Abstract
In eukaryotes, eRF1 and eRF3 are associated in a complex that mediates translation termination. The regulation of the formation of this complex in vivo is far from being understood. In mammalian cells, depletion of eRF3a causes a reduction of eRF1 level by decreasing its stability. Here, we investigate the status of eRF3a when not associated with eRF1. We show that eRF3a forms altered in their eRF1-binding site have a decreased stability, which increases upon cell treatment with the proteasome inhibitor MG132. We also show that eRF3a forms altered in eRF1 binding as well as wild-type eRF3a are polyubiquitinated. These results indicate that eRF3a is degraded by the proteasome when not associated with eRF1 and suggest that proteasomal degradation of eRF3a controls translation termination complex formation by adjusting the eRF3a level to that of eRF1.
Keywords: release factor eRF3a, GSPT1, eRF1, translation termination, proteasome, eukaryote
INTRODUCTION
In eukaryotes, two interacting polypeptides, eRF1 and eRF3, are involved in translation termination. eRF1 recognizes stop codons and activates the peptidyl transferase center of the ribosome that triggers the release of the nascent polypeptide. eRF3, which belongs to the small GTPase family, stimulates eRF1 release activity in a GTP-dependant manner (Kisselev et al. 2003). The activation of eRF3 GTPase activity strictly requires the presence of both eRF1 and the ribosome (Frolova et al. 1996). Recent data suggest that GTP hydrolysis by eRF3 couples stop codon recognition by eRF1 with peptidyl-tRNA hydrolysis by the peptidyl transferase center (Salas-Marco and Bedwell 2004; Alkalaeva et al. 2006).
eRF1 and eRF3 form a stable complex through interaction of their C-terminal domains (Ito et al. 1998; Ebihara and Nakamura 1999; Merkulova et al. 1999). Studies of the interaction of guanine nucleotides with eRF3 alone and with the eRF3•eRF1 complex show that GTP and eRF1 bind to eRF3 with strong positive cooperativity (Hauryliuk et al. 2006; Pisareva et al. 2006) and that the stimulatory effect of eRF1 on binding of eRF3 to GTP strongly depends on the presence of Mg2+. At physiological Mg2+ concentration, the affinity of the eRF1•eRF3 complex to GTP is slightly higher than to GDP, and the dissociation of GTP from the complex is much slower than of GDP. Thus, it is postulated that, in eukaryotic cells, eRF3 is predominantly bound to GTP in a relatively long-lived eRF1•eRF3•GTP ternary complex, and eRF3 that is not associated with eRF1 is likely to be in the GDP-bound form (Hauryliuk et al. 2006; Pisareva et al. 2006). These studies, together with the data obtained from the crystal structure of yeast eRF3 (Kong et al. 2004), indicate that the mechanism of nucleotide binding/exchange on eRF3 is very distinct among GTP-binding proteins: Mg2+ does not stabilize GDP binding to eRF3 alone or to its complex with eRF1, and a GTP/GDP exchange factor (GEF) is not required for rapid GDP dissociation from any form of the factor (Kong et al. 2004; Pisareva et al. 2006).
Whereas most of the parameters that govern eRF1–eRF3 association are now well characterized, the regulation of the translation termination complex formation is far from being understood in vivo. In mammals, two distinct proteins, eRF3a and eRF3b, are able to function as a translation termination factor in vivo (Chauvin et al. 2005). Depletion of eRF3a, which is likely to be the main factor acting in translation termination, causes a significant reduction of the eRF1 level by decreasing its stability (Chauvin et al. 2005). In budding yeast, some reports showed that a reduced level of either eRF1 or eRF3 did not affect the level of its partner (Chabelskaya et al. 2004; Salas-Marco and Bedwell 2004), whereas Valouev et al. (2002) reported that depletion of either of the release factors is accompanied by a reduction in the level of the other.
