Abstract
Junctional epidermolysis bullosa (JEB) is an autosomal recessive skin blistering disease with both lethal and nonlethal forms, with most patients shown to have defects in laminin-5. We analyzed the location of mutations, gene expression levels, and protein chain assembly of the laminin-5 heterotrimer in six JEB patients to determine how the type of genetic lesion influences the pathophysiology of JEB. Mutations within laminin-5 genes were diversely located, with the most severe forms of JEB correlating best with premature termination codons, rather than mapping to any particular protein domain. In all six JEB patients, the laminin-5 assembly intermediates we observed were as predicted by our previous work indicating that the α3β3γ2 heterotrimer assembles intracellularly via a β3γ2 heterodimer intermediate. Since assembly precedes secretion, mutations that disrupt protein–protein interactions needed for assembly are predicted to limit the secretion of laminin-5, and likely to interfere with function. However, our data indicate that typically the most severe mutations diminish mRNA stability, and serve as functional null alleles that block chain assembly by resulting in either a deficiency (in the nonlethal mitis variety) or a complete absence (in lethal Herlitz-JEB) of one of the chains needed for laminin-5 heterotrimer assembly.
Epidermolysis bullosa (EB)1 is a family of genetic blistering skin diseases with three major forms dependent upon the exact layer of the skin where the split occurs (1, 2). In epidermolysis bullosa simplex defects in keratin 5 and 14 that make up tonofilaments within the basal keratinocytes cause cell fragility and result in separation from the suprabasilar layers (3, 4). In dystrophic forms of EB cleavage occurs just below the basement membrane zone due to defects in anchoring filaments made of collagen VII (5, 6). In the junctional forms of EB (JEB) blistering occurs at the lamina lucida (1) and is associated with defects in the protein components of hemidesmosomes that attach basal keratinocytes to the basement membrane zone (BMZ). JEB is an autosomal recessive disease, with six variants currently recognized by the National Epidermolysis Registry (1), ranging from the mild, localized JEB inversa form to the severe, generalized JEB Herlitz form (H-JEB), which typically results in death early in infancy. In our study, we focused on the individual polypeptide chains of laminin-5, the primary protein component of the anchoring filaments shown to be most often defective in JEB. The anchoring filaments connect the hemidesmosomes located along the basal surface of basal keratinocytes to the underlying basement membrane zone. Microscopy data have shown these filaments to be absent or reduced in numbers in JEB (7, 8).
The first realization that laminin-5 is defective in JEB came from immunofluorescence studies using the monoclonal antibody GB3, which illustrated reduced or absent staining for the basement membrane of JEB patients (7, 9). It was later shown that GB3 recognizes a large glycoprotein (10) termed nicein (7), kalinin (11, 12), or epiligrin (13), by different investigators, but is now known as the epithelium-specific laminin variant, laminin-5 (14). Laminin-5 is an isoform of the classical laminin, laminin-1(reviewed in reference 15) and is also a heterotrimer that is presumed to form a cruciform structure of disulfide linked chains (11). There are three separate genes for the chains of laminin-5 (14), LAMA3 (16), LAMB3 (17), and LAMC2 (18, 19). These code for three polypeptides (12): α3 of 200 kD, β3 of 145 kD, and γ2 of 155 kD. The latter contains the antigenic determinant recognized by GB3 (20), the immunoreagent most often used in the diagnosis of JEB.
Extensive mutation detection analysis has been conducted in numerous JEB patients for each of the laminin-5 chain genes and mutations have been mapped to each of the chains, including γ2 (21–24), β3 (25–27), and α3 (28). Such studies, designed to pinpoint the exact location of genetic mutations in specific patients provide a potential framework for correlations to be drawn between the site of mutation and clinical phenotypes. For example, in JEB patients containing mutations in laminin-5 chains with the most severe, lethal form of JEB (H-JEB) it was hoped that there might be a clustering of mutations at certain locations within the genes, reflecting protein domains most crucial to laminin-5 function. Instead, the site of the genetic lesions appears to vary widely.
We sought to further clarify how specific types of mutations in laminin-5 chains result in the observed differences in clinical phenotypes. An emphasis on understanding the downstream effects of the mutations and comparing multiple patient samples necessitated brevity in the presentation of the mutation data. We biopsied JEB patients and cultured keratinocytes because they provided us with the unique opportunity of evaluating gene expression at both the mRNA and protein levels. Here we analyze JEB patient keratinocytes with defects in each of the three chains of the laminin-5 heterotrimer. The clearest correlation we found was on the protein level, where the H-JEB patients had a complete absence of any one of the three chains. In contrast, the less severe mitis forms of JEB had normal, or somewhat reduced, levels of one of the chains. The protein analysis also provided a test for our hypothesis, described in our previous studies, showing that the α3β3γ2 heterotrimer assembles intracellularly via a β3γ2 intermediate (29). Since each of the H-JEB patients had one expressed chain missing, the mutant genes for these chains performed as functional null alleles, and allowed us to look at assembly intermediates when each of the three chains was missing. The assembly intermediates present in each of the H-JEB patients were as predicted by our assembly model.
In general, the mRNA levels for the laminin-5 chains corresponded to the observed protein levels, indicating that the mutations are causing a net decrease in the mRNA levels. Thus, mRNA levels for the mutant chains are often drastically reduced, or undetectable in H-JEB patients as compared with more normal levels in nonlethal JEB patients. These observations agree with both our data and data from other laboratories that have shown that H-JEB patients often have premature termination codons (PTCs) in at least one allele (22, 30). PTCs have been shown to give rise to unstable mRNA transcripts (31, 32), thereby explaining the often undetectable mRNA levels. We conclude that it is the near, or complete absence of laminin-5 heterotrimer, typically caused by mutations that destabilize one of the transcripts for laminin-5, that is the hallmark of lethal H-JEB.
Materials and Methods
Patients.
Skin biopsies were obtained from six unrelated JEB patients, D, G, J, L, M, and V (Table I). In all cases, JEB was diagnosed according to the revised criteria for subtyping EB (1). Four patients (G, J, L, and M, all died shortly after birth) had the severe Herlitz form of JEB, one patient had an intermediate form of JEB (patient V alive at age 5), and the final patient had the mild, generalized “mitis” form of JEB patient (patient D, died at age 44). All patients had generalized blisters. mAb GB 3 did not react with the skin biopsies from patients D, G, J, L, and M and reacted only weakly with that of patient V. Transmission electron microscopic analysis showed a dermal–epidermal split in the lamina lucida and a reduced number of hemidesmosomes (data not shown).
Cell Culture and Immortalization.
Keratinocytes were cultured from neonatal foreskin for use as the normal control sample, and from JEB patient skin biopsies. All gene expression experiments were conducted using immortalized keratinocyte cell cultures, because of the advantage of a more uniform and readily available source of cells. Immortalized patient L keratinocytes were obtained as described (33). Transfections of all of the other primary cells were accomplished by lipofectin (GIBCO BRL, Gaithersburg, MD) after one or two passages. Cells were grown in serum-free media (SFM; GIBCO BRL) and transfected with p18ccB (34, 35), containing a subgenomic fragment of HPV-18 encoding intact open-reading frames of E6 and E7. These genes are sufficient for inducing the immortalization of keratinocytes. At 2 d after transfection, selection by G418 (100 mg/ml; Sigma Chemical Co., St. Louis, MO) was applied for a total of 3–4 d. Individual colonies were cloned and passaged once a week at a split ratio of 1:6 or 1:8. The cell lines that survived >20 passages in culture were considered immortalized and used for study.
Antibodies.
Polyclonal anti–laminin-5 antisera (11) was kindly provided by Dr. Peter Marinkovich (Stanford University, Stanford, CA). Polyclonal anti-β3 antisera was raised against a glutathione/S-transferase/laminin-5 β3 fusion protein as previously described (29).
Biosynthetic Labeling and Immunoprecipitation.
