Abstract
In contrast to conventional T cells, natural killer (NK) 1.1+ T cell receptor (TCR)-α/β+ (NK1+T) cells, NK cells, and intestinal intraepithelial lymphocytes (IELs) bearing CD8-α/α chains constitutively express the interleukin (IL)-2 receptor (R)β/15Rβ chain. Recent studies have indicated that IL-2Rβ/15Rβ chain is required for the development of these lymphocyte subsets, outlining the importance of IL-15. In this study, we investigated the development of these lymphocyte subsets in interferon regulatory factor 1–deficient (IRF-1−/−) mice. Surprisingly, all of these lymphocyte subsets were severely reduced in IRF-1−/− mice. Within CD8-α/α+ intestinal IEL subset, TCR-γ/δ+ cells and TCR-α/β+ cells were equally affected by IRF gene disruption. In contrast to intestinal TCR-γ/δ+ cells, thymic TCR-γ/δ+ cells developed normally in IRF-1−/− mice. Northern blot analysis further revealed that the induction of IL-15 messenger RNA was impaired in IRF-1−/− bone marrow cells, and the recovery of these lymphocyte subsets was observed when IRF-1−/− cells were cultured with IL-15 in vitro. These data indicate that IRF-1 regulates IL-15 gene expression, which may control the development of NK1+T cells, NK cells, and CD8-α/α+ IELs.
In addition to the conventional lymphocyte subsets, other lineages have been identified as NK1.1+TCR-α/β+ (NK1+T) cells, NK cells, and intestinal intraepithelial lymphocytes (IELs). NK1+T cells have been recently classified as a lymphocyte subset that shares common features with both NK cells and conventional T cells. This lineage expresses NK markers including NKR-P1, Ly-49, and IL-2Rβ/15Rβ as well as an invariant Vα14J281TCR-α chain in combination with Vβ8, Vβ7, or Vβ2 (1, 2). Expression of these TCRs is required for NK1+ T cell development (3, 4). They are positively selected by MHC class I–related CD1 or thymic leukemia (TL) molecules (5–7). The majority of TCR-α/β+ or TCR-γ/δ+ intestinal IEL expresses CD8-α/α homodimers. Both NK1+T cells and CD8-α/α+ intestinal IELs can develop through either extrathymic or alternative thymic pathways (1, 2, 8). Notably, the IL-2Rβ/15Rβ chain is required for the development of NK1+T cells, NK cells, and CD8-α/α+ intestinal IELs (9, 10), and IL-15 preferentially promotes the proliferation of these lymphocyte subsets (10–12).
IFN regulatory factor 1 (IRF-1), an IFN-inducible transcriptional activator, was initially identified as a protein that binds cis-acting DNA elements in the IFN-β promoter (13–15) and the IFN-stimulated response element of IFN-α/β–stimulated genes (16, 17). Recent studies with IRF-deficient (IRF-1−/−) mice demonstrated a reduction of CD8+TCR-α/β+ cells and decreased MHC class I levels as a consequence of reduced expression of transporter associated with antigen processing 1 (TAP-1) and low molecular weight protein 2 (LMP-2; 18, 19).
Since IRF-1 deficiency has been related to T cell maturation, we examined the development of NK1+T cells, NK cells, and IELs in IRF-1−/− mice. Data indicated that these lymphocyte subsets were selectively reduced and IL-15 messenger RNA (mRNA) was barely detectable in IRF-1−/− mice. Therefore, IRF-1 regulates the IL-15 gene that is required for survival and/or expansion of these lymphocyte subsets in vivo.
Materials and Methods
Mice.
Mice deficient in IRF-1 (18) were backcrossed five times with C57BL/6 mice. Homozygous IRF-1−/− mice were bred and identified by staining blood with anti-CD8 and -CD4 mAb. Wild-type or heterozygous mice were used as controls. All mice were maintained in our animal facility according to institutional guidelines, and experiments were done between 8 and 14 wk of age.
Cell Preparation and Culture.