In this study, we investigate the status of human eRF3a when not associated with eRF1. We show that alteration of the eRF1-binding site by deletion or mutation decreases the stability of eRF3a. Using GFP-eRF3a fusion proteins, we show that eRF3a carrying mutations in the eRF1-binding site and, to a lesser extent, wild-type eRF3a are ubiquitinated and degraded by the proteasome. We discuss a model describing the regulation of translation termination complex formation.
RESULTS
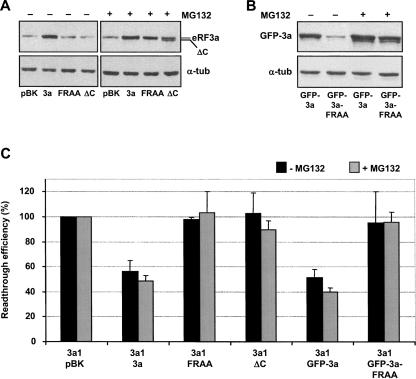
The C-terminal region of eRF3 has been found to be the primary binding site for eRF1 (Ebihara and Nakamura 1999; Merkulova et al. 1999). In this region, the highly conserved GRFTLRD motif plays a crucial role in mediating eRF1 binding (Kong et al. 2004). eRF3 variants with either substitutions in the GRFTLRD motif or C-terminal truncation show reduced interaction with eRF1 (Kong et al. 2004). To examine the influence of eRF1 binding disruption on eRF3a stability, human eRF3a variants carrying either a deletion of the last 31 C-terminal amino acids (eRF3a-ΔC) or mutations in the GRFTLRD motif (eRF3a-FRAA) were created and overexpressed in HEK293 cells. When compared to wild-type eRF3a overexpression, surprisingly, eRF3a-ΔC and eRF3a-FRAA overexpression was not detected by Western blotting (Fig. 1A), suggesting that these proteins had an increased sensitivity to cellular proteases; the band observed in lanes pBK, FRAA, and ΔC corresponds to endogenous eRF3a. Proteolytic degradation was confirmed by treatment of electroporated cells with MG132, a specific inhibitor of the proteasome. As shown in Figure 1A, the presence of MG132 increased the level of overexpressed eRF3a-FRAA and eRF3a-ΔC proteins. The same stabilization by MG132 treatment was observed in HEK293 cells electroporated with plasmids expressing wild-type eRF3a and the eRF3a-FRAA variant in fusion with the green fluorescent protein (GFP), GFP-3a and GFP-3a-FRAA, respectively (Fig. 1B). These fusion proteins allowed us to discriminate between the overexpressed variant of eRF3a and the endogenous wild-type eRF3a. These results suggested that eRF3a variants altered in eRF1 binding were unstable and degraded by the proteasome.
FIGURE 1.
Stabilization of eRF3a forms mutated in eRF1-binding site by MG132. Extracts of HEK293 cells electroporated with (A) the empty vector pBK-CMV (lanes pBK), plasmid expressing wild-type human eRF3a (lanes 3a), plasmids expressing eRF3a variants with either a double mutation in eRF1-binding site (lanes FRAA), or a deletion of the last 31 amino acids containing the eRF1-binding site (lanes ΔC); (B) plasmids expressing GFP fusion with wild-type eRF3a (lanes GFP-3a), and with eRF3a variant carrying the double mutation in the eRF1-binding site (lanes GFP-3a-FRAA) were analyzed by Western blotting with antibodies to human eRF3a; equal protein loading was verified by the detection of α-tubulin (α-tub). Cells were treated or not with MG132 as indicated above each lane. (C) Translation termination activity of eRF3a forms mutated in the eRF1-binding site. 559C cells stably expressing a lacZ gene containing a premature stop codon were depleted in eRF3a by electroporation of a plasmid expressing a siRNA (si-3a1) directed against eRF3a mRNA and re-electroporated 3 d later with either the empty vector pBK-CMV (3a1/pBK) or plasmid expressing wild-type eRF3a (3a1/3a), or plasmids expressing the variants eRF3a-FRAA (3a1/FRAA), eRF3a-ΔC (3a1/ΔC), GFP-eRF3a (3a1/GFP-3a), and GFP-3a-FRAA (3a1/GFP-3a-FRAA). Cells were treated or not with MG132 as indicated, and the extracts performed 3 d after the second electroporation were used for the β-galactosidase assay. The results of readthrough assays were normalized to the level of 3a1/pBK-CMV electroporated cells, which served as the standard for the readthrough level promoted by eRF3a depletion (lane 3a1/pBK). Thus, for each experiment and for each cell culture condition, the percent readthrough level relative to the readthrough level of cells electroporated with plasmid expressing si-3a1 and re-electroporated with pBK-CMV (3a1/pBK) was calculated and expressed as the readthrough efficiency. Results were expressed as the mean of three experiments; error bars show the standard error of the mean.
