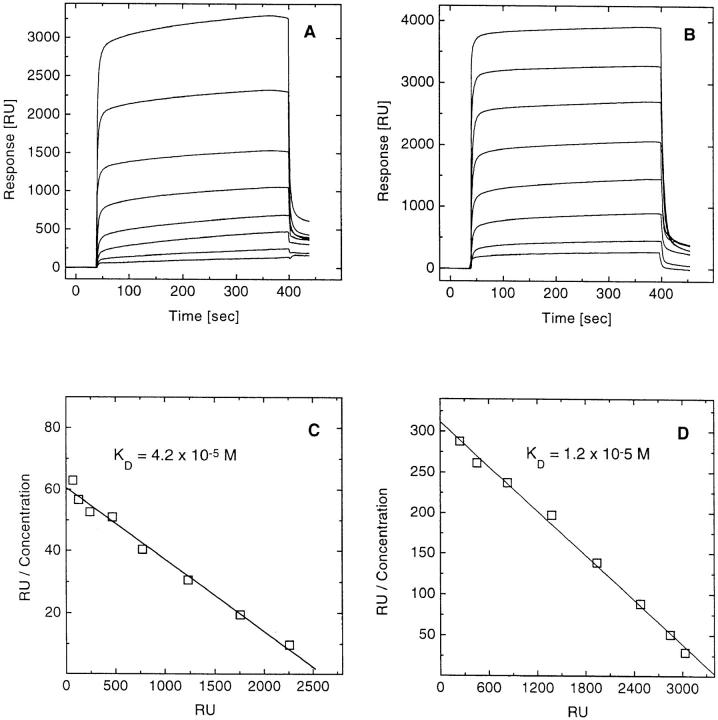
We coupled most of the SEC3 and SEB mutants described above directly to the dextran matrix of sensor chips through the primary amino groups of the proteins. Unglycosylated 14.3.d β chain in increasing concentrations (1–128 μM) was injected and concentration-dependent surface plasmon resonance profiles such as those shown in Fig. 2 for SEC3 N60A (panel A) and SEB V26Y (panel B) were recorded. As in the case of the wild-type proteins, the binding of all SEC3 and SEB mutants tested was characterized by very fast on- and off-rates. In fact, these kinetic rates were too fast to accurately measure, although the association and dissociation rate constants may be estimated at >105 M−1s−1 and >0.1 s−1, respectively. Therefore, affinities were determined under equilibrium binding conditions, in which we took report points for Scatchard analysis 20–40 s after injection. Scatchard plots for SEC3 N60A and SEB V26Y, after correction for nonspecific binding, are shown in Fig. 2, C and D. The plots are linear, with correlation coefficients of 0.99; K ds were calculated directly from the slopes. The predicted maximum specific binding, calculated from the x-intercept assuming a linear relationship between mass of bound protein and measured RU (27), indicated that nearly 90% of immobilized SAG molecules retained their binding activity. Similar results were observed for the other mutants tested; Table 1 lists the K d values measured by BIAcore. Comparison of the affinities obtained by sedimentation equilibrium and BIAcore showed reasonable agreement, with values for K d differing by not more than a factor of two. These variations are probably attributable to intrinsic differences between the two methods. In the sedimentation equilibrium both binding partners are in solution, whereas in BIAcore one of the ligands is chemically coupled to a solid support, which may result in small changes in the conformation and/or accessibility of the immobilized molecule. However, we do not observe any systematic difference between these two very different physical methods. In some cases, the affinities obtained from BIAcore were lower (SEC3 T20A, N60A, and K103A) than those from sedimentation equilibrium, whereas in others the reverse was true (SEC3 wild-type, Y26A, and SEB V26Y). Further, we found that both methods gave very reproducible results, with K d values deviating by not more than 30% in each case. We therefore believe that these two methods combined give an accurate picture of the functional importance of individual SEC3 or SEB contact residues in complex formation with the TCR-β chain.
Figure 2.