Liver mononuclear cells (MNCs) and IELs were prepared as previously described (20). In some experiments, liver MNCs or IELs obtained from IRF-1−/− mice were cultured with 100 ng/ml mouse IL-15 (provided by Immunex Co., Seattle, WA) for 7 d.
Antibodies and Flow Cytometric Analysis.
The following mAb conjugates were purchased from PharMingen (San Diego, CA) and used in this study: M1/69-FITC (anti-HSA), 53-5.8-FITC (anti-CD8β), H57-597-FITC and -PE (anti–TCR-β), TM-β1-PE (anti–IL-2Rβ), GL-3-PE (anti–TCR-δ), 53-6.7-PE (anti-CD8), PK136-PE and -biotin (anti-NK1.1), 1B1-PE (anti-CD1), 27D-biotin (anti–LFA-1), IM7-biotin (anti-CD44), and KJ16-biotin (anti-Vβ8.1,8.2). B22-purified mAb (anti–H-2Db) was prepared in our laboratory. Biotinylated mAbs were detected with streptavidin red 670 (GIBCO BRL, Gaithersburg, MD) and purified mAbs were detected with goat anti–mouse IgG-FITC or goat anti–rat IgM-FITC; 106 cells were stained in 2% FCS PBS, washed, and analyzed by FACScan® using the Lysis II program (Becton Dickinson, Mountain View, CA).
Analysis for IL-15 mRNA Expression.
Bone marrow (BM) cells were isolated and stimulated by 30 μg/ml LPS and 100 U/ml IFN-γ for 6 h. Total cellular RNA was isolated with TRIZOL (GIBCO BRL) according to the manufacturer's protocol. 10 μg of total RNA were subjected to electrophoresis in a denaturing 1.0% agarose gel containing 2% formaldehyde and transferred to Hybond N+ nylon membrane (Amersham Corp., Arlington Heights, IL). The filter was hybridized with mouse IL-15 cDNA probe radioactively labeled with [32P]dCTP. The mouse IL-15 cDNA used as a probe was obtained by polymerase chain reaction using specific primers: sense primer 5′-GCC AGC TCA TCT TCA ACA-3′ and antisense primer 5′-TAA GTC TGA GAC GAG CTC TTT-3′. Radioactivity was assessed using phosphorimager (Molecular Dynamics, Sunnyvale, CA). The filter was stripped and rehybridized with a β-actin cDNA probe.
Results and Discussion
Impaired NK1+T Cell and NK Cell Development in IRF-1−/− Mice.
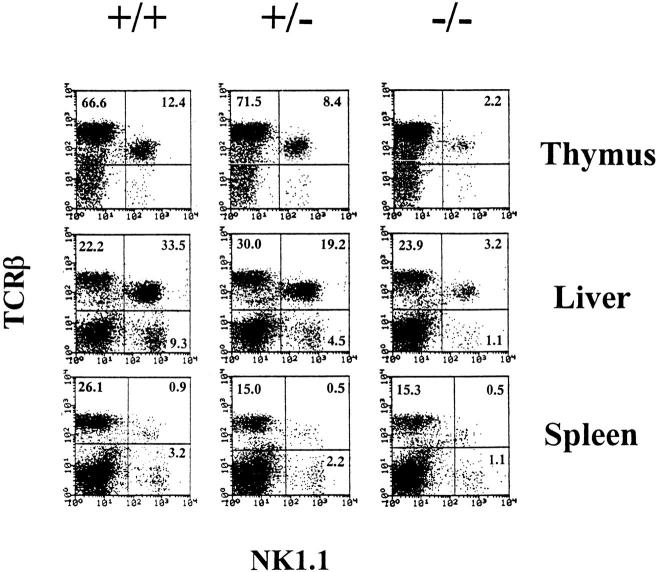
Mouse NK1+T cells are generally either CD4+8− or CD4−8− cells that are primarily found in the thymus, liver, and BM (1, 2). We examined the NK1+T cell subset in mice deficient for IRF-1−/−. Surprisingly, the percentages of thymic and liver NK1+T cells were decreased by 4–5 fold and 8–10 fold, respectively, in IRF-1−/− mice. The total number of thymic NK1+T cells obtained from IRF-1−/− mice was 10-fold lower than in wild-type control mice. Interestingly, a partial reduction of NK1+T cells was also seen in IRF-1+/− mice (Fig. 1, Table 1). The IL-2Rβ/15Rβ+TCR-α/β+ cells were also decreased, suggesting that the pronounced reduction of NK1+T cells detected in IRF-1−/− mice was not simply due to the loss of NK1.1 molecules from the cell surface (data not shown). The small number of NK1+T cells detected in IRF-1−/− mice expressed the IL-2Rβ/15Rβ chain and preferentially expressed Vβ8+ TCR as seen in control mice (data not shown). In addition, analysis of the thymus, liver, and spleen using IRF-1+/+, IRF-1+/−, and IRF-1−/− mice clearly demonstrated a reduction of NK cells (TCR-β−NK1.1+) in IRF-1−/− mice (Fig. 1). This is consistent with the lack of NK cell function previously reported in IRF-1−/− mice (21). Interestingly, IRF-1+/− mice consistently showed an intermediate phenotype, reflecting the dose-dependent requirement for genes regulated by IRF-1. These analysis showed that IRF-1 is important for NK cell and NK1+T cell development.