We then analyzed the translation termination activity of eRF3a variants by complementation experiments in eRF3a-depleted cells (Chauvin et al. 2005). Parallel cultures of 559C cells, a cell line expressing a lacZ reporter gene interrupted by a premature stop codon, were depleted in eRF3a by electroporation of a plasmid expressing a siRNA, si-3a1, specifically targeting eRF3a mRNA. Three days after electroporation, cells were re-electroporated with the plasmids expressing wild-type and variants eRF3a. Two days later, half of the cell plates were treated with MG132 for 16 h, and the effect on stop codon readthrough was analyzed by measuring the β-galactosidase activity. Cells electroporated with the empty vector pSuper and re-electroporated with plasmid pBK-CMV served as a negative control. Cells electroporated with plasmid expressing si-3a1 and re-electroporated with plasmid pBK-CMV served as the standard for the readthrough level promoted by eRF3a depletion (Fig. 1C, lane 3a1/pBK). For each electroporation experiment, the amount of endogenous eRF3a and overexpressed proteins was verified by Western blot analysis (data not shown). As shown in Figure 1C, wild-type eRF3a alone (Fig. 1C, lane 3a1/3a) or in fusion with GFP (Fig. 1C, lane 3a1/GFP-3a) alleviates the effect of eRF3a depletion on translational readthrough with identical efficiencies. This indicated that the GFP-3a fusion protein had an activity in translation termination comparable to that of wild-type eRF3a. Moreover, MG132 treatment did not modify the translation termination activity of these proteins. Overexpression of eRF3a-FRAA, eRF3a-ΔC, and GFP-3a-FRAA (Fig. 1C, lanes 3a1/FRAA, 3a1/ΔC, and 3a1/GFP-3a-FRAA) did not modify the standard readthrough level induced by eRF3a depletion (Fig. 1C, lane 3a1/pBK), even when these proteins were stabilized by MG132 treatment. This result indicated that eRF3a-FRAA, eRF3a-ΔC, and GFP-3a-FRAA were not active in translation termination and was consistent with the fact that these forms of eRF3a had altered eRF1-binding capacities and were not able to form an active translation termination complex.
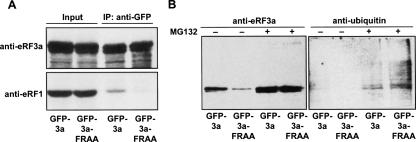
The absence of eRF1 binding to GFP-3a-FRAA was further confirmed by coimmunoprecipitation experiments. Extracts of cells expressing GFP-3a and GFP-3a-FRAA and treated with MG132 were subjected to immunoprecipitation with anti-GFP antibodies. Immunoprecipitates were analyzed by Western blotting with either anti-eRF3a or anti-eRF1 antibodies. As shown in Figure 2A, endogenous cellular eRF1 was coimmunoprecipitated with GFP-3a but not with GFP-3a-FRAA. This result confirmed that the two point mutations, F615 → A and R618 → A, introduced in the eRF3 C-terminal domain, prevented eRF1 binding to eRF3a. The moderate level of eRF1 coimmunoprecipitated with GFP-3a is likely due to the association of a part of eRF1 with endogenous eRF3a.
FIGURE 2.