Immortalized keratinocytes from both normal and JEB patients were cultured in SFM. Dissociated cells were seeded at 105 cells per 60-mm dish, allowed to attach in complete growth medium for 24 h, and then incubated in methionine- and cysteine-deficient SFM for 2 h. Cells were then cultured in deficient SFM containing 50 mCi/ml of protein labeling mix ([35S]methionine and [35S]cysteine; Dupont-NEN, Boston, MA) for 24 h. Cell and medium fractions were processed for immunoprecipitation, which was performed as previously described (20). In brief, cell layers were harvested with a cell scraper and ice-cold RIPA buffer (10 mM Tris-HCl, pH 7.4, 150 mM NaCl, 2 mM EDTA, 250 mM PMSF, 1 mM n-methylmaleimide, 2 mM l-methionine, 2 mM l-cysteine, 0.3% NP-40, 0.05% triton X-100, 0.3% sodium deoxycholate, and 0.1% BSA containing 0.1% SDS). 250 mM PMSF was added to the culture medium which was then centrifuged at 12,000 rpm to remove cells and debris. TCA-precipitable radioactivity of the medium and cell fractions was counted. Both the medium (2.0 × 106 TCA-precipitable cpm) and the cell lysate (2.0 × 107 TCA-precipitable cpm) were cleared by the addition of preimmunized rabbit serum, anti-rabbit IgG-conjugated agarose beads (Sigma Chemical Co.), and gelatin sepharose (Pharmacia Biotechnology, Inc., Piscataway, NJ) for 1 h at 4°C. Immunologically reactive laminin-5 was precipitated from precleared medium and cell fraction by 16 h incubation with a mixture of anti-rabbit IgG-conjugated agarose beads and anti–laminin-5 antisera. After incubation, precipitates were pelleted by centrifugation at 2,500 rpm for 10 min and washed with RIPA buffer (medium sample) or RIPA buffer containing 0.1% SDS (for cell extract). After five washes, the pellets were mixed with SDS sample buffer, heated to 95°C for 3 min and analyzed by SDS-PAGE.
Gel Electrophoresis.
Two-dimensional SDS-PAGE was performed as previously described (36). After soaking the gels in Amplify (Amersham Pharmacia Biotechnology, Buckinghamshire, England), fluorography was used to visualize radiolabeled proteins in the acrylamide gels followed by drying and exposure to film.
Northern Blot Analysis.
Total RNA was prepared from patient and normal cells using RNAeasy (QIAGEN Inc., Chatsworth, CA). The RNA was subjected to electrophoresis on a 1.2% denaturing agarose gel containing 1.2 M formaldehyde and transferred onto nylon membranes (Hybond N; Amersham Pharmacia Biotechnology). Membranes were subsequently hybridized at high stringency with 32P-labeled cDNA probes for the laminin-5 chains as follows; Na3 for the α3 chain (37), PCR1.3 for the γ2 chain (19), and kal 85 for the β3 chain (17), using RapidHyb (Amersham Pharmacia Biotechnology). Blots were exposed to x-ray film using intensifying screens.
DNA Isolation and PCR Amplification.
Genomic DNA was isolated from peripheral blood lymphocytes by standard procedures and used as a template for amplification of individual exons within the LAMB3 gene. Oligonucleotide primers spanning each of the gene's 23 exons were synthesized on the basis of intronic sequences to generate PCR products. For amplification by the PCR, ∼50 ng of genomic DNA was used as the template in a reaction mixture containing 5 μl of Gene Amp 10× PCR buffer with MgCl2 (Perkin-Elmer Corp., Norwalk, CT), 2.5 mM of each nucleotide, 10 pmol of each primer and 1.25 U of AmpliTaq DNA Polymerase (Perkin-Elmer Corp.) in a total volume of 50 μl. The PCR primers used to identify only those mutations shown here are listed below with 5′ identifying numbers and using R to indicate sense strand and L to indicate antisense: patient J in exon 3, −130 R gcagcgagtggaagcttg, +130 L gtataaggccagatcctagcc; patient V in exon 7, −98 R ttcaagtgtgaggcatataatgg, +133 L gatttgatacttgccctgcttg; patient M in exon 15, −183 R gattgtgctggtcgccaagcc, +202 L tcgcgccttttggaccagct; patient M in exon 20, −158 R ggggtcctgggacatcaaagctac, +341 L gttcagcaggtactgcggcc; and patient D in exon 17, −64 R cgtgtggggtggattggttatt, +105 L acactaaagtccagagattttaggtga.
The primers correspond to flanking intronic sequences. In the case of sense primers, the number indicates the position upstream from the intron–exon border and in the case of antisense primers, to the position downstream from the exon–intron border. The amplification conditions for the typical primer were 94°C for 7 min followed by 40 cycles of 94°C for 45 s; 50–66°C for 45 s; 72°C for 1 min in a 9600 GeneAmp PCR thermocycler (Perkin-Elmer Corp.). 5-μl aliquots of the PCR products were analyzed on a 3% agarose gel and visualized by ethidium bromide staining.
Heteroduplex Analysis.
10 μl of the PCR sample was prepared for heteroduplex analysis according to the Mutation Detection Enhancement (MDE) protocol supplied by the manufacturers (JT Baker Inc. Phillipsburg, NJ). Samples were run both using PCR products amplified from patient DNA alone, or were mixed with PCR products amplified from normal DNA, to detect heteroduplexes. Staining with ethidium bromide was used to visualize the heteroduplexes (data not shown).
Genomic DNA Sequencing.
When a heteroduplex was detected, the PCR product was sequenced by cycle sequencing (Sequitherm cycle sequencing; Epicentre Technologies Corp., Madison, WI) and run on a 6% denaturing polyacrylamide gel using the Genomyx LR system (Genomyx, Foster City, CA). Mutations were confirmed by separate rounds of PCR amplification followed by cycle sequencing, repeated at least three times to diminish the possibility of PCR generated mutations. Mutations from three previously uncharacterized JEB patients are presented here, patients D, J, and V. Family members were not readily available for patients D or V, so no familial analysis or verification was performed. Patient J's mutations were confirmed by familial analysis (data not shown).
Results
Clinical Phenotypes and Location of Mutations in Junctional Epidermolysis Bullosa Patients.
Six unrelated JEB patients were selected for investigation in this study (Table I), including four patients with the H-JEB and two patients with the less severe mitis variety. These six patients were chosen for presentation because defects in each of the three laminin-5 chains of the heterotrimer are represented. Specifically, we sought to evaluate the effect on laminin-5 chain assembly when each of the three chains of the heterotrimer is defective. Because the highest percentage of JEB patients contain mutations in the β3 chain (30), we also present four patients with mutations in the β3 chain (patients D, V, J, and M). These patients demonstrate a correlation between the type of mutation, its effect on laminin-5 expression, and the severity of the clinical phenotype.
Two patients with β3 chain mutations had the less severe mitis form of JEB (patient D and V), and a single allele mutation for each patient is presented. The mutation identified in patient D is a PTC resulting from a nonsense mutation. In patient V we identified a splice site mutation. Two patients (J and M) with H-JEB who had β3 chain mutations were also characterized. Patient J is homozygous for a nonsense mutation at a position located early in the transcript for the β3 chain. The location corresponds to a mutational hotspot previously identified (30) but characterized for the first time here in this patient. Patient M has a single-bp deletion on one allele that creates a frame shift resulting in a PTC 94 bp downstream, and a splice site mutation on the second allele (35). We also present patient G with a defect in the α3 chain as judged by protein and mRNA data, but for whom the exact location of mutation has not yet been determined. Finally, patient L with a γ2 chain defect is presented containing a homozygous nonsense mutation that has been previously identified (22). By collecting protein assembly data for patient L, we further clarify the impact of these γ2 chain mutations.
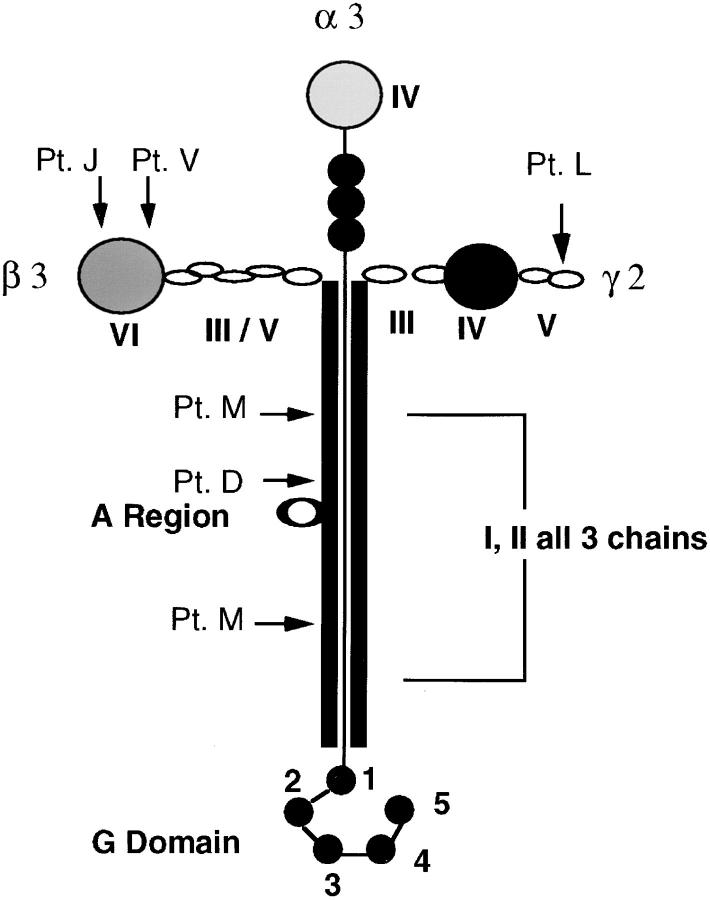
The locations of laminin-5 mutations in these patients are summarized in Fig. 1. The diverse locations of these mutations exemplifies the lack of correlation between clinical severity and localization of mutations to specific domains. Some of the mutations in these patients were localized to the NH2-terminal ends of the chains that form the short arms of laminin-5, as indicated for patients L, J, and V. Other mutations were contained within the COOH-terminal domains I/II that form the long arm, shown for patients D and M. This region is presumed to form a triple α-helical coiled-coil (14, 38) based on homology to laminin-1, and is important for the assembly of the three chains into a heterotrimer.
