Abstract
Escherichia coli contains five members of the DEAD-box RNA helicase family, a ubiquitous class of proteins characterized by their ability to unwind RNA duplexes. Although four of these proteins have been implicated in RNA turnover or ribosome biogenesis, no cellular function for the RhlE DEAD-box protein has been described as yet. During an analysis of the cold-sensitive growth defect of a strain lacking the DeaD/CsdA RNA helicase, rhlE plasmids were identified from a chromosomal library as multicopy suppressors of the growth defect. Remarkably, when tested for allele specificity, RhlE overproduction was found to exacerbate the cold-sensitive growth defect of a strain that lacks the SrmB RNA helicase. Moreover, the absence of RhlE exacerbated or alleviated the cold-sensitive defect of deaD or srmB strains, respectively. Primer extension and ribosome analysis indicated that RhlE regulates the accumulation of immature ribosomal RNA or ribosome precursors when deaD or srmB strains are grown at low temperatures. By using an epitope-tagged version of RhlE, the majority of RhlE in cell extracts was found to cosediment with ribosome-containing fractions. Since both DeaD and SrmB have been recently shown to function in ribosome assembly, these findings suggests that rhlE genetically interacts with srmB and deaD to modulate their function during ribosome maturation. On the basis of the available evidence, I propose that RhlE is a novel ribosome assembly factor, which plays a role in the interconversion of ribosomal RNA-folding intermediates that are further processed by DeaD or SrmB during ribosome maturation.
Keywords: RNA helicase, ribosome, Escherichia coli, DEAD-box protein, ribosome assembly
INTRODUCTION
One of the most important cellular activities is translation, a series of steps that encompasses the transfer of genetic information from messenger RNA to yield the corresponding polypeptide or protein. Protein synthesis is carried out inside the ribosome, a macromolecular complex composed of several dozen protein and several RNA constituents. Despite its complexity, the formation of ribosomes in the cell is an efficiently regulated process, as might be expected based upon the requirements of actively growing cells to mediate high levels of protein synthesis. The reconstitution of prokaryotic ribosomes from its individual constituents has been successfully demonstrated, but it requires multiple treatment steps and much longer times as compared with the rates with which they are assembled in vivo (Nomura et al. 1984; Nierhaus 1991).
Apart from the ribosomal components, a number of cellular factors have been implicated in ribosome assembly. These factors are believed to be responsible for promoting the fidelity and speed of ribosome formation in vivo. Studies in Saccharomyces cerevisiae have suggested that nearly 200 proteins function as ribosome assembly factors (Venema and Tollervey 1999; Harnpicharnchai et al. 2001; Grandi et al. 2002; Saveanu et al. 2003). In contrast, only 10–15 assembly factors have been identified in E. coli (Alix and Guerin 1993; Bylund et al. 1998; El Hage et al. 2001; Charollais et al. 2003, 2004; Inoue et al. 2003; Gutgsell et al. 2005; Bharat et al. 2006; Hwang and Inouye 2006; Jiang et al. 2006), suggesting either a lower degree of ribosomal complexity in prokaryotes, or that additional ribosome assembly factors remain to be discovered (Fromont-Racine et al. 2003; Hage and Tollervey 2004).
Among the known ribosome assembly factors, a prominent class belongs to the DEAD-box family of proteins. DEAD-box proteins are characterized by the presence of nine characteristic motifs, including the eponymous amino acid sequence D-E-A-D. Many of these proteins possess RNA helicase activity, the ability to dissociate RNA duplexes using nucleotide triphosphates as a source of energy, as well as other functions such as RNA chaperone, RNA annealing, or RNA–protein reorganization ability (Rocak and Linder 2004; Cordin et al. 2006; Iost and Dreyfus 2006; Linder 2006). At least 14 of the 26 DEAD-box proteins in S. cerevisiae are classified as ribosome assembly factors, suggesting that the biogenesis of ribosomes in this organism involves numerous structural reorganization steps that could include the dissociation of base-pairing interactions (de la Cruz et al. 1999). However, the precise molecular details regarding helicase function in ribosome biogenesis have yet to be delineated.
The prokaryotic organism, E. coli, contains five DEAD-box proteins, each of which functions as a bona fide RNA helicase in vitro (Kalman et al. 1991; Iost and Dreyfus 2006). Two of these proteins, DeaD and SrmB, are implicated in ribosome assembly, since each factor associates with ribosomal subunits, and the absence of either protein causes ribosome biogenesis defects (Charollais et al. 2003, 2004). Both factors were initially identified as multicopy suppressors of ribosomal protein mutants (Nishi et al. 1988; Toone et al. 1991). Among the remaining three DEAD-box proteins, one, DbpA has been shown biochemically to interact with 23S ribosomal RNA (rRNA), the major RNA component of large ribosomal subunit (Fuller-Pace et al. 1993). In particular, the ATPase and helicase activities of DbpA are specifically stimulated by interaction with helix 92 of 23S rRNA (Fuller-Pace et al. 1993; Diges and Uhlenbeck 2001; Tsu et al. 2001). A second protein, RhlB, appears to play a specialized role in RNA turnover, as it is a major component of the “degradosome,” an RNA degrading complex involved in RNA turnover and processing reactions (Py et al. 1996; Coburn et al. 1999). The third protein, RhlE, is arguably the least well understood of the E. coli DEAD-box proteins. Biochemically, RhlE functions as an ATPase and a RNA helicase, and limited studies on this protein have indicated that it can associate with RNA degradation and modification proteins (Raynal and Carpousis 1999; Bizebard et al. 2004; Khemici et al. 2004). However, no function for this enzyme has been described so far.
A common property of strains defective in ribosome or ribosome assembly factors is cold-sensitive growth. Among the E. coli DEAD-box proteins, strains lacking DeaD or SrmB display such a phenotype (Iost and Dreyfus 2006). To further investigate the functions of these proteins, a genetic screen was performed to identify multicopy suppressors of the cold-sensitive growth defect of a deaD strain. Interestingly, rhlE was identified as a multicopy suppressor of this defect. Additional experiments indicated that rhlE also interacts with srmB genetically, but these interactions have opposite effects on growth and ribosome maturation as compared with its interactions with deaD. By fractionation of cell extracts on a sucrose density gradient, RhlE was found to be primarily associated with ribosomes. On the basis of these observations, I propose that RhlE is a novel ribosome assembly factor that regulates DeaD and SrmB function during ribosome biogenesis.