Ubiquitination of GFP-3a and GFP-3a-FRAA. (A) HEK293 cells expressing GFP-3a (lanes GFP-3a) and GFP-3a-FRAA (lanes GFP-3a-FRAA) fusion proteins were treated with MG132, and proteins (200 μg) were immunoprecipitated with mouse monoclonal anti-GFP antibodies. Protein input (20 μg) and immunoprecipitates (IP:GFP) were analyzed by Western blotting using anti-eRF3a (top panel) and anti-eRF1 (bottom panel) rabbit polyclonal antibodies. (B) HEK293 cells expressing GFP-3a (lanes GFP-3a) and GFP-3a-FRAA (lanes GFP-3a-FRAA) fusion proteins were treated or not with MG132 as indicated above each lane. Cell extracts (200 μg of protein) were immunoprecipitated with mouse monoclonal anti-GFP antibodies, and immunoprecipitates were analyzed by Western blotting using anti-eRF3a (left panel) and anti-ubiquitin (right panel) antibodies.
We next examined the polyubiquitination status of GFP-3a and GFP-3a-FRAA overexpressed in cells treated or not with MG132. Polyubiquitinated forms of GFP-3a-FRAA were observed in MG132-treated cells in which proteasome inhibition enhanced polyubiquitin chain accumulation (Fig. 2B, lane GFP-3a-FRAA). As it has been shown that GFP is not a substrate for ubiquitin–proteasome-dependent proteolysis (Dantuma et al. 2000), the target for polyubiquitination was likely the eRF3a moiety of GFP-3a-FRAA. Of course, we cannot rule out the possibility that eRF3a forms containing mutations in the eRF1-binding site were misfolded and targeted to ubiquitin–proteasome-dependent proteolysis. However, in yeast, eRF3 variants harboring C-terminal truncation that included the eRF1-binding site were expressed at almost the same level as that of wild-type eRF3, suggesting that these truncated proteins did not have increased sensitivity to proteolysis (Chabelskaya et al. 2004). Moreover, we also observed polyubiquitinated forms of GFP-3a in MG132-treated cells, although at a lower level than that of GFP-3a-FRAA (Fig. 2B, lane GFP-3a). The presence of polyubiquitinated forms of GFP-3a suggested that a fraction of GFP-3a was also targeted to the proteasome degradation pathway. Interestingly, these observations not only confirmed that eRF3a forms mutated in eRF1-binding site were degraded by the proteasome but also indicated that a fraction of wild-type eRF3a underwent the same degradation process.
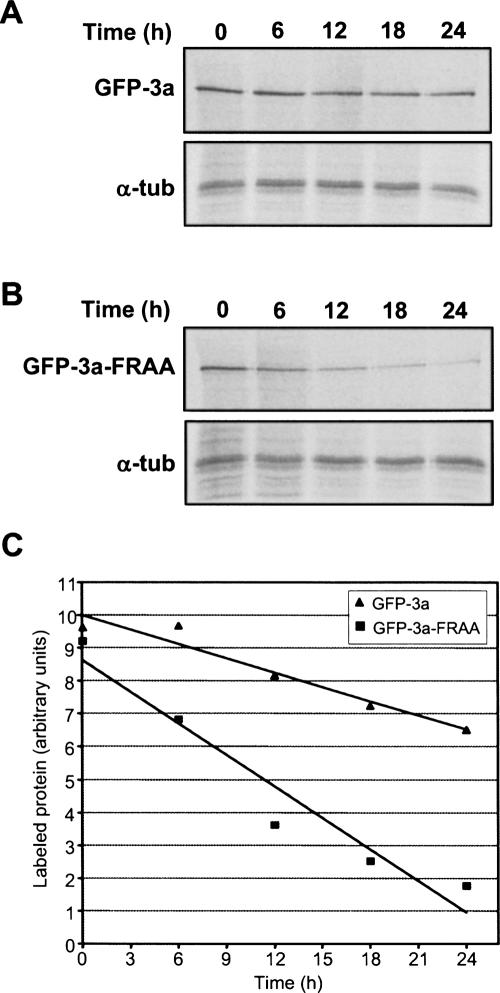
