Abstract
Polynucleotide phosphorylase (PNPase) is a diverse enzyme, involved in RNA polyadenylation, degradation, and processing in prokaryotes and organelles. However, in human mitochondria, PNPase is located in the intermembrane space (IMS), where no mitochondrial RNA (mtRNA) is known to be present. In order to determine the nature and degree of its involvement in mtRNA metabolism, we stably silenced PNPase by establishing HeLa cell lines expressing PNPase short-hairpin RNA (shRNA). Processing and polyadenylation of mt-mRNAs were significantly affected, but, to different degrees in different genes. For instance, the stable poly(A) tails at the 3′ ends of COX1 transcripts were abolished, while COX3 poly(A) tails remained unaffected and ND5 and ND3 poly(A) extensions increased in length. Despite the lack of polyadenylation at the 3′ end, COX1 mRNA and protein accumulated to normal levels, as was the case for all 13 mt-encoded proteins. Interestingly, ATP depletion also altered poly(A) tail length, demonstrating that adenylation of mtRNA can be manipulated by indirect, environmental means and not solely by direct enzymatic activity. When both PNPase and the mitochondrial poly(A)-polymerase (mtPAP) were concurrently silenced, the mature 3′ end of ND3 mRNA lacked poly(A) tails but retained oligo(A) extensions. Furthermore, in mtPAP-silenced cells, truncated adenylated COX1 molecules, considered to be degradation intermediates, were present but harbored significantly shorter tails. Together, these results suggest that an additional mitochondrial polymerase, yet to be identified, is responsible for the oligoadenylation of mtRNA and that PNPase, although located in the IMS, is involved, most likely by indirect means, in the processing and polyadenylation of mtRNA.
Keywords: RNA polyadenylation, PNPase, poly(A)-polymerase, mitochondria, human
INTRODUCTION
Although progress has been made toward understanding the mechanisms of processing and polyadenylation of human mitochondrial RNA (mtRNA), much is yet to be deciphered. In comparison to human mitochondria, the RNA degradation pathways in other organelles and prokaryotes, such as plant mitochondria and chloroplasts, Escherichia coli, and other bacteria, are much better defined (Regnier and Arraiano 2000; Dreyfus and Regnier 2002; Condon 2003; Bollenbach et al. 2004; Gagliardi et al. 2004; Kushner 2004; Slomovic et al. 2006). The mechanism by which RNA is metabolized in these systems is generally termed “polyadenylation-stimulated RNA degradation.” The basic scheme of this pathway consists of three sequential stages, commencing with endonucleolytic cleavage of the full-length transcript. Polyadenylation plays a central role in the second step, as it stimulates the degradation of the intermediate cleavage products, which occurs in the final stage. Due to the rapidity of the degradation of these molecules, they are of very low abundance, relative to the full-length transcript, and their detection and isolation, therefore, depend on multiple stages of highly specific amplification. The presence of truncated polyadenylated molecules is considered a tell-tale sign, hinting that degradation-stimulating polyadenylation exists in the studied system (Slomovic et al. 2005).
Unlike RNA encoded by the genomes of other organelles and prokaryotes, human mt-mRNAs are known to undergo stable polyadenylation at their mature 3′ ends, similar to the stable polyadenylation that occurs in the nuclei of eukaryotic cells (Ojala et al. 1981; Fernandez-Silva et al. 2003). This polyadenylation occurs once the polycistronic RNA, transcribed from the mt genome, is cleaved upstream of and downstream from each of the tRNAs, which “punctuate” between the mRNA genes. One known function of the stable poly(A) tails of human mt-mRNA is to complete the partial stop codons of U or UA that occur in a number of the mitochondrial genes, as a result of extreme genomic condensation, thereby allowing efficient protein translation (Ojala et al. 1981). There is still controversy surrounding the question of additional roles for the stable polyadenylation of mt-mRNA, especially concerning its influence upon RNA stability (Tomecki et al. 2004; Nagaike et al. 2005).
Recently, by bioinformatic and biological means, truncated, nonabundant, polyadenylated mitochondrial mRNA, tRNA, and rRNA in human cells were detected and isolated (Slomovic et al. 2005). This was one of the first instances in which stable and unstable polyadenylation were shown to coexist in the same system and suggested that a polyadenylation-stimulated RNA degradation pathway is present in human mitochondria. However, of the enzymes involved in the polyadenylation of the mature 3′ ends of mRNA and the truncated transcripts, as well as the ribonuclease responsible for mt-RNA degradation, only one mitochondrial poly(A)-polymerase (mtPAP) was identified (Tomecki et al. 2004; Nagaike et al. 2005). This mtPAP was reported to be responsible for producing poly(A) tails at the mature 3′ ends of mt-mRNA, by extending the oligo(A) tails present there, the source of which remains unclear.
One of the major enzymes involved in the RNA polyadenylation-stimulated degradation pathway is polynucleotide phosphorylase (PNPase) (Grunberg-Manago 1999; Littauer and Grunberg-Manago 1999). In spinach chloroplasts (cp) and cyanobacteria, this phosphorolytic processive enzyme is responsible for both the polymerization and degradation of cpRNA by using nucleotide diphosphate (NDP) and inorganic phosphate (Pi), respectively (Kudla et al. 1996; Lisitsky et al. 1996; Yehudai-Resheff et al. 2001; Rott et al. 2003). In E. coli, PAP is active in polyadenylation; while PNPase is mostly active in 3′ to 5′ phosphorolysis during RNA degradation and 3′ end processing (Kushner 2002). However, a substantial degree of activity in the polymerization of heteropolymeric tails has also been shown for PNPase (Mohanty and Kushner 2000; Mohanty et al. 2004). Also, PNPase bares a strong structural resemblance to the archaeal exosome and in human cells, contains a mitochondrial target peptide (Piwowarski et al. 2003; Buttner et al. 2006; Houseley et al. 2006). It would, therefore, be natural to choose this enzyme as a major candidate for either polyadenylation, degradation, or both, of human mtRNA. However, the human PNPase, originally identified in an overlapping pathway screen to discover genes displaying coordinated expression as a consequence of terminal differentiation and senescence of melanoma cells, was recently localized to the intermembrane space (IMS) in mitochondria and not to the matrix (Leszczyniecka et al. 2002; Chen et al. 2006). In addition, PNPase was purified and reported to interact with TCL1, a nonenzymatic oncoprotein that promotes B- and T-cell malignancies (French et al. 2007). Assuming that this enzyme, unlike the homologous plant mitochondrial and chloroplast PNPases that exist in the matrix and stroma, respectively, is indeed exclusively localized outside of the matrix, it would have no obvious RNA substrate. Nonetheless, previous RNA interference (RNAi) studies of human PNPase have reported that this enzyme's silencing can influence polyadenylation of mt-mRNA while mRNA accumulation levels remain normal (Nagaike et al. 2005; Chen et al. 2006).