RESULTS
Suppression of the deaD cold-sensitive growth defect by rhlE overexpression
Many DEAD-box proteins are known to be important for cellular function at low temperatures—conditions under which RNA can become trapped in incorrect conformations and could require helicase function to resolve misfolding (Noble and Guthrie 1996; Mohr et al. 2002). Compatible with this viewpoint, the absence of two E. coli DEAD-box proteins, DeaD and SrmB, causes progressive growth defects at reduced growth temperatures (Charollais et al. 2003, 2004). Furthermore, ribosomes extracted from srmB and deaD strains grown at low temperatures display a number of defects, including the accumulation of 40S particles that lack a subset of the proteins found in the 50S large ribosomal subunit and incompletely processed 23S ribosomal RNA (rRNA) (Charollais et al. 2003, 2004). Presumably, these defects are a consequence of misfolded regions within the rRNA that are normally corrected by DeaD or SrmB. On this basis, both DeaD and SrmB have been categorized as ribosome assembly factors. However, the molecular basis by which they promote ribosome assembly is not known.
One question I wanted to address is whether the low-temperature growth defect of deaD or srmB strains could be alleviated by extragenic suppressors. The identification of such suppressors can provide valuable insights, especially if they restore an important function carried out by the absent protein. Initial experiments with mutagenized deaD or srmB strains plated at low temperatures indicated that genomic suppressors can be readily identified (data not shown). However, as the identification of the responsible chromosomal mutation(s) can be laborious, further characterization was not pursued. Instead, I decided to focus on a search for plasmid-borne multicopy suppressors, which should function via gene overexpression and be easier to characterize.
To identify such suppressors, a multicopy E. coli chromosomal library was transformed into a strain deleted for deaD (CJ1825ΔdeaD) and plated on LB-agar plates supplemented with chloramphenicol. The library contains random segments of chromosomal DNA inserted into a pACYC184-based plasmid, which increases the copy number of cloned DNA 20-fold (Jain and Belasco 1995; Sarkar et al. 2002). The plates were incubated for several days at 20°C, until small colonies could be visualized. Among the background of small colonies, a few larger colonies, which could harbor potential multicopy growth suppressors, were identified. Plasmid DNA was isolated from such colonies and retransformed into CJ1825ΔdeaD to confirm a plasmid-specific suppressor phenotype. One set of suppressors that alleviated the CJ1825ΔdeaD low-temperature growth defect contained the gene for the RhlE RNA helicase (Fig. 1A). Sequencing of two such plasmids indicated that each contained the entire rhlE coding region, as well as 30 or 355 base pairs (bp) of sequences upstream of the coding region and 1106 or 470 bp of downstream sequences, respectively. Although the expression signals for rhlE have not been defined, the latter plasmid (pCJ1078), which is more likely to contain an intact promoter and ribosome binding site, was used for the growth experiments described below. In addition, based on restriction polymorphism or partial sequence analysis, four other rhlE clones were identified, but these were not characterized further.
FIGURE 1.
Effect of RhlE overexpression on the cold-sensitive growth of ΔdeaD and ΔsrmB strains. (A) Suppression of the ΔdeaD cold-sensitive growth defect by RhlE overexpression. A control plasmid (pACYC184) or a RhlE plasmid (pCJ1078) were transformed into MG1655ΔdeaD. The transformed strains were streaked on a LB-agar-chloramphenicol plate, followed by incubation at 20°C. (B) MG1655 or the indicated derivative strains were transformed with pACYC184 or pCJ1078 and grown in LB supplemented with chloramphenicol at 16°C or 20°C. The strain doubling time was derived from measurements of cell culture density versus time for three to four independent cultures. The mean cell doubling time and standard deviation are indicated. (n.d.) Not determined. (C) RhlE overexpression exacerbates the ΔsrmB cold-sensitive growth defect. MG1655ΔsrmB was transformed with either pACYC184 or pCJ1078, streaked on a LB-agar-chloramphenicol plate, and grown at 16°C.
Growth rate measurements of strains, transformed with either a control plasmid or pCJ1078, were performed to corroborate the plate phenotypes (Fig. 1B). Whereas the rhlE plasmid had a minimal or a slightly detrimental effect on growth of a wild-type strain, it decreased the cell-doubling time for a ΔdeaD strain by 42%. As no physiological role for RhlE has been defined, additional studies to characterize RhlE function were pursued.
rhlE overexpression exacerbates the srmB cold-sensitive growth defect
To further characterize the effect of rhlE overproduction, the RhlE plasmid was transformed into MG1655 or MG1655ΔsrmB, and the transformed cells were plated at 16°C. Remarkably, the RhlE plasmid had a deleterious effect on growth of MG1655ΔsrmB (Fig. 1C). Additional growth rate measurements in liquid medium at 16°C corroborated the phenotype seen on plates (Fig. 1B). Thus, the RhlE plasmid, when transformed into MG1655ΔsrmB, dramatically increased the culture doubling time by over threefold. In contrast, transformation of this plasmid into MG1655 increased the doubling time slightly, which might reflect a nonspecific consequence of increased RhlE production. Thus, the detrimental expression of RhlE overexpression appears to be specific to the ΔsrmB strain. In summary, RhlE overexpression has opposite effects on the growth of ΔdeaD and ΔsrmB strains at low temperatures.
Growth of ΔdeaDΔrhlE and ΔsrmBΔrhlE strains at low temperatures
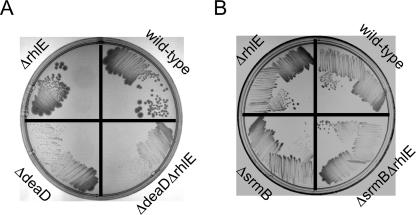
If the effects of RhlE overexpression on the growth of deaD or srmB strains are physiologically relevant, one prediction would be that deletion of rhlE should confer opposite effects on the growth of ΔdeaD and ΔsrmB strains as compared with the effects of RhlE overexpression. To determine if that is the case, double helicase-deficient strains MG1655ΔdeaDΔrhlE or MG1655ΔsrmBΔrhlE were streaked on LB-agar plates, along with the corresponding single-mutant and the wild-type strains, at 16°C or 20°C, respectively (Fig. 2). At 20°C, the ΔrhlEΔdeaD strain grew more slowly as compared with the isogenic ΔdeaD strain (Fig. 2A). This phenotype is the opposite of the enhanced growth of a ΔdeaD strain that harbors an RhlE plasmid (Fig. 1A). Similarly, a ΔsrmBΔrhlE strain grew better than the ΔsrmB strain at 16°C (Fig. 2B), which contrasts with the poorer growth of a ΔsrmB strain that contained an RhlE plasmid (Fig. 1C). These observations indicate that the phenotypes observed with RhlE overexpression are not a nonspecific consequence of gene overexpression, but instead reflect genuine functional interactions between these sets of helicase-encoding genes.