Binding of 14.3.d. β chain to immobilized SEC3 N60A or SEB V26Y. The β chain was injected at eight different concentrations ranging from 1 to 128 μM over surfaces coupled with (A) SEC3 N60A (2,700 RU) or (B) SEB V26Y (3,600 RU). Buffer flow rates were 5 μl/ min and the equilibrium was typically reached within 10 s. Scatchard analysis of the binding of the β chain to SEC3 N60A (C) with data derived from A after correction for nonspecific binding, and of the binding of the β chain to SEB V26Y (D) with data derived from (B), yielded linear plots with correlation coefficients of 0.99 in both cases. The apparent K ds for the β chain–SEC3 N60A and the β chain–SEB V26Y reactions were 42 and 12 μM, respectively. Similar binding profiles and Scatchard plots were obtained with the others SEC3 and SEB mutants.
The Biological Activity of SEC3 and SEB Mutants Correlates with their Affinity for the TCR-β chain.
To mimic normal physiological conditions as closely as possible, we used resting lymph node T cells bearing the 14.3.d TCR from RAG-2−/− TCR transgenic mice (23) to measure the stimulatory effects of different SAGs on BALB/c spleen cells expressing I-Ed (Fig. 3). The results are summarized in Table 1 as the dose of each SAG required to induce T cell proliferation 100-fold above background. Dose-response curves for most of the mutant SAGs did not reach a plateau, even at the highest SAG concentrations tested (Fig. 3). Thus, it is very difficult to estimate either the maximal stimulatory capacity (total proliferation in the assays), or the concentration of SAG that is required to obtain 50% of the maximal effect. Nevertheless, we believe that our definition of the stimulatory capacity gives an accurate picture of the relative biological activities of the different SEB and SEC3 mutants. A comparison with the results from the binding experiments reveals a direct correlation between the affinity and the biological activity of SEC3 and SEB mutants: the higher the affinity of a particular mutant for the TCR-β chain, the greater its mitogenic potency. For instance, SEC3 mutants G102A, K103A, and G106A, all located on the flexible disulfide loop of this SAG, bind to the β chain with K ds similar to that of wild-type SEC3 and also show similar biological activity. On the other hand, SEC3 mutants N23A and Q210A, with K ds ⩾150 μM, exhibit no measurable stimulatory effects whatsoever. However, another very low-affinity mutant, SEC3 Y90A, is still able to activate T cells, although ∼50-fold higher concentrations are needed to achieve the same effect as with wild-type. This difference between no apparent biological activity and low biological activity is reflected in the results from BIAcore measurements, in which SEC3 Y90A yielded a K d of 240 μM, whereas SEC3 N23A and Q210A did not show any detectable binding (Table 1). Except for F176A, the mutants that have intermediate affinities (i.e., SEC3 T20A, Y26A, N60A, V91A) consistently stimulated T cells transgenic for the 14.3.d TCR less efficiently than wild-type SEC3, but significantly better than the three mutants with the weakest binding. A very similar picture emerged from experiments with the various SEB mutants: SEB V26Y, with its increased affinity, also exhibited a slightly enhanced mitogenic potency compared to wild-type SEB, whereas the triple mutant, with no detectable binding to the 14.3.d β chain, only barely stimulated T cells. However, an apparent exception to this simple affinity-activity rule was the finding that SEB stimulated transgenic T cells ∼10-fold better than SEC3, even though the affinity of SEB for the TCR-β chain is ∼35-fold lower than that of SEC3 (Table 1). This effect was observed using either BALB/c spleen cells or MHC class II-negative mouse fibroblasts expressing HLA-DR1 as APCs (data not shown).
Figure 3.
Functional characterization of mutant SAGs. The mitogenic potencies of wild-type SEC3 and SEC3 mutants are shown in A and of wild-type SEB and SEB mutants in B. Proliferation of lymph node T cells from RAG-2−/− TCR transgenic mice was measured using irradiated BALB/c spleen cells in the presence of various SAGs. The SAG concentrations used are indicated with the different symbols. Proliferation results at 72 h are shown here. The results were qualitatively the same at 24 h.
SEB Binds HLA-DR1 More Tightly than SEC3.