In order to determine the nature of this enzyme's involvement in human mtRNA metabolism, we established stable clonal PNPase knockdown human cell lines. Additionally, these PNPase-silenced cells were subjected to transient siRNA silencing of mtPAP as well. Here we show that, although PNPase is located in the IMS, it is still able to significantly influence the 5′ and 3′ processing and stable 3′ end polyadenylation of various mitochondrial transcripts. Furthermore, by indirect means, such as ATP depletion, poly(A) tail length can be altered, and not solely by direct enzymatic intervention. Also, PNPase silencing did not impair the polyadenylation of truncated molecules. However, although they are present in mtPAP-silenced cells as well, these truncated transcripts had significantly shorter tails. Moreover, although mature 3′ end polyadenylation was abolished in concurrent silencing of PNPase and mtPAP, oligoadenylation remained. Therefore, we also propose that an additional, unidentified enzyme is present within the matrix and is involved in the oligoadenylation of mtRNA.
RESULTS
Stable shRNA-mediated silencing of PNPase in HeLa cells
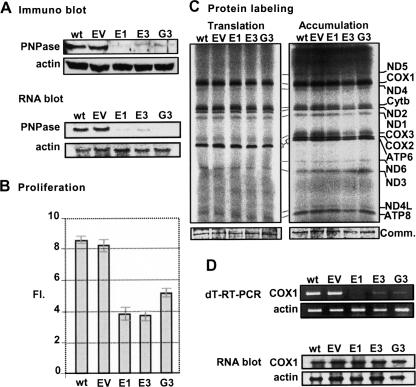
In order to determine whether PNPase plays a role in the processing or polyadenylation of human mitochondrial RNA, we established PNPase-silenced HeLa cell lines. This was achieved by transfecting cells with pSUPER.retro.puro (Oligoengine), an H1 promoter-driven RNAi retroviral vector constitutively expressing short hairpin RNA (shRNA), targeting the PNPase mRNA in one of two chosen sites, schematically termed here “E” and “G.” A number of colonies were isolated and PNPase levels were assessed with both Western and Northern blots. Three clonal cell lines with substantial silencing of PNPase were thereby established; two stably expressed shRNA directed toward site “E,” E1 and E3, and one, toward site “G,” G3 (Fig. 1A). A negative control line was achieved by transfecting cells with an empty pSUPER.retro.puro vector, termed “EV,” which allowed antibiotic resistance but lacked expression of the shRNA. Figure 1A shows substantial silencing of PNPase in E1, E3, and G3 lines, compared to the non-transfected cells (wild type [wt]) and the negative control (EV). Quantitative analysis in which the amounts of PNPase protein were compared to serial dilutions of the wt and EV cells proteins revealed <10% of PNPase in E1 and <15% in E3, and G3 cells (data not shown).
FIGURE 1.
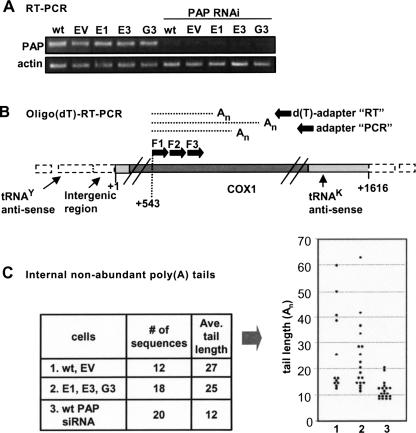
Characterization of HeLa cell lines with stable shRNA-mediated PNPase-silencing. (A) Western and Northern blots showed substantial silencing of PNPase in three PNPase-shRNA clonal cell lines; E1, E3, and G3, compared to non-transfected HeLa cells (wt), and a negative control line termed “empty vector” (EV). (B) Proliferation assay in which identical amounts of cells from each line were plated, counted after 96 h, and the resulting number divided by the initial amount, yielding the “fold increase” rate (FI). (C) Mitochondrion-encoded proteins were labeled by incubating cells with [35S]methionine–[35S]cysteine in the presence of the 80S ribosome inhibitor, emetine. Following radioactive labeling for 25 min to measure protein translation and 120 min to measure accumulation, in left and right gel pictures, respectively, equal amounts of extracted proteins were fractionated on SDS-PAGE, dried, and analyzed by autoradiography. Identification of the 13 mitochondrion-encoded proteins is presented to the right. A panel from the gels stained with Coomassie (Comm.) is included below. (D) Oligo(dT) RT-PCR of the COX1 mRNA in the different cell lines is presented in the upper part. Relative amounts of the transcript were determined by nonsaturating PCR amplification of oligo(dT)-primed cDNA with gene-specific primers and fractionation on agarose gel. Quantitative control was achieved by β-actin PCR amplification using randomly primed cDNA. RNA blots analysis is presented in the lower part. Total RNA extracted from the cell lines was fractionated by denaturing agarose gels, transferred to nylon membrane, and hybridized with 32P-specific oligo-DNA probe for the COX1 and β-actin (used as loading control) transcripts.
PNPase knockdown cells had a lower proliferation rate but normal mitochondrial protein translation and accumulation
A proliferation assay was applied in which an equal number of cells were plated for each line, cells were counted after 96 h, and the final number was divided by the initial, yielding the fold increase (FI) rate. This assay showed that PNPase silencing resulted in an FI rate as much as 50% lower than that of the wt and EV cells (Fig. 1B). Also, the growth medium turned yellow more rapidly than that of the control cells, presumably because of lactate accumulation due to mitochondrial dysfunction (data not shown). These results were consistent with previous work on PNPase RNAi (Chen et al. 2006). Since previous studies have analyzed the effects of PNPase overexpression in human cells and reported various effects on cell growth and, perhaps, improper localization in the cell, we did not complement the RNAi down-regulation with overexpression of PNPase (Leszczyniecka et al. 2002; Sarkar et al. 2005; Sarkar and Fisher 2006).
In order to determine if the translation rate (25-min pulse labeling) and accumulation levels (120-min labeling) of mitochondrial proteins were affected by the silencing of PNPase, cells were incubated with a [35S]methionine–cysteine mix in the presence of emetine, which inhibited the translational activity of 80S ribosomes in the cytoplasm while allowing labeling of proteins translated within the mitochondria. Figure 1C shows that the translation rate is not affected (left picture) nor are the levels of accumulation (right picture) of all 13 mitochondrion-encoded proteins, by the knockdown of PNPase. We note that not all 13 proteins are visible in the translation gel picture, due to the short pulse time.