FIGURE 2.
Growth of wild-type and helicase-deficient strains at low temperatures. (A) Strain MG1655 (wild type) and isogenic derivatives containing ΔrhlE, ΔdeaD, or ΔdeaDΔrhlE mutations were streaked on LB-agar plates and incubated at 20°C. (B) MG1655 and isogenic derivatives containing ΔrhlE, ΔsrmB, or ΔsrmBΔrhlE deletions were streaked on LB-agar plates and incubated at 16°C.
Characterization of rRNA processing in helicase-deficient strains
One characteristic ribosomal defect of deaD or srmB strains is the accumulation of 23S rRNA precursors. The mature rRNA is generated by initial cleavage of the ribosomal operon RNA by RNase III, followed by additional processing events at the 5′ and 3′ ends to generate the final product (Deutscher 2006). In deaD and srmB strains, however, elevated levels of the 23S RNase III cleaved precursor are observed (Charollais et al. 2003, 2004). Such precursors contain three to seven extra nucleotides at the 5′ end and seven to nine extra nucleotides at the 3′ end.
To evaluate whether the effect of rhlE deletion on the growth of deaD or srmB strains correlates with 23S rRNA processing efficacy, RNA was analyzed from wild-type and mutant strains. First, RNA was extracted from ΔsrmB or ΔsrmBΔrhlE strains grown at 16°C, or from wild-type and ΔrhlE strains as controls, and analyzed by primer extension (Fig. 3A). The extent of 23S rRNA 5′ end processing was evaluated by quantifying products that correspond to the mature rRNA end and to a prominent precursor that contains a 7-nucleotide (nt) extension. This analysis indicated that the ratio of unprocessed rRNA to mature RNA in a wild-type strain was 0.56. The relatively high levels of unprocessed rRNA under this condition are primarily due to inefficient processing at low growth temperatures (data not shown). A similar level of precursor was observed in the ΔrhlE strain, indicating that RhlE by itself does not influence 23S rRNA processing. In the ΔsrmB strain, a much higher precursor to mature ratio of 2.5 was observed, indicating a processing defect, which is consistent with the known role of SrmB in ribosome maturation (Charollais et al. 2003). However, the corresponding ratio in the ΔsrmBΔrhlE double-mutant strain was reduced to 0.84. Therefore, the introduction of a ΔrhlE deletion into a ΔsrmB strain background reduces the rRNA processing defect, which also correlates with restored growth (Fig. 2B).
FIGURE 3.
23S rRNA processing defects in helicase-deficient strains. RNA was extracted from the indicated strains, annealed with a labeled oligonucleotide probe (5′-CCTTCATCGCCTCTGACTGCC-3′) that hybridizes to 23S rRNA, reverse transcribed, and fractionated on a denaturing gel. The reverse transcription products that correspond to the mature 5′ end (M), and to a precursor containing seven additional nucleotides (P), are indicated. An intermediate product corresponding to the +3 product can also be visualized. The assignment of the precursor and mature products are based on additional experiments (data not shown). The ratios of the +7 precursor to mature RNA were quantified by phosphorimaging. (A) RNA was analyzed from isogenic MG1655 derivatives grown at 16°C: lane 1, wild type; lane 2, ΔrhlE; lane 3, ΔsrmB; lane 4, ΔsrmBΔrhlE. (B) RNA was analyzed from MG1655 derivatives grown at 22°C: lane 1, wild type; lane 2, ΔrhlE; lane 3, ΔdeaD; lane 4, ΔdeaDΔrhlE.
Similarly, RNA was extracted from wild-type, ΔrhlE, ΔdeaD, or ΔdeaDΔrhlE strains grown at 22°C, and the extent of 23S rRNA processing was determined in each case (Fig. 3B). As expected from their growth phenotypes (Fig. 2A), low levels of unprocessed rRNA were found to accumulate in wild-type and ΔrhlE strains, and substantially higher levels in the ΔdeaD strain. Surprisingly, the levels of unprocessed rRNA in the ΔdeaDΔrhlE strain, as compared with the ΔdeaD strain, were not significantly different. Although these negative results could indicate that RhlE has no effect on rRNA processing in this context, it was possible that defects could be subtler and might require a more sensitive analysis. Therefore, a direct visualization of ribosomal profiles by ultracentrifugation was undertaken.
Ribosome profiles in helicase-deficient strains
To investigate possible differences between ribosomes in ΔdeaD and ΔdeaDΔrhlE strains, ribosomal profiles were generated by sucrose density gradient ultracentrifugation of extracts derived from cells grown at 22°C. Peaks corresponding to 70S ribosomes and the 30S small subunit were observed in each case (Fig. 4A). However, the 50S peak was substantially altered in comparison with the profile observed using a wild-type strain (data not shown), and a significant accumulation of particles at ∼40S was observed. Such particles correspond to precursors of the 50S large ribosomal subunit, and their presence is diagnostic of ribosome biogenesis defects (Charollais et al. 2003, 2004). In the ΔdeaD strain, the 40S particles and 50S subunits were present in nearly equal amounts (Fig. 4A, left panel), but increased amounts of 40S particles were observed in the ΔdeaDΔrhlE strain (Fig. 4A, right panel). To provide a semiquantitative measure of the ribosome assembly defect, the peak height of 40S particles was compared with the height of the 70S ribosomes. In the ΔdeaD strain, this ratio was 0.28, whereas in the ΔdeaDΔrhlE strain the 40S/70S height ratio was increased to 0.51. Therefore, the poorer growth of the double-mutant strain correlates with higher levels of 40S ribosomal particles, suggesting that the absence of RhlE negatively influences ribosome biogenesis in a ΔdeaD strain background.
FIGURE 4.
Ribosome profiles of mutant strains and RNA analysis. Cell lysates were fractionated by ultracentrifugation on a 14%–32% sucrose density gradient and ribosomal profiles were generated. (A) Ribosomal profiles derived from MG1655ΔdeaD (left) or MG1655ΔdeaDΔrhlE (right) grown at 22°C. Sedimentation is from right to left. The positions of the 70S ribosome, the 50S and 30S subunits, and a 40S precursor that accumulates in these strains are shown. The designation of these peaks is based on parallel ultracentrifugation studies on a wild-type strain in which 30S, 50S, and 70S peaks were well separated (data not shown). The numbers below the sedimentation values correspond to 12 fractions, starting at the 70S ribosomal peak and ending at the 30S subunit peak, which were collected for RNA analysis (see below). (B) RNA was extracted from the fractions underlined in A and analyzed by primer extension using a 23S rRNA probe (Materials and Methods). The migration of the mature (M) and two prominent precursors (+3) and (+7) are indicated. The abundance of the mature and precursor products were quantified by phosphorimaging, and the percentage of RNA that contains a mature end is denoted. (C) Ribosomal profiles derived from MG1655ΔsrmB (left) and MG1655ΔsrmBΔrhlE strains (right) grown at 16°C and analyzed as described in A.