Figure 1.
Structural model of laminin-5 and location of analyzed mutations. Protein domains in the α3, β3, and γ2 chains are labeled with roman numerals by homology to laminin-1. The short arms contain globular domains flanked by EGF-like repeats as depicted by the small circles. The long arms of the three chains are associated in an α-helical coiled-coil of approximately the same length as in laminin-1, 600 AA. The heptad repeats of the long arm are interrupted at one region in the β3 chain, termed the A region. The α3 chain contains a COOH-terminal end with a globular extension containing five similar regions (G domain). Arrows with single letter designations denote approximate locations of mutations for the JEB patients discussed.
Identification of Mutations in Laminin-5 Chain Genes.
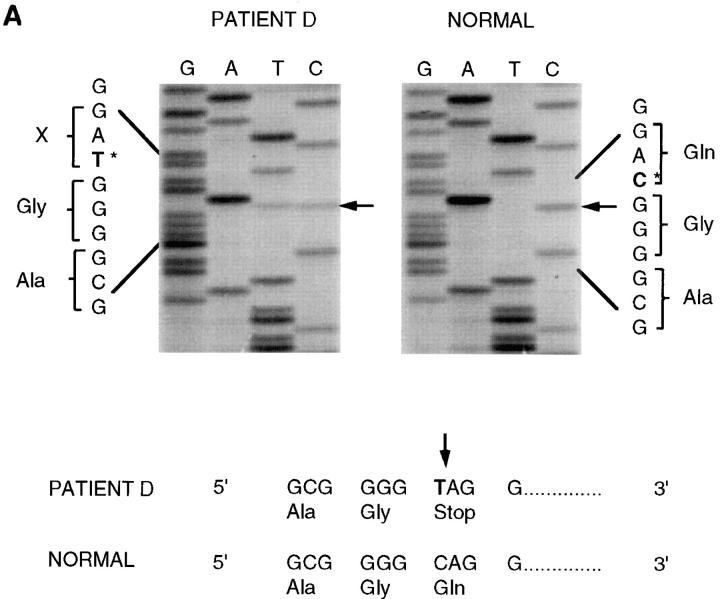
Two of the patients studied have previously published mutations (patient L [22] and our earlier work on patient M [Pereira, P., C. Tzou, L. Pan, T. Kutzkey, K. Hultquist, S. Cobberly, L. Nall, J. McGuire, M.P. Marinkovich, E.A. Bauer, G.S. Herron, W.K. Hoeffler, and E.A. Welsh, manuscript submitted for publication]), whereas three of the other patients were previously uncharacterized for their mutations in the β3 chain (patients D, V, and J). We show direct DNA sequencing of PCR products amplified from genomic DNA from these three patients. To screen for mutations, a series of PCR products spanning the entire LAMB3 gene were resolved on MDE gels to distinguish heteroduplex mismatched DNA strands indicative of polymorphic sequence variations (data not shown). Direct DNA sequencing of a polymorphic PCR product using primers spanning exon 17 for patient D revealed a C-to-T transition at position 2621 of the cDNA (Fig. 2 A). A comparison of the normal sequence on the right with that of the proband on the left shows that the C position signal drops to 50% intensity at this position and that a T also appears at this same position. A nonsense mutation is created on one of the alleles where the T appears but is normal at this position in the other allele.
Figure 2.
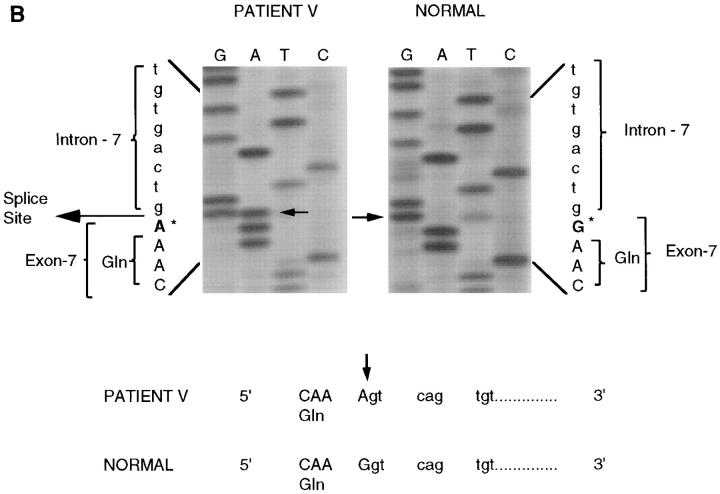
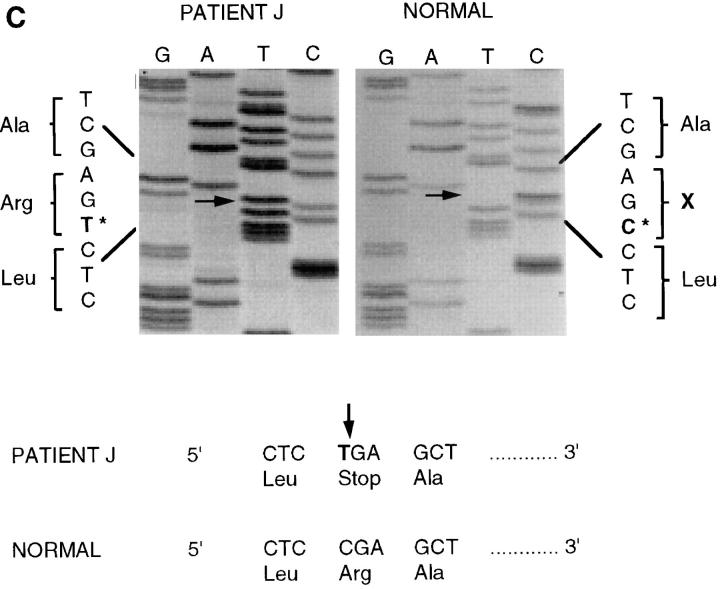
DNA sequencing of mutations in JEB patients. (A) Direct DNA sequencing of patient D's PCR amplified genomic DNA revealed a heterozygous C-to-T transition (arrow) at position 2621 of the cDNA resulting in a stop codon substituted for a glutamine residue designated as Q832X. Shown is the sense strand reading 5′ to 3′ from bottom to top. Both the cytosine normally present and the substituted thymine are visible at the location. An unrelated normal is analyzed in the right panel and shows only a cytosine at this position. (B) Direct DNA sequencing of patient V's PCR amplified genomic DNA showed a heterozygous G-to-A transition (arrow) at position 754 of the cDNA resulting in a splice site mutation at the exon 7, intron 7 junction. Shown is the sense strand reading 5′ to 3′ from bottom to top. Both the guanine normally present and the substituted adenine are visible at this location. An unrelated normal is analyzed in the right panel and shows only a guanine at this position. (C) Direct DNA sequencing of patient J's PCR amplified genomic DNA showed a homozygous C-to-T transition (arrow) at position 250 of the cDNA resulting in a nonsense mutation in exon 3, designated R42X. An unrelated normal is analyzed in the right panel and shows only a guanine at this position.
Direct DNA sequencing of a polymorphic PCR product using primers spanning exon 7 of patient V revealed a G-to-A transition at position 754 of the cDNA. A comparison of the normal sequence on the right with that of the proband on the left shows the presence of an A only in the proband (Fig. 2 B, arrow), as well as a lighter band in the G lane for the same position. Both the G and A bands are visible because both alleles of the β3 chain gene (normal and mutant) are equally represented in the PCR product. The transition of a G-to-A occurs within the consensus splice donor site AG/GT. Although, this is the first characterization of a mutation in this patient, interestingly the same location of mutations was reported previously for two other unrelated, nonlethal JEB patients (25, 39).