PNPase silencing caused drastic changes in stable 3′ adenylation as well as 5′ and 3′ processing of the COX1 mRNA
In previous studies, mt-mRNA levels were assessed at or beyond 7 d post-RNAi transfection to determine the effect of PNPase silencing (Nagaike et al. 2005; Chen et al. 2006). In these studies, COX1 and other mt-mRNA levels were not affected. Here, when oligo(dT)-primed RT-PCR was applied to total RNA purified from the stable PNPase knockdown lines, COX1 levels appeared to have decreased drastically, while for three control cytoplasmic mRNAs, B2M, GAPDH, and HRPT, levels remained normal (Fig. 1D and data not shown). In contrast, an RNA blot with a COX1 oligo probe disclosed normal COX1 mRNA levels (Fig. 1D, bottom). This assay was repeated multiple times with different COX1 primers and showed consistent results. We hypothesized that the apparent decrease in the COX1 signal, according to oligo(dT)-RT-PCR, may have been caused by inefficient priming of the oligo(dT) during reverse transcription, due to a possible effect of PNPase silencing on the poly(A) tails known to exist at the mature 3′ ends of mt-mRNA.
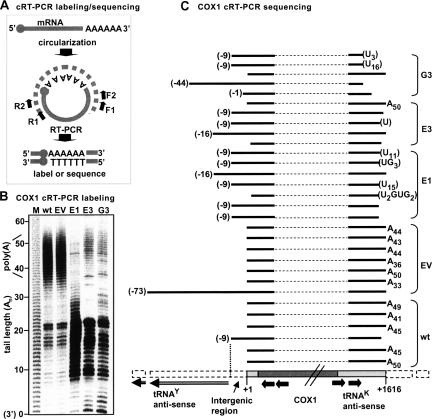
To analyze the status of COX1 stable 3′ polyadenylation in the three PNPase knockdown lines, and thereby determine whether indeed a lack of 3′ polyadenylation was responsible for the apparent decrease in the COX1 signal during oligo(dT)-RT-PCR, circularized-reverse transcription PCR (cRT-PCR) assays were carried out. The cRT-PCR labeling and sequencing assays used here are a modified combination of assays applied in other works (Temperley et al. 2003; Perrin et al. 2004) and are described in Figure 2A. Briefly, in COX1 cRT-PCR labeling, the 5′ and 3′ extremities of total RNA were joined by ligation, reverse transcribed with a COX1 primer, and PCR amplified and 32P labeled. The resulting products consisted of the adjoined 5′ and 3′ ends along with any 3′ (or 5′) extensions. The lengths of the extensions were derived by subtracting from the product size, as determined by resolving with 10% denaturing PAGE and autoradiography, the expected size of a properly processed molecule bearing a naked 3′ end. As presented in Figure 2B, in wt and empty vector (EV) control cells, signals related to COX1 molecules harboring poly(A) tails averaging ∼44 adenosines in length, were obtained. However, in the three PNPase knockdown lines, this poly(A) fraction was barely present. In these lines there were, instead, signals that could have been related to molecules with short extensions of 10–23 nucleotides (nt), at the 3′ or 5′ end. Mitochondrial mRNA molecules are processed from a polycistronic transcript and therefore, a second possibility was that these signals originated from COX1 molecules, not necessarily adenylated but, due to improper processing, retained 5′ or 3′ regions from the polycistronic RNA, and therefore, appeared larger in the gel.
FIGURE 2.
Improper polyadenylation and processing of the COX1 transcript in PNPase-silenced cell lines. (A) cRT-PCR labeling and sequencing methods, used to investigate the 5′ and 3′ ends of a target mRNA, are described. Both start with the circularization of total RNA which contains the target mRNA, with T4 RNA ligase. Next, a gene-specific reverse oligo, generally termed R1, is used to prime reverse transcription, initiated ∼100 nt downstream of the 5′ end. Afterward, two consecutive PCR reactions with F1+R2 and F2+R2 oligos, respectively, amplify the adjoined 5′ and 3′ extremities and simultaneously increase specificity. At this point, there are two options: For sequencing, the products are cloned to T/A vectors, PCR-screened, and sequenced, in order to analyze individual clone sequences (cRT-PCR sequencing). To obtain a more global view of the target mRNA population instead, a third PCR reaction, similar to the second, can be applied, in which either the R2 or F2 oligo is labeled with [γ-32P]ATP. Products are resolved in 10% acrylamide gel, followed by autoradiography (cRT-PCR labeling). The 3′ poly(A) tail lengths can be calculated by subtracting the expected length of a properly processed naked 3′ end molecule from that of the actual product as compared to a nucleotide ladder. (B) The 3′ and 5′ ends of COX1 were analyzed in control (wt and EV) and PNPase-silenced (E1, E3, and G3) cells using the cRT-PCR labeling technique (as described above for A). Products were resolved by 10% denaturing PAGE, followed by autoradiography, and product size was determined by comparison to a nucleotide ladder produced by alkaline hydrolysis of a [32P]RNA (lane M). Assuming proper processing of the mRNA, the product size represents the length of the poly(A) tail added to the 3′ end, a naked 3′ end marked as “0.” However, products could also originate from molecules with impaired processing. In order to differentiate between these two possibilities, cRT-PCR sequencing was performed as shown in part C of the figure. (C) cRT-PCR sequencing of COX1 is shown. The region of the human mitochondrial genome containing the COX1 gene is schematically displayed at the bottom. The first nucleotide of the COX1 transcript at the 5′ end is marked as +1. The translation initiation codon starts at number +4, and the amino acid coding region is colored in dark gray with the two diagonal lines indicating that it is not drawn to scale. The 5′ and 3′ UTRs, composed of 3 nt and the tRNAK antisense, respectively, are shown in light gray. The flanking sequences, including the 9-nt intergenic region and tRNAY antisense located upstream of the COX1 gene, are marked with a dashed white line. Four black arrows represent the R2, R1, F2, and F1 primers used in cRT-PCR. Above the gene scheme, individually sequenced COX1 clones are shown for each cell line. A dashed line symbols the inferred internal part of the COX1 mRNA that was not physically isolated, as only the transcript extremities were amplified (as described above for A). Black lines show the sequenced segments of the 5′ and 3′ ends with the relative position aligned to the scheme below. The 5′ end sites, initiating at positions other than the proper +1, are labeled in parentheses. At the 3′ end of the transcript, either the number of adenosines is indicated or, in parentheses, the post-transcriptionally added nonadenosine extensions that could be located either at the 3′ or at the 5′ end of the transcript.