Because primer extension analysis of 23S rRNA using total RNA preparations from ΔdeaD and ΔdeaDΔrhlE strains showed no significant differences (Fig. 3B), a more detailed analysis was carried out using RNA purified from selected ribosomal fractions (Fig. 4A). Thus, RNA from four fractions (numbers 7–10), which sediment between 40S and 50S, were purified and analyzed by primer extension (Fig. 4B). As a control, RNA from the 70S peak (fraction number 1) was also analyzed. In each case, the 40S–50S fractions contained a substantial amount of unprocessed 23S RNA, with only 2%–13% of the RNA containing a mature 5′ end. Most of the immature RNA contained 3–7 additional nucleotides at the 5′ end. As expected, the percentage of immature RNA was lower in fractions that contained the 50S subunit and higher in fractions that contained 40S particles. However, higher levels of mature RNA were observed in the ΔdeaD strain as compared with analogous fractions from the ΔdeaDΔrhlE strain. For example, in fraction number 7, 13% of the 23S rRNA was mature in the ΔdeaD strain, whereas the corresponding number for the ΔdeaDΔrhlE strain was 8%. Similarly, in fraction number 10, 4% of the 23S rRNA derived from the ΔdeaD strain and 2% of the 23S rRNA from ΔdeaDΔrhlE strain was mature. In contrast, RNA from 70S ribosomes contained similar ratios of mature to immature RNA in each strain, which is consistent with the observation that final 23S rRNA maturation occurs only when 30S and 50S subunits associate to form 70S ribosomes (Srivastava and Schlessinger 1988). Overall, the primer extension analysis revealed differences in 23S rRNA processing within 40S–50S particles that correlate with the growth characteristics of ΔdeaD and ΔdeaDΔrhlE strains.
In addition, ribosomal profiles were generated from extracts derived from ΔsrmB or ΔsrmBΔrhlE strains grown at 16°C (Fig. 4C). In the ΔsrmB strain (Fig. 4C, left panel), there were higher levels of 40S particles as compared with 50S subunits. In contrast, in the ΔsrmBΔrhlE strain, the amounts of 40S particles were significantly reduced and were equivalent to the levels of the 50S subunits (Fig. 4C, right panel). The lower levels of 40S particles in the latter case is consistent with a reduction in the amount of unprocessed rRNA and improved growth in this strain (Figs. 2B, 3A).
RhlE is a ribosome-associated factor
The ability of rhlE function to regulate ribosome biogenesis suggests that RhlE may function via direct associations with the ribosome. To test this possibility, a plasmid encoding a Flag-tag epitope appended to the RhlE C terminus was constructed and transformed into E. coli. The transformed cells were grown in LB medium, and cell extracts were used for sucrose density gradient ultracentrifugation and fraction collection (Fig. 5). The fractions were analyzed by Western blot using antibodies to the Flag-tag to investigate interactions between tagged RhlE and the ribosome.
FIGURE 5.
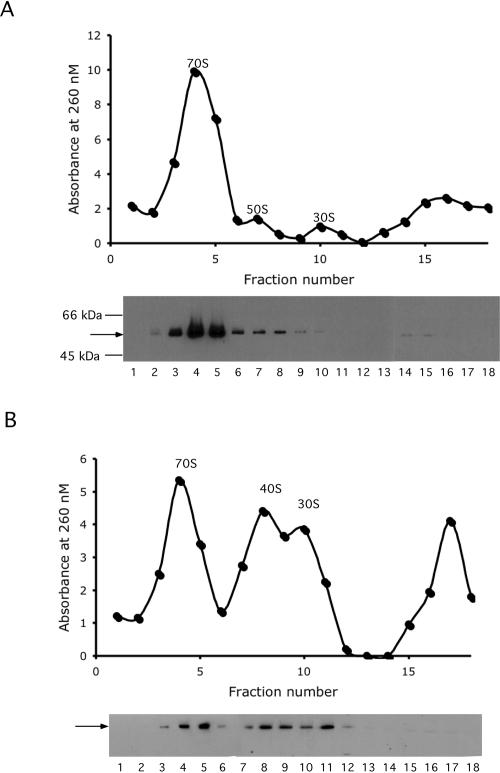
Association of RhlE with ribosomes. A Flag-tagged RhlE plasmid construct, pCJ1086, was transformed into MG1655ΔrhlE (A) or MG1655ΔrhlEΔdeaD (B). Transformed cells were grown in LB-ampicillin at 20°C, induced with arabinose, and harvested by centrifugation. To better discriminate between 70S ribosomes and polysomes, an extended sucrose concentration range (14%–40%) was used, as compared with 14%–32% used in the previous set of experiments, and 18 2-mL fractions were collected. (Top) The absorbance of each fraction at 260 nM was measured. The positions of the 70S ribosomes and the ribosomal subunits are indicated. (Bottom) Equal volumes of the fractions were used for Western blot analysis to detect Flag-tagged RhlE. The migration of Flag-RhlE is indicated by an arrow, and the positions of molecular weight markers are indicated.
In one set of studies, the tagged construct was expressed in MG1655ΔrhlE (Fig. 5A). The ΔrhlE derivative was used to reduce competition from endogenous RhlE; however, similar results were obtained when these experiments were performed in the MG1655 background (data not shown). Cells were analyzed by ultracentrifugation in buffer containing 100 mM NH4Cl and 10 mM MgCl2, similar to conditions that have been used to analyze the association of assembly factors with the ribosome (Sato et al. 2005; Bharat et al. 2006; Hwang and Inouye 2006). Western blot analysis of the individual fractions indicated that the majority of tagged RhlE (molecular weight=53 kDa) were found in fractions 4 and 5. As these fractions coincided with the 70S ribosome peak, this suggested that RhlE associates with 70S ribosomes.
