Abstract
MicroRNAs are small noncoding RNAs that regulate many cellular processes in a post-transcriptional mode. MicroRNA knockdown by antisense oligonucleotides is a useful strategy to explore microRNA functionality and as potential therapeutics. MicroRNA-122 (miR-122) is a liver-specific microRNA, the main function of which has been linked with lipid metabolism and liver homeostasis. Here, we show that lipofection of an antisense oligonucleotide based on a Locked Nucleic Acids (LNA)/2′-O-methyl mixmer or electroporation of a Peptide Nucleic Acid (PNA) oligomer is effective at blocking miR-122 activity in human and rat liver cells. These oligonucleotide analogs, evaluated for the first time in microRNA inhibition, are more effective than standard 2′-O-methyl oligonucleotides in binding and inhibiting microRNA action. We also show that microRNA inhibition can be achieved without the need for transfection or electroporation of the human or rat cell lines, by conjugation of an antisense PNA to the cell-penetrating peptide R6-Penetratin, or merely by linkage to just four Lys residues, highlighting the potential of PNA for future therapeutic applications as well as for studying microRNA function.
Keywords: microRNA, miR-122, PNA, antisense, peptide, targeting
INTRODUCTION
MicroRNAs are a novel class of small noncoding RNAs that are implicated in post-transcriptional gene expression regulation. Initially found to regulate diverse developmental events in Caenorhabditis elegans (Lee et al. 1993), they are now known to be involved in many other important processes, including cell metabolism (Xu et al. 2003; Esau et al. 2006), inflammation (Moschos et al. 2007; O'Connell et al. 2007), pathogen infection (Jopling et al. 2006; Triboulet et al. 2007), and cancer (Cimmino et al. 2005; He et al. 2005; O'Donnell et al. 2005; Garzon et al. 2006; O'Connell et al. 2007). It is thus not surprising that the development of sequence-specific microRNA antagonists is so appealing (Weiler et al. 2006; Ebert et al. 2007). The importance of such reagents is twofold. First, use of microRNA inhibitors is a rapid and inexpensive way to assign and characterize microRNA function, in contrast to the time-consuming and more difficult strategy of creating gene knockouts. Secondly, microRNA targeting represents a novel and still undeveloped approach toward potential therapeutic applications.
Oligonucleotide (ON) analogs inhibit microRNA function essentially by a steric block, RNase H-independent and RISC-independent, antisense mechanism through complementary binding of the ON to the microRNA sequence. The cellular outcome of such binding is still unclear, with reports arguing either in favor of a mechanism based on simple sequestration by stoichiometric complex formation between the mature microRNA and the ON inhibitor (Chan et al. 2005), or in favor of a yet unknown mechanism by which complex formation leads to degradation of the target microRNA (Krutzfeldt et al. 2005, 2007; Esau et al. 2006). A number of ON analog types have been proposed that provide both metabolic stability as well as good RNA binding, two fundamental requirements for microRNA inhibition. Early literature reports showed that well established 2′-O-methyl (OMe) ONs were in principle capable of sequence-specific microRNA inhibition (Meister et al. 2004), and these have become the standard choice for studies involving microRNA function (Chang et al. 2004; Cheng et al. 2005; Jopling et al. 2005; O'Donnell et al. 2005; Bhattacharyya et al. 2006a; Saetrom et al. 2007). Although somewhat resistant against nuclease degradation, such OMe inhibitors show only moderately strong RNA binding power, especially when combined with phosphorothioate (PS) linkages. More recently, other types of materials with improved properties have been described, for example, 2′-O-methoxyethyl (MOE), 2′-fluoro, their mixmer combinations with DNA, and those with PS backbones (Esau et al. 2004, 2006; Jopling et al. 2005; Davis et al. 2006).
ONs based on locked nucleic acids (LNAs) have been shown to have extremely strong RNA binding ability (Koshkin et al. 1998), which has led recently to the application to microRNA inhibition of LNA/DNA mixmers, which have higher sequence specificity than all LNA ONs (Naguibneva et al. 2006; Ørom et al. 2006), and such reagents are now available commercially. In order to facilitate tissue and cell uptake and produce long-lasting effects in vivo, Krutzfeldt and colleagues developed “antagomirs,” a particular type of microRNA ON inhibitor containing a cholesterol moiety conjugated to the 3′-end of an OMe oligonucleotide that contains partial PS linkage modifications at the termini (Krutzfeldt et al. 2005, 2007). Although the cholesterol-modified ON was far superior to the unconjugated construct, it does not seem to be strictly necessary for in vivo use, since Esau and colleagues showed in a similar model that unconjugated MOE PS ONs were also very effective at microRNA target knockdown in mice (Esau et al. 2006).
In earlier work, we showed that steric block antisense ONs composed of LNA/OMe mixmers directed to the trans-activation-responsive RNA TAR were very effective at blocking HIV-1 Tat-dependent trans-activation in a HeLa cell-based luciferase reporter model system when delivered by lipofection, and also showed antiviral activity (Arzumanov et al. 2001; Turner et al. 2005a; Brown et al. 2006). These ON mixmers were designed to combine the very high binding affinity and biostability provided by LNA units together with the greater nuclease resistance and RNA binding strength of OMe nucleotides as compared with DNA.
Synthetic ONs with electrically neutral backbones have also shown great promise as steric block antisense agents. For example, phosphorodiamidate morpholino oligonucleotides (PMOs) or their conjugates with a cell-penetrating peptide (CPP) have been applied very effectively for inhibition of RNA function by blocking mRNA translation (Ahn et al. 2002), redirection of splicing (Suwanmanee et al. 2002), and more recently as microRNA antagonists (Kloosterman et al. 2007), and they are being taken toward clinical applications such as Duchenne Muscular Dystrophy (Fletcher et al. 2007). Peptide nucleic acids (PNAs) are ON analogs where the negatively charged deoxyribose phosphate backbone is replaced by an electroneutral peptide-like backbone consisting of N-(2-aminoethyl)glycine units directly linked to the DNA bases (Nielsen et al. 1994). We and others have shown that PNAs, especially as conjugates of CPPs, have high efficiency in many RNA targeting applications, for example in inhibition of bacterial mRNA translation (Xue-Wen et al. 2007), inhibition of HIV-1 Tat-dependent trans-activation (Turner et al. 2005b) and RNA reverse transcription (Koppelhus et al. 1997), and as splicing redirection agents (Turner et al. 2005b; Abes et al. 2007).
miR-122 is a liver-specific microRNA, initially discovered in cloning studies of small RNAs of mouse origin (Lagos-Quintana et al. 2002). This microRNA is of particular therapeutic interest, since it has been shown not only to facilitate Hepatitis C RNA replication (Jopling et al. 2005), but also to be up-regulated in HIV-1 infected cells (Triboulet et al. 2007). Here, we show that an LNA/OMe mixmer delivered by lipofection and a PNA delivered by electroporation are efficient microRNA blocking agents in human and rat liver cells, with superior activity compared with standard OMe oligonucleotides. miR-122 inhibition was evaluated by Northern blot and by the up-regulation effect upon various negatively regulated messenger RNAs (mRNAs). We also show that substantial microRNA blocking activity is obtained in the absence of transfection agents or electroporation for a disulfide conjugate of anti-miR-122 PNA with the CPP R6-Penetratin or for PNA containing just four additional Lys residues.