Figure 1.
IRF-1 is important for NK1+T cell and NK cell maturation. Thymocytes, liver, and spleen MNCs from indicated strains were stained with M1/69-FITC (anti-HSA), H57-597-PE (anti–TCR-β), and PK136-biotin (anti-NK1.1) plus streptavidin 670. HSA− cells are shown.
Table 1.
Impaired Maturation of Thymic NK1+T Cells in IRF-1− /− Mice
| Mice | Total thymocytes (× 106) | HSA− cell | NK1+T cell | Total NK1+T cell (× 104) | ||||
|---|---|---|---|---|---|---|---|---|
| % | % | |||||||
| IRF-1+/+ | 81.7 ± 4.7 | 3.4 ± 0.4 | 11.9 ± 1.4 | 39.5 ± 8.8 | ||||
| IRF-1+/− | 87.3 ± 10.8 | 3.2 ± 0.6 | 8.1 ± 0.6 | 22.6 ± 7.5 | ||||
| IRF-1−/− | 86.0 ± 11.8 | 1.6 ± 0.4 | 2.4 ± 0.4 | 3.3 ± 0.6 |
Four to six mice from each group were individually analyzed. Thymocytes were stained with M1/69-FITC (anti-HSA), H57-PE (anti– TCR-β), and PK136-biotin (anti-NK1.1) plus streptavidin 670. The percentage of NK1+T cells was calculated in the HSA− thymocyte population.
Previous reports have shown that CD4−8+TCRα/β+ cells were selectively reduced in thymus and periphery of IRF-1−/− mice (18). The data demonstrated a crucial role for IRF-1 in T cell development for the first time. A recent paper suggested that IRF-1 controls MHC class I expression through the regulation of transporter associated with antigen 1 and low molecular weight protein (19). Since mouse NK1+T cells require β2-microgloblin–associated CD1 and TL molecules for development (5–7, 20, 22, 23), we examined CD1 expression on thymocytes from IRF-1−/− mice. Consistent with a recent paper (19), Fig. 2 showed that the lack of the IRF-1 gene clearly resulted in reduced H-2Db expression. However, the mean intensities of CD1 on IRF-1−/− thymocytes was comparable to littermate controls, suggesting that the IRF-1 gene does not control NK1+T cell development through CD1 expression. In addition, we can further exclude the role of the TL antigen in NK1+T cell development, since both IRF-1−/− and control mice are of the C57B1/6 background and do not express TL.
Figure 2.
Normal CD1 expression on IRF-1−/− thymocytes. Thymocytes from the indicated strains were stained with 1B1-FITC (anti-CD1), 57.6.7-PE (anti-CD8), and L3T4-biotin (anti-CD4) plus streptavidin 670, and double-positive CD4+8+ thymocytes were analyzed for CD1 expression. For H-2Db expression, total thymocytes were stained with B22 (anti–H-2Db) plus goat anti–mouse Ig-FITC.