Patient J was analyzed by direct DNA sequencing of a polymorphic PCR product using primers spanning exon 3, and revealed a C-to-T transition at nucleotide position 250 of the cDNA. The normal sequence on the right shows the presence of a C on both alleles at this position, in agreement with the published sequence for the β3 chain (Fig. 2 C, arrow in left panel). The same position in patient J contains only a T. Because both alleles of the β3 chain gene are equally represented in the PCR product, patient J is homozygous for a T at position 250. This point mutation results in the change of an arginine (CGA) to a PTC (TGA), represented as R42X. Although this is the first characterization of this particular patient, the location of this mutation was previously identified in other patients as a mutational hotspot (30). The repeat findings of mutations in additional unrelated patients will eventually facilitate screening procedures, since mutational hotspots can be probed for directly. Considering our current emphasis on understanding the downstream effects of underlying mutations by characterizing mRNA levels and determining effects on laminin-5 chain assembly, the more common mutations are preferred examples since they are representative of more patients.
Northern Analysis of mRNA Levels of All Three Laminin-5 Chains in JEB Patients.
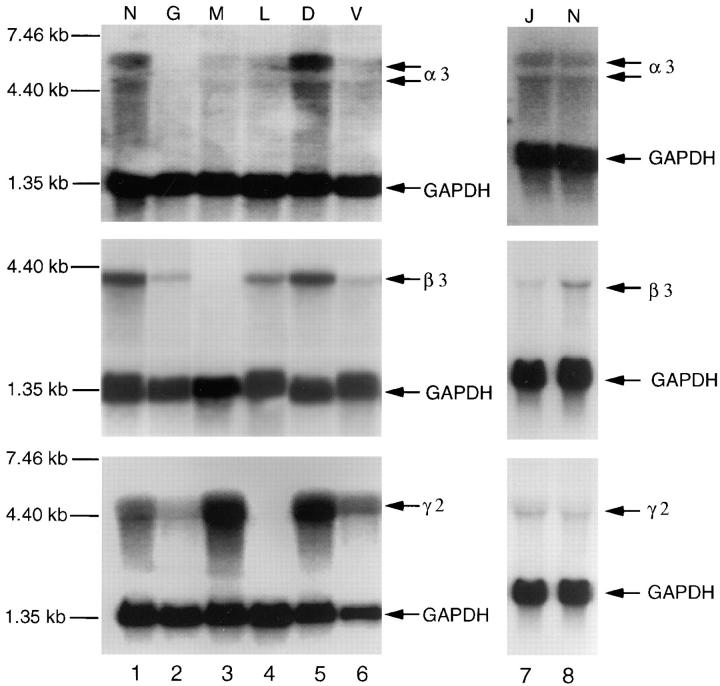
Total RNA was isolated from keratinocyte cultures from either normal or JEB patients. Transcripts of the expected size were observed for each of the laminin-5 chains using normal keratinocyte RNA (Fig. 3, lanes N on both sides). Patient G, who does not produce α3 chain protein, gave no detectable signal for α3 mRNA. However, mRNA was detectable for both the β3 and γ2 chains, although at somewhat reduced levels compared with normal (lane G). Patient M, who we previously characterized as containing mutations in the β3 chain did not produce detectable mRNA for the β3 chain, whereas α3 mRNA was produced at a reduced level, and γ2 was produced at normal, or perhaps greater levels (lane M). The other previously characterized JEB patient, patient L, had no detectable mRNA for the γ2 chain, but did produce mRNA for the α3 and β3 chains. Three of the four JEB patients containing β3 chain mutations produced detectable levels of all three chains. Patient D produced normal levels of all three chains, whereas patient V had reduced levels of β3 chain mRNA, and somewhat reduced levels of α3 chain mRNA as well. Patient J, with homozygous PTC mutations in the β3 chain, had detectable, but somewhat reduced, levels of the β3 transcript. The levels for the mRNAs of the α3 and γ2 chains were normal. The presence of mRNA for the β3 chain is noteworthy because other patients with PTC mutations often showed no detectable mRNA for the chain containing the mutation (compare to patients M and L).
Figure 3.
H-JEB patients' mRNA levels for the mutant chains are generally lower than in nonlethal JEB. Northern analysis of total RNA isolated from keratinocytes from either an unrelated normal control (N, lanes 1 and 8), lethal H-JEB (patients G, M, and L, lanes 2–4, and J, lane 7), or nonlethal JEB (patients D, V, lanes 5 and 6) hybridized with probes specific for laminin-5 chains, α3 (top panel), β3 (middle panel), γ2 (bottom panel), and for GAPDH as a control for approximately equal loadings of RNA. Film exposure for the α3 chain on patients V,G, and L was fivefold longer than for the other lanes because of a lower α3 signal in these patients. Position of RNA size markers shown on the right. Northern for patient J was done separately, lane 7, and is compared with a normal control, lane 8.
Laminin-5 Subunit Assembly in Patient Keratinocytes.
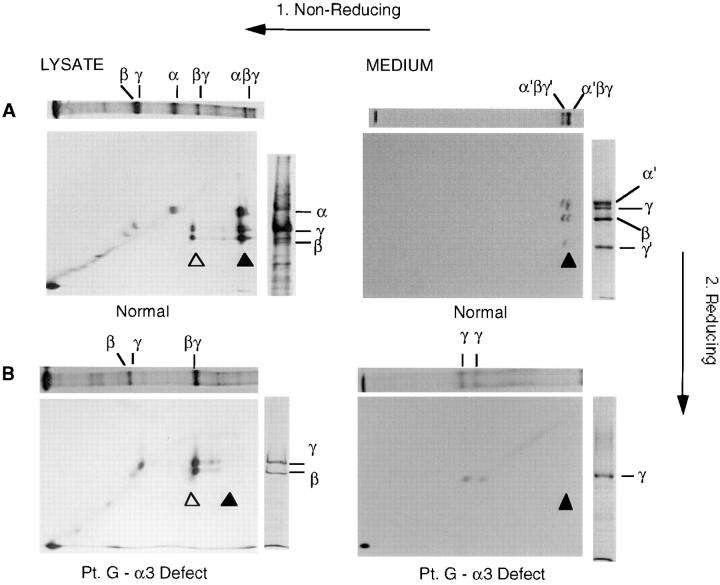
Laminin-5 immunoprecipitates from normal and patient keratinocytes were examined by two-dimensional (2-D) SDS-PAGE. In the normal cell lysate the 460-kD complex (Fig. 4 A, top panel, αβγ) observed under nonreducing conditions dissociated under reducing conditions into an equimolar mixture of α3, β3, and γ2 subunits (larger panel, three spots directly below band, solid triangle). The second specific band (σ) from the origin dissociates under nonreducing conditions into two protein bands of 145 and 155-kD, indicative of the disulfide-linked β3γ2 heterodimer we identified previously (Fig. 4 A, larger panel, two spots directly below band, open triangle; reference 29). Monomers of the three subunits, run under reducing conditions, are shown as markers in the right side panels (Fig. 4, A–E) and appear as spots along a diagonal (larger panels).
Figure 4.
Assembly of laminin-5 chains in JEB patient keratinocytes. Cells were radiolabeled for either 2 h (lysates-derived from whole cells) or 24 h (medium-secreted proteins in tissue culture) and immunoprecipitated with anti–laminin-5 antiserum. The samples were subjected to 2-D SDS-PAGE as described in Materials and Methods. Electrophoresis in the first dimension was performed under nonreducing conditions, and in the second dimension under reducing conditions. Duplicate nonreducing (top panels) and reducing (side panels) gels are shown along each dimension to allow alignment of the component chains seen in the 2-D gel. Open triangle points to position of β3γ2 heterodimers, when present. Solid triangle points to position of α3β3γ2 heterotrimers. Gels show the result when each of the three laminin-5 chains is missing due to mutation in a particular JEB patient. (A) Normal keratinocyte lysate (left panel) and tissue culture medium (right panel), all chains present. (B) Patient G keratinocyte lysate (left panel) and tissue culture medium (right panel), α3 chain missing. (C) Patient D keratinocyte lysate (left panel) and tissue culture medium (right panel), truncated β3 chain. (D) Patient V keratinocyte lysate (left panel) and tissue culture medium (right panel), β3 chain deficiency. (E) Patient M keratinocyte lysate (left panel) and tissue culture medium (right panel), β3 chain missing. (F) Patient L keratinocyte lysate (left panel) and tissue culture medium (right panel), γ2 chain missing. (α) unprocessed α3 subunit; (β) β3 subunit, (βt) truncated β3 subunit; (γ) unprocessed γ2 subunit; (α′ ) processed α3 subunit; (γ′ ) processed γ2 subunit; (FN) fibronectin.