However, it cannot be excluded that RhlE forms an oligomeric complex that fortuitously cofractionates with ribosomes. Also, because of the low concentration of the 30S and 50S ribosomal subunits in the preceding experiment, it was not clear whether RhlE can also bind the individual subunits. To address these issues, similar analyses were carried out in MG1655ΔrhlEΔdeaD. In this strain, 70S ribosomes are significantly reduced in amount, whereas the abundance of the ribosomal subunits and the 40S particle becomes pronounced (Fig. 5B). Western blot analysis of the fractions indicated that there was a significant increase of RhlE in fractions 7–11, which correspond to the ribosomal subunits. These results support the notion that RhlE is ribosome associated, and can either bind to the intact 70S ribosome or to individual ribosomal subunits. This pattern of association is different from that of SrmB and DeaD, each of which binds to ribosomal subunits but not to 70S ribosomes (Charollais et al. 2003, 2004).
The presence of RhlE in fractions 7–9 is not unexpected, since these contain 40S–50S particles (Fig. 5A,B), which are potential targets of RhlE function. However, fractions 10 and 11, which should contain mainly 30S subunits, also had appreciable amounts of RhlE (Fig. 5B). This suggested a possibility that RhlE might possess high affinity for 30S subunits as well. Alternatively, the presence of RhlE could be due to binding to naked 23S rRNA or to early precursors of the large ribosomal subunit. To distinguish between these possibilities, RNA was extracted from fractions 7–11 and analyzed by gel electrophoresis. 23S rRNA was found in fractions 7–9, as expected, but not in fractions 10 and 11, which contained only 16S rRNA (data not shown). These results indicate that the presence of RhlE in the latter two fractions is most likely due to association with the small ribosomal subunit or its precursors. Whether this association has any consequences for the assembly or function of this subunit remains to be determined.
Another point of interest is that very little RhlE was found in fractions corresponding to free protein. Measurement of β-galactosidase activity indicated that peak activity was in fraction 14 (data not shown). As β-galactosidase forms a tetramer with a molecular weight of 5 × 105 Da, uncomplexed RhlE would be expected in fractions 14–18. The significant absence of RhlE in these fractions suggests that RhlE preferentially associates with ribosomes and very little is present in the free form or bound to low molecular weight nucleic acids. To investigate whether RhlE can be dissociated from ribosomes by changing salt or magnesium ion concentration, ultracentrifugation was performed at 150 mM NH4Cl with either 1 or 10 mM MgCl2. When the collected fractions were analyzed, again, very little RhlE was found in fractions 14–18 (data not shown). Therefore, RhlE is primarily ribosome bound, and the interactions between ribosomes and RhlE appear to be relatively specific. Overall, these data support the hypothesis that RhlE mediates its molecular functions via direct interactions with the ribosome.
DISCUSSION
Nearly all organisms contain multiple members of the DEAD-box protein family, which have been shown to regulate a variety of processes in RNA metabolism. E. coli contains five DEAD-box proteins, which appear to play multiple roles in RNA turnover and ribosome biogenesis, but whose precise molecular functions have yet to be elucidated. In particular, very little is known about RhlE, apart from the fact that it associates with proteins involved in RNA degradation and displays helicase activity in vitro (Raynal and Carpousis 1999; Bizebard et al. 2004; Khemici et al. 2004). No phenotype with the loss of rhlE has been observed, indicating an enigmatic role for this protein in the cell.
During the course of investigations into the cold-sensitive phenotype of a ΔdeaD strain, rhlE plasmids were identified as multicopy suppressors of this defect. Very recently, similar findings were reported, but additional studies on this topic have not been described (Awano et al. 2007). Further analysis led to the observation that overproduction or the absence of RhlE have opposite effects in deaD and srmB strains. Whereas RhlE overproduction suppresses the ΔdeaD cold-sensitive growth defect, these defects are instead exacerbated in a ΔsrmB strain. In contrast, loss of RhlE accentuates growth defects in a ΔdeaD strain background, but diminishes the defects of a ΔsrmB strain. Similar trends were observed when 23S rRNA processing or 50S ribosomal subunit maturation were assessed. Furthermore, by expressing an epitope-tagged RhlE, most of the protein was found to be ribosome associated. Collectively, these findings suggest that RhlE has some role in ribosome assembly that is related to the function of SrmB and DeaD.
Strains lacking SrmB or DeaD display many similar defects; therefore, the opposite effects of RhlE in these strains were unanticipated. However, it has recently been shown that the 40S particles that accumulate in srmB or deaD strains contain some notable differences. In srmB strains, the 40S particles lack several ribosomal proteins that are normally present in the 50S subunit, including L13, which is important for an early step of 50S assembly in vitro, and several proteins that are incorporated late in the assembly process (L28, L34, L35, and L36) (Charollais et al. 2003). In contrast, 40S particles found in the ΔdeaD strain have a low abundance of proteins involved in the late step only (L6, L16, L25, L28, L32, and L33) (Charollais et al. 2004). Since both SrmB and DeaD are believed to be important for assisting conformational changes during 50S assembly, these differences suggest that they act on distinct rRNA conformations.
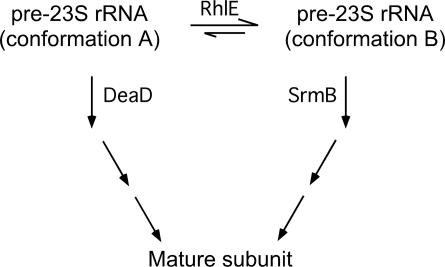
Based on the results described above, a model for RhlE function during ribosome maturation is proposed (Fig. 6). By this model, 50S maturation proceeds via multiple intermediates, DeaD and SrmB act on a nonoverlapping set of intermediates to promote further maturation, and substrates for DeaD and SrmB are interconvertible via RhlE. When RhlE is overproduced, the equilibrium shifts toward intermediates that require SrmB (conformation B). As a result, ribosome biogenesis defects are greater in cells that lack SrmB and lesser in cells that lack DeaD, as compared with strains that do not overproduce RhlE. Similarly, the absence of RhlE is expected to shift the equilibrium to favor processing via the deaD pathway (conformation A). Consequently, when RhlE is absent, cells lacking DeaD are expected to display an enhanced defect, but cells lacking SrmB should show reduced defects. In addition, a fraction of the preribosomes may undergo maturation via SrmB- and DeaD-independent pathways, since a strain lacking both factors retains viability (data not shown). Although the proposed model is speculative, future studies to test its validity could include identifying unique features of the proposed rRNA intermediates and demonstrating that RhlE can stimulate the interconversion between such intermediates.
FIGURE 6.