Maturation of Intestinal IELs Is Reduced in IRF-1− /− Mice.
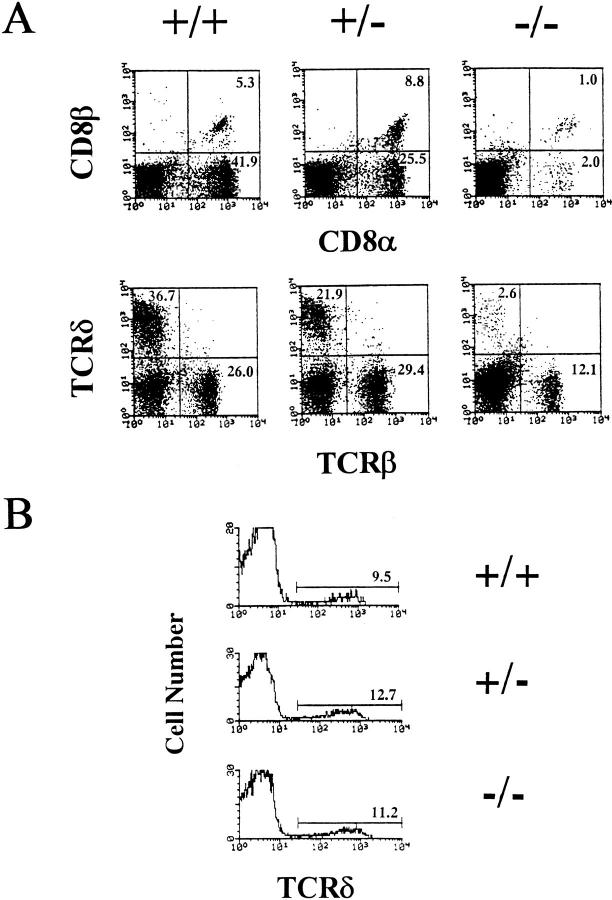
The majority of IELs express CD8 and can be divided into two subsets. One population bears CD8-α/β+ heterodimers and expresses TCR-α/β+, whereas the other expresses CD8-α/α+ homodimers consisting of TCR-α/β+ and TCR-γ/δ+ cells. Using thymectomized recombinase activating gene (RAG)-deficient mice reconstituted with BM cells from athymic (nude) mice, thymus-independent development of CD8-α/α+ IELs has been clearly demonstrated to occur (8). Surprisingly, in IRF-1−/− mice, the percentage of intestinal CD8-α/α+ IELs was approximately eight- to ninefold less than in wild-type control mice. As seen with NK1+T cells, mice heterozygous for IRF-1+/− showed altered CD8-α/α+ IEL development. TCR-γ/δ+ IELs were profoundly reduced by IRF gene disruption (Fig. 3 A, Table 2). In addition, CD8-α/β+T cells were also reduced as seen in periphery. The total cell numbers of IELs from IRF-1−/− mice (0.4 ± 0.1 × 106) were three- to fourfold lower than those from littermate controls (1.5 ± 0.3 × 106). Therefore, IRF-1 controls the expression of genes important for IEL T cell development. It is likely that the reduced development of intestinal γ/ δ+T cells is controlled by other mechanisms unrelated to MHC class I expression in IRF-1−/− mice. Previous studies using β2-microglobulin–deficient (MHC class I−/−) mice showed a reduction in TCR-α/β+ IELs, but not TCR-γ/ δ+ IELs (24), demonstrating that TCR-α/β+ and TCR-γ/ δ+ IELs have differential requirements for β2-microglobulin dependent selection.
Figure 3.
IRF-1 controls intestinal IEL development. (A) Intestinal IELs were obtained from either IRF-1+/+ mice, IRF-1+/− mice, or IRF-1−/− mice and stained with H57-597 (anti–TCR-β) and GL-3-PE (anti– TCR-δ), or 53.6.7-FITC (anti–CD8α) and Lyt3-PE (anti-CD8β). (B) Thymocytes were stained with L3T4-FITC (anti-CD4), GL-3-PE (anti– TCR-δ), and 53.6.7-FITC (anti-CD8α). Histograms are gated on double-negative CD4−8− thymocytes and TCR-δ expression is shown.