RESULTS
Genes regulated by miR-122
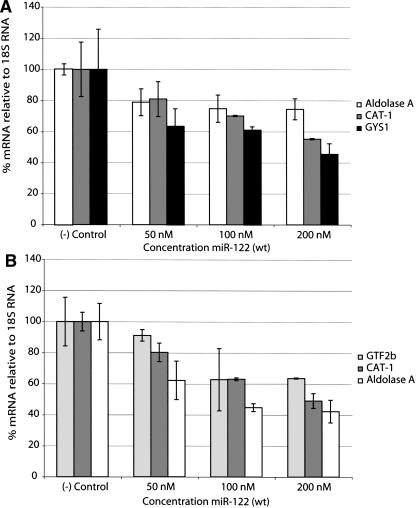
In order to evaluate the efficiency of microRNA blocking agents, we first set out to validate possible mRNA targets that are subject to miR-122 negative regulation in liver cells. Thus, we decided to transfect human hepatocellular carcinoma cells, Huh7, and primary rat hepatocytes with the corresponding synthetic double-stranded miR-122 mimic. A previous study showed the direct reduction in mRNA levels upon binding of miR-122 to the mRNAs of GYS1, SLC7A1 (CAT-1), Aldolase A, CCNG1, and P4HA1 in mouse AML12 cells (Esau et al. 2006). Here, we transfected the human or rat cells with the miR-122 mimic by lipofection at different concentrations, followed by 24 h of incubation, lysis, and Trizol RNA extraction. A number of predicted (TargetScan) (Griffith-Jones et al. 2006) or mouse pre-validated mRNA targets were quantified for each species by real-time RT-PCR. Figure 1A shows that enhancement of endogenous miR-122 activity by external addition of the synthetic microRNA mimic leads to mRNA knockdown in a sequence-specific and concentration-dependent manner in human Huh7 cells. Our results show that all three predicted regulated genes in humans (and pre-validated in mouse [Esau et al. 2006]), Aldolase A, CAT-1, and GYS1, are targets for miR-122 negative regulation.
FIGURE 1.
Transfection of a synthetic double-stranded miR-122 mimic leads to mRNA destabilization in both Huh7 (A) and primary rat hepatocytes (B). ([–] Control) Nontransfected cells.
Similar experiments in primary rat hepatocytes showed that the rat miR-122 mimic regulates the corresponding rat mRNAs of CAT-1 and Aldolase A, also by direct reduction in mRNA levels (Fig. 1B). A novel predicted target for miR-122 regulation, GTF2b (transcription initiation factor IIB), was also found to be down-regulated in miR-122-supplemented rat hepatocytes (Fig. 1B). These results validate the use of these target mRNAs, respectively, within human Huh7 cells and primary rat hepatocytes for evaluation of miR-122 inhibitors.
miR-122 inhibition by complementary LNA/OMe mixmer ONs
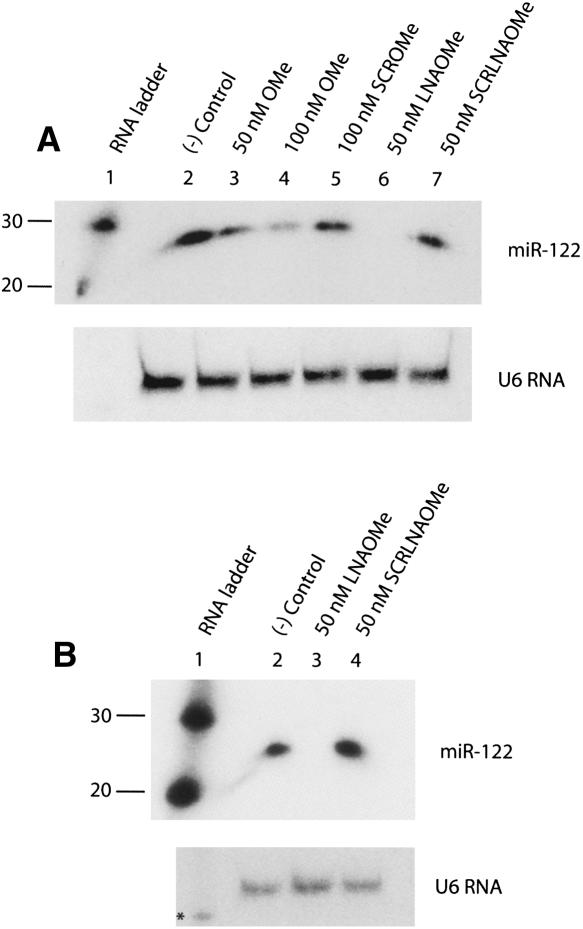
In order to evaluate the potential as inhibitors of microRNA action of LNA/OMe mixmers in comparison with standard OMe oligonucleotides, we transfected Huh7 cells with either miR-122-complementary or scrambled (scr) 23-mer LNA/OMe ONs. Northern blot analysis of total RNA from transfected cells is shown in Figure 2. Samples treated with 50 nM or 100 nM standard 31-mer OMe ON showed dose-dependent reductions in levels of endogenous miR-122 compared with a nontransfected control, but the miR-122 decrease was significant only at 100 nM (Fig. 2A, lanes 2–4). By contrast, Huh7 transfection with 50 nM LNA/OMe ON reduced the miR-122 RNA signal to undetectable levels (Fig. 2A, lane 6). Scrambled OMe and LNA/OMe ON controls did not affect miR-122 levels at the concentrations tested (Fig. 2A, lanes 5,7). Similar results were also seen with transfection of 50 nM LNA/OMe ON into primary rat hepatocytes, where the treatment also led to a complete loss of miR-122 signal (Fig. 2B, lane 3), but not for scrambled control (Fig. 2B, lane 4). By contrast, neither OMe nor LNA/OMe ONs were able to affect pre-miR-122 levels in Huh7 cells or primary rat hepatocytes (data not shown).
FIGURE 2.
Northern blot of total RNA from Huh7 cells (A), or primary rat hepatocytes (B), transfected by lipofection with miR-122 inhibitors and controls. U6 RNA was used as loading control. ([–] Control) Nontransfected cells, (*) RNA ladder band corresponding to 100 nucleotides.
Further analysis of Northern blot membranes with a radiolabeled miR-122 probe for antisense inhibitor detection suggested that after Trizol purification only LNA/OMe ONs, but not their OMe counterparts, were still present in the RNA samples analyzed (Supplemental Fig. S1A). This observation suggests that the superior stability of LNA/OMe ONs toward cellular degradation and their higher RNA binding strength compared with OMe ONs confers a greater ability to maintain microRNA target binding over a longer time period. To eliminate the possibility of the OMe ON being removed during the RNA purification step, a control in vitro experiment was carried out, showing that although the OMe ON can potentially be lost during RNA extraction (Supplemental Fig. S1B, lanes 2,3, top panel), it is retained if both miR-122 and carrier RNA are present, conditions similar to those expected in extracts from cells (Supplemental Fig. S1B, lanes 8,9, top panel).
miR-122 inhibition by antisense oligonucleotides leads to mRNA accumulation of miR-122-regulated genes
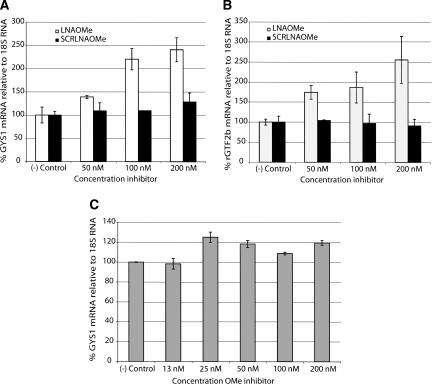
It is clear from Figure 1 that endogenous miR-122 negatively regulates the mRNAs corresponding to CAT-1, GYS1, and Aldolase A in human Huh7 cells, and those of GTF2b, Aldolase A, and CAT-1 in primary rat hepatocytes. Therefore, if inhibition of miR-122 is achieved by complementary ONs, this should lead to an intracellular accumulation of the negatively regulated mRNAs. To confirm this hypothesis, we transfected Huh7 cells and primary rat hepatocytes with LNA/OMe mixmers at different concentrations and measured in each case the levels of one of the down-regulated mRNAs (GYS1 and GTF2b, respectively). The results (Fig. 3) show that in both cell lines there is a dose-dependent increase in the intracellular concentration of the regulated mRNA obtained as the amount of transfected LNA/OMe ON is increased, but not for scrambled LNA/OMe ON (Fig. 3A,B). Further, we found that lipofection of the standard OMe ON in Huh7 cells did not produce a significant increase in GYS1 mRNA levels, even at higher doses (Fig. 3C), which correlates with the poor binding observed to miR-122, seen by Northern blotting (Fig. 2A).