Proposed function of RhlE in ribosome assembly. RhlE is proposed to play a role in the interconversion of two sets of 23S rRNA conformational forms within pre-50S ribosomes that arise during the ribosome maturation process. Conformational form (A) denotes substrates that require DeaD for further processing, whereas form (B) denotes substrates that require SrmB. The absence of RhlE shifts the population toward form (A), increasing the dependence for DeaD, and alleviating the requirement for SrmB. The overproduction of RhlE has the opposite effect.
Among the requirements for a detailed understanding of the ribosome is a definition of the processes through which ribosomes assemble in the cell. A key first step is to identify the set of molecular components that are responsible for imparting fidelity and speed to the assembly process (Williamson 2003). The studies described here examine RhlE, the least well understood of the E. coli DEAD-box RNA helicases, and provide a first glimpse into its cellular functions, implicating this protein as a ribosome assembly factor. Although the precise molecular function of this protein remains to be elucidated, the finding that it has a role in ribosome biogenesis will help to clarify its function in this process, to complete the list of components in the ribosome biogenesis pathway, and ultimately, to recapitulate a biochemical system for swift and faithful ribosome assembly in vitro.
MATERIALS AND METHODS
Strains and plasmids
Strains CJ1825 and MG1655 have been described previously (Jain and Belasco 1995; Blattner et al. 1997) and are wild type for the DEAD-box helicase genes. Strain derivatives containing deletions within DEAD-box genes were individually created using λRed-mediated recombination of linear DNA in CJ1825, resulting in the replacement of deaD, srmB, or rhlE with cassettes that confer resistance to tetracycline, kanamycin, and chloramphenicol, respectively (Datsenko and Wanner 2000). The incorporation of the linear DNA at the correct locus was confirmed by PCR. The strain derivatives containing a deletion of the deaD, srmB, and rhlE genes are denoted as CJ1825ΔdeaD, CJ1825ΔsrmB, and CJ1825ΔrhlE, respectively. Details of the gene deletions are available upon request. These alleles were transduced to the MG1655 background by P1 transduction to generate MG1655ΔdeaD, MG1655ΔsrmB, MG1655ΔrhlE, MG1655ΔdeaDΔrhlE, and MG1655ΔsrmBΔrhlE. The multicopy suppressors were identified in the CJ1825 background and subsequent experiments were carried out in the more commonly used MG1655 strain background.
A tagged RhlE plasmid, pCJ1086, was constructed by subcloning a PCR product into pMPM-A4, a multicopy plasmid that contains a PBAD arabinose-inducible promoter (Mayer 1995). RhlE was amplified from E. coli chromosomal DNA using primers with the sequences 5′-AAAAAGCTTGTCATGGCAGGATTATTCATCG-3′ and 5′-TTGGATCCTACTTGTCATCGTCATCCTTGTAGTCGATGTCATGATCTTTATAATCACCGTCATGGTCTTTGTAGTCCTGCGCAGCGGCAGGTTTACGCGG-3′, which include cleavage sequences for HindIII and BamHI restriction enzymes, respectively. The latter primer also contains an overhang sequence coding for three tandem copies of the Flag-tag epitope (DYKDHDGDYKDHDIDYKDDDDK) that immediately follows the last rhlE sense codon. The PCR product was digested with BamHI and HindIII, purified, and ligated with BglII and HindIII digested pMPM-A4 DNA to yield the plasmid, pCJ1086.
Genetic screening for multicopy suppressors
A previously described pACYC184-based E. coli multicopy plasmid library was transformed into CJ1825ΔdeaD (Sarkar et al. 2002). Transformants were plated on LB-agar plates supplemented with chloramphenicol and incubated at 20°C until a large number of small colonies (>10,000) and a few larger colonies could be visualized. Plasmid DNA was isolated from the latter set of colonies and retransformed into CJ1825ΔdeaD to confirm the growth-suppression effect. DNA was reisolated from 14 transformed colonies that recapitulated the suppressor effect and sequenced to identify the chromosomal regions present in the plasmids.
RNA analysis
RNA was extracted from exponentially growing cells using the hot-phenol method (Aiba et al. 1981) or from ribosomal fractions (see below). Primer extension reactions to assay 23S rRNA 5′ end maturation were carried out using a radioactively labeled oligonucleotide primer that anneals 35–55 nt from the 5′ end of mature 23S rRNA (Slagter-Jäger et al. 2007). The primer extension products were separated on a 6% polyacrylamide–8M urea sequencing gel, dried, and visualized using a Molecular Dynamics Storm 840 PhosphorImager.
Ribosome and protein analysis
Cells were grown in 80 mL at LB medium, harvested at OD600 = 0.5 − 0.7, pelleted by centrifugation, washed, and resuspended in 0.8 mL of buffer A (10 mM Tris at pH 7.5, 60 mM KCl, and 10 mM MgCl2). The concentrated cells were lysed by sonication, adjusted to 2.2 mL with buffer A, and the cell debris was pelleted by centrifugation at 14,000 rpm for 15 min in a microfuge at 4°C. A total of 2 mL of the supernatant (40 A260 units) was layered on a 14%–32% sucrose gradient in buffer containing 10 mM Tris (pH 7.5), 50 mM NH4Cl, 10 mM MgCl2, and 1 mM DTT. The lysate was centrifuged at 22,000 rpm for 19 h at 4°C in a Beckman L8-70M ultracentrifuge using a SW28 rotor. For analysis of RNA from ribosomal fractions, 12 fractions were collected, RNA was purified by phenol extraction and ethanol precipitation and analyzed by primer extension as described above.
For Western blot analysis, cells transformed with pCJ1086 were grown in 250 mL of LB medium supplemented with ampicillin at 20°C and arabinose to 0.2% was added at early log-phase. After 1 h, cells were pelleted and lysed. A total of 140 A260 units were fractionated by sucrose density gradient ultracentrifugation in buffer containing 10 mM Tris (pH 7.5), 100 mM NH4Cl, 10 mM MgCl2, and 1 mM DTT, as described above. Eighteen 2.0 mL fractions were collected and nucleic acid absorbance was measured at 260 nM. A total of 15 μL volumes of each fraction were separated on NuPage Novex Bis-Tris 4%–12% gradient polyacrylamide gels (Invitrogen) and analyzed for the presence of RhlE-Flag protein by Western blot analysis using anti-Flag primary antibody (Sigma) and anti-mouse horseradish peroxidase conjugated antibody (GE Healthcare). The immunoreactive bands were visualized using ECL-Plus detection reagents (GE Healthcare).
ACKNOWLEDGMENTS
I thank Dr. Nancy Gutgsell for assistance with sucrose density gradient ultracentrifugation. This work was supported by start-up funds from the Lucille P. Markey Foundation.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.800308.