In the culture medium (Fig. 4 A, right-hand panels) the two forms of extracellularly processed laminin-5, 440-kD (α′βγ) and 400-kD (α′βγ′) were observed under nonreducing conditions. Under reducing conditions the 440-kD form (right-most band in top panel) dissociated into a mixture of processed 165-kD α3 and 140-kD β3, and unprocessed 155-kD γ2 subunits. The 400-kD laminin-5 (the second band from the origin under nonreducing conditions) also dissociated into processed α3 and β3, but displayed a complete substitution of the processed 105-kD γ2 chain for unprocessed 155-kD γ2 (larger panel, two sets of three spots directly below each of two bands, solid triangle). To simplify discussions only the main larger panels containing the 2-D gels for each figure are referred to in the following descriptions, and not the side panels containing 1-D gels.
In the cell lysate from patient G (Fig. 4 B), the largest complex observed under nonreducing conditions is heterodimeric β-γ (Fig. 4 B, bottom left panel two spots above open triangle). Monomers of β3 and γ2 subunits were also present, but because the α3 subunit was completely absent, no heterotrimer assembly involving this subunit was found (no spots visible above solid triangle). Likewise, in the medium from patient G (Fig. 4 B, bottom right panel) the absence of assembly resulted in the near absence of secreted forms (no spots visible above solid triangle). Only γ2 chain monomers were observed; other subunits were not detectable even after longer exposure of the gel (data not shown).
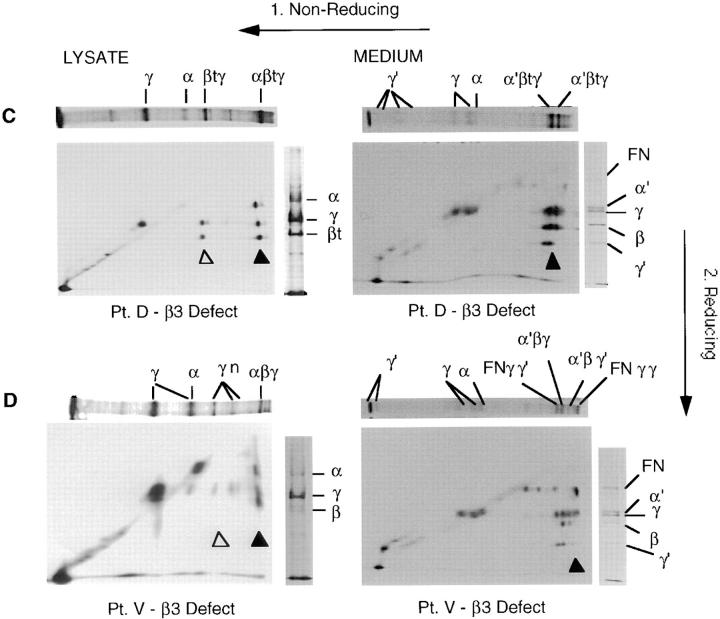
The following three JEB patients showed defects in the β3 chain but display unique characteristics (Fig. 4, C–E). The assembly pattern of laminin-5 is essentially normal for patient D, except that a truncated form of β3 (βt estimated 120-kD molecular mass) substitutes for the normal β3 chain. Thus, in the cell lysate from patient D (Fig. 4 C), all three chains of the heterotrimer assemble (top left panel, three spots above solid triangle). The heterodimer also forms normally (two spots above open triangle). These data show that the truncated β3 protein from patient D is capable of heterodimer and heterotrimer formation. The medium from patient D (Fig. 4 C, top right panel) again shows the normal pattern of assembled heterotrimer (three spots above solid triangle), but incorporates the truncated β3 chain. Interestingly, monomeric α3 and γ2 chains were also present and migrated to a central position along the diagonal in the 2-D gels, with degradation products of these monomers seen along the diagonal on the lower left. The larger molecular mass band of fibronectin is also seen, although none of the detectable forms appear to be associated with the laminin-5 complexes.
A somewhat greater deviation from the normal pattern was observed in the cell lysate from patient V (Fig. 4 D). Although the heterotrimer was detectable, the amount was more than fivefold less than in normal cells (the autoradiograph was exposed five times longer than for the normal keratinocyte sample). No heterodimer formation was visible under reducing conditions (no doublet spot seen above open triangle). Instead, multiple high molecular mass bands (γn) were present which lined up entirely with the 155-kD γ2 chain upon reduction. Disulfide-linked multimers (spots directly below γn bands) of the γ2 subunit are present. We previously reported multimer formation of recombinant γ2 subunits (29). Monomers of the α3 and γ2 subunits are present, whereas the β3 monomer is absent. The severely reduced level of β3 subunit appears to be the limiting subunit for heterotrimer assembly in this patient. Likewise, in the medium from patient V (Fig. 4 D, bottom right panel) less assembly resulted in lower levels of secreted heterotrimer (three spots visible above solid triangle). Monomeric α3 and γ2 chains were also present and migrated to a central position along the diagonal of this 2-D gel. The larger band of fibronectin is also seen, and appears to be associated with the γ2 chain.
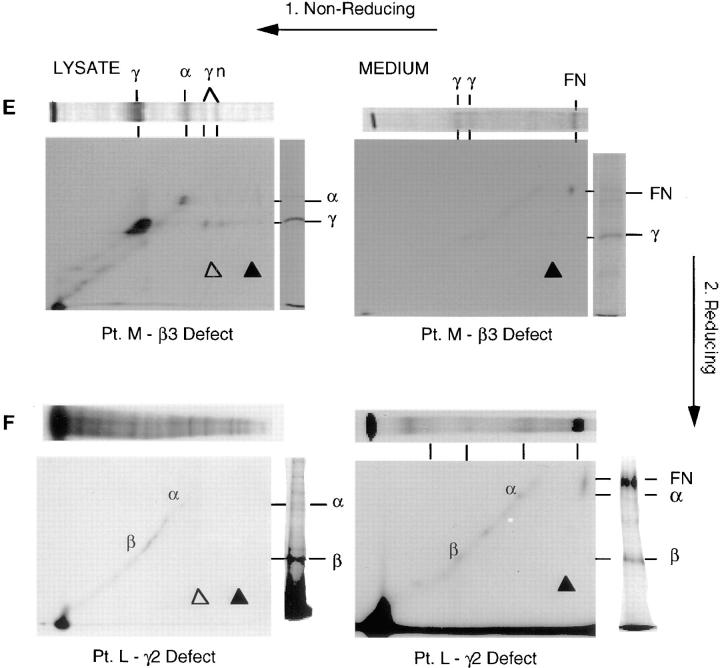
The cell lysate from patient M displays a complete lack of β3 chain expression and the consequent absence of larger complexes (Fig. 4 E, top left panel, no spots above solid triangle). Some multimerization of the γ2 chain is visible, whereas no βγ heterodimer forms (no spots above open triangle). In the medium from patient M (Fig. 4 E, top right panel) the lack of assembly resulted in the absence of secreted forms (no spots above solid triangle), although some γ2 chain is present. Likewise, patient J has a complete absence of β3 chain expression as analyzed on Western blots (data not shown), and we conclude patient J has an analogous pattern of expression to patient M.
A similar situation is observed in the cell lysate from patient L (Fig. 4 F). Again, there is an absence of larger complexes (no spots above open or solid triangles) resulting from the lack of γ2 chain. Only α3 and β3 monomers are present. Similarly, in the medium from patient L (Fig. 4 F, bottom right panel) the lack of assembly resulted in the absence of secreted forms (no spots above solid triangle).
Discussion
We present information on the synthesis of mutant gene products in junctional epidermolysis bullosa patients with laminin-5 defects, coupled with mutation site mapping data, to provide insight into ways that genetic lesions effect gene product function. We found that clinical severity corresponded to the extent of laminin-5 heterotrimer assembly. The hallmark of the most severe form of JEB, H-JEB, is the lack of one of the protein chains needed to assemble laminin-5, causing an absence of the heterotrimer, as shown here for patients G, M, J, and L. The lack of expression could often be traced to the near absence of RNA transcripts (Fig. 3) in the patients we analyzed, several of whom had PTCs. Others have shown that PTCs can cause instability of the RNA (31, 32, 40), and the importance of this mechanism in creating the downstream effects of many human genetic diseases needs to be more fully recognized. The H-JEB patients presented have functional null alleles for one of the laminin-5 chains. In contrast, we found that in the nonlethal generalized atrophic benign, or mitis varieties, expression of the mutant chains' mRNA (Fig. 3) and protein (Fig. 4, A–F) were reduced, but not absent, as shown here for patients D and V.