Table 2.
Intestinal and Thymic T Cell Subsets in IRF-1− /− Mice
| Mice | Intestine | Thymus* | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CD8-α/α+ | CD8-α/β+ | TCR-α/β+ | TCR-γ/δ+ | TCR-γ/δ+ | ||||||
| IRF-1+/+ | 42.5 ± 4.3 | 7.5 ± 3.0 | 26.7 ± 2.6 | 34.7 ± 4.0 | 9.3 ± 0.5 | |||||
| IRF-1+/− | 27.0 ± 2.6 | 10.5 ± 2.3 | 29.5 ± 3.6 | 21.0 ± 4.4 | 10.7 ± 1.8 | |||||
| IRF-1−/− | 5.0 ± 3.4 | 3.2 ± 1.7 | 15.8 ± 8.9 | 4.6 ± 2.8 | 9.6 ± 0.8 | |||||
Four mice in each group were individually analyzed.
Total thymocytes were stained with GK1.5-FITC (anti-CD4), GL-3-PE (anti–TCR-γ/δ), and 53.6.7-biotin (anti-CD8) plus streptavidin 670. TCR-γ/δ+ cells were analyzed on gated double-negative (CD4−8−) thymocytes.
Since the majority of thymus-independent intestinal TCR-γ/δ+ cells were absent in IRF-1−/− mice, we also examined whether thymic TCR-γ/δ+ cells were present in these mice. Although the number of intestinal TCR-γ/δ+ cells were decreased by 10-fold in IRF-1−/− mice, thymic TCR-γ/δ+ cells were normal (Fig. 3 B, Table 2). Thus, IRF-1 selectively affected the development of intestinal TCR-γ/δ+ cells.
IL-15 mRNA Expression Is Impaired in IRF-1− /− BM Cells.
As certain cytokines are crucial for lymphocyte development, it is possible that a reduction in the expression of cytokine receptors or cytokines may result in poor selection, survival, or expansion of NK1+T cells, NK cells, and intestinal IEL subsets in IRF-1−/− mice. IL-15 is one of the most likely targets because NK1+T cells, NK cells and intestinal IEL subsets are severely reduced in IL-2Rβ/ 15Rβ−/− mice (9, 10), while present in normal numbers in IL-2, IL-7Rα, or IL-7–deficient mice (10, 25–27). Interestingly, IL-15 preferentially promotes the proliferation of these T cell subsets (10–12). Thus, we examined IL-15 mRNA expression by Northern blot analysis (Fig. 4 A). Wild-type BM cells cultured in the presence of LPS and IFN-γ for 6 h, clearly increased IL-15 mRNA levels. In contrast, IL-15 mRNA remained undetectable in IRF-1−/− BM cells, even after induction with LPS and IFN-γ. These data demonstrate that IRF-1 regulates the expression of IL-15.
Figure 4.

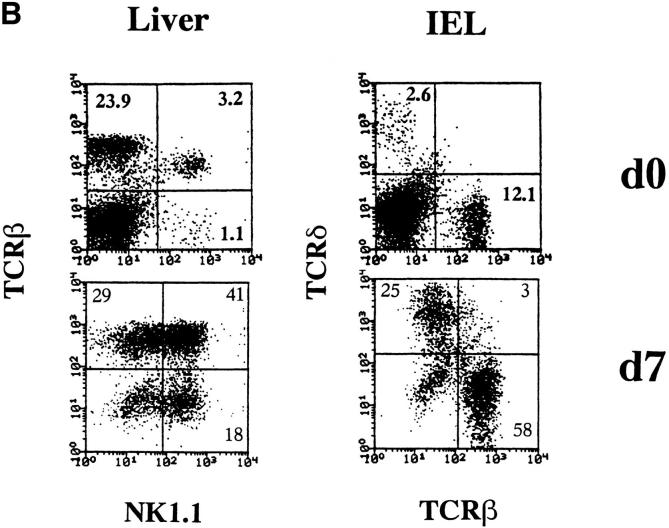
Impaired lineage development correlates with the absence of IL-15. (A) Limited IL-15 expression in the absence of IRF-1. BM cells were isolated from IRF-1−/− mice or control wild-type (WT) mice. Total RNA was extracted from untreated BM cells or BM cells cultured for 6 h in the presence of LPS (30 μg/ml) and IFN-γ (100 U/ml). Northern blot analysis was performed using IL-15 cDNA and β-actin probes. (B) IL-15 induces the expansion of NK1+T cells, NK cells, and IEL subsets. Liver MNCs and intestinal IELs were isolated from IRF-1−/− mice and cultured with 100 ng/ml mouse IL-15 for 7 d.