FIGURE 3.
miR-122 inhibition by antisense oligonucleotides leads to intracellular accumulation of negatively regulated mRNAs. (A) GYS1 mRNA levels in Huh7 cells. (B) GTF2b mRNA levels in primary rat hepatocytes. (C) GYS1 mRNA levels in Huh7 cells transfected by lipofection with increasing amounts of OMe ON. ([–] Control) Nontransfected cells.
Peptide nucleic acid (PNA) analogs and their peptide conjugates for microRNA inhibition
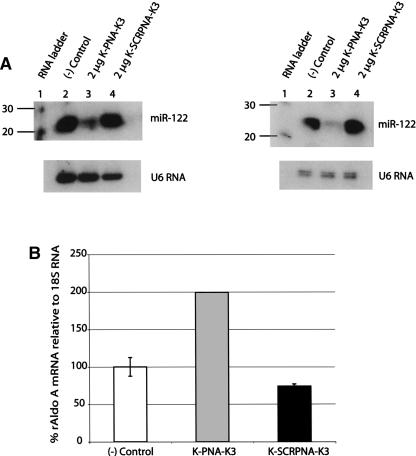
In order to evaluate the potential of PNAs for microRNA inhibition in cells, we synthesized the corresponding PNA 23-mer antisense to the mature miR-122. These PNAs contained one additional Lys residue at the N terminus and three Lys residues at the C terminus to aid solubility, as has been our common practice (Turner et al. 2005b; Abes et al. 2007). Electroporation of the K-PNA-K3 ON into Huh7 or primary rat hepatocytes led to a sequence-specific reduction in intensities of the miR-122 band in Northern blots of total RNA, but not for a scrambled K-PNA-K3 (Fig. 4A,B). Since our Northern blot analysis suggested that electroporated K-PNA-K3 may not be present in the RNA fraction isolated from the cells (Supplemental Fig. S2), we quantified the levels of mature miR-122 in the RNA fractions by real-time RT-PCR. These data (Supplemental Fig. S3) support our above finding, that RNA extracts from cells treated with K-PNA-K3 become highly depleted in mature miR-122. As shown before for LNA/OMe mixmer oligonucleotides, the K-PNA-K3 ON binds to miR-122 and induces an increase in the intracellular levels of a regulated mRNA, Aldolase A (Fig. 4C).
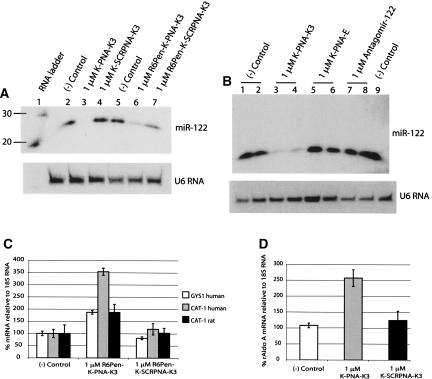
FIGURE 4.
MicroRNA inhibitors based on peptide nucleic acids (PNAs). (A) Northern blots from total RNA for samples electroporated with K-PNA-K3 and K-SCRPNA-K3 in either Huh7 cells (left) or primary rat hepatocytes (right). (B) Aldolase A mRNA levels in primary rat hepatocytes electroporated with 2 μg of the indicated ONs. ([–] Control) Nontransfected cells.
In previous publications aimed toward potential therapeutic applications, we have synthesized conjugates of PNA with cell-penetrating peptides (CPP) in attempts to improve their cell uptake in the absence of transfection agents or electroporation. For such purposes, we developed a cell-penetrating peptide, R6-Penetratin, which when conjugated to antisense PNA ONs showed a remarkable ability to enter HeLa cells in model systems and transfer into cell nuclei. Such CPP–PNA conjugates were shown to inhibit Tat-dependent trans-activation at the TAR RNA target in a luciferase reporter system with integrated plasmids (Turner et al. 2005b) or to redirect splicing in a luciferase up-regulation system involving an aberrant inserted β-globin intron (Abes et al. 2007). We therefore decided to synthesize an N-terminal conjugate of K-PNA-K3 to the C terminus of the CPP R6-Penetratin through a disulfide linkage afforded by added Cys residues, in a similar way to that previously described (Turner et al. 2005b, 2006; Abes et al. 2007). Incubation of the resultant R6Pen-K-PNA-K3 with Huh7 cells in the absence of any transfection agent led to only a low level detected of endogenous miR-122, compared with scrambled control, as observed by Northern blot (Fig. 5A, lanes 6,7). Surprisingly, transfection of the unconjugated K-PNA-K3, also in the absence of any transfection agent, led to a complete disappearance of the miR-122 signal, but not for scrambled K-PNA-K3 (Fig. 5A, lanes 3,4).
FIGURE 5.
Cell delivery of K-PNA-K3 and R6Pen-K-PNAK3 oligonucleotides into Huh7 cells in the absence of transfecting agents. (A) Northern blot of total RNA from cells incubated with 1 μM ONs. (B) Northern blot of total RNA from cells incubated with 1 μM electroneutral K-PNA-E, Antagomir-122, and controls. (C) mRNA levels of miR-122-regulated genes in Huh7 cells and primary rat hepatocytes incubated with the R6-Penetratin conjugate. (D) Aldolase A mRNA levels in primary rat hepatocytes incubated with K-PNA-K3 in the absence of transfecting agent, at 1 μM. ([–] Control) Nontransfected cells.
We wondered whether the additional four Lys residues included in the PNA oligomer might be partially responsible for the observed activity. We have previously observed that Lys residues added to PNA ONs enabled them to be taken up in HeLa cells where they become trapped in endosomal compartments (Turner et al. 2005b; Abes et al. 2006), the uptake presumably driven by electrostatic interactions of the Lys residues with cell-surface proteoglycans (Richard et al. 2005). Although such additional charges were not sufficient to obtain biological activity in either of two model systems that require nuclear delivery (Turner et al. 2005b; Abes et al. 2007), perhaps there is a sufficient “proton sponge” effect of the primary amine groups of the Lys residues to afford some endosomal leakage, at least sufficient for microRNA inhibition. Thus, to test whether the presence of the additional four Lys residues in the PNA ON is responsible for uptake and subsequent activity, we synthesized an electrically neutral PNA, K-PNA-E, where the 3′-(Lys)3 was substituted by a 3′-glutamic acid residue. Figure 5B (lanes 5,6) shows that the electroneutral PNA was not able to reduce miR-122 levels in Huh7 cells. As a comparison, we also incubated the corresponding 3′-cholesterol OMe/PS antagomir ON, which has been developed for in vivo delivery (Krutzfeldt et al. 2005), with Huh7 cells in the absence of transfection agent and found that this ON was not active under these conditions (Fig. 5B, lanes 7,8). As shown above for the LNA/OMe mixmer (Fig. 3), addition of R6Pen-K-PNA-K3 (Fig. 5C) induced an increase in the abundances of negatively regulated mRNA in both Huh7 and primary rat hepatocytes in the absence of a transfection agent. The parent K-PNA-K3 also showed a similar effect on a negatively regulated gene when incubated with primary rat hepatocytes (Fig. 5D).