To resolve this question, cRT-PCR sequencing was applied (Fig. 2C). This assay, described in Figure 2A, allowed a more particular view of the 3′ extension status as individual clones were analyzed. The sequenced wt and EV clones supported the cRT-PCR labeling results as all but two wt/EV clones, which bore naked 3′ ends, displayed stable 3′ end poly(A) tails together averaging ∼44 nt in length. This tail length is consistent with similar results from previous research (Tomecki et al. 2004). One of the two nonadenylated clones (third from bottom) terminated 2 nt before the expected mature 3′ end and contained an additional 9 nt at the 5′ end (see below). As for the cRT-PCR sequencing of the PNPase silenced cells, in contrast to the wt/EV sequences, all but one of the sequenced clones did not have post-transcriptionally added poly(A) tails. Furthermore, most of the sequenced COX1 transcripts from these cell lines terminated slightly upstream of the proper 3′ end site and contained nonadenosine extensions (using the applied method, it is impossible to say if these extensions are located at the 5′ or 3′ end). Interestingly, these extensions, comprised mostly of uridines, were detected in eight out of the 17 PNPase-RNAi COX1 clones. The addition of post-transcriptional added poly(U) tails in trypanosome mitochondria and a recently discovered family of poly(U) polymerases, present in many organisms, were previously reported (Ryan and Read 2005; Kwak and Wickens 2007). However, the type of extension reported here has not been described before in human mitochondria.
For wt/EV clones, the COX1 5′ end was mapped to 3 nt upstream of the ATG translation initiator codon, producing a very short 5′ untranslated region for the COX1 transcript, as previously shown (Montoya et al. 1981). As mentioned above, of the 13 sequenced wt/EV clones, only two bore naked 3′ ends and the rest were polyadenylated. The 5′ end of one of these clones retained 9 nt, which are encoded upstream to the COX1 transcription initiation point. As shown in Figure 2C, the 9 nt comprise an intergenic space separating between the 5′ end of COX1, encoded on the heavy (H) strand and the 5′ end of the tyrosine tRNA gene (tRNAY), encoded on the complementary light (L) strand. This intergenic 9-nt retention was found in nine of the 17 clones obtained from the PNPase-RNAi cell lines (Fig. 2C). A possible scenario for the processing of the COX1 5′ end is suggested, in which first a cleavage of the polycistronic RNA at position −9 takes place, followed by removal of these 9 nt to produce the mature 5′ end. In the stable PNPase RNAi lines, the later step is impaired and the correct 5′ end is not produced. Therefore, the signals that appeared in the COX1 cRT-PCR labeling assay (Fig. 2B) for the RNAi cell lines did not correspond to fully processed oligoadenylated COX1 transcripts, rather to nonadenylated, deficiently processed molecules, most including non-A extensions. Additionally, a cRT-PCR labeling assay applied to PNPase-silenced HEK 293 cells (expressing shRNA targeting the PNPase “E” site) showed a similar loss in stable 3′ COX1 polyadenylation (data not shown). Together, these results revealed that reduction of PNPase resulted in impaired processing and adenylation of COX1 while accumulation of COX1 mRNA and translation of the corresponding protein remained unaffected.
Analyses of additional mt-mRNAs revealed varied effects resulting from PNPase silencing
The mature COX1 transcript is encoded on the heavy strand of the mitochondrial genome and includes a 71-nt 3′ UTR which is the antisense of the tRNASer1. Therefore, the COX1 mRNA does not require 3′ polyadenylation in order to complete a translation stop codon, as is the case with several other transcripts (Ojala et al. 1981). If the complete lack of 3′ adenylation is a general phenomenon for mitochondrial transcripts in PNPase silenced cell lines, including those with incomplete stop codons, it could not account for the results presented in Figure 1C, which resolved that all 13 mitochondrion-encoded proteins are translated and accumulate to normal levels. In order to check whether the effect of PNPase silencing on stable 3′ adenylation correlates with the necessity to complete a stop codon via adenylation of a certain mt-mRNA, we chose three additional genes for cRT-PCR labeling and sequencing analysis; ND3, COX3, and ND5. Both ND3 and COX3 require stable 3′ adenylation to produce a functional stop codon, while ND5 does not, as it contains a 598-nt 3′ untranslated region. RNA was purified from the wt, EV control, and the three PNPase knockdown lines and was subjected to cRT-PCR labeling assays for these genes (Fig. 3A).
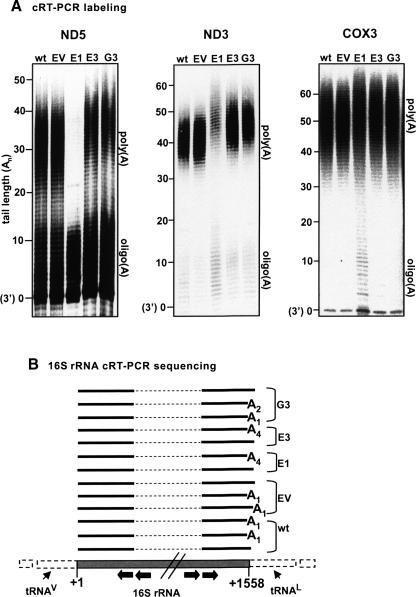
FIGURE 3.
Varied effects of PNPase-silencing on the stable polyadenylation of several mitochondrial transcripts. (A) The 3′ ends of ND5, ND3, and COX3 were analyzed by cRT-PCR labeling in control and PNPase-silenced cells. Labels are the same as in Fig. 2B. The oligo(A) and poly(A) fractions are indicated to the right of each gel. Note that, for ND5 and ND3, the poly(A) fractions in E3 and G3 cells were extended by several nucleotides and in E1, significantly diminished. (B) cRT-PCR sequencing of 16S rRNA did not detect an effect of PNPase knockdown on the nature of 3′ oligoadenylation of this transcript. Gene scheme labels are the same as in Fig. 2C.
In the wt and control lanes of the cRT-PCR labeling assay of ND5, two populations of ND5 mRNA were clearly observed: signals originating from ND5 molecules with oligo(A) extensions (up to ∼13 adenosines) and molecules with poly(A) tails (∼25–42 adenosines). Also, a discrete band derived from ND5 transcripts with naked 3′ ends could be seen. In clone E1, the poly(A) fraction was barely detectable and only the oligo(A) fraction was clearly visible. In E3 and G3, the poly(A) fraction was slightly reduced compared to wt and was shifted ∼5 nt upward, disclosing a lengthening in the poly(A) tails. A similar result was reported previously for transient silencing of PNPase (Nagaike et al. 2005). This slight elongation in poly(A) length was consistent throughout all repetitions of this assay. Furthermore, the fact that the same phenotype was observed in two clonal lines, E3 and G3, each expressing a separate PNPase shRNA directed toward a different site in the PNPase mRNA, strongly supports that this was indeed the result of PNPase silencing.
For ND3 (Fig. 3A, center), the oligo(A) fraction was less visible for all cell lines than in the case of ND5, but the population of molecules with poly(A) tails could be clearly observed. As in ND5, a similar shift of several adenosines in the poly(A) tail length of ND3 transcripts was detected, in the PNPase-silenced cell lines. In the E1 cell line, although lengthened, the poly(A) tail fraction appeared to be substantially diminished and consequently, molecules with oligo(A) tails slightly accumulated.