NK1+T Cells, NK Cells, and Intestinal IELs were Recovered by IL-15 In Vitro.
To further examine the importance of IL-15 for maturation of NK1+T cells, NK cells, and intestinal IEL subsets in IRF-1−/− mice, liver MNCs and intestinal IELs were isolated from these mice and cultured with 100 ng/ml mouse IL-15 for 7 d (Fig. 4 B). Recovery of these lymphocyte subsets was observed. This suggested that IL-15 is essential for the survival or expansion of NK1+T cells, NK cells, and intestinal IELs, and not early development or commitment.
NK1+T cells, NK cells, and intestinal IELs share cell surface markers and other common features during development. In addition to the expression and developmental requirement of IL-2Rβ/15Rβ chain, they also express the NK complex that encompasses NKR-P1 and Ly-49 (1, 2, 28, 29). In contrast, conventional T cells do not express these products. Although the majority of T cells develops in the thymus, NK cells develop normally in athymic nude mice. The developmental origin of NK1+T cells can be either thymus dependent or independent (2, 30, 31). Thymus-independent development of intestinal CD8-α/α+ T cells has been clearly demonstrated to occur (8). Thus, NK1+T cells and intestinal CD8-α/α+ T cells are related to the NK lineage and can be distinguished from mainstream T cells. Our results demonstrate that IRF-1 controls the expression of IL-15, which is likely to be important for the maturation of the related NK1+T cell, NK cell, and CD8-α/α+ IEL lineages.
Acknowledgments
We wish to thank Dr. Hans-Willi Mittrucker (Amgen Institute, Toronto, Ontario, Canada) for providing the C57BL/6 background IRF-1−/− mice, Dr. Yutaka Tagaya (National Cancer Institute, Bethesda, MD) for helpful discussion, and Arsen Zakarian (Ontario Cancer Institute, Toronto, Ontario, Canada) for technical assistance.
This work was supported by the Medical Research Council of Canada. P.S. Ohashi is a recipient of a Medical Research Council scholarship.
References
- 1.Bendelac A. Mouse NK1+T cells. Curr Opin Immunol. 1995;7:367–374. doi: 10.1016/0952-7915(95)80112-x. [DOI] [PubMed] [Google Scholar]
- 2.MacDonald HR. NK1.1+ T cell receptor-α/β+cells: new clues to their origin, specificity, and function. J Exp Med. 1995;182:633–638. doi: 10.1084/jem.182.3.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taniguchi M, Koseki H, Tokuhisa T, Masuda K, Sato H, Kondo E, Kawano T, Cui J, Perkes A, Koyasu S, Makino Y. Essential requirement of an invariant Vα14 T cell antigen receptor expression in the development of natural killer T cells. Proc Natl Acad Sci USA. 1996;93:11025–11028. doi: 10.1073/pnas.93.20.11025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ohteki T, MacDonald HR. Stringent Vβ requirement for the development of NK1.1+ T cell receptor-α/β+cells in mouse liver. J Exp Med. 1996;183:1277–1282. doi: 10.1084/jem.183.3.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bendelac A, Lantz O, Quimby ME, Yewdell JW, Bennink JR, Brutkiewicz RR. CD1 recognition by mouse NK1+T lymphocytes. Science. 1995;268:863–865. doi: 10.1126/science.7538697. [DOI] [PubMed] [Google Scholar]
- 6.Smiley ST, Kaplan MH, Grusby MJ. Immunoglobulin E production in the absence of interleukin-4– secreting CD1-dependent cells. Science. 1997;275:977–979. doi: 10.1126/science.275.5302.977. [DOI] [PubMed] [Google Scholar]