MicroRNA fate after binding to antisense ON inhibitor
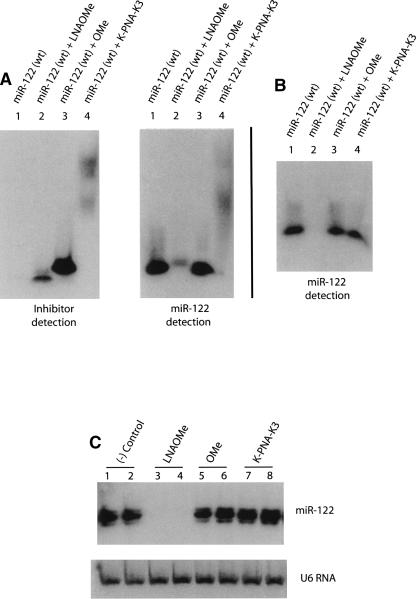
An important issue regarding microRNA inhibition by synthetic ONs is the final fate of the targeted microRNA. We addressed this question by carrying out Northern blot analysis. We first asked if the stringent denaturing conditions used in the PAGE gels (samples incubated 5 min at 95°C and immediately loaded onto a 14% polyacrylamide/20% formamide/8 M urea gel) are sufficient to dissociate the potential complexes formed between miR-122 and the corresponding inhibitor. We therefore tested this in an in vitro experiment (Fig. 6). Not surprisingly, we found that the LNA/OMe ON formed a complex with high thermal stability, which was not dissociated completely under the denaturing conditions used for gel separation, as shown by the lower intensity and retarded mobility of the detected miR-122 (Fig. 6A, cf. lane 2 of both panels). Similarly, PNA formed stable complexes with miR-122, which are seen in this case as broad bands of high molec-ular mass (Fig. 6A, lane 4, both panels). In the case of addition of the OMe ON, strong single bands seen at very similar positions for both the inhibitor and miR-122 probe (Fig. 6A, lane 3, both panels) make it difficult to prove unequivocally the presence/absence of a complex, but such a complex would be expected to be significantly weaker than for the other inhibitors.
FIGURE 6.
Northern blots of wild-type miR-122 and of its equimolar mixes with all ON inhibitors tested. (A) Fifty picomoles of the indicated ONs were mixed, diluted in loading buffer containing 20% formamide/8 M urea, and incubated 5 min at 95°C before loading gels. (B) ONs were mixed as described above and extracted with 150 μL of Trizol prior to PAGE and Northern blot analysis. (C) Inhibitor amounts corresponding to 50 nM (or 1 μM for K-PNA-K3) transfections were spiked into a Huh7 cell lysate (one lysate equals one well on a six-well plate) before Trizol extraction (1 mL). Total RNA was subjected to PAGE and Northern blot. ([–] Control) Total RNA not spiked with ONs.
Northern blots of RNA cell extracts shown in Figures 2, 4, and 5 were negative for complex detection for each of the ON types, which did not seem to correlate with the results shown in Figure 6 for the in vitro experiments where complexes were seen. Therefore, we carried out further Northern blot experiments to determine if such complexes can be found in the RNA fraction following Trizol purification. Figure 6B indeed shows that after Trizol RNA purification, which involves denaturing conditions (phenol, guanidine isothiocyanate, chloroform), for synthetic miR-122 (wt) mixed with either OMe ON or K-PNA-K3, there is a complete recovery of the microRNA signal, supporting the notion that any microRNA present in RNA cell extracts is likely to be released from either of these two miR-122-ON complexes and recovered in the RNA fraction. By contrast, the sample corresponding to miR-122 complex with LNA/OMe ON did not show evidence of miR-122 presence (Fig. 6B, lane 2).
We therefore tested whether the miR-122:LNA/OMe ON complex could be recovered from a cell extract. Thus, we added to a Huh7 cell Trizol lysate an amount of ON inhibitor equivalent to a standard transfection (50 nM for OMe and LNA/OMe, 1 μM for K-PNA-K3) and carried out Northern blots following total RNA extraction by the standard procedure (Fig. 6C). Just as in the case of Figure 6B, the miR-122 signal is fully restored for both OMe and K-PNA-K3 treated samples (Fig. 6C, lanes 5–8), but once again the LNA/OMe-treated sample did not produce a band corresponding to miR-122 (Fig. 6C, lanes 3,4). These results confirm that the PNA and OMe complexes with the targeted microRNA are disrupted during Trizol RNA purification and thus allow visualization of intact microRNA bands. Since in Figures 2 and 5 ablation of the miR-122 signal was seen by PNA or OMe ON inhibitor treatment of cells, the effective blocking of the microRNA seen in the presence of complementary ONs here is strong evidence for intracellular RNA target degradation by a pathway still as yet undiscovered. By contrast, in the case of miR-122:LNA/OMe ON complexes, the microRNA is not directed into the RNA fraction during Trizol purification. Hence, the up-regulation of miR-122-regulated mRNAs must be taken as the major evidence for inhibitor anti-microRNA activity. In this particular case, the disappearance of the miR-122 band in Northern blots cannot be used as evidence of a microRNA degradation pathway, even though the complex formation with LNA/OMe oligomer is clearly extremely strong (Fig. 6).
DISCUSSION
The standard ON type used in many microRNA inhibition studies is OMe (Meister et al. 2004). For example, Chang et al. (2004) used a 22-mer OMe ON at high concentration (30 μM) for long time periods (3-d treatment) to inhibit miR-122 in liver cells, and even then protein expression of a miR-122-targeted gene could not be recovered completely. A second study used a lower concentration (50 nM) of a 31-mer anti-miR-122 OMe ON for 48 h to study the miR-122 requirements of HCV replication (Jopling et al. 2005). Although 2′-O-methoxyethyl (MOE) ONs have been shown to have improved miR-122 inhibition properties in mouse cells over OMe ONs (Davis et al. 2006; Esau et al. 2006), such materials are not available commercially. Current commercial suppliers of microRNA inhibitors (e.g., Dharmacon, Exiqon) utilize a number of nucleotide modifications. In the case of Exiqon, their microRNA probes consist of LNA/DNA mixmers, based on an original publication from this company (Válóczi et al. 2004), and LNA/DNA mixmers have been used also for microRNA function inhibition (Naguibneva et al. 2006; Ørom et al. 2006). We have used LNA/OMe oligomers extensively for other steric block applications with excellent sequence specificity (Arzumanov et al. 2001; Brown et al. 2005) and here found that a transfected 23-mer LNA/OMe led to complete removal of the miR-122 signal by Northern blot in two model liver cell systems (Fig. 2) and up-regulated a corresponding miR-122-regulated mRNA, whereas a standard 31-mer OMe ON [21] was ineffective (Fig. 3). The mixmer LNA/OMe ON we designed had 10/23 LNA approximately alternating with OMe residues. It was not the purpose of this study to optimize the ratio of the two analogs or their placement within mixmers, but we have shown previously that ∼40% LNA distributed evenly throughout an OMe oligomer showed best results in another steric block application (Arzumanov et al. 2003). Apart from their lower binding strength, OMe ONs are degradable within cells (Baker et al. 1997), which might also reflect their lower activity and explain why we have not observed the OMe ON in a Northern blot when probing with an anti-inhibitor ON, in contrast to a LNA/OMe-treated sample, where it is clearly visible (Supplemental Fig. S1A).
Davis et al. (2006) showed a loose correlation between the melting temperature of the hybrid between 22-mer ONs and complementary RNA, but this was not absolute, since the strongest binding ON type, consisting of LNA/MOE units with all PS linkages, was not the best in target microRNA knockdown (uniform MOE). However, LNA/OMe or LNA/MOE with diester linkages were not reported. Our results suggest that LNA/OMe are worthy of more detailed investigation for microRNA target knockdown both in cells and in vivo.
We also showed that a 23-mer K-PNA-K3 complementary to miR-122 and electroporated into Huh7 cells or rat primary hepatocytes reduced miR-122 expression and up-regulated a miR-122-regulated mRNA (Fig. 4). This is the first time that a non-negatively charged ON analog has been shown to knock down microRNA function in a cell line. Another type of well-known electroneutral analog, PMO, has been injected into zebrafish embryos to modulate miR-375 activity to demonstrate its role in pancreatic islet development (Kloosterman et al. 2007), but there have been no previous reports of PMO or PNA targeting microRNAs in cell lines.