Unlike the clear phenotypes observed in the cRT-PCR labeling assays of ND5 and ND3, analysis of COX3 did not disclose a marked effect of PNPase RNAi (Fig. 3A, right). In all cell lines, including wt and EV, a fraction of COX3 molecules lacking any 3′ extensions was visible. In cell line E1, a slight accumulation of molecules with oligo(A) tails is evident, but no shift in poly(A) tail length relative to the wt and control is apparent in the PNPase-RNAi cells.
cRT-PCR sequencing was performed for ND5 and ND3 (see Fig. 5C, below, for ND5 results; ND3 data are not shown). However, this method could not mirror the phenotypes observed in the cRT-PCR labeling assays for these genes; instead, molecules of varying adenosine extension lengths, evident in the labeling assay, were among those sequenced. This is due to the fact that the shift in poly(A) tail length in the PNPase knockdown cells, as disclosed in cRT-PCR labeling, although consistent, was minor, and the sequencing method provides a very localized view of each individual clone. Therefore, in order to detect this slight shift while sequencing individual clones, a statistical amount of clones would need to be sequenced. It did, however, confirm that the signals observed in the labeling assay were related to properly processed ND5 and ND3 molecules and the difference in size depended solely upon tail length.
FIGURE 5.
cRT-PCR labeling and sequencing assays disclosed the effect of mtPAP silencing on the polyadenylation of the mature 3′ ends of ND5 and ND3. (A) Transient mtPAP RNAi was applied to wt cells as well as PNPase-silenced lines, as shown in Fig. 4A. RNA purified from these cells and also from a non-transfected control was then subjected to cRT-PCR labeling to analyze ND3. The oligo(A) and poly(A) fractions are indicated to the right of the gel. The poly(A) fraction was barely detectable. However, the oligo(A) fraction intensified substantially, even in cells with concurrent stable PNPase and transient mtPAP silencing. (B) ND5 cRT-PCR labeling as described for A. The poly(A) fraction was hardly detectable, and, unlike ND3, no oligo(A) tails accumulated; instead, only molecules with naked 3′ ends were present. (C) Results of cRT-PCR sequencing for ND5 in control (wt and EV), and PNPase knockdown lines (E1, E3, and G3) are shown along with those of transient silencing of mtPAP in wt and E3 cells (upper 16 lines). Statistically, no difference could be seen using the cRT-PCR sequencing method, between control (wt and EV) and the PNPase-silenced lines, despite the effect revealed in the corresponding cRT-PCR labeling assay (Fig. 3A). However, as disclosed in the cRT-PCR labeling assay (see B), the majority of transcripts from mtPAP siRNA-transfected cells (upper 16 lanes) lacked oligo(A) extensions. The ND5 gene scheme and sequenced clone labels are the same as for COX1, as in Fig. 2C.
Previous studies have shown that rRNAs encoded in the mitochondrial genome undergo 3′ oligoadenylation (Dubin et al. 1982). In order to determine if PNPase silencing affects the oligoadenylation of 16S rRNA, we performed cRT-PCR sequencing and analyzed the sequences of a number of clones for each cell line (Fig. 3B). This assay did not disclose an effect of PNPase knockdown on the oligoadenylation of mt-rRNA.
In conclusion, the nature and intensity of the effects of PNPase RNAi on different mt-mRNAs varied. Loss of polyadenylation, improper processing of both 5′ and 3′ ends, and the addition of nonadenosine nucleotides were found in the case of COX1. While no major change was noted for COX3, in ND5 and ND3, the poly(A) fraction was reduced to different degrees but consistently increased in length and no processing deficiency was detected. Furthermore, the nature of the effect of PNPase-silencing on the adenylation of mt-mRNA did not depend upon their necessity to gain complete stop codons through 3′ adenylation. Together, these results showed that the accumulation of PNPase to normal levels in human cells is required for correct processing and polyadenylation of mitochondrial transcripts.
The role of poly(A)-polymerase (mtPAP) and PNPase in polyadenylation of truncated polyadenylated mtRNA, considered to be degradation intermediates
In previous work, by both biological and bioinformatic means, we detected the presence of truncated polyadenylated molecules corresponding to mitochondrial tRNA, mRNA, and rRNA in human cells (Slomovic et al. 2005). This type of polyadenylation, termed “internal” polyadenylation, as it appears not at the mature 3′ end of the RNA sequence but at sites within the transcript, is believed to stimulate degradation of these fragments, as seen in prokaryotes and other organelles (Dreyfus and Regnier 2002; Kushner 2004; Slomovic et al. 2005). The enzymes involved in the polyadenylation and degradation of these RNA fragments in human mitochondria are yet to be identified, but as mtPAP has been shown to be involved in the stable 3′ end polyadenylation of mitochondrial mRNA (Tomecki et al. 2004; Nagaike et al. 2005), it may also be involved in the internal polyadenylation process. Likewise, PNPase may influence this activity, and therefore, we decided to investigate the effect of either transient mtPAP RNAi or stable PNPase RNAi on the internal polyadenylation of mitochondrial RNA. To silence mtPAP, cell lines wt, EV, E1, E3, and G3 were transfected with siRNA duplex targeting the mtPAP mRNA, and total RNA was purified 72 h post initial transfection. Levels of mtPAP mRNA, pre- and post-transfection, were assessed with semiquantitative random-primed RT-PCR (Fig. 4A). Pre-transfection lanes showed that mtPAP levels in the stable PNPase knockdown cells were not affected. Therefore, PNPase silencing did not affect mtPAP levels, and any phenotypes observed to this point in E1, E3, and G3 cell lines were not due to decreased mtPAP expression. The post-transfection lanes showed efficient siRNA-silencing of mtPAP in all cell lines, with remaining mtPAP transcript levels of less than 5%. Analyses of mature 3′ end stable polyadenylation in cells with concurrent stable PNPase-silencing and transient mtPAP RNAi are discussed in the following section, but to study the influence of either stable PNPase silencing or transient mtPAP RNAi on internal adenylation, we used RNA from wt, EV, E1, E3, and G3 prior to mtPAP transfection as well as wt cells transfected with mtPAP siRNA.
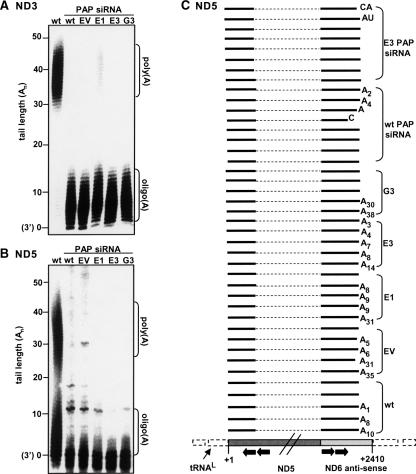
FIGURE 4.