We then measured the half-life of GFP-3a and GFP-3a-FRAA overexpressed in HEK293 cells by pulse-chase 35S-labeling of cellular proteins. Samples collected at 6-h time points were analyzed by immunoprecipitation with anti-GFP and anti-α-tubulin antibodies (Fig. 3). First, we observed that, after the 2-h labeling period, the amount of GFP-3a-FRAA was almost identical to that of GFP-3a, suggesting that the two proteins were translated with the same efficiency (Fig. 3, time 0). Second, there was a significant difference in the decrease of GFP-3a-FRAA and GFP-3a as a function of time. The half-lives calculated from the linear regression lines (Fig. 3C) were ∼14 h for GFP-3a-FRAA and ∼34 h for GFP-3a. Wild-type GFP was found to be stable in mammalian cells with a half-life of 26 h but can be destabilized by the fusion with proteins with shorter half-lives (Corish and Tyler-Smith 1999; Kant et al. 2006). Furthermore, it has been shown that, in HEK293 cells, the stability of ERK3 protein that is degraded by the proteasome was independent of the presence of the N-terminal GFP tag (Kant et al. 2006). Thus, the half-lives calculated for GFP-3a and GFP-3a-FRAA are probably closely related to the half-lives of wild-type eRF3a and eRF3a-FRAA, respectively.
FIGURE 3.
Pulse-chase labeling of HEK293 cells expressing (A) GFP-3a and (B) GFP-3a-FRAA. Labeled proteins were immunoprecipitated with anti-GFP (top panel) and anti-α-tubulin antibodies (bottom panel). (C) [35S]-Labeled GFP-3a and GFP-3a-FRAA bands were quantified using the BAS1000 Fuji image plate program V 2.0, and the value at each time point was plotted against time: (filled triangles) GFP-3A, (filled squares) GFP-3a-FRAA. The regression line is presented for each curve.
DISCUSSION
As a starting point for these experiments, there is the surprising observation that eRF1 abundance strongly correlates to that of eRF3a in a wide range of human cell lines (Chauvin et al. 2005). This raises the possibility that the relative abundances of eRF1 and eRF3a are tightly adjusted by a mechanism that controls the formation of the translation termination complex and the efficiency of termination. The first clue was provided by the fact that eRF3a depletion induces a decrease in eRF1 stability in human cells, suggesting that eRF1 is proteolytically degraded when not complexed with eRF3a (Chauvin et al. 2005). To go further in the understanding of this regulation, we examined the status of eRF3a when not associated with eRF1. Using eRF3a variants with altered capacities in eRF1 binding, we clearly show that, symmetrically to what was observed for eRF1, eRF3a is proteolytically degraded when not associated with eRF1. However, we show that eRF3a degradation involves the ubiquitin–proteasome-degradation pathway, whereas, in contrast, eRF1 degradation in eRF3a-depleted cells is not sensitive to MG132 proteasome inhibitor and probably not dependent on proteasome (data not shown).