An advantage of PNA is that it can be readily conjugated with amino acids or peptides to modify its cell uptake properties. We previously reported that a K-PNA-K3 ON disulfide conjugated to an R6-Penetratin peptide was able to enter HeLa cells and show substantial steric block RNA targeting activity in two model systems that require nuclear delivery (Turner et al. 2005b; Abes et al. 2007). Here, we found that a similar R6-Penetratin disulfide conjugate to K-PNA-K3 showed substantial and sequence-dependent inhibition of miR-122 in Huh7 cells (Fig. 5A) or rat primary hepatocytes (data not shown) and corresponding up-regulation of miR-122-regulated mRNA (Fig. 5C). Remarkably, incubation of Huh7 cells with 1 μM of the unconjugated K-PNA-K3 ON was also able to effect the same miR-122 inhibition (Fig. 5A,B) and increase the abundance of a miR-122-regulated mRNA (Fig. 5D). Since this activity was not achieved by a PNA that maintained overall electroneutrality (Fig. 5B), it seems likely that, at least in hepatocytes, the four added Lys residues in K-PNA-K3 are sufficient to allow both cell entry and to reach a compartment where mature miR-122 is present. Although mRNAs targeted by microRNAs reside, at least for a time, in cytosolic RNA P-bodies (Liu et al. 2005; Bhattacharyya et al. 2006b), it has been shown recently that antagomir to miR-122 does not colocalize with P-bodies when hepatocytes taken from mouse liver were examined (Krutzfeldt et al. 2007).
Our experiments do not address the issue of the subcellular site of action of ON inhibitors of microRNAs, but cationic PNAs, such as those containing multiple Lys residues or conjugated to the HIV-1 Tat basic region, are taken up in HeLa cells through an endocytotic mechanism and are sequestered in endosomes (Richard et al. 2005; Abes et al. 2006). Whether such endosomes then release the PNA into the cytosol, because of a “proton sponge” effect of the Lys residues, in order to travel to another cytosolic compartment, or whether there is a fusion of PNA-containing endosomes with the microRNA-containing cytosolic compartments must be the subject of further experimentation. MicroRNA may be a more susceptible cytosolic target than mRNA, since a PNA complementary to human caveolin-1 mRNA conjugated to a Lys-rich signal peptide was unable to reduce gene expression after 56 h incubation in HeLa cells or primary endothelial cells (Kaihatsu et al. 2004), and additional Lys residues were shown also not to be sufficient to obtain nuclear activity for PNA (Turner et al. 2005b; Abes et al. 2007).
Use of microRNA-targeted mRNAs levels as an important measure of inhibitor effectiveness has been well validated in mouse studies of miR-122 (Krutzfeldt et al. 2005, 2007; Esau et al. 2006), but few studies have described mRNA targets of miR-122 in cell lines. We showed that three mRNAs, CAT-1, GYS1, and Aldolase A, are targets in Huh7 cells and three mRNAs, CAT-1, Aldolase A, and GTF2b, are targets in rat hepatocytes (Fig. 1). The amino acid transporter CAT-1 mRNA was shown by OMe ON inhibitor transfection to be a miR-122 target in Huh7 cells a few years ago (Chang et al. 2004), but another similar study also in Huh7 cells did not observe significant up-regulation of CAT-1 mRNA levels by Northern blot analysis, but concluded instead that the mRNA was released from cytoplasmic processing P-bodies and recruited into polysomes (Bhattacharyya et al. 2006a,b). We did not see significant up-regulation of miR-122-regulated mRNAs GYS1 (Fig. 3C) or CAT-1 (data not shown) by 31-mer OMe ON transfected into either Huh7 cells or rat hepatocytes. By contrast, we did see significant up-regulation of GYS1 in Huh7 cells using LNA/OMe ON transfection (Fig. 3A), which agrees with data seen in mice for a MOE ON (Esau et al. 2006). With CAT-1 mRNA, no significant up-regulation was seen with the LNA/OMe ON in Huh7 cells, but we did observe an increase in CAT-1 mRNA in rat hepatocytes (data not shown). Similarly, R6Pen-K-PNA-K3 also showed CAT-1 mRNA up-regulation when incubated with either Huh7 cells or rat hepatocytes (Fig. 5). Thus, overall LNA/OMe and PNA ONs appear more effective than the corresponding OMe ON for studying microRNA regulation of target mRNAs.
The fate of microRNAs targeted by antisense ONs within cells has been unclear to date. Our LNA/OMe inhibition data do not allow us to draw conclusions on this aspect, since the complex is not partitioned into the Trizol fraction during RNA isolation from cells (Fig. 6). However, the loss of miR-122 signals in Northern blots using K-PNA-K3 (Figs. 2, 4, 5) strongly suggests that miR-122 targeted by the ON within cells is degraded in some way. These results are consistent with conclusions from in vivo experiments where microRNA degradation products were detected in mice treated with OMe/PS-cholesterol antagomirs (Krutzfeldt et al. 2005) and with Northern blot experiments of extracts from HeLa cells treated with MOE/PS antisense ONs (Davis et al. 2006). This degradation cannot be due to RNase H, as proposed from experiments with LNA/DNA ONs (Naguibneva et al. 2006), since PNA ONs do not direct RNase H cleavage of RNA (Knudsen and Nielsen 1996), nor is it likely to be due to Ago2, the nuclease component of the RISC complex implicated in siRNA action (Krutzfeldt et al. 2007). Thus, the pathway of degradation is yet to be established.
The 23-mer LNA/OMe and PNA ONs have been designed to be complementary to the mature form of miR-122 that is active in the cytosol. Recently, electroneutral PMO ONs have been designed to target cytosolic pre-miRNA-375 and nuclear pri-mRNA-375 precursors by microinjection into developing zebrafish embryos (Kloosterman et al. 2007). Our results with K-PNA-K3 inhibition of miR-122 suggests that it may be possible to design PNA probes for cell studies to distinguish the cytoplasmic mature and/or pre-microRNA forms (using a Lys-modified PNA probe) from the nuclear pri-miRNA or blocking microRNA gene expression at the transcription level (using a R6-Pen-K-PNA-K3 probe). We are currently investigating also whether our K-PNA-K3 microRNA inhibition results obtained in human Huh7 cells and rat primary hepatocytes can be extended to a range of other cell types.
We have shown that LNA/OMe and PNA ONs provide powerful new types of microRNA inhibitory reagents. It is already clear that different ON chemistries result in somewhat different characteristics of microRNA inhibition, and further experience will help establish where different ON types may best find application. As far as we are aware, LNA/OMe mixmers have not been evaluated in vivo for any application hitherto, but steric block LNA/DNA mixmers containing all phosphorothioate linkages gave efficient splice redirection in mice (Roberts et al. 2006). By contrast, PNA and its peptide conjugates have been evaluated in a number of in vivo studies toward splicing redirection (Sazani et al. 2002; Albertshofer et al. 2005) and cancer (Boffa et al. 2005) and have been shown to have wide tissue distribution. Our promising results in cell culture for K-PNA-K3 suggest that relatively simple PNA constructs would be worth evaluating in vivo for microRNA inhibition applications.