- 7.Joyce S, Negishi I, Boesteanu A, DeSilva AD, Sharma P, Chorney MJ, Loh DY, Kaer LV. Expansion of natural (NK1+) T cells that express αβ T cell receptors in transporters associated with antigen presentation–1 null and thymic leukemia antigen positive mice. J Exp Med. 1996;184:1579–1584. doi: 10.1084/jem.184.4.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guy-Grand D, Vassalli P. Gut intraepithelial T lymphocytes. Curr Opin Immunol. 1993;5:247–252. doi: 10.1016/0952-7915(93)90012-h. [DOI] [PubMed] [Google Scholar]
- 9.Suzuki H, Duncan GS, Takimoto H, Mak TW. Abnormal development of intestinal intraepithelial lymphocytes and peripheral natural killer cells in mice lacking the IL-2 receptor β chain. J Exp Med. 1997;185:499–505. doi: 10.1084/jem.185.3.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohteki T, Shirley H, Suzuki H, Mak TW, Ohashi PS. Role for IL-15/IL-15Rβ chain in NK1.1+ T cell receptor–αβ+cell development. J Immunol. 1997;159:5931–5935. [PubMed] [Google Scholar]
- 11.Inagaki-Ohara K, Nishimura H, Mitani A, Yoshikai Y. Interleukin-15 preferentially promotes the growth of intestinal intraepithelial lymphocytes bearing γδ T cell receptor in mice. Eur J Immunol. 1997;27:2885–2891. doi: 10.1002/eji.1830271121. [DOI] [PubMed] [Google Scholar]
- 12.Mrozek E, Anderson P, Caliguiri MA. Role of interleukin-15 in the development of human CD56+ natural killer cells from CD34+hematopoietic progenitor cells. Blood. 1996;87:2632–2640. [PubMed] [Google Scholar]
- 13.Pine R, Decker T, Kessler DS, Levy DE, Darnell JE., Jr Purification and cloning of interferon-stimulated gene factor 2 (ISGF2): ISGF2 (IRF-1) can bind to the promoters of both β interferon– and interferon–stimulated genes but is not a primary transcriptional activator of either. Mol Cell Biol. 1990;10:2448–2457. doi: 10.1128/mcb.10.6.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miyamoto M, Fujita T, Kimura Y, Maruyama M, Harada H, Sudo Y, Miyata T, Taniguchi T. Regulated expression of a gene encoding a nuclear factor, IRF-1, that specifically binds to IFN-β gene regulatory elements. Cell. 1988;54:903–913. doi: 10.1016/s0092-8674(88)91307-4. [DOI] [PubMed] [Google Scholar]
- 15.Fujita T, Sakakibara J, Sudo Y, Miyamoto M, Kimura Y, Taniguchi T. Evidence for a nuclear factor(s), IRF-1, mediating induction and slicing properties to human IFN-β gene regulatory elements. EMBO (Eur Mol Biol Organ) J. 1988;7:3397–3405. doi: 10.1002/j.1460-2075.1988.tb03213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parrington J, Rogers NC, Gewert DR, Pine R, Veals SA, Levy DE, Stark GR, Kerr IM. The interferon-stimulable response elements of two human genes detect overlapping sets of transcription factors. Eur J Biochem. 1993;214:617–626. doi: 10.1111/j.1432-1033.1993.tb17961.x. [DOI] [PubMed] [Google Scholar]
- 17.Levy DE, Kessler DS, Pine R, Reich N, Darnell JE., Jr Interferon-induced nuclear factors that bind a shared promotor element correlate with positive and negative transcriptional control. Genes Dev. 1988;2:383–393. doi: 10.1101/gad.2.4.383. [DOI] [PubMed] [Google Scholar]
- 18.Matsuyama T, Kimura T, Kitagawa M, Pfeffer K, Kawakami T, Watanabe N, Kundig TM, Amakawa R, Kishihara K, Wakeham A, et al. Targeted disruption of IRF-1 or IRF-2 results in abnormal type I IFN gene induction and aberrent lymphocyte development. Cell. 1993;75:83–97. [PubMed] [Google Scholar]