To determine whether the surprisingly strong mitogenic potency of SEB relative to SEC3 could be attributed to tighter binding to MHC class II on APCs, we examined the binding of SEB and SEC3 to soluble recombinant I-Ed loaded with HA 110–120 (28), as well as to HLA-DR1 loaded with HA 306–318. Both peptide–MHC class II complexes behaved well in separate sedimentation equilibrium runs, which yielded apparent molecular weights of 49 kD for I-Ed and 50 kD for DR1, in close agreement with expected values (52 kD for both I-Ed and DR1). However, we could not detect any binding of I-Ed to either SEB or SEC3, in agreement with BIAcore experiments reported by Kozono et al. using I-Ek (29). In contrast, we could easily measure a K d of 14 μM for the interaction of SEB with HLA-DR1 and a corresponding value of 48 μM for SEC3. This is also consistent with the observation that DR1 is a much more efficient presenting element for SEB and SEC3 than I-E in T cell stimulation assays (30). Therefore, the unexpectedly high mitogenic potency of SEB relative to SEC3 could be at least partially explained by the tighter binding of SEB to MHC class II. This observation is supported by earlier studies that also suggested a correlation between affinity and biological activity, since SAGs with weaker binding to MHC also showed decreased mitogenic potency (31, 32).
We also determined the affinities of two SEC3 mutants, G102A and V91A, for HLA-DR1. SEC3 G102A had the same affinity for the 14.3.d β chain as the wild-type SAG, whereas the affinity of SEC3 V91A was decreased by ∼30-fold. In contrast, both mutants bound to HLA-DR1 with similar affinities (K d = 54 μM for V91A and K d = 59 μM for G102A), and about equally as well as wild-type SEC3. These results demonstrate that mutations in at least these particular TCR-contacting residues of SEC3 have no influence on affinity for MHC, in agreement with the finding that the corresponding residues of SEB are not in contact with MHC in the crystal structure of the SEB–DR1 complex (12).
Discussion
While the crystal structure of the 14.3.d β chain–SEC3 complex (6) provides detailed information on the molecular architecture of the TCR–SAG interface, it does not tell us what the actual functional contribution of individual residues to complex stabilization is. To answer this question, SEC3 residues in contact with the β chain were subjected to alanine-scanning mutagenesis (24) in order to analyze their relative importance to binding. The free energy of binding was calculated from the equation ΔG = −RT ln (1/K d), where R is the univeral gas constant and T is the absolute temperature. In this way, we constructed a “thermodynamic map” of the TCR binding site of SEC3 that shows the relative loss of binding-free energy for individual alanine mutants compared to wild-type SEC3 (ΔΔG = ΔGmutant − ΔGwild-type). As shown in Table 1, the most destabilizing alanine substitutions are located at positions N23, Y90, and Q210 with a ΔΔG ⩾2.5 kcal/mol. Significant contributions (1.3 to 2.1 kcal/mol) can also be observed for contact residues T20, Y26, N60, V91, and F176. In contrast, replacement of G102, K103, and G106 by alanine has little or no effect (<0.5 kcal/mol). In agreement with this result, these four interface residues form part of the 93–110 disulfide loop of SEC3 that was only poorly visible in the crystal structure of the β–SEC3 complex, suggesting high intrinsic mobility (6). Therefore, 8 out of 11 SEC3 contact residues are energetically important in binding the TCR-β chain such that stabilization of the β–SEC3 complex is achieved by the accumulation of many productive interactions of varying strength over a large portion of the interface with the TCR. This finding that a majority of contact residues significantly contribute to binding is similar to the case of an anti–hen eggwhite lysozyme antibody binding to an antiidiotypic antibody (22), but different from the binding of human growth hormone to its receptor, where only a few contact residues appear to be responsible for the formation of a stable complex (33).