MATERIALS AND METHODS
Oligonucleotides, miRNA mimics, and PNA peptides
Human/rat miR-122 mimic duplex sequences were obtained from the miRBase Sequence Database (Release 9.2) and were purchased from Dharmacon; human/rat sense strand, 5′-UGGAGUGUGACAAUGGUGUUUGU-3′; human antisense strand, 5′-AAACGCCAUUAUCACACUAAAUA-3′; rat antisense strand, 5′-AAACGCCAUCAUCACACUAAACA-3′. LNA/OMe ONs were synthesized as previously described (Turner et al. 2006). Synthesized sequences contained approximately alternating LNA and 2′-OMe residues, with an LNA/OMe ratio of ∼0.4. These ON sequences corresponded either to the full-complement of wild type, mature miR-122, 5′-aCaAaCaCcAuuGuCaCaCuCca-3′ (LNA/OMe), or to a scrambled sequence, 5′-uCaGcCauAaCuAaCcCauAcCa-3′. PNA ONs were synthesized on an APEX 396 robotic peptide synthesizer as described previously (Turner et al. 2006). PNA sequences contained an N-terminal Cys for post-synthetic modification and four Lys residues for improved solubility. K-PNA-K3: Cys-K-5′-ACAAACACCATTGTCACACTCCA-3′-KKK, K-SCRPNA-K3: Cys-K-5′-TCAGCCATAACTAACCCATACCA-3′-KKK. An additional PNA ON was synthesized where the three Lys residues at the 3′-end were replaced by one glutamic acid residue, K-PNA-E, Cys-K-5′-ACAAACACCATTGTCACACTCCA-3′-E. Antagomir-122 was purchased from Dharmacon, 5′-A*C*AAACACCAUUGUCACACU*C*C*A*-Chol-3′ (uppercase, OMe; *, PS linkage).
The sequence of the peptide R6-Penetratin is RRRRRRRQIKIWFQNRRMKWKKGGC, and this was synthesized and characterized as previously described (Turner et al. 2005a). PNA oligonucleotides were conjugated to R6-Pen(Cys) as described previously (Turner et al. 2006). Briefly, 10 nmol of the corresponding PNA (0.5–1 mM) were diluted in 6 μL of NH4Ac 1 M and 30 μL of formamide. After addition of Pys-R6-Pen (3 eq, 10 mM stock), the solution was incubated for 1 h at room temperature and purified by RP-HPLC (0.1% TFA-containing buffers). Freeze-dried fractions were dissolved in water and repurified through an ion-exchange column for counter-ion replacement, from trifluoroacetate to acetate. Purified fractions were freeze-dried three times against water/ethanol (80:20).
Cell culture and transfections
For Northern blot or real-time RT-PCR experiments, Huh7 cells were plated in either a six- or 96-well plate format and maintained in standard cell culture medium, ∼20 h before transfection. For primary rat hepatocytes cultures (Dominion Pharmakyn), plates were initially treated with 0.3 mg/mL fibronectin for 5 min and air dried. Hepatocytes were then plated at ∼5 × 105 cells/mL (HCB media, Cambrex) in a six-well plate format. Wells were carefully washed after 1.5 h and further incubated for 14 h in fresh media. For lipid-based transfections, ONs were mixed with Lipofectamine 2000 (Invitrogen) following the manufacturer's instructions and incubated with cells in serum-free media for 4 h. Cells were washed once with PBS and incubated for 20 h in full media containing antibiotics, before RNA extraction. Total RNA was purified using the TRI reagent (Sigma) following the manufacturer's instructions. RNA was quantified using the Quant-iT RiboGreen RNA assay kit (Invitrogen).
Polyacrylamide gel electrophoresis (PAGE) and Northern blots
Fifteen to 20 μg of total RNA from each experimental condition was loaded onto a 14% acrylamide (acrylamide:bis-acrylamide 19:1)/20% formamide/8 M urea gel and run at 10 W for 1.2 h at room temperature. Gels were transferred onto a GeneScreen Plus hybridization membrane (PerkinElmer) for 1 h at 400 mA at room temperature in a semi-dry transfer unit (Hoefer Scientific Instruments). Blotted membranes were cross-linked with a UV dose of 2400 × 100 μJoules (UV Stratalinker 1800, Stratagene) and baked for 1 h at 80°C. Prehybridization was performed for 30 min in ULTRA-hyb Oligo solution (Ambion) at 42°C. For microRNA detection, a subsequent incubation with 250 pmol of a specific [γ-32P]ATP-labeled miRCURY LNA detection probe (Exiqon) was carried out (or with a DNA probe for U6 RNA control, 5′-TTGCGTGTCATCCTTGCGCAGG-3′). Membranes were sequentially washed with 2× SSC/0.1% SDS (30 min), 1× SSC/0.1% SDS (30 min), and 0.2× SSC/0.1% SDS (10 min) at 42°C.
Real-time RT-PCR
Messenger RNAs of regulated genes were quantified relative to 18S RNA, in a 7900 HT Fast Real-Time PCR System (Applied Biosystems) using a one-step RT-PCR approach (QuantiTect Probe RT-PCR, Qiagen). Briefly, the 25 μL reactions contained 25 ng of total RNA, 0.25 μL of reverse transcriptase mix, 12.5 μL of master mix, 1.25 μL of Taqman probe/primers (Taqman Gene Expression Assays, Applied Biosystems), and 11 μL of water. The one-step RT-PCR was performed for 30 min at 50°C, followed by an inactivation/denaturation step (15 min, 95°C) and 40 amplification cycles (15 sec at 94°C and 1 min at 60°C). MicroRNA-122 was quantified relative to RNU6B by real-time PCR using a two-step approach (Taqman MicroRNA Transcription kit, Applied Biosystems). Initially, total RNA (25 ng) was reverse-transcribed using specific primers for either miR-122 or RNU6B in a 15 μL reaction volume (0.15 μL of dNTP mix [100 mM], 1 μL of MultiScribe RT [50 U/μL], 0.19 μL of RNase inhibitor [20 U/μL], 1.5 μL of 10× RT buffer, 3 μL of 5× RT primer, 4.16 μL of H2O, and 5 μL of RNA [25 ng]). Real-time PCR reactions were performed in 20 μL reaction volumes, containing 10 μL of Taqman 2× PCR master mix (No AmpErase UNG, Applied Biosystems), 1 μL of Taqman assay 20×, 9 μL of H2O, and 2 μL of cDNA. The RT step consisted of two 30-min incubations at 16°C and 42°C, followed by 5 min of inactivation at 85°C. The PCR step consisted of an initial deactivation/denaturation step of 15 min at 95°C, followed by 40 amplification cycles (15 sec at 94°C and 1 min at 60°C).
SUPPLEMENTAL DATA
Supplemental Materials can be found at http://www2.mrc-lmb.cam.ac.uk/groups/mg/papers.html.
ACKNOWLEDGMENTS
We thank Donna Williams, Matthew Watson, and David Owen for PNA, LNA/OMe ONs, and peptide syntheses. We also thank Ramon Mayoral for technical assistance and Chris Goldring (University of Liverpool) for kindly providing Huh7 cells.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.844108.