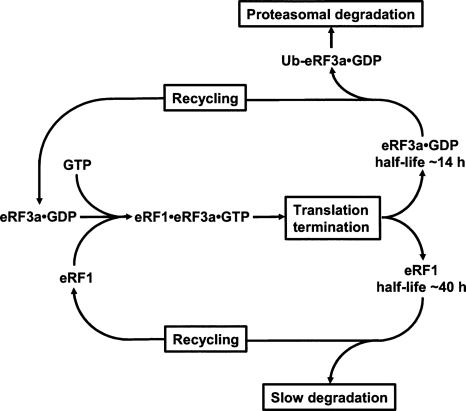
Protein degradation may constitute a primary mechanism by which cells regulate translation termination complex formation (Fig. 4). Taking the hypothesis that the half-life of ∼14 h calculated for the GFP-3a-FRAA fusion protein (Fig. 3) roughly corresponds to that of eRF3a when not associated with eRF1 and according to the results of kinetic studies indicating that in cells eRF3 is in the GDP-bound form when not associated with eRF1 (Hauryliuk et al. 2006; Pisareva et al. 2006), eRF3a•GDP is likely to be the substrate for polyubiquitination and proteasome degradation. Our results also suggest that after the dissociation of eRF3a•GDP from the post-termination complex, the degradation of eRF3a is approximately three times faster than the degradation of eRF1 alone, which half-life is ∼40 h (Chauvin et al. 2005). In addition, the long half-life of GFP-3a fusion protein (∼34 h), which likely corresponds to the global half-life of eRF3a (alone and associated with eRF1), tends to confirm the view of a long-lived eRF1•eRF3•GTP ternary complex deduced from kinetic studies (Pisareva et al. 2006). Taken together, these results explain the strong correlation between the abundances of eRF1 and eRF3a observed in human cell lines.
FIGURE 4.
A model for the regulation of the translation termination complex formation (see description in the Discussion).
However, the relatively long half-life of either release factor, eRF1 and eRF3a, suggests that the formation of the translation termination complex mainly depends on the recycling of the release factors and that renewal and proteasomal degradation of eRF3a allow cells to adjust the amount of eRF3a needed for efficient translation termination to the variations of the global translation rate. When cells move from proliferation to the resting state, the decrease of translation rate will decrease the requirement for translation termination factors, and free eRF3a will be first degraded by the proteasome, followed by a slow decrease in eRF1 level. Conversely, in cells undergoing transition from rest to proliferation, the need for eRF3a increases with the rate of protein synthesis. Given that the eRF3a/GSPT1 gene is expressed in a proliferation-dependent manner in mammalian cells (Hoshino et al. 1989), we can speculate that newly synthesized eRF3a mRNAs will provide for eRF3a renewal.
MATERIALS AND METHODS
Plasmids
Plasmids expressing small interfering RNA si-3a1 targeting eRF3a mRNA and plasmid pCMV-heRF3a expressing the entire human eRF3a have been previously described (Chauvin et al. 2005). Plasmid pCMV-heRF3a-FRAA carrying the double mutation F615 → A and R618 → A in the GRFTLRD motif, and plasmid pCMV-heRF3a-ΔC, in which the last 31 codons of the eRF3a open reading frame (ORF) were deleted, were obtained using the ExSite PCR-based site-directed mutagenesis kit (Stratagene), plasmid pCMV-heRF3a as template, and the appropriate oligonucleotides. Plasmid pGFP-heRF3a, in which the human eRF3a open reading frame (ORF) was fused to the 3′ end of green fluorescent protein ORF, was obtained by ligating the BamHI-Klenow filled-PstI short fragment of pCMV-heRF3a in EcoRI-Klenow filled-PstI fragment of pEGFP-C1 vector (Clontech). The BamHI site of the eRF3a coding sequence is located immediately downstream from the first initiator ATG codon of the eRF3a ORF so that the GFP-eRF3a fusion ORF contains the entire eRF3a coding sequence. An identical strategy was used for pGFP-heRF3a-FRAA construction.
Cell culture and electroporation
The 559C cell line is a derivative of the human HEK293 cell line (ATCC No. CRL-1573) stably expressing a lacZ gene in which the coding sequence is interrupted by a TAG stop codon (Chauvin et al. 2005). Cell cultures and electroporation was performed as previously described (Chauvin et al. 2005). For MG132 treatment, MG132 (Sigma) diluted in DMSO was added to cell culture medium at 10 μM final concentration for 16 h. Control cells were treated with an equivalent amount of DMSO.
Antibodies, Western blot analysis, and readthrough assay
The antibodies against human eRF3a and eRF1 were previously described (Chauvin et al. 2005). The mouse monoclonal anti-GFP and anti-ubiquitin antibodies were from Santa Cruz Biotechnologies; the rabbit polyclonal anti-GFP antibody was from Clontech; and the anti-α-tubulin (DM1A) and anti-rabbit IgG and anti-mouse IgG peroxydase-linked antibodies were from Amersham Biosciences. Western blotting and readthrough assays were performed as previously described (Chauvin et al. 2005).