Laminin-5 is a primary component of anchoring filaments, which through their association with hemidesmosomes, are responsible for attachment of basal epithelial cells to the BMZ. Four of the six patients had H-JEB, and were chosen for presentation because each was defective in one of the three chains of the heterotrimer, the α3 chain (patient G), the β3 chain (patients M and J), and the γ2 chain (patient L). We analyzed these patients for synthesis and secretion of the individual chains of the laminin-5 heterotrimer by immunoprecipitation and resolution on 2-D gels. The same methodology was used in our earlier study on laminin-5 chain assembly conducted in SCC25 cells (29). This allowed us to determine both the amounts of the component chains expressed, and the extent of assembly of chain monomers into larger complexes. We concluded earlier that the assembly of laminin-5 proceeds from a β3γ2 heterodimer to the α3β3γ2 heterotrimer. These observations provided the basis for our interpretation of any deviations from normal laminin-5 assembly found in JEB patients. In normal keratinocytes (Fig. 4 A) both the β3γ2 heterodimer (spots above open triangle) and the α3β3γ2 heterotrimer (spots above solid triangle) are seen in the lysate (left panel), whereas in the medium (right panel) only the heterotrimer is secreted (spots above solid triangle). For patient G with H-JEB (Fig. 4 B) there was an absence of detectable α3 chain, but the β3γ2 heterodimer was still evident as an assembly intermediate (spots above open triangle). This observation is consistent with our earlier finding of β3γ2 heterodimer formation in the absence of the α3 chain (29). The heterodimer was not secreted into the culture medium, although some γ2 chain monomeric complexes were secreted in the absence of complete heterotrimer assembly. In patient D with nonlethal JEB (Fig. 4 C) the β3 chain was truncated (βt), but was still able to associate normally into heterodimers and heterotrimers (spots above open and solid triangles). This is the first description of a JEB patient with normal levels of mRNA for all three chains, and a nearly normal pattern of assembly of the individual chains into laminin-5. Patient V with nonlethal JEB (Fig. 4 D) produced limiting amounts of β3, so that no β3 was found as monomers or heterodimers but all was incorporated into heterotrimers (spots above solid triangle). In contrast, in patient M with H-JEB (Fig. 4 E) a frame shift PTC abrogated production of the β3 chain resulting in an absence of β3γ2 heterodimer (no heterodimer spot above open triangle) and a complete absence of heterotrimers (no spots above solid triangles). Another H-JEB patient, patient J, also had a complete absence of β3 protein (data not shown) due to the presence of homozygous PTCs positioned early in the transcripts. Similarly, H-JEB patient L (Fig. 4 F) shows a complete absence of the γ2 chain, resulting in no heterodimer or heterotrimer formation (no spots above either open or solid triangle). This illustrates again that H-JEB patients have a total absence of one of the chains of laminin-5, and therefore possess a functional null allele phenotype.
Complete assembly does not occur in the absence of any one of the chains of the heterotrimer, and therefore no significant amount of laminin-5 is secreted. We found that the γ2 chain could multimerize in the absence of one of the other chains, and was secreted to a limited extent. H-JEB keratinocytes provided natural ‘knockout' genes for laminin-5 chains and were thus valuable reagents for understanding assembly, helping to confirm our earlier findings of an ordered assembly from a β3γ2 heterodimer to the heterotrimer (29). Our results also explain why chain specific antibodies have not been particularly informative in discerning which chain contains the gene mutation (41), since defects in any of the three chains can limit assembly and secretion of the complete heterotrimer. In the patients shown here a null allele phenotype for any of the chains correlated with lethal H-JEB, whereas nonlethal JEB patients exhibited some expression of the mutant alleles.
The variable levels of functional laminin-5 chains seen in JEB patients is a reflection of both the type and location of mutation. By bringing together data identifying mutations and characterizing laminin-5 expression, we can improve our ability to predict how certain mutations will effect expression. The predictive power of mutational data for guessing clinical phenotypes will improve if we can discern the set of rules governing final expression levels from mutant alleles. An earlier effort to characterize expression levels in JEB patients was conducted before mutation data was readily available, so no correlations between expression levels and mutant alleles were drawn. That study also focused only on H-JEB patients, and did not include an analysis of less severely effected JEB patients (42). In addition to establishing the correlation between the extent of laminin-5 expression and the severity of JEB, we also show that the laminin-5 protein levels are reflected in the mRNA levels. Thus, the absence of laminin-5 protein can often be traced to the absence of mRNA transcript (Fig. 3). The preponderance of PTC mutations among H-JEB patients is noted in our data (Table I) and in published work that first identified the hotspot mutation R42X (30), that we also found to be present in our patient J. We show for the first time in H-JEB that the presence of PTCs typically cause a dramatic reduction in the mRNA levels from the mutant alleles. A smaller decrease in mRNA from a patient with the milder generalized atrophic benign form of JEB containing a PTC has been previously noted (25). Others have pointed out the high probability of PTCs being present in H-JEB patients (43). Our work is independent confirmation of this observation, and importantly, we add protein data and RNA analysis to show the functional significance of the PTCs. In other systems PTCs have been shown to destabilize mRNA transcripts (31, 32). Our data may also suggest a functional difference in the ability of different types of PTCs to trigger so-called ‘nonsense mediated mRNA decay'(38, 44). Patient M keratinocytes have a PTC that results from a frameshift located downstream of a deletion, and the mRNA levels for that transcript are dramatically reduced. In contrast, patient J with homozygous nonsense PTCs has less of a reduction in message levels, and therefore the effect of all PTCs may not be equivalent in terms of mRNA stability. Perhaps if the location of the PTC in patient J were further downstream a stable truncated product could be produced, since clearly the mRNA transcript is present. Patient D may illustrate this phenomenon, since no reduction in mRNA level is seen for the mutant transcript, despite the presence of a nonsense PTC on one allele in this patient. Patient L may be an exception, where nonsense PTCs resulted in undetectable mRNA levels of the mutant transcripts (but in this case the cells were obtained from a different laboratory and were immortalized using a different protocol). Also in agreement with our observations is previously published data on a JEB patient with a nonsense PTC where only a small reduction in the mutant transcript was noted. However, the significance of the slight reduction in mRNA levels as being characteristic of less severe disease had not been noted (25). Our data from patient J also points out that H-JEB may afflict patients with less significant reductions in mRNA levels if both alleles contain nonsense mutations early in the transcript that result in a total absence of translated protein. The current understanding among EB researchers is that all PTCs within laminin-5 have identical consequences. However, we believe that there may be possible differences in mRNA stability between nonsense PTCs and frameshift PTCs that could result in clinical differences. Experiments further addressing these observations are currently ongoing in our laboratory.
The molecular characterization of JEB patients is an important step toward future targeting of this disease for gene therapy. There are two possible methods of introduction of the therapeutic gene. The first is simply the addition of the therapeutic gene in the patient host cells that still contains the defective version. The second involves the removal of the defective copy of the gene and replacement with a normal copy through homologous recombination. The attempts at conducting gene therapy using the first method require a determination of the extent to which the endogenous defective gene products may interfere with the function of the therapeutic gene in the patient host cells. Many genetic diseases, including JEB, are recessive and therefore the assumption is that the addition of a normal gene copy will be dominant and restore function. Yet considerations, such as relative levels of expression of the therapeutic gene versus endogenous gene, may complicate the results. Furthermore, it remains possible that not all patient backgrounds will respond equivalently to gene therapy efforts. In some JEB patients the transcripts from the mutant alleles are unstable, as in patients M, L, and G, whereas in others they are more stable, as in patient J, and perhaps patients D and V. From our results, it is not clear whether the therapeutic gene transcript would be equally stable in these two categories of patient host cells. Further work is required to determine how various patient backgrounds respond to the addition of a therapeutic gene.
Table 1.