REFERENCES
- Abes, S., Williams, D., Prevot, P., Thierry, A.R., Gait, M.J., Lebleu, B. Endosome trapping limits the efficiency of splicing correction by PNA–oligolysine conjugates. J. Control. Release. 2006;110:595–604. doi: 10.1016/j.jconrel.2005.10.026. [DOI] [PubMed] [Google Scholar]
- Abes, S., Turner, J.J., Ivanova, G.D., Owen, D., Williams, D., Arzumanov, A., Clair, P., Gait, M.J., Lebleu, B. Efficient splicing correction by PNA conjugation to an R6-Penetratin delivery peptide. Nucleic Acids Res. 2007;35:4495–4502. doi: 10.1093/nar/gkm418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn, D.-G., Kourakis, M.J., Rohde, L.A., Silver, L.M., Ho, R.K. T-box gene tbx5 is essential for formation of the pectoral limb bud. Nature. 2002;417:754–758. doi: 10.1038/nature00814. [DOI] [PubMed] [Google Scholar]
- Albertshofer, K., Siwkowski, A.M., Wancewicz, E.V., Esau, C.C., Watanabe, T., Nishihara, K.C., Kinberger, G.A., Malik, L., Eldrup, A.B., Manoharan, M., et al. Structure–activity relationship study on a simple cationic peptide motif for cellular delivery of antisense peptide nucleic acid. J. Med. Chem. 2005;48:6741–6749. doi: 10.1021/jm050490b. [DOI] [PubMed] [Google Scholar]
- Arzumanov, A., Walsh, A.P., Rajwanshi, V.K., Kumar, R., Wengel, J., Gait, M.J. Inhibition of HIV-1 Tat-dependent trans-activation by steric block chimeric 2′-O-methyl/LNA oligoribonucleotides. Biochemistry. 2001;40:14645–14654. doi: 10.1021/bi011279e. [DOI] [PubMed] [Google Scholar]
- Arzumanov, A., Stetsenko, D.A., Malakhov, A.D., Reichelt, S., Sørensen, M.D., Babu, B.R., Wengel, J., Gait, M.J. A structure–activity study of the inhibition of HIV-1 Tat-dependent trans-activation by mixmer 2′-O-methyl oligoribonucleotides containing locked nucleic acid (LNA), α-LNA or 2′-thio-LNA residues. Oligonucleotides. 2003;13:435–453. doi: 10.1089/154545703322860762. [DOI] [PubMed] [Google Scholar]
- Baker, B.F., Lot, S.S., Condon, T.P., Cheng-Flourney, S., Lesnik, E.A., Sasmor, H.M., Bennett, C.F. 2′-O-(2-methoxy)ethyl-modified anti-intercellular adhesion molecule 1 (ICAM-1) oligonucleotides selectively increase the ICAM-1 mRNA level and inhibit formation of the ICAM-1 translation initiation complex in human umbilical vein endothelial cells. J. Biol. Chem. 1997;272:11994–12000. doi: 10.1074/jbc.272.18.11994. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya, S.N., Habermacher, R., Martine, U., Closs, E.I., Filipowicz, W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006a;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya, S.N., Habermacher, R., Martine, U., Closs, E.I., Filipowicz, W. Stress-induced reversal of microRNA repression and mRNA P-body localization in human cells. Cold Spring Harb. Symp. Quant. Biol. 2006b;71:513–521. doi: 10.1101/sqb.2006.71.038. [DOI] [PubMed] [Google Scholar]
- Boffa, L.C., Cutrona, G., Cilli, M., Mariani, M.R., Matis, S., Pastorini, M., Damonte, G., Millo, E., Roncella, S., Ferrarini, M. Therapeutically promising PNA complementary to a regulatory sequence for c-myc: Pharmacokinetics in an animal model of human Burkitt's lymphoma. Oligonucleotides. 2005;15:85–93. doi: 10.1089/oli.2005.15.85. [DOI] [PubMed] [Google Scholar]
- Brown, D., Arzumanov, A.A., Turner, J.J., Stetsenko, D.A., Lever, A.M., Gait, M.J. Antiviral activity of steric-block oligonucleotides targeting the HIV-1 trans-activation response and packaging signal stem–loop RNAs. Nucleosides Nucleotides Nucleic Acids. 2005;24:393–396. doi: 10.1081/ncn-200059813. [DOI] [PubMed] [Google Scholar]
- Brown, D.E., Arzumanov, A., Syed, S., Gait, M.G., Lever, A.M. Inhibition of HIV-1 replication by oligonucleotide analogues directed to the packaging signal and trans-activating response region. Antivir. Chem. Chemother. 2006;17:1–9. doi: 10.1177/095632020601700101. [DOI] [PubMed] [Google Scholar]
- Chan, J.A., Krichevsky, A.M., Kosik, K.S. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- Chang, J., Nicolas, E., Marks, D., Sander, C., Lerro, A., Buendia, M.A., Xu, C., Mason, W.S., Moloshok, T., Bort, R., et al. miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol. 2004;1:106–113. doi: 10.4161/rna.1.2.1066. [DOI] [PubMed] [Google Scholar]
- Cheng, A.M., Byrom, M.W., Shelton, J., Ford, L.P. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 2005;33:1290–1297. doi: 10.1093/nar/gki200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimmino, A., Calin, G.A., Fabbri, M., Iorio, M.V., Ferracin, M., Shimizu, M., Wojcik, S.E., Aqeilan, R.I., Zupo, S., Dono, M., et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl. Acad. Sci. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, S., Lollo, B., Freier, S., Esau, C. Improved targeting of miRNA with antisense oligonucleotides. Nucleic Acids Res. 2006;34:2294–2304. doi: 10.1093/nar/gkl183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert, M.S., Neilson, J.R., Sharp, P.A. MicroRNA sponges: Competitive inhibitors of small RNAs in mammalian cells. Nature. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esau, C., Kang, X., Peralta, E., Hanson, E., Marcusson, E.G., Ravichandran, L.V., Sun, Y., Koo, S., Perera, R.J., Jain, R., et al. MicroRNA-143 regulates adipocyte differentiation. J. Biol. Chem. 2004;279:52361–52365. doi: 10.1074/jbc.C400438200. [DOI] [PubMed] [Google Scholar]
- Esau, C., Davis, S., Murray, S.F., Yu, X.X., Pandey, S.K., Pear, M., Watts, L., Booten, S.L., Graham, M., McKay, R., et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Fletcher, S., Honeyman, K., Fall, A.M., Harding, P.L., Johnsen, R.D., Steinhaus, J.P., Moulton, H.M., Iversen, P.L., Wilton, S.D. Morpholino oligomer-mediated exon skipping averts the onset of dystrophic pathology in the mdx mouse. Mol. Ther. 2007;151:587–592. doi: 10.1038/sj.mt.6300245. [DOI] [PubMed] [Google Scholar]
- Garzon, R., Fabbri, M., Cimmino, A., Calin, G.A., Croce, C.M. MicroRNA expression and function in cancer. Trends Mol. Med. 2006;12:580–587. doi: 10.1016/j.molmed.2006.10.006. [DOI] [PubMed] [Google Scholar]
- Griffith-Jones, S., Grocock, R.J., van Dongen, S., Bateman, A., Enright, A.J. miRBase: MicroRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, L., Thomson, J.M., Hemann, M.T., Hernando-Monge, E., Mu, D., Goodson, S., Powers, S., Cordon-Cardo, C., Lowe, S.W., Hannon, G.J., et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jopling, C.L., Yi, M., Lancaster, A.M., Lemon, S.M., Sarnow, P. Modulation of Hepatitis C Virus RNA abundance by a liver-specific microRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- Jopling, C.L., Norman, K.L., Sarnow, P. Positive and negative modulation of viral and cellular mRNAs by liver-specific microRNA miR-122. Cold Spring Harb. Symp. Quant. Biol. 2006;71:369–376. doi: 10.1101/sqb.2006.71.022. [DOI] [PubMed] [Google Scholar]
- Kaihatsu, K., Huffman, K.E., Corey, D.R. Intracellular uptake and inhibition of gene expression by PNAs and PNA–peptide conjugates. Biochemistry. 2004;43:14340–14347. doi: 10.1021/bi048519l. [DOI] [PubMed] [Google Scholar]