Cell protein labeling and immunoprecipitation
Pulse-chase labeling of cellular proteins and immunoprecipitation were previously described (Chauvin et al. 2005) except that [35S]-labeled GFP-eRF3a fusion proteins were immunoprecipitated with 5 μL of rabbit polyclonal anti-GFP antibodies.
For coimmunoprecipitation of eRF1, 2 d after electroporation, cells were collected by scraping and pelleted. Cell pellets were resuspended in 250 μL of co-IP buffer (10 mM sodium phosphate buffer at pH 7.5, 100 mM NaCl, 5 mM MgCl2, 1 mM GTP, 0.01% Triton X-100), lysed by sonication in the presence of protease inhibitor cocktail (Roche), and centrifuged at 12,000g for 30 min at 4°C. An aliquot fraction of the resulting supernatants (∼200 μg of proteins) was incubated with mouse monoclonal anti-GFP antibodies fixed on protein G-Sepharose beads in 350 μL of co-IP buffer for 2 h at 4°C. Immunoprecipitates were washed with co-IP buffer and then Western blotted with indicated antibodies. For the detection of ubiquitinylated forms of eRF3a, cell pellets were lysed by sonication in 250 μL of RIPA buffer (10 mM sodium phosphate buffer at pH 7.5, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) containing 5 mM N-ethyl maleimide (NEM) to block deubiquitinating enzymes and protease inhibitor cocktail (Roche) and centrifuged at 12,000g for 30 min at 4°C. An aliquot fraction of the resulting supernatants (∼200 μg of proteins) was incubated with anti-GFP monoclonal antibodies fixed on protein G-Sepharose beads in 350 μL of RIPA buffer containing 5 mM NEM for 2 h. Immunoprecipitates were washed four times with RIPA buffer, boiled in SDS loading buffer, and then Western blotted with indicated antibodies.
ACKNOWLEDGMENTS
This work was supported by the Association pour la Recherche sur le Cancer (grant no. 3784). C.C. held fellowships from the Fondation pour la Recherche Médicale and from the Société Française du Cancer.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.728608.
REFERENCES
- Alkalaeva, E.Z., Pisarev, A.V., Frolova, L.Y., Kisselev, L.L., Pestova, T.V. In vitro reconstitution of eukaryotic translation reveals cooperativity between release factors eRF1 and eRF3. Cell. 2006;125:1125–1136. doi: 10.1016/j.cell.2006.04.035. [DOI] [PubMed] [Google Scholar]
- Chabelskaya, S., Kiktev, D., Inge-Vechtomov, S., Philippe, M., Zhouravleva, G. Nonsense mutations in the essential gene SUP35 of Saccharomyces cerevisiae are nonlethal. Mol. Genet. Genomics. 2004;272:297–307. doi: 10.1007/s00438-004-1053-1. [DOI] [PubMed] [Google Scholar]
- Chauvin, C., Salhi, S., Le Goff, C., Viranaicken, W., Diop, D., Jean-Jean, O. Involvement of human release factors eRF3a and eRF3b in translation termination and regulation of the termination complex formation. Mol. Cell. Biol. 2005;25:5801–5811. doi: 10.1128/MCB.25.14.5801-5811.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corish, P., Tyler-Smith, C. Attenuation of green fluorescent protein half-life in mammalian cells. Protein Eng. 1999;12:1035–1040. doi: 10.1093/protein/12.12.1035. [DOI] [PubMed] [Google Scholar]
- Dantuma, N.P., Lindsten, K., Glas, R., Jellne, M., Masucci, M.G. Short-lived green fluorescent proteins for quantifying ubiquitin/proteasome-dependent proteolysis in living cells. Nat. Biotechnol. 2000;18:538–543. doi: 10.1038/75406. [DOI] [PubMed] [Google Scholar]