- 19.White LC, Wright KL, Felix NJ, Ruffner H, Reis LF, Pine R, Ting JP-Y. Regulation of LMP2 and TAP1 genes by IRF-1 explains the paucity of CD8+ T cells in IRF-1−/−mice. Immunity. 1996;5:365–376. doi: 10.1016/s1074-7613(00)80262-9. [DOI] [PubMed] [Google Scholar]
- 20.Ohteki T, MacDonald HR. Major histocompatibility complex class I related molecules control the development of CD4+8− and CD4−8− subsets of natural killer 1.1+ T cell receptor–α/β+cells in the liver of mice. J Exp Med. 1994;180:699–704. doi: 10.1084/jem.180.2.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gordon SD, Mittrucker H-W, Kagi D, Matsuyama T, Mak TW. The transcriptional factor interferon regulatory factor-1 is essential for natural killer cell function in vivo. J Exp Med. 1996;184:2043–2048. doi: 10.1084/jem.184.5.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bendelac A, Killeen N, Littman DR, Schwartz RH. A subset of CD4+thymocytes selected by MHC class I molecules. Science. 1994;263:1774–1778. doi: 10.1126/science.7907820. [DOI] [PubMed] [Google Scholar]
- 23.Coles MC, Raulet D. Class I dependence of the development of CD4+CD8−NK1.1+thymocytes. J Exp Med. 1994;180:395–399. doi: 10.1084/jem.180.1.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Correa I, Bix M, Liao N-S, Zijlstra M, Jaenisch R, Raulet D. Most γδ T cells develop normally in β2-microglobulin-deficient mice. Proc Natl Acad Sci USA. 1992;89:653–657. doi: 10.1073/pnas.89.2.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He Y-W, Malek TR. Interleukin-7 receptor α is essential for the development of γδ+T cells, but not natural killer cells. J Exp Med. 1996;184:289–293. doi: 10.1084/jem.184.1.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sadlack B, Merz H, Schorle H, Schimpl A, Feller AC, Horak I. Ulcerative colitis-like disease in mice with a disrupted interleukin-2 gene. Cell. 1993;75:253–261. doi: 10.1016/0092-8674(93)80067-o. [DOI] [PubMed] [Google Scholar]
- 27.Vicari AP, Herbelin A, Leite-de-Moraes MDC, von Freeden-Jeffry U, Murray R, Zlotnik A. NK1.1+T cells from IL-7–deficient mice have a normal distribution and selection but exhibit impaired cytokine production. Int Immunol. 1996;8:1759–1766. doi: 10.1093/intimm/8.11.1759. [DOI] [PubMed] [Google Scholar]
- 28.Yokoyama WM, Seamen WE. The Ly-49 and NKR-P1 gene families encoding lectin-like receptors on natural killer cells: the NK gene complex. Annu Rev Immunol. 1993;11:613–635. doi: 10.1146/annurev.iy.11.040193.003145. [DOI] [PubMed] [Google Scholar]
- 29.Guy-Grand D, Cuenod-Jabri B, Malassis-Seris M, Selz F, Vassalli P. Complexity of the mouse gut T cell immune system: identification of two distinct natural killer T cell intraepithelial lineages. Eur J Immunol. 1996;26:2248–2256. doi: 10.1002/eji.1830260942. [DOI] [PubMed] [Google Scholar]
- 30.Hashimoto W, Takeda K, Anzai R, Ogasawara K, Sakihara H, Sugiura K, Seki S, Kumagai K. Cytotoxic NK1.1 Ag+αβ T cells with intermediate TCR induced in the liver of mice by IL-12. J Immunol. 1995;154:4333–4340. [PubMed] [Google Scholar]
- 31.Shimamura M, Ohteki T, Launois P, Garcia A-M, MacDonald HR. Thymus-independent generation of NK1+ T cells in vitrofrom fetal liver precursors. J Immunol. 1997;158:3682–3689. [PubMed] [Google Scholar]