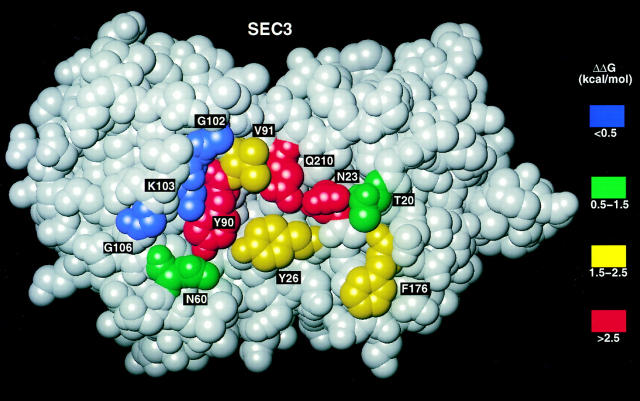
Fig. 4 shows the functional epitope of SEC3 involved in complex formation with the 14.3.d β chain mapped onto its three-dimensional structure. With the exception of F176, which lies at the periphery, the most energetically important SEC3 residues (N23, Y26, Y90, V91, and Q210) are clustered at the center of the cleft between the small and large domains of the enterotoxin molecule. All five of these SEC3 residues interact with a continuous stretch of amino acids (residues 50–55) in the CDR2 loop of the Vβ domain. Hydrophilic residues N23 and Q210 form hydrogen bonds with backbone atoms of this part of CDR2, whereas hydrophobic residues Y26, Y90, and V91 make a number of van der Waals contacts with main chain and side chain atoms in the CDR2 loop (6). The finding that mutations in the highly mobile 93–110 disulfide loop of SEC3 have little or no effect on binding affinity is in agreement with the high sequence variability in this region present in SEC1-3, SEB, and streptococcal pyrogenic exotoxin A (SPEA) (Fig. 5), all of which bind the 14.3.d β chain and activate Vβ8.2-bearing T cells (15). On the other hand, “hot spot” residues making the greatest energetic contribution to stabilization of the β–SEC3 complex (N23, Y90, and Q210), as well as the less important N60, are strictly conserved in SEC1-3, SEB, and SPEA. Even in SAGs such as SEA, SED, and SEE, which do not recognize Vβ8.2, position 23 is also occupied by an asparagine; this residue has proven critical for T cell stimulation in earlier mutagenesis experiments (32). These observations emphasize the functional importance of each of the conserved interface residues, and makes it understandable why SAGs having other residues at these positions show different Vβ-binding specificities.
Figure 4.
Space-filling model of the surface of SEC3 in contact with the 14.3.d TCR-β chain in the crystal structure. Residues are color coded according to the loss of binding free energy (ΔΔG) upon alanine substitution: red, >2.5 kcal/mol; yellow, 1.5-2.5 kcal/mol; green, 0.5-1.5 kcal/mol; blue, <0.5 kcal/mol.
Figure 5.
Sequence alignment of bacterial superantigens (SEC1-3, SEB, SPEA, SEA, SED, and SEE). Residues of SEC3 in contact with the TCR-β chain are boxed in colors according to the loss of binding-free energy (ΔΔG) upon alanine substitution. The color code is the same as that used in Fig. 4. The homologous residues in the other SAGs are only boxed in color if they are identical with those in SEC3. SEB residues contacting MHC in the crystal structure of the SEB–HLA–DR1 complex (12), and the corresponding residues of SEC3, are boxed in cyan if identical in SEB and SEC3 and in magenta if different.
The results obtained with the SEB–SEC chimeras were quite surprising, because only SEB mutant V26Y had a higher affinity (K d = 19 μM, taken as an average of sedimentation equilibrium and BIAcore measurements) than wild-type SEB (K d = 130 μM). The two other SEB single mutants, L20T and Y91V, did not show improved binding and the triple mutant did not show any detectable binding to the 14.3.d β chain. Since alanine-scanning mutagenesis of SEC3 implicated Y26 as functionally important, it is reasonable that replacement of the valine that is present in wild-type SEB by the bulkier tyrosine should increase the number of van der Waals contacts to the β chain and hence improve the affinity of that mutant. Other studies also revealed a key role of residue 26 in SEC 1-3 and SEB in the specificity of these SAGs for different human TCRs. Thus, SEB and SEC1 react strongly with human Vβ3 but not with Vβ13.1, whereas SEC3 and SEC2 bind well to Vβ13.1 but only weakly with Vβ3 (8). Site-directed mutagenesis has shown that these reactivity differences can be fully attributed to the amino acid at position 26, which is valine in SEB and SEC1 but tyrosine in SEC2 and SEC3. However, it is quite puzzling that the SEB single mutant Y91V and the SEB triple mutant, with an almost completely grafted SEC3 binding site, react with the 14.3.d β chain only very weakly, or not at all. This behavior could be explained by unanticipated conformational changes in SEB induced by these mutations, or by subtle structural differences between the β–SEB and β–SEC3 complexes that are not apparent at the current resolution of the x ray structures (∼3.5 Å). For instance, small differences in the relative orientation of SAG and TCR in the two complexes may lead to unanticipated effects on binding when interface residues are mutated. Furthermore, attempts to rationally design better-binding SEC3 mutants (e.g., G19K, Y26W, Y90W, V91I, and F176E) were unsuccessful, since in all cases the affinity was the same as that of wild-type SEC3, or even up to 10-fold lower. It is therefore entirely possible that the TCR binding site of SEC3 is already optimized and that the introduction of charged (G19K, F176E) or bulkier (Y90W) residues leads to less favorable interactions and a consequent decrease in affinity.