Truncated nonabundant COX1 transcripts with adenosine tails were isolated from cells with either PNPase or mtPAP silencing. (A) Transient mtPAP RNAi was applied to wild-type cells as well as PNPase-silenced lines, and mtPAP mRNA levels were measured with semiquantitative RT-PCR pre- (five left lanes) and post- (five right lanes) mtPAP siRNA transfection. PNPase silencing did not affect mtPAP levels, but application of mtPAP siRNA successfully lowered mtPAP mRNA levels to less than 10%. (B) The oligo(dT)-RT-PCR method was used to isolate nonabundant, truncated, polyadenylated mitochondrial transcripts, assumed to be degradation intermediates. RT was primed with an oligo(dT)-adapter and product cDNA was subjected to a PCR reaction with a gene-specific forward primer, generally termed F1, paired with the adapter oligo. Resulting products were used as templates for a second round of PCR with a nested forward primer, termed F2, paired with the adapter oligo and resulting products were then cloned. Primer F3 was paired with the adapter-oligo to isolate the positive clones which were then sequenced. The schematic presentation of the COX1 gene is as in Fig. 2C. (C) The left table summarizes the effect of either PNPase or mtPAP RNAi-mediated silencing on the adenylation of truncated COX1 fragments, believed to be degradation intermediates. These molecules were isolated with the oligo(dT)-RT-PCR method, as described in the scheme (see B). PNPase silencing did not affect tail length, while knockdown of mtPAP resulted in a decrease in tail length. In the graph to the right, the tail lengths of each individual sequenced clone are presented and compared between assays 1, 2, and 3, as described in the table.
Total RNA was subjected to oligo(dT)-primed RT followed by three consecutive PCR reactions with nested COX1 forward primers paired with the adapter oligo, as described in Figure 4B. The nested primers enhanced specificity and efficiency in isolation of the truncated adenylated COX1 molecules as they are nonabundant compared to the full-length species (Slomovic et al. 2005). Due to oligo(dT) annealing flexibility, tail lengths may be altered from their original length. However, the degradation rapidity of these polyadenylated intermediates does not allow their efficient isolation via cRT-PCR techniques, and a drastic difference in tail length would be expected to be detectable using the oligo(dT) method. Therefore, a total of 50 individual clones were sequenced in order to determine if any difference could be detected between wt cells and those in which either PNPase or mtPAP were silenced. The results, presented in Figure 4C, disclosed that truncated adenylated COX1 molecules were obtained in all the examined cell lines, suggesting the involvement of an additional mitochondrial polymerase. However, differences were observed in the length of the tails. While the average tail length obtained from PNPase silenced cells was similar to that found in wt cells, 25 and 27 adenosines, respectively, with tails reaching >60 adenosines in length, those obtained from mtPAP silenced cells displayed oligo(A) tails averaging only 12 adenosines. This outcome could be explained by a scenario which resembles stable adenylation at the mature 3′ end, in which mtPAP extends oligo(A) tails synthesized by an additional mt-polymerase, thereby producing poly(A) tails (Tomecki et al. 2004).
Concurrent stable PNPase RNAi and transient mtPAP RNAi disclosed that neither is responsible for the oligoadenylation of mtRNA 3′ ends
Previous studies have described the involvement of mtPAP in the elongation of oligo(A) tails at the mature 3′ ends of mitochondrial RNA (Tomecki et al. 2004; Nagaike et al. 2005). Here, cRT-PCR labeling assays for ND3 and ND5 were applied to RNA from wt cells, as a control, and from wt, EV, E1, E3, and G3 lines in which mtPAP was silenced with transient RNAi (Fig. 5A,B). The ND3 cRT-PCR labeling assay showed that silencing of mtPAP resulted in depletion of the poly(A) fraction while the oligo(A) fraction, barely detectable in wt, markedly accumulated. Interestingly, in E1, E3, and G3 cells, in which PNPase had been stably silenced and mtPAP was then transiently knocked down, the oligo(A) fraction remained. This suggests that, in this case, neither mtPAP nor PNPase was responsible for the synthesis of these extensions rather, an additional polymerase.
In contrast to ND3, the cRT-PCR labeling assay of ND5, showed an accumulation of molecules with naked 3′ ends and neither the poly(A) nor the oligo(A) fraction accumulated (Fig. 5B). In order to gain a more localized view of the 3′ end, cRT-PCR sequencing of ND5 was applied (Fig. 5C). 16 ND5 clones, consisting of eight from wt cells transfected with mtPAP siRNA and eight from E3 cells transfected with mtPAP siRNA as well, were sequenced. The results supported the cRT-PCR labeling assay as only two clones were properly processed and contained oligo(A) tails of A2 or A4. Of the remaining clones, the majority showed naked 3′ ends and the rest either terminated upstream of the normal 3′ site or contained short, non-A extensions of 1 or 2 nt. In conclusion, in some genes mtPAP silencing may cause accumulation of oligoadenylated mt-mRNA, possibly produced by an additional, mitochondrial polymerase, but at least in the case of ND5, this silencing resulted in the loss of both the oligo(A) and poly(A) fractions.
PNPase silencing affected ATP levels and membrane potential, and ATP depletion altered the length of poly(A) tails
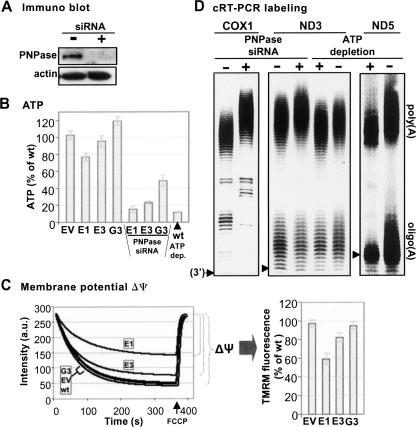
Previous reports have suggested that PNPase silencing indirectly caused a decrease in cellular ATP (Chen et al. 2006). We measured ATP levels in E1, E3, and G3, and the results (see Fig. 6B, left four columns) show that the effect varied between the cell lines. Only clone E1 showed a marked decrease in ATP levels, while E3 was only slightly lower than wt, and in G3, the amount of ATP was slightly increased. When transient PNPase siRNA transfection was added to the stable silencing already present in the cells and ATP was measured, ATP levels significantly decreased (Fig. 6B). The relatively minor ATP decrease observed in the stably silenced cells before the addition of transient PNPase siRNA transfection may have been due to the fact that initial PNPase silencing results in a substantial ATP decrease while stable cell lines with constitutive PNPase silencing may adapt and recover almost normal ATP levels. The efficiency of transient PNPase silencing with siRNA duplexes, targeting site E within the PNPase mRNA, is demonstrated in the Western blot in Figure 6A, in which HeLa cells were either mock transfected (Fig. 6B, “−” lane) or transfected with PNPase-siRNA (Fig. 6B, “+” lane).