REFERENCES
- Aiba, H., Adhya, S., de Crombrugghe, B. Evidence for two functional gal promoters in intact Escherichia coli cells. J. Biol. Chem. 1981;256:11905–11910. [PubMed] [Google Scholar]
- Alix, J.H., Guerin, M.F. Mutant DnaK chaperones cause ribosome assembly defects in Escherichia coli . Proc. Natl. Acad. Sci. 1993;90:9725–9729. doi: 10.1073/pnas.90.20.9725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awano, N., Xu, C., Ke, H., Inoue, K., Inouye, M., Phadtare, S. Complementation analysis of the cold-sensitive phenotype of the Escherichia coli csdA deletion strain. J. Bacteriol. 2007;189:5808–5815. doi: 10.1128/JB.00655-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharat, A., Jiang, M., Sullivan, S.M., Maddock, J.R., Brown, E.D. Cooperative and critical roles for both G domains in the GTPase activity and cellular function of ribosome-associated Escherichia coli EngA. J. Bacteriol. 2006;188:7992–7996. doi: 10.1128/JB.00959-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bizebard, T., Ferlenghi, I., Iost, I., Dreyfus, M. Studies on three E. coli DEAD-box helicases point to an unwinding mechanism different from that of model DNA helicases. Biochemistry. 2004;43:7857–7866. doi: 10.1021/bi049852s. [DOI] [PubMed] [Google Scholar]
- Blattner, F.R., Plunkett, G., Bloch, C.A., Perna, N.T., Burland, V., Riley, M., Collado-Vides, J., Glasner, J.D., Rode, C.K., Mayhew, G.F., et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- Bylund, G.O., Wipemo, L.C., Lundberg, L.A., Wikstrom, P.M. RimM and RbfA are essential for efficient processing of 16S rRNA in Escherichia coli . J. Bacteriol. 1998;180:73–82. doi: 10.1128/jb.180.1.73-82.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charollais, J., Pflieger, D., Vinh, J., Dreyfus, M., Iost, I. The DEAD-box RNA helicase SrmB is involved in the assembly of 50S ribosomal subunits in Escherichia coli . Mol. Microbiol. 2003;48:1253–1265. doi: 10.1046/j.1365-2958.2003.03513.x. [DOI] [PubMed] [Google Scholar]
- Charollais, J., Dreyfus, M., Iost, I. CsdA, a cold-shock RNA helicase from Escherichia coli, is involved in the biogenesis of 50S ribosomal subunit. Nucleic Acids Res. 2004;32:2751–2759. doi: 10.1093/nar/gkh603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coburn, G.A., Miao, X., Briant, D.J., Mackie, G.A. Reconstitution of a minimal RNA degradosome demonstrates functional coordination between a 3′ exonuclease and a DEAD-box RNA helicase. Genes & Dev. 1999;13:2594–2603. doi: 10.1101/gad.13.19.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordin, O., Banroques, J., Tanner, N.K., Linder, P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- Datsenko, K.A., Wanner, B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz, J., Kressler, D., Linder, P. Unwinding RNA in Saccharomyces cerevisiae: DEAD-box proteins and related families. Trends Biochem. Sci. 1999;24:192–198. doi: 10.1016/s0968-0004(99)01376-6. [DOI] [PubMed] [Google Scholar]
- Deutscher, M.P. Degradation of RNA in bacteria: Comparison of mRNA and stable RNA. Nucleic Acids Res. 2006;34:659–666. doi: 10.1093/nar/gkj472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diges, C.M., Uhlenbeck, O.C. Escherichia coli DbpA is an RNA helicase that requires hairpin 92 of 23S rRNA. EMBO J. 2001;20:5503–5512. doi: 10.1093/emboj/20.19.5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Hage, A., Sbai, M., Alix, J.H. The chaperonin GroEL and other heat-shock proteins, besides DnaK, participate in ribosome biogenesis in Escherichia coli . Mol. Gen. Genet. 2001;264:796–808. doi: 10.1007/s004380000369. [DOI] [PubMed] [Google Scholar]
- Fromont-Racine, M., Senger, B., Saveanu, C., Fasiolo, F. Ribosome assembly in eukaryotes. Gene. 2003;313:17–42. doi: 10.1016/s0378-1119(03)00629-2. [DOI] [PubMed] [Google Scholar]
- Fuller-Pace, F.V., Nicol, S.M., Reid, A.D., Lane, D.P. DbpA: A DEAD box protein specifically activated by 23s rRNA. EMBO J. 1993;12:3619–3626. doi: 10.1002/j.1460-2075.1993.tb06035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi, P., Rybin, V., Bassler, J., Petfalski, E., Strauss, D., Marzioch, M., Schafer, T., Kuster, B., Tschochner, H., Tollervey, D., et al. 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell. 2002;10:105–115. doi: 10.1016/s1097-2765(02)00579-8. [DOI] [PubMed] [Google Scholar]
- Gutgsell, N.S., Deutscher, M.P., Ofengand, J. The pseudouridine synthase RluD is required for normal ribosome assembly and function in Escherichia coli . RNA. 2005;11:1141–1152. doi: 10.1261/rna.2550105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hage, A.E., Tollervey, D. A surfeit of factors: Why is ribosome assembly so much more complicated in eukaryotes than bacteria? RNA Biol. 2004;1:10–15. [PubMed] [Google Scholar]
- Harnpicharnchai, P., Jakovljevic, J., Horsey, E., Miles, T., Roman, J., Rout, M., Meagher, D., Imai, B., Guo, Y., Brame, C.J., et al. Composition and functional characterization of yeast 66S ribosome assembly intermediates. Mol. Cell. 2001;8:505–515. doi: 10.1016/s1097-2765(01)00344-6. [DOI] [PubMed] [Google Scholar]
- Hwang, J., Inouye, M. The tandem GTPase, Der, is essential for the biogenesis of 50S ribosomal subunits in Escherichia coli . Mol. Microbiol. 2006;61:1660–1672. doi: 10.1111/j.1365-2958.2006.05348.x. [DOI] [PubMed] [Google Scholar]
- Inoue, K., Alsina, J., Chen, J., Inouye, M. Suppression of defective ribosome assembly in a rbfA deletion mutant by overexpression of Era, an essential GTPase in Escherichia coli . Mol. Microbiol. 2003;48:1005–1016. doi: 10.1046/j.1365-2958.2003.03475.x. [DOI] [PubMed] [Google Scholar]