Our comparison between affinity and biological activity clearly shows that SAG mutants that bind the TCR-β chain more tightly stimulate T cells more efficiently than do SAG mutants with lower affinities. This observation complements recent studies of T cell activation by peptide– MHC ligands, which strongly suggest that the affinity of the TCR–peptide–MHC interaction plays a critical role in determining the nature of the T cell response (34). For example, it has been shown that peptide–MHC antagonists that induce T cell anergy may act by binding the TCR with affinities that are below those of peptide–MHC agonists. Furthermore, the so-called “serial triggering” model of T cell activation argues that the rather low affinities and fast off-rates observed for many peptide–MHC–TCR interactions are in fact necessary to enable a single peptide– MHC complex to serially engage a large number of TCRs (35, 36). Indeed, ligands with too high of an affinity for the TCR are predicted to be actually less efficient at triggering T cells. We have shown that, like peptide–MHC, the binding of SAGs to the TCR is characterized by relatively low affinities and fast dissociation rates. This suggests that SAGs “mimic” the interaction of peptide–MHC complexes with the TCR in terms of affinities and kinetics, a conclusion also drawn by Viola and Lanzavecchia (36). However, it remains to be established whether there is indeed an optimum affinity for T cell activation by SAGs, such that SAG variants with either higher or lower affinities than this optimum value exhibit decreased ability to stimulate T cells. Alternatively, mutant SAGs with progressively higher affinities for the TCR relative to the wild-type would stimulate T cells increasingly well, until some plateau of maximum stimulation is reached. Unfortunately, our current data do not permit us to distinguish between the two possibilities since we were unable to engineer SAG variants with markedly enhanced binding to the TCR-β chain. In the best case, SEB V26Y bound to the β chain only approximately sevenfold more tightly than did the wild-type SAG.
The only notable discrepancy in the observed correlation of activity with affinity is that SEB exhibited an ∼10-fold higher potency than SEC3 in T cell stimulation assays, even though its affinity for the TCR-β chain (K d = 130 μM) is much lower than that of SEC3 (K d = 3.8 μM). Since the activation of T cells by SAGs requires simultaneous binding of the SAG to TCR and to MHC on the APCs, this discrepancy could be reasonably explained by a tighter binding of SEB than SEC3 to MHC molecules. The crystal structure of the complex between HLA-DR1 and SEB (12) reveals that MHC-contacting residues of SEB are different in SEC3 (Fig. 5). These sequence differences may well result in different affinities of SEB and SEC3 for MHC class II and thereby exert a significant effect on the mitogenic potency of these SAGs. Indeed, we found that SEB binds to HLA-DR1 with a K d of 14 μM, whereas the corresponding value for SEC3 was 48 μM. This threefold higher affinity of SEB for MHC appears to compensate for its 35-fold lower affinity for the TCR. However, it is somewhat puzzling that this rather modest difference in SAG–MHC affinity has such a large impact on biological activity, especially given that the effects of changes in TCR–SAG affinity on SAG activity are proportionately much less pronounced (Table 1). One explanation may be that the tighter binding of SEB to HLA-DR1 is mainly the result of a slower off-rate leading to a considerably longer residence time of SEB molecules on the MHC and therefore to much more efficient presentation of the SAG to the T cell. Another could be that some minor structural differences between the trimolecular MHC–SEB–TCR and MHC–SEC3–TCR complexes may result in altered binding properties that cannot be easily discerned by examining either the MHC–SAG or TCR–SAG complexes individually. Another important question regarding T cell activation by SAGs remains as well: how can a SAG efficiently cross-link the APC and T cell if its affinity for both MHC and TCR is quite weak? This would appear to be less of an issue for conventional T cell activation by peptide–MHC complexes because only one low-affinity interaction is involved. One possibility is that accessory molecules such as CD4 help stabilize the TCR–SAG–MHC complex sufficiently for activation to occur. Another is that the overall stability of the MHC–SAG–TCR complex is greater than one would expect from considering the TCR–SAG and MHC–SAG interactions independently. In other words, the binding of SAGs to TCR and MHC may be a cooperative process such that the SAG–MHC complex binds the TCR with a higher affinity than does SAG alone. Indeed, surface plasmon resonance experiments have suggested a tighter binding of an immobilized TCR to SEB complexed with HLA-DR1 than to SEB alone (37). Furthermore, our current model of the MHC–SAG–TCR complex (6) suggests that the Vα domain of the TCR can contact the MHC β1 domain, perhaps helping to stabilize the overall complex. However, only an actual crystal structure determination of an MHC–SAG–TCR complex, combined with further binding experiments with all three components, will reveal whether cooperative effects in the trimolecular complex in fact occur, and if so, how exactly they act to determine T cell stimulation by SAGs.