FIGURE 6.
PNPase silencing caused changes in cellular ATP level and membrane potential in PNPase-silenced cells, and ATP depletion manipulated poly(A)-tail length. (A) Immunoblot analysis, of proteins obtained from control (mock transfected) cells and cells transiently transfected with PNPase siRNA duplexes, is shown. (B) ATP levels were determined in the stable PNPase-silenced cell lines before and after additional transient PNPase siRNA transfection. Levels were normalized by total protein and are presented as percentages of wt ATP. The measurement for cells in which ATP had been depleted with azide and deoxyglucose is shown at the far right. (C) Mitochondrial membrane potential (Δψ) was measured in the five cell lines using the cationic indicator tetramethylrhodamine methyl ester (TMRM). The Δψ was determined by subtracting the TMRM fluorescence at mitochondrial saturation (minimum) from the fluorescence detected upon addition of the uncoupler, carbonylcyanide m-chlorophenylhydrazone (CCCP) (maximum). The results of one representative experiment are shown to the left and a table summarizing multiple measurements in which the results are presented as percentages of wt Δψ is shown to the right. (D) cRT-PCR labeling assays for COX1 and ND3 were applied to RNA from mock siRNA control and transient PNPase siRNA cells to compare the effect of transient silencing to that of stable (four left lanes). Note the elongation of the COX 1 and ND3 poly(A) fraction in transient PNPase RNAi. To assess the effect of ATP depletion on mtRNA adenylation, cRT-PCR labeling was applied to control and ATP depletion cells, for ND3 and ND5 (four right lanes). Although ATP levels decreased in PNPase-silenced cells, as shown in B, and poly(A) tails lengthened, ATP depletion with azide and deoxyglucose caused the opposite effect; shortening of the poly(A) tail fraction in both genes and, in ND5, shortening of the oligo(A) tails as well.
To determine whether mitochondrial membrane potential (Δψ) may have been partially collapsed in the PNPase-silenced lines, as has been shown in previous work, Δψ was measured and compared in the five cell lines using tetramethylrhodamine methyl ester (TMRM) (Fig. 6C). As shown, the Δψ in E1 was 59% of wt, and in E3, 82%. These decreases coincided with the decrease in cellular ATP levels in these two lines (Fig. 6B).
To compare the effects of stable versus transient PNPase silencing, concerning alteration of poly(A) tail length, COX1 and ND3 cRT-PCR labeling assays were applied to RNA from mock siRNA cells and from wt cells transfected with PNPase siRNA (Fig. 6D, four left lanes). Both for COX1 and ND3, the transient PNPase silencing caused a lengthening in the poly(A) tail fraction. In the case of COX1, this result was similar to that observed during an assay applied to cells with transiently silenced PNPase in previous work but differed from the COX1 cRT-PCR labeling assay applied here to stably silenced cells (Fig. 2B; Nagaike et al. 2005). For ND3, this result was identical to that witnessed in the stable PNPase-silenced lines, confirming that this phenotype was indeed due to PNPase knockdown and not resulting from or compensating the prolonged silencing of PNPase in stable knockdown cell lines.
As described, PNPase silencing altered both ATP levels (especially upon transient silencing) as well as the stable poly(A) tail length of mt-mRNA. To determine whether any correlation could be drawn between these effects, cellular ATP was depleted in wt cells by treatment with azide and deoxyglucose; depletion was validated (see Fig. 6B, last lane on the right).
For ND3 and ND5, cRT-PCR labeling assays were applied to RNA from control cells and from cells in which ATP was depleted (Fig. 6D, four right lanes). Interestingly, ATP depletion caused a slight but consistent decrease in poly(A) length in both genes. For ND5, a decrease was also detected in the length of the oligo(A) fraction. This phenotype is opposite to that observed in PNPase silencing. However, it demonstrates that poly(A) tail length can be indirectly altered by environmental and conditional means. This is supported by previous works that reported that ATP concentrations can influence polyadenylation, as well as the ratio of mt-rRNA transcription compared to mt-mRNA transcription, within the matrix (Gaines et al. 1987).
DISCUSSION
PNPase is recognized as a central enzyme in RNA processing, polyadenylation, and degradation in numerous systems, such as bacteria, cyanobacteria, plant mitochondria, and chloroplasts (Kudla et al. 1996; Lisitsky et al. 1996; Deutscher and Li 2001; Yehudai-Resheff et al. 2001; Kushner 2002; Condon 2003; Rott et al. 2003; Gagliardi et al. 2004; Deutscher 2006; Yehudai-Resheff et al. 2007). In plant mitochondria and chloroplasts, PNPase is located in the matrix and stroma, respectively, in clear correlation with its role in the metabolism of RNA encoded in the genomes of these organelles (Yehudai-Resheff et al. 2001; Walter et al. 2002; Perrin et al. 2004). However, it was recently discovered that the human PNPase is not located in the mitochondrial matrix, but instead, in the IMS (Chen et al. 2006; Rainey et al. 2006). Due to lack of knowledge of any other exoribonuclease known to be located in the matrix of human mitochondria, the question arose whether PNPase is involved in mtRNA metabolism. In this work, we established clonal HeLa cell lines with stable shRNA expression targeting PNPase. The results revealed a significant effect upon the processing and polyadenylation of mitochondrial mRNA.
How does PNPase modulate mtRNA processing and polyadenylation?
Considering the IMS location of the human PNPase, how could the results of this work, which demonstrate that PNPase affects processing and polyadenylation of mtRNA, be interpreted? We consider three alternatives to explain this discrepancy. The first explanation could be that, although PNPase is mostly located in the IMS, a minute amount enters the mitochondrial matrix and is directly involved in the processing and polyadenylation of mtRNA. In this case, it is possible that the observed varied effect that PNPase silencing has on different mt-mRNAs may be due to residual enzyme and varying binding affinities to its mRNA substrates. Although it is difficult to absolutely exclude this possibility, we consider it unlikely because of the following reasons: (1) The considerably different effects of the PNPase RNAi treatment on the different mt-mRNAs do not support a direct enzymatic involvement of this enzyme in processing, degradation, and/or polyadenylation, despite possible varying binding affinities. (2) Recent biochemical analysis of the human PNPase disclosed that, unlike the bacterial and chloroplast PNPases, which bind poly(A) with high affinity, the human enzyme displayed low binding affinity to poly(A)-RNA (Hayakawa and Sekiguchi 2006; Portnoy et al. 2008). In addition, the optimal degradation activity was detected in the presence of 0.1 mM Pi, in contrast to other PNPases, which degrade RNA optimally at 10 mM Pi, the physiological Pi concentration in bacteria and organelles (Littauer and Grunberg-Manago 1999; Yehudai-Resheff et al. 2001). (3) When polymerizing RNA, PNPase and the archaeal exosome, its homolog, produce heteropolymeric poly(A)-rich tails, rather than homopolymeric poly(A) tails, in all systems analyzed as of today (Bollenbach et al. 2004; Slomovic et al. 2006). In this and previous work, hundreds of isolated and analyzed human mitochondrial polyadenylated transcripts, whether full-length or truncated, harbored only homopolymeric, not heteropolymeric, poly(A) tails (Tomecki et al. 2004; Slomovic et al. 2005). The only exceptions are the poly(U)-rich tails revealed in this work for COX1 (Fig. 2C). However, these were observed in the PNPase-RNAi cell lines, and therefore, most likely were not produced by PNPase. Taken together, human PNPase does not seem to be directly involved in poly(A) metabolism and processing in the mitochondrial matrix.