Clinical Phenotype and Location of Mutations in JEB Patients
| Patient | G | D | V | J | M | L | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diagnosis | H-JEB | JEB mitis | JEB mitis | H-JEB | H-JEB | H-JEB | ||||||
| Age & status | Dead at 9 wk | Dead at 44 yr | Alive at 7 yr | Dead at 8 wk | Dead at 1 yr | Dead at 8 wk | ||||||
| Defective chain | α3 | β3 | β3 | β3 | β3 | γ2 | ||||||
| Type of mutation | ND | PTC | Splice site | PTC | PTC | PTC | ||||||
| Location (cDNA) | ND | C2621T | G754A | C250T | 3112delC | C283T | ||||||
| A.A. location | ND | Q832X | E210K | R42X | V1027X | R95X | ||||||
| Protein domain | ND | Dom-I/II | Dom-VI | Dom-VI | Dom-I | Dom-V | ||||||
| Type of mutation | ND | ND | ND | PTC | PTC | |||||||
| Location (cDNA) | ND | ND | ND | C250T | G2257(+)T* | C283T | ||||||
| A.A. location | ND | ND | ND | R42X | (In intron) | R95X | ||||||
| Protein domain | ND | ND | ND | Dom-VI | Dom-II | Dom-V |
Comparison of clinical data from six JEB patients shows two distinct groups, one group with lethal H-JEB (patients G, M, J, and L, all died within the first year), and the other group with less severe JEB mitis (patient D died as an adult and patient V is alive at age 7). Defects were detected in each one of the three laminin-5 chains, α3, β3, or γ2. Genetic information for patients M1 and L (22) were from a previous analysis, and include both alleles. Identification of single allele mutations for patients D and V are presented in Fig. 2, A and B. Patient J contained a homozygous PTC and is presented in Fig. 2 C. PTC, premature termination codon, location in cDNA indicates nucleotide normally at the position, followed by the numbered nucleotide position (numbering from the first nucleotide of the transcript including a 128-bp leader preceding the AUG), and ended by the nucleotide substitution.
Indicates mutation located one base past nucleotide position 2257 as indicated by + in the first base of intronic splice consensus sequence. A.A., amino acid location indicates amino acid normally at the position, followed by the numbered A.A. position, and ended by the amino acid substituted, or stop codon, at that position.
Acknowledgments
We thank Dr. Guerrino Meneguzzi for the immortalized cell line LSV5. We also thank Dr. Schiller for the HPV-18 E6-E7 immortalizing plasmid 18CCB, Dr. David Woodley for keratinocytes from patient D that we immortalized, and Dr. Peter Marinkovich who provided anti-laminin-5 polyclonal antibody. This study would not have been possible without our access to JEB biopsies obtained through the National Epidermolysis Bullosa Registry. Undergraduate student Alonzo Sexton contributed to early versions of the Northerns for laminin-5.
This work was supported by NIAMS Grants AR41045-03 and AR19537.
Footnotes
1 Abbreviations used in this paper: 1-D, 1-dimensional; BMZ, basement membrane zone; EB, epidermolysis bullosa; JEB, junctional EB; PTC, premature termination codon; SFM, serum-free media.
References
- 1.Fine JD, Bauer EA, Briggaman RA, Carter DM, Eady RAJ, Esterly NB, Holbrook KA, Hurwitz S, Johnson L, Lin A, Pearson R, Sybert VP. Revised clinical and laboratory criteria for subtypes of inherited epidermolysis bullosa. J Am Acad Dermatol. 1991;24:119–135. doi: 10.1016/0190-9622(91)70021-s. [DOI] [PubMed] [Google Scholar]
- 2.Smith LT. Ultrastructural findings in epidermolysis bullosa. Arch Dermatol. 1993;129:1578–1584. [PubMed] [Google Scholar]
- 3.Fuchs E, Coulombe PA. Of mice and men: genetic skin diseases of keratin. Cell. 1992;69:899–902. doi: 10.1016/0092-8674(92)90607-e. [DOI] [PubMed] [Google Scholar]
- 4.Epstein EH., Jr Molecular genetics of epidermolysis bullosa. Science. 1992;256:799–804. doi: 10.1126/science.1375393. [DOI] [PubMed] [Google Scholar]
- 5.Sakai LY, Keene DR, Morris NP, Burgeson RE. Type VII collagen is a major structural component of anchoring fibrils. J Cell Biol. 1986;103:1577–1586. doi: 10.1083/jcb.103.4.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ryynanen M, Knowlton RG, Parente MG, Chung LC, Chu ML, Uitto J. Human type VII collagen: genetic linkage of the gene (COL7A1) on chromosome 3 to dominant dystrophic epidermolysis bullosa. Am J Hum Genet. 1991;49:797–803. [PMC free article] [PubMed] [Google Scholar]
- 7.Verrando P, Hsi BL, Yeh CJ, Pisani A, Serieys N, Ortonne JP. Monoclonal antibody GB3, a new probe for the study of human basement membranes and hemidesmosomes. Exp Cell Res. 1987;170:116–128. doi: 10.1016/0014-4827(87)90121-2. [DOI] [PubMed] [Google Scholar]
- 8.Schofield OM, Fine JD, Verrando P, Heagerty AH, Ortonne JP, Eady RA. GB3 monoclonal antibody for the diagnosis of junctional epidermolysis bullosa: results of a multicenter study. J Am Acad Dermatol. 1990;23:1078–1083. doi: 10.1016/0190-9622(90)70336-g. [DOI] [PubMed] [Google Scholar]
- 9.Hsi BL, Yeh CJ. Monoclonal antibodies to human amnion. J Reprod Immunol. 1986;9:11–21. doi: 10.1016/0165-0378(86)90021-5. [DOI] [PubMed] [Google Scholar]
- 10.Verrando P, Pisani A, Ortonne JP. The new basement membrane antigen recognized by the monoclonal antibody GB3 is a large size glycoprotein: modulation of its expression by retinoic acid. Biochim Biophys Acta. 1988;942:45–56. doi: 10.1016/0005-2736(88)90273-8. [DOI] [PubMed] [Google Scholar]
- 11.Rousselle P, Lunstrum GP, Keene DR, Burgeson RE. Kalinin: an epithelium-specific basement membrane adhesion molecule that is a component of anchoring filaments. J Cell Biol. 1991;114:567–576. doi: 10.1083/jcb.114.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marinkovich MP, Lunstrum GP, Burgeson RE. The anchoring filament protein kalinin is synthesized and secreted as a high molecular weight precursor. J Biol Chem. 1992;267:17900–17906. [PubMed] [Google Scholar]
- 13.Domloge-Hultsch N, Gammon WR, Briggaman RA, Gil SG, Carter WG, Yancey KB. Epiligrin, the major human keratinocyte integrin ligand, is a target in both an acquired autoimmune and an inherited subepidermal blistering skin disease. J Clin Invest. 1992;90:1628–1633. doi: 10.1172/JCI116033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burgeson RE, Chiquet M, Deutzmann R, Ekblom P, Engel J, Kleinman H, Martin G, Meneguzzi G, Paulsson M, Sanes J, et al. A new nomenclature for laminins. Matrix Biology. 1994;14:209–215. doi: 10.1016/0945-053x(94)90184-8. [DOI] [PubMed] [Google Scholar]
- 15.Engel J. Laminins and other strange proteins. Biochemistry. 1992;31:10643–10651. doi: 10.1021/bi00159a001. [DOI] [PubMed] [Google Scholar]
- 16.Ryan MC, Tizard R, VanDevanter DR, Carter WG. Cloning of the gene encoding the α3 chain of the adhesive ligand epiligrin. Expression in wound repair. J Biol Chem. 1994;269:22779–22787. [PubMed] [Google Scholar]
- 17.Gerecke DR, Wagman DW, Champliaud MF, Burgeson RE. The complete primary structure for a novel laminin chain, the laminin B1k chain. J Biol Chem. 1994;269:11073–11080. [PubMed] [Google Scholar]