Acknowledgments
We thank Claudia Beck for technical assistance and Cynthia V. Stauffacher (Purdue University, West Lafayette, IN) for providing coordinates of SEC3.
Abbreviations used in this paper
- HA
hemaglutinin peptide
- HBS
Hepes-buffered saline
- Kd
dissociation constant
- RU
resonance unit
- SAG
superantigen
- SEA
staphylococcal enterotoxin A
- SEB
staphylococcal enterotoxin B
- SEC
staphylococcal enterotoxin C
- SED
staphylococcal enterotoxin D
- SEE
staphylococcal enterotoxin E
- SPEA
streptococcal pyrogenic exotoxin A
Footnotes
This research was supported by National Institutes of Health (NIH) grant 36900 and National Multiple Sclerosis Society grant RG2747 (R.A. Mariuzza); NIH grant HL-36611 (P.M. Schlievert); NIH grant AI-28401 and USDA grant 9402399 (G.A. Bohach); and grants from the National Cancer Institute of Canada (R.-P. Sékaly). Support from the Lucille P. Markey Charitable Trust is also gratefully acknowledged. The Basel Institute for Immunology was founded and is supported by F. Hoffmann-LaRoche Ltd., Basel, Switzerland. L. Leder is a Fellow of the Swiss National Science Foundation. R.-P. Sékaly holds a Medical Research Council of Canada Scientist Award.
References
- 1.Bohach GA, Fast DJ, Nelson RD, Schlievert PM. Staphylococcal and streptococcal pyrogenic toxins involved in toxic shock syndrome and related illnesses. Crit Rev Microbiol. 1990;17:251–272. doi: 10.3109/10408419009105728. [DOI] [PubMed] [Google Scholar]
- 2.Kotzin BL, Leung DYM, Kappler J, Marrack P. Superantigens and their potential role in human disease. Adv Immunol. 1993;54:99–106. doi: 10.1016/s0065-2776(08)60534-9. [DOI] [PubMed] [Google Scholar]
- 3.Conrad B, Weissmahr RN, Boni J, Arcari R, Schupbach J, Mach B. A human endogeneous retroviral superantigen as candidate autoimmune gene in type I diabetes. Cell. 1997;90:303–313. doi: 10.1016/s0092-8674(00)80338-4. [DOI] [PubMed] [Google Scholar]
- 4.Brocke S, Gaur A, Piercy C, Gautam A, Gijebls K, Fathman CG, Steinman L. Induction of relapsing paralysis in experimental autoimmune encephalomyelitis by bacterial superantigen. Nature. 1993;365:642–644. doi: 10.1038/365642a0. [DOI] [PubMed] [Google Scholar]
- 5.Cole BC, Griffiths MM. Triggering and exacerbation of autoimmune arthritis by the mycoplasma arthritidis superantigen MAM. Arthritis Rheum. 1993;36:994–1002. doi: 10.1002/art.1780360717. [DOI] [PubMed] [Google Scholar]
- 6.Fields BA, Malchiodi EL, Li H, Ysern X, Stauffacher CV, Schlievert PM, Karajalainen K, Mariuzza RA. Crystal structure of a T cell receptor β-chain complexed with a superantigen. Nature. 1996;384:188–192. doi: 10.1038/384188a0. [DOI] [PubMed] [Google Scholar]
- 7.Patten PA, Rock EP, Sonoda T, Fazekas de St B, Groth, Jorgensen JL, Davis MM. Transfer of putative complementarity-determining region loops of T cell receptor V domains confers toxin reactivity but not peptide/MHC specificity. J Immunol. 1993;150:2281–2294. [PubMed] [Google Scholar]