- Kloosterman, W.P., Agendijk, A.K., Ketting, R.F., Moulton, J.D., Plasterk, R.H.A. Targeted inhibition of miRNA maturation with morpholinos reveals a role for miR-375 in pancreatic islet development. PLoS Biol. 2007;5:e203. doi: 10.1371/journal.pbio.0050203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen, H., Nielsen, P. Antisense properties of duplex- and triplex-forming PNAs. Nucleic Acids Res. 1996;24:494–500. doi: 10.1093/nar/24.3.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppelhus, U., Zachar, V., Nielsen, P., Liu, X., Eugen-Olsen, J., Ebbesen, P. Efficient in vitro inhibition of HIV-1 gag reverse transcription by peptide nucleic acid (PNA) at minimal ratios of PNA/RNA. Nucleic Acids Res. 1997;25:2167–2173. doi: 10.1093/nar/25.11.2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshkin, A.A., Singh, S.K., Nielsen, P., Rajwanshi, V.K., Kumar, R., Meldgaard, M., Olsen, C.E., Wengel, J. LNA (locked nucleic acids): Synthesis of the adenine, cytosine, guanine, 5-methylcytosine, thymine and uracil bicyclonucleoside monomers, oligomerisation, and unprecedented nucleic acid recognition. Tetrahedron. 1998;54:3607–3630. [Google Scholar]
- Krutzfeldt, J., Rajewsky, N., Braich, R., Rajeev, K.G., Tuschl, T., Manoharan, M., Stoffel, M. Silencing of microRNAs in vivo with “antagomirs.”. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- Krutzfeldt, J., Kuwajima, S., Braich, R., Rajeev, K.G., Pena, J., Tuschl, T., Manoharan, M., Stoffel, M. Specificity, duplex degradation and subcellular localization of antagomirs. Nucleic Acids Res. 2007;35:2885–2892. doi: 10.1093/nar/gkm024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana, M., Rauhut, R., Yalcin, A., Meyer, J., Lendeckel, W., Tuschl, T. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- Lee, R.C., Feinbaum, R.L., Ambros, V. The C. elegans heterochromic gene lin-4 encodes small RNAs with antisense complementary to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Liu, J., Valencia-Sanchez, M.A., Hannon, G.J., Parker, R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 2005;7:719–723. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister, G., Landthaler, M., Dorsett, Y., Tuschl, T. Sequence-specific inhibition of microRNA- and siRNA-induced RNA silencing. RNA. 2004;10:544–550. doi: 10.1261/rna.5235104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moschos, S.A., Williams, A.E., Perry, M.M., Birrell, M.A., Belvisi, M.G., Lindsay, M.A. Expression profiling in vivo demonstrates rapid changes in lung microRNA levels following lipopolysaccharide-induced inflammation but not in the anti-inflammatory action of glucocorticoids. BMC Genomics. 2007;8:240. doi: 10.1186/1471-2164-8-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naguibneva, I., Ameyar-Zazoua, M., Nonne, N., Polesskaya, A., Ait-Si-Ali, S., Groisman, R., Souidi, M., Pritchard, L.L., Harel-Bellan, A. An LNA-based loss-of-function assay for micro-RNAs. Biomed. Pharmacother. 2006;60:633–638. doi: 10.1016/j.biopha.2006.07.078. [DOI] [PubMed] [Google Scholar]
- Nielsen, P.E., Egholm, M., Buchardt, O. Peptide nucleic acid (PNA). A DNA mimic with a peptide backbone. Bioconjug. Chem. 1994;5:3–7. doi: 10.1021/bc00025a001. [DOI] [PubMed] [Google Scholar]
- O'Connell, R.M., Taganov, K.D., Boldin, M.P., Cheng, G., Baltimore, D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc. Natl. Acad. Sci. 2007;104:1604–1609. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell, K.A., Wentzel, E.A., Zeller, K.I., Dang, C.V., Mendell, J.T. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- Ørom, U.A., Kauppinen, S., Lund, A.H. LNA-modified oligonucleotides mediate specific inhibition of microRNA function. Gene. 2006;372:137–141. doi: 10.1016/j.gene.2005.12.031. [DOI] [PubMed] [Google Scholar]
- Richard, J.P., Melikov, K., Brooks, H., Prevot, P., Lebleu, B., Chernomordik, L.V. Cellular uptake of unconjugated TAT peptide involves clathrin-dependent endocytosis and heparan sulfate receptors. J. Biol. Chem. 2005;280:15300–15306. doi: 10.1074/jbc.M401604200. [DOI] [PubMed] [Google Scholar]
- Roberts, J., Palma, E., Sazani, P., Ørum, H., Cho, M., Kole, R. Efficient and persistent splice switching by systemically delivered LNA oligonucleotides in mice. Mol. Ther. 2006;14:471–475. doi: 10.1016/j.ymthe.2006.05.017. [DOI] [PubMed] [Google Scholar]
- Saetrom, P., Heale, B.S.E., Snove O., Jr, Aagaard, L., Alluin, J., Rossi, J.J. Distance constraints between microRNA target sites dictate efficacy and cooperativity. Nucleic Acids Res. 2007;35:2333–2342. doi: 10.1093/nar/gkm133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sazani, P., Gemignani, F., Kang, S.-H., Maier, M.A., Manoharan, M., Persmark, M., Bortner, D., Kole, R. Systemically delivered antisense oligomers upregulate gene expression in mouse tissues. Nat. Biotechnol. 2002;20:1228–1233. doi: 10.1038/nbt759. [DOI] [PubMed] [Google Scholar]
- Suwanmanee, T., Sierakowska, H., Fucharoen, S., Kole, R. Repair of a splicing defect in erythroid cells from patients with β-thalassemia/HbE disorder. Mol. Ther. 2002;6:718–726. doi: 10.1006/mthe.2002.0805. [DOI] [PubMed] [Google Scholar]
- Triboulet, R., Mari, B., Lin, Y.-L., Chable-Bessia, C., Bennasser, Y., Lebrigand, K., Cardinaud, B., Maurin, T., Barbry, P., Baillat, V., et al. Suppression of microRNA-silencing pathway by HIV-1 during virus replication. Science. 2007;315:1579–1582. doi: 10.1126/science.1136319. [DOI] [PubMed] [Google Scholar]
- Turner, J.J., Arzumanov, A.A., Gait, M.J. Synthesis, cellular uptake and HIV-1 Tat-dependent trans-activation inhibition activity of oligonucleotide analogues disulphide-conjugated to cell-penetrating peptides. Nucleic Acids Res. 2005a;33:27–42. doi: 10.1093/nar/gki142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, J.J., Ivanova, G.D., Verbeure, B., Williams, D., Arzumanov, A.A., Abes, S., Lebleu, B., Gait, M.J. Cell-penetrating peptide conjugates of peptide nucleic acids (PNA) as inhibitors of HIV-1 Tat-dependent trans-activation in cells. Nucleic Acids Res. 2005b;33:6837–6849. doi: 10.1093/nar/gki991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, J.J., Williams, D., Owen, D., Gait, M.J. Current protocols in nucleic acid chemistry. Wiley; New York: 2006. Disulfide conjugation of peptides to oligonucleotides and their analogs; pp. 4.28.21–24.28.21. [DOI] [PubMed] [Google Scholar]
- Válóczi, A., Hornyik, C., Varga, N., Burgyán, J., Kauppinen, S., Havelda, Z. Sensitive and specific detection of microRNAs by northern blot analysis using LNA-modified oligonucleotide probes. Nucleic Acids Res. 2004;32:e175. doi: 10.1093/nar/gnh171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiler, J., Hunziker, J., Hall, J. Anti-miRNA oligonucleotides (AMOs): Ammunition to target miRNAs implicated in human disease? Gene Ther. 2006;13:496–502. doi: 10.1038/sj.gt.3302654. [DOI] [PubMed] [Google Scholar]
- Xu, P., Vernooy, S.Y., Guo, M., Hay, B.A. The Drosophila microRNA miR-14 suppresses cell death and is required for normal fat metabolism. Curr. Biol. 2003;13:790–795. doi: 10.1016/s0960-9822(03)00250-1. [DOI] [PubMed] [Google Scholar]
- Xue-Wen, H., Jie, P., Xian-Yuan, A., Hong-Xiang, Z. Inhibition of bacterial translation and growth by peptide nucleic acids targeted to domain II of 23S rRNA. J. Pept. Sci. 2007;13:220–226. doi: 10.1002/psc.835. [DOI] [PubMed] [Google Scholar]