- 18.Kallunki P, Sainio K, Eddy R, Byers M, Kallunki T, Sariola H, Beck K, Hirvonen H, Shows TS, Tryggvason KA. A truncated laminin chain homologous to the B2 chain: structure, spacial expression, and chromosomal assignment. J Cell Biol. 1992;119:679–694. doi: 10.1083/jcb.119.3.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vailly J, Verrando P, Champliaud M-F, Gerecke D, Wagman DW, Baudoin C, Aberdam D, Burgeson RE, Bauer EA, Ortonne J-P. The 100-kDa chain of nicein/kalinin is a laminin B2 chain variant. Eur J Biochem. 1994;219:209–218. doi: 10.1111/j.1432-1033.1994.tb19932.x. [DOI] [PubMed] [Google Scholar]
- 20.Matsui C, Nelson CF, Hernandez GT, Herron GS, Bauer EA, Hoeffler WK. Gamma 2 chain of laminin-5 is recognized by monoclonal antibody GB3. J Invest Dermatol. 1995;105:648–652. doi: 10.1111/1523-1747.ep12324108. [DOI] [PubMed] [Google Scholar]
- 21.Pulkkinen L, Christiano AM, Airenne T, Haakana H, Tryggvason K, Uitto J. Mutations in the γ2 chain gene (LAMC2) of kalinin/laminin 5 in the junctional forms of epidermolysis bullosa. Nat Genet. 1994;6:293–297. doi: 10.1038/ng0394-293. [DOI] [PubMed] [Google Scholar]
- 22.Aberdam D, Galliano MF, Vailly J, Pulkkinen L, Bonifas J, Christiano AM, Tryggvason K, Uitto J, Epstein EH, Jr, Ortonne JP, Meneguzzi G. Herlitz's junctional epidermolysis bullosa is linked to mutations in the gene (LAMC2) for the gamma 2 subunit of nicein/kalinin (LAMININ-5) Nat Genet. 1994;6:299–304. doi: 10.1038/ng0394-299. [DOI] [PubMed] [Google Scholar]
- 23.Vailly J, Pulkkinen L, Christiano AM, Tryggvason K, Uitto J, Ortonne JP, Meneguzzi G. Identification of a homozygous exon-skipping mutation in the LAMC2 gene in a patient with Herlitz's junctional epidermolysis bullosa. J Invest Dermatol. 1995;104:434–437. doi: 10.1111/1523-1747.ep12666027. [DOI] [PubMed] [Google Scholar]
- 24.Baudoin C, Miquel C, Gagnoux-Palacios L, Pulkkinen L, Christiano AM, Uitto J, Tadini G, Ortonne JP, Meneguzzi G. A novel homozygous nonsense mutation in the LAMC2 gene in patients with the Herlitz junctional epidermolysis bullosa. Hum Mol Genet. 1994;3:1909–1910. doi: 10.1093/hmg/3.10.1909. [DOI] [PubMed] [Google Scholar]
- 25.McGrath JA, Pulkkinen L, Christiano AM, Leigh IM, Eady RA, Uitto J. Altered laminin 5 expression due to mutations in the gene encoding the beta 3 chain (LAMB3) in generalized atrophic benign epidermolysis bullosa. J Invest Dermatol. 1995;104:467–474. doi: 10.1111/1523-1747.ep12605904. [DOI] [PubMed] [Google Scholar]
- 26.Pulkkinen L, Christiano AM, Gerecke D, Wagman DW, Burgeson RE, Pittelkow MR, Uitto J. A homozygous nonsense mutation in the beta 3 chain gene of laminin 5 (LAMB3) in Herlitz junctional epidermolysis bullosa. Genomics. 1994;24:357–360. doi: 10.1006/geno.1994.1627. [DOI] [PubMed] [Google Scholar]
- 27.Vailly J, Pulkkinen L, Miquel C, Christiano AM, Gerecke D, Burgeson RE, Uitto J, Ortonne JP, Meneguzzi G. Identification of a homozygous one-basepair deletion in exon 14 of the LAMB3 gene in a patient with Herlitz junctional epidermolysis bullosa and prenatal diagnosis in a family at risk for recurrence. J Invest Dermatol. 1995;104:462–466. doi: 10.1111/1523-1747.ep12605898. [DOI] [PubMed] [Google Scholar]
- 28.Kivirikko S, McGrath JA, Baudoin C, Aberdam D, Ciatti S, Dunnill MG, McMillan JR, Eady RA, Ortonne JP, Meneguzzi G, Uitto J. A homozygous nonsense mutation in the alpha 3 chain gene of laminin 5 (LAMA3) in lethal (Herlitz) junctional epidermolysis bullosa. Hum Mol Genet. 1995;4:959–962. doi: 10.1093/hmg/4.5.959. [DOI] [PubMed] [Google Scholar]
- 29.Matsui C, Wang CK, Nelson CF, Bauer EA, Hoeffler WK. The assembly of laminin 5 subunits. J Biol Chem. 1995;270:23496–23503. doi: 10.1074/jbc.270.40.23496. [DOI] [PubMed] [Google Scholar]
- 30.Kivirikko S, McGrath JA, Pulkkinen L, Uitto J, Christiano AM. Mutational hotspots in the LAMB3 gene in lethal (Herlitz) type of junctional epidermolysis bullosa. Hum Mol Genet. 1996;5:231–237. doi: 10.1093/hmg/5.2.231. [DOI] [PubMed] [Google Scholar]
- 31.Urlaub G, Mitchell PJ, Ciudad CJ, Chasin LA. Nonsense mutations in the dihydrofolate reductase gene affect RNA processing. Mol Cell Biol. 1989;9:2868–2880. doi: 10.1128/mcb.9.7.2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheng J, Fogel-Petrovic M, Maquat LE. Translation to near the distal end of the penultimate exon is required for normal levels of spliced triosephosphate isomerase mRNA. Mol Cell Biol. 1990;10:5215–5225. doi: 10.1128/mcb.10.10.5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miquel C, Gagnoux-Palacios L, Durand-Clement M, Marinkovich MP, Ortonne JP, Meneguzzi G. Establishment and characterization of cell line LSV5 that retains the altered adhesive properties of human junctional epidermolysis bullosa keratinocytes. Exp Cell Res. 1996;224:279–290. doi: 10.1006/excr.1996.0138. [DOI] [PubMed] [Google Scholar]
- 34.Barbosa MS, Schlegel R. The E6 and E7 genes of HPV-18 are sufficient for inducing two-stage in vitro transformation of human keratinocytes. Oncogene. 1989;4:1529–1532. [PubMed] [Google Scholar]
- 35.Villa LL, Schlegel R. Differences in transformation activity between HPV-18 and HPV-16 map to the viral LCR-E6-E7 region. Virology. 1991;181:374–377. doi: 10.1016/0042-6822(91)90507-8. [DOI] [PubMed] [Google Scholar]
- 36.Peters BP, Hartle RJ, Krzesicki RF, Kroll TG, Perini F, Balun JE, Goldstein IJ, Ruddon RW. The biosynthesis, processing, and secretion of laminin by human choriocarcinoma cells. J Biol Chem. 1985;260:14732–14742. [PubMed] [Google Scholar]
- 37.Vidal F, Baudoin C, Miquel C, Galliano MF, Christiano AM, Uitto J, Ortonne JP, Meneguzzi G. Cloning of the laminin α3 chain (LAMA3) and identification of a homozygous deletion in a patient with Herlitz junctional epidermolysis bullosa. Genomics. 1995;30:273–280. doi: 10.1006/geno.1995.9877. [DOI] [PubMed] [Google Scholar]
- 38.Hunter I, Schulthess T, Engel J. Laminin chain assembly by triple and double stranded coiled-coil structures. J Biol Chem. 1992;267:6006–6011. [PubMed] [Google Scholar]
- 39.McGrath JA, Christiano AM, Pulkkinen L, Eady RA, Uitto J. Compound heterozygosity for nonsense and missense mutations in the LAMB3 gene in nonlethal junctional epidermolysis bullosa. J Invest Dermatol. 1996;106:1157–1159. doi: 10.1111/1523-1747.ep12340210. [DOI] [PubMed] [Google Scholar]
- 40.McIntosh I, Hamosh A, Dietz HC. Nonsense mutations and diminished mRNA levels. Nat Genet. 1993;4:219. doi: 10.1038/ng0793-219. [DOI] [PubMed] [Google Scholar]
- 41.McMillan JR, McGrath JA, Pulkkinen L, Kon A, Burgeson RE, Ortonne J-P, Meneguzzi G, Ortonne JP, Eady RAJ. Immunohistochemical analysis of the skin in junctional epidermolysis bullosa using laminin-5 chain specific antibodies is of limited value in predicting the underlying gene mutation. Br J Dermatol. 1997;136:817–822. [PubMed] [Google Scholar]
- 42.Baudoin C, Miquel C, Blanchet-Bardon C, Gambini C, Meneguzzi G, Ortonne JP. Herlitz junctional epidermolysis bullosa keratinocytes display heterogeneous defects of nicein/kalinin gene expression. J Clin Invest. 1994;93:862–869. doi: 10.1172/JCI117041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pulkkinen L, McGrath JA, Christiano AM, Uitto J. Detection of sequence variants in the gene encoding the β3 chain of laminin-5 (LAMB3) Hum Mutat. 1995;6:77–84. doi: 10.1002/humu.1380060115. [DOI] [PubMed] [Google Scholar]
- 44.Peltz SW, Jacobson A. mRNA stability: in trans-it. Curr Opin Cell Biol. 1992;4:979–983. doi: 10.1016/0955-0674(92)90129-z. [DOI] [PubMed] [Google Scholar]