- 8.Deringer JR, Ely RJ, Stauffacher CV, Bohach GA. Subtype-specific interactions of type C staphylococcal enterotoxins with the T-cell receptor. Mol Microbiol. 1996;22:523–534. doi: 10.1046/j.1365-2958.1996.1381506.x. [DOI] [PubMed] [Google Scholar]
- 9.Woodland DL, Blackman MA. How do T cell receptors, MHC molecules and superantigens get together? . Immunol Today. 1993;14:208–212. doi: 10.1016/0167-5699(93)90164-G. [DOI] [PubMed] [Google Scholar]
- 10.Webb SR, Gascoigne NRJ. T-cell activation by superantigens. Curr Opin Immunol. 1994;6:467–475. doi: 10.1016/0952-7915(94)90129-5. [DOI] [PubMed] [Google Scholar]
- 11.Fields BA, Ober EL, Lebedeva MI, Braden BC, Ysern X, Kim JK, Shao X, Ward ES, Mariuzza RA. Crystal structure of the Vα domain of T cell antigen receptor. Science. 1995;270:1821–1824. doi: 10.1126/science.270.5243.1821. [DOI] [PubMed] [Google Scholar]
- 12.Jardetzky TS, Brown JH, Gorga JC, Stern LJ, Urban RG, Chi Y, Stauffacher CV, Strominger JL, Wiley DC. Three-dimensional structure of a human class II histocompability antigen complexed with superantigen. Nature. 1994;368:711–718. doi: 10.1038/368711a0. [DOI] [PubMed] [Google Scholar]
- 13.Bentley GA, Boulot G, Karjalainen K, Mariuzza RA. Crystal structure of the β chain of a T cell antigen receptor. Science. 1995;267:1984–1987. doi: 10.1126/science.7701320. [DOI] [PubMed] [Google Scholar]
- 14.Kubo RT, Born W, Kappler JW, Marrack P, Pigeon M. Characterization of a monoclonal antibody which detects all murine αβ T cell receptors. J Immunol. 1989;142:2736–2742. [Google Scholar]
- 15.Malchiodi EL, Eisenstein E, Fields BA, Ohlendorf DH, Schlievert PM, Karajalainen K, Mariuzza RA. Superantigen binding to a T cell receptor β chain of known three-dimensional structure. J Exp Med. 1995;182:1833–1845. doi: 10.1084/jem.182.6.1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones CL, Kahn SA. Nucleotide sequence of the enterotoxin B gene from Staphylococcus aureus. . J Bacteriol. 1986;166:29–33. doi: 10.1128/jb.166.1.29-33.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hovde CJ, Hackett SP, Bohach GA. Nucleotide sequence of the staphylococcal enterotoxin C3: sequence comparison of all three type C staphyloccal enterotoxins. Mol Gen Genet. 1990;220:329–333. doi: 10.1007/BF00260504. [DOI] [PubMed] [Google Scholar]
- 18.Ward ES. Secretion of T-cell receptor fragments from recombinant Escherichia colicells. J Mol Biol. 1992;224:885–888. doi: 10.1016/0022-2836(92)90455-s. [DOI] [PubMed] [Google Scholar]
- 19.Passalacqua EF, Brehm RD, Acharya KR, Tranter HS. Crystallization and preliminary X-ray analysis of a microbial superantigen staphylococcal enterotoxin C2. J Mol Biol. 1993;233:170–172. doi: 10.1006/jmbi.1993.1493. [DOI] [PubMed] [Google Scholar]
- 20.Summers, M.D., and G.E. Smith. 1988. A manual of methods for baculovirus and insect cell culture procedures. In Texas Agricultural Experiment Station Bulletin No. 1555. College Station, TX.