- Ebihara, K., Nakamura, Y. C-terminal interaction of translational release factors eRF1 and eRF3 of fission yeast: G-domain uncoupled binding and the role of conserved amino acids. RNA. 1999;5:739–750. doi: 10.1017/s135583829998216x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolova, L., Le Goff, X., Zhouravleva, G., Davydova, E., Philippe, M., Kisselev, L. Eukaryotic polypeptide chain release factor eRF3 is an eRF1- and ribosome-dependent guanosine triphosphatase. RNA. 1996;2:334–341. [PMC free article] [PubMed] [Google Scholar]
- Hauryliuk, V., Zavialov, A., Kisselev, L., Ehrenberg, M. Class-1 release factor eRF1 promotes GTP binding by class-2 release factor eRF3. Biochimie. 2006;88:747–757. doi: 10.1016/j.biochi.2006.06.001. [DOI] [PubMed] [Google Scholar]
- Hoshino, S., Miyazawa, H., Enomoto, T., Hanaoka, F., Kikuchi, Y., Kikuchi, A., Ui, M. A human homolog of the yeast GST1 gene codes for a GTP-binding protein and is expressed in a proliferation-dependent manner in mammalian cells. EMBO J. 1989;8:3807–3814. doi: 10.1002/j.1460-2075.1989.tb08558.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, K., Ebihara, K., Nakamura, Y. The stretch of C-terminal acidic amino acids of translational release factor eRF1 is a primary binding site for eRF3 of fission yeast. RNA. 1998;4:958–972. doi: 10.1017/s1355838298971874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant, S., Schumacher, S., Singh, M.K., Kispert, A., Kotlyarov, A., Gaestel, M. Characterization of the atypical MAPK ERK4 and its activation of the MAPK-activated protein kinase MK5. J. Biol. Chem. 2006;281:35511–35519. doi: 10.1074/jbc.M606693200. [DOI] [PubMed] [Google Scholar]
- Kisselev, L., Ehrenberg, M., Frolova, L. Termination of translation: Interplay of mRNA, rRNAs and release factors? EMBO J. 2003;22:175–182. doi: 10.1093/emboj/cdg017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, C., Ito, K., Walsh, M.A., Wada, M., Liu, Y., Kumar, S., Barford, D., Nakamura, Y., Song, H. Crystal structure and functional analysis of the eukaryotic class II release factor eRF3 from S. pombe . Mol. Cell. 2004;14:233–245. doi: 10.1016/s1097-2765(04)00206-0. [DOI] [PubMed] [Google Scholar]
- Merkulova, T.I., Frolova, L.Y., Lazar, M., Camonis, J., Kisselev, L.L. C-terminal domains of human translation termination factors eRF1 and eRF3 mediate their in vivo interaction. FEBS Lett. 1999;443:41–47. doi: 10.1016/s0014-5793(98)01669-x. [DOI] [PubMed] [Google Scholar]
- Pisareva, V.P., Pisarev, A.V., Hellen, C.U., Rodnina, M.V., Pestova, T.V. Kinetic analysis of interaction of eukaryotic release factor 3 with guanine nucleotides. J. Biol. Chem. 2006;281:40224–40235. doi: 10.1074/jbc.M607461200. [DOI] [PubMed] [Google Scholar]
- Salas-Marco, J., Bedwell, D.M. GTP hydrolysis by eRF3 facilitates stop codon decoding during eukaryotic translation termination. Mol. Cell. Biol. 2004;24:7769–7778. doi: 10.1128/MCB.24.17.7769-7778.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valouev, I.A., Kushnirov, V.V., Ter-Avanesyan, M.D. Yeast polypeptide chain release factors eRF1 and eRF3 are involved in cytoskeleton organization and cell cycle regulation. Cell Motil. Cytoskeleton. 2002;52:161–173. doi: 10.1002/cm.10040. [DOI] [PubMed] [Google Scholar]