- Iost, I., Dreyfus, M. DEAD-box RNA helicases in Escherichia coli . Nucleic Acids Res. 2006;34:4189–4197. doi: 10.1093/nar/gkl500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain, C., Belasco, J.G. RNase E autoregulates its synthesis by controlling the degradation rate of its own mRNA in Escherichia coli: Unusual sensitivity of the rne transcript to RNase E activity. Genes & Dev. 1995;9:84–96. doi: 10.1101/gad.9.1.84. [DOI] [PubMed] [Google Scholar]
- Jiang, M., Datta, K., Walker, A., Strahler, J., Bagamasbad, P., Andrews, P.C., Maddock, J.R. The Escherichia coli GTPase CgtAE is involved in late steps of large ribosome assembly. J. Bacteriol. 2006;188:6757–6770. doi: 10.1128/JB.00444-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalman, M., Murphy, H., Cashel, M. rhlB, a new Escherichia coli K-12 gene with an RNA helicase-like protein sequence motif, one of at least five such possible genes in a prokaryote. New Biol. 1991;3:886–895. [PubMed] [Google Scholar]
- Khemici, V., Toesca, I., Poljak, L., Vanzo, N.F., Carpousis, A.J. The RNase E of Escherichia coli has at least two binding sites for DEAD-box RNA helicases: Functional replacement of RhlB by RhlE. Mol. Microbiol. 2004;54:1422–1430. doi: 10.1111/j.1365-2958.2004.04361.x. [DOI] [PubMed] [Google Scholar]
- Linder, P. Dead-box proteins: A family affair—Active and passive players in RNP-remodeling. Nucleic Acids Res. 2006;34:4168–4180. doi: 10.1093/nar/gkl468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer, M.P. A new set of useful cloning and expression vectors derived from pBlueScript. Gene. 1995;163:41–46. doi: 10.1016/0378-1119(95)00389-n. [DOI] [PubMed] [Google Scholar]
- Mohr, S., Stryker, J.M., Lambowitz, A.M. A DEAD-box protein functions as an ATP-dependent RNA chaperone in group I intron splicing. Cell. 2002;109:769–779. doi: 10.1016/s0092-8674(02)00771-7. [DOI] [PubMed] [Google Scholar]
- Nierhaus, K.H. The assembly of prokaryotic ribosomes. Biochimie. 1991;73:739–755. doi: 10.1016/0300-9084(91)90054-5. [DOI] [PubMed] [Google Scholar]
- Nishi, K., Morel-Deville, F., Hershey, J.W., Leighton, T., Schnier, J. An eIF-4A-like protein is a suppressor of an Escherichia coli mutant defective in 50S ribosomal subunit assembly. Nature. 1988;336:496–498. doi: 10.1038/336496a0. [DOI] [PubMed] [Google Scholar]
- Noble, S.M., Guthrie, C. Identification of novel genes required for yeast pre-mRNA splicing by means of cold-sensitive mutations. Genetics. 1996;143:67–80. doi: 10.1093/genetics/143.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura, M., Gourse, R., Baughman, G. Regulation of the synthesis of ribosomes and ribosomal components. Annu. Rev. Biochem. 1984;53:75–117. doi: 10.1146/annurev.bi.53.070184.000451. [DOI] [PubMed] [Google Scholar]
- Py, B., Higgins, C.F., Krisch, H.M., Carpousis, A.J. A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature. 1996;381:169–172. doi: 10.1038/381169a0. [DOI] [PubMed] [Google Scholar]
- Raynal, L.C., Carpousis, A.J. Poly(A) polymerase I of Escherichia coli: Characterization of the catalytic domain, an RNA binding site and regions for the interaction with proteins involved in mRNA degradation. Mol. Microbiol. 1999;32:765–775. doi: 10.1046/j.1365-2958.1999.01394.x. [DOI] [PubMed] [Google Scholar]
- Rocak, S., Linder, P. DEAD-box proteins: The driving forces behind RNA metabolism. Nat. Rev. Mol. Cell Biol. 2004;5:232–241. doi: 10.1038/nrm1335. [DOI] [PubMed] [Google Scholar]
- Sarkar, N., Cao, G.J., Jain, C. Identification of multicopy suppressors of the pcnB plasmid copy number defect in Escherichia coli . Mol. Genet. Genomics. 2002;268:62–69. doi: 10.1007/s00438-002-0723-0. [DOI] [PubMed] [Google Scholar]
- Sato, A., Kobayashi, G., Hayashi, H., Yoshida, H., Wada, A., Maeda, M., Hiraga, S., Takeyasu, K., Wada, C. The GTP binding protein Obg homolog ObgE is involved in ribosome maturation. Genes Cells. 2005;10:393–408. doi: 10.1111/j.1365-2443.2005.00851.x. [DOI] [PubMed] [Google Scholar]
- Saveanu, C., Namane, A., Gleizes, P.E., Lebreton, A., Rousselle, J.C., Noaillac-Depeyre, J., Gas, N., Jacquier, A., Fromont-Racine, M. Sequential protein association with nascent 60S ribosomal particles. Mol. Cell. Biol. 2003;23:4449–4460. doi: 10.1128/MCB.23.13.4449-4460.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slagter-Jäger, J.G., Puzis, L., Gutgsell, N.S., Belfort, M., Jain, C. Functional defects in transfer RNAs lead to the accumulation of ribosomal RNA precursors. RNA. 2007;13:597–605. doi: 10.1261/rna.319407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava, A.K., Schlessinger, D. Coregulation of processing and translation: Mature 5′ termini of Escherichia coli 23S ribosomal RNA form in polysomes. Proc. Natl. Acad. Sci. 1988;85:7144–7148. doi: 10.1073/pnas.85.19.7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toone, W.M., Rudd, K.E., Friesen, J.D. deaD, a new Escherichia coli gene encoding a presumed ATP-dependent RNA helicase, can suppress a mutation in rpsB, the gene encoding ribosomal protein S2. J. Bacteriol. 1991;173:3291–3302. doi: 10.1128/jb.173.11.3291-3302.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsu, C.A., Kossen, K., Uhlenbeck, O.C. The Escherichia coli DEAD protein DbpA recognizes a small RNA hairpin in 23S rRNA. RNA. 2001;7:702–709. doi: 10.1017/s1355838201010135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venema, J., Tollervey, D. Ribosome synthesis in Saccharomyces cerevisiae . Annu. Rev. Genet. 1999;33:261–311. doi: 10.1146/annurev.genet.33.1.261. [DOI] [PubMed] [Google Scholar]
- Williamson, J.R. After the ribosome structures: How are the subunits assembled? RNA. 2003;9:165–167. doi: 10.1261/rna.2164903. [DOI] [PMC free article] [PubMed] [Google Scholar]