- 21.Stern LJ, Wiley DC. The human class II MHC protein HLA-DR1 assembles as empty αβ heterodimers in the absence of antigenic peptide. Cell. 1992;68:465–477. doi: 10.1016/0092-8674(92)90184-e. [DOI] [PubMed] [Google Scholar]
- 22.Dall'Aqua W, Goldman ER, Eisenstein E, Mariuzza RA. A mutational analysis of the binding of two different proteins to the same antibody. Biochemistry. 1996;35:9667–9776. doi: 10.1021/bi960819i. [DOI] [PubMed] [Google Scholar]
- 23.Kirberg J, Baron A, Jakob S, Rolink A, Karjalainen K, von Boehmer H. Thymic selection of CD8+single positive cells with a class II major histocompatibility complex–restricted receptor. J Exp Med. 1994;180:25–34. doi: 10.1084/jem.180.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wells JA. Systematic mutational analysis of protein-protein interfaces. Methods Enzymol. 1991;202:390–411. doi: 10.1016/0076-6879(91)02020-a. [DOI] [PubMed] [Google Scholar]
- 25.Swaminathan S, Furey W, Pletcher J, Sax M. Crystal structure of staphylococcal entertoxin B, a superantigen. Nature. 1992;359:801–806. doi: 10.1038/359801a0. [DOI] [PubMed] [Google Scholar]
- 26.Schlievert PM, Bohach GA, Ohlendorf DH, Stauffacher CV, Leung DY, Murray DL, Prassad GS, Earhart CA, Jablonski LM, Hoffmann ML, Chi YI. Molecular structure of staphylococcus and streptococcus superantigens. J Clin Immunol. 1995;15:4–10. doi: 10.1007/BF01540887. [DOI] [PubMed] [Google Scholar]
- 27.Granzow R, Reed R. Interactions in the fourth dimension. Biotechnology. 1992;10:390–393. doi: 10.1038/nbt0492-390. [DOI] [PubMed] [Google Scholar]
- 28.Wallny HJ, Sollami G, Karjalainen K. Soluble major histocompatibility complex class II molecules produced in Drosophilacells. Eur J Immunol. 1995;25:1262–1266. doi: 10.1002/eji.1830250520. [DOI] [PubMed] [Google Scholar]
- 29.Kozono H, Parker D, White J, Marrack P, Kappler J. Multiple binding sites for bacterial superantigens on soluble class II MHC molecules. Immunity. 1995;3:187–196. doi: 10.1016/1074-7613(95)90088-8. [DOI] [PubMed] [Google Scholar]
- 30.Herman A, Croteau G, Sékaly RP, Kappler J, Marrack P. HLA-DR alleles differ in their ability to present staphyloccal enterotoxins to T cells. J Exp Med. 1990;172:709–717. doi: 10.1084/jem.172.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chintagumpala MM, Mollick JA, Rich RR. Staphylococcal toxins bind to different sites on HLA-DR. J Immunol. 1991;147:3876–3881. [PubMed] [Google Scholar]
- 32.Kappler JW, Herman A, Clements J, Marrack P. Mutations defining functional regions of the superantigen staphylococcal enterotoxin B. J Exp Med. 1992;175:387–396. doi: 10.1084/jem.175.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cunningham BC, Wells JA. Comparison of a structural and functional epitope. J Mol Biol. 1993;234:554–563. doi: 10.1006/jmbi.1993.1611. [DOI] [PubMed] [Google Scholar]
- 34.Sloan-Lancaster J, Allen PM. Altered peptide-ligand induced partial T cell activation: molecular mechanisms and role in T cell biology. Annu Rev Immunol. 1996;14:1–27. doi: 10.1146/annurev.immunol.14.1.1. [DOI] [PubMed] [Google Scholar]
- 35.Valitutti S, Muller S, Cella M, Padovan E, Lanzavecchia A. Serial triggering of many T-cell receptors by a few peptide-MHC complexes. Nature. 1995;375:148–151. doi: 10.1038/375148a0. [DOI] [PubMed] [Google Scholar]
- 36.Viola A, Lanzavecchia A. T cell activation determined by number and tunable thresholds. Science. 1996;273:104–106. doi: 10.1126/science.273.5271.104. [DOI] [PubMed] [Google Scholar]
- 37.Seth A, Stern LJ, Ottenhoff THM, Engel I, Owen MJ, Lamb JR, Klausner RD, Wiley DC. Binary and ternary complexes between T cell receptor, class II MHC and superantigen in vitro. Nature. 1994;369:324–327. doi: 10.1038/369324a0. [DOI] [PubMed] [Google Scholar]