The second possible explanation, concerning the effect of PNPase silencing on the processing and polyadenylation of mtRNA, is that phosphorolytic activity of PNPase in the IMS plays an important function in the equilibrium of the NDP/Pi concentrations inside the mitochondrial matrix, which may influence the NTP concentrations as well. Indeed, human PNPase displays phosphorolytic activity, degrading and polymerizing various analyzed RNA substrates (Leszczyniecka et al. 2002; Chen et al. 2006; Hayakawa and Sekiguchi 2006; Portnoy et al. 2008). By maintaining correct nucleotide concentrations by phosphorolytic activity, PNPase may affect poly(A) tail length within the mitochondrial matrix, from the IMS. This effect could be a direct purpose of the phosphorolytic activity of PNPase in the IMS, as a means of poly(A) tail length modulation. However, PNPase may play a more generalized role in maintaining proper NDP/Pi concentrations in the mitochondrion which, among other aspects, influences poly(A) tails. Likewise, it could be suggested that the processing of COX1 mRNA, and perhaps additional mt-mRNAs, is sensitive to changes in the nucleotides/Pi concentrations and, therefore, impaired in PNPase-silenced cells. Interestingly, transient PNPase-RNAi leads to a significant decrease in ATP concentration, although, in our stable cell lines, pretransient PNPase siRNA duplex transfection, these changes were less prominent, perhaps resulting from acclimation of the cells after prolonged PNPase silencing (Fig. 6B; Chen et al. 2006). In addition, the experiments in which ATP was depleted resulted in changes in the lengths of oligo and poly(A) tails, disclosing that proper ATP levels are essential for normal polyadenylation (Fig. 6). Indeed, the influence of ATP levels on mtRNA transcription and polyadenylation was reported 20 years ago by the Attardi group (Gaines et al. 1987). This type of activity, as a modulator of NDP/Pi concentrations, has recently been proposed for the Chlamydomonas chloroplast PNPase, explaining the effect of Pi starvation on the stability of chloroplast transcripts (Yehudai-Resheff et al. 2007).
If indeed PNPase displays phosphorolytic activity in the IMS, thereby maintaining proper NDP/Pi concentrations in the matrix, RNA molecules, serving as substrates for either polymerization or degradation, could be expected to be present in the IMS.
A third explanation for the effect of PNPase-RNAi on mt-mRNA is that PNPase has no designated activity within the IMS and is merely stored in this compartment to be released to the cytoplasm upon apoptotic stimuli (Chen et al. 2006). Perhaps, when PNPase is lacking in the IMS, the general disruption of mitochondrial homeostasis, which was shown here to impair mitochondrial electrochemical membrane potential, and was reported to decrease respiratory chain activity, alter mitochondrial morphology, and impair ATP generation, includes mt-mRNA processing and polyadenylation deficiencies, as well. In this case, the partial collapse in membrane potential could be the cause of the coinciding partial decrease in ATP shown in Figure 6.
Processing of the mature 5′ end of COX1 and nonadenosine 3′ extensions
In the PNPase knockdown cells, the COX1 mRNA was most drastically affected as, unlike the other examined mRNAs, it experienced defects in both 5′ and 3′ processing as well as abolishment of its stable poly(A) tail. Concerning 5′ processing, sequencing of COX1 transcripts, isolated from these cells, showed that the majority of molecules retained a 9-nt sequence which occurs immediately upstream of the COX1 gene, between its 5′ end and the 5′ end of the tRNAY gene, which is encoded on the opposite strand (the L-strand) of the mitochondrial genome (see Fig. 2C, map, bottom). Similar to the endocleavage of the mt precursor RNA, guided by secondary structures of the tRNA genes, the antisense of the tRNAY gene, located upstream of COX1, most likely creates such a structure which guides the cleavage of the H-strand polycistronic RNA, exactly 9 nt upstream of COX1. Next, to produce the mature 5′ end of COX1, these 9 nt must be subsequently removed. The silencing of PNPase impaired this trimming activity, evident by the accumulation of molecules retaining the 9-nt sequence, and thereby revealed it. Additionally, in PNPase-silenced cells, many isolated COX1 molecules contained uridine-rich extensions. As revealed by the cRT-PCR sequencing method, these additions could be either located at the 5′ or 3′ end of the transcript. However, a favorable explanation could be that, once lacking poly(A) tails, the 3′ ends were exposed to polymerization of additional nucleotides. This type of nonadenosine post-transcriptional addition has not been reported before for human mt-mRNA, and it would be interesting to investigate it further.
Is polyadenylation related to stability and translation of mt-RNA?
Generation of functional stop codons at the 3′ ends of certain mt-mRNAs is considered a major function of mature 3′ end adenylation in mitochondria (Ojala et al. 1981). Whether these extensions also grant stability to the mt-mRNA, akin to the stable polyadenylation which occurs in the nuclei of eukaryotic cells, is not definite (Temperley et al. 2003; Tomecki et al. 2004; Nagaike et al. 2005). In the case of COX1, as detected in the PNPase knockdown cells, this transcript does not gain stability by means of poly(A); despite the loss of stable polyadenylation, COX1 mRNA accumulated to normal levels.
The COX1 gene encodes a complete and functional stop codon followed by a 3′ UTR and therefore, does not rely on 3′ adenylation as a means to acquire a functional stop codon. However, polyadenylation of nucleus-encoded mRNA, which also do not require poly(A) tails for stop-codon generation, is nonetheless essential for efficient translation. Here, in PNPase-RNAi cells, the COX1 protein was translated and accumulated to normal levels, as for the remaining 12 mitochondria-encoded proteins, despite the polyadenylation impairment.
Is there a second PAP in the mitochondria?
The stable polyadenylation which occurs at the mature 3′ ends of mt-mRNAs is considered to be a two-step process in which the 3′ ends are oligoadenylated, followed by extension of this short tail by mtPAP, to form a long poly(A) tail (Tomecki et al. 2004). Here, too, when RNA, purified from cells treated with mtPAP siRNA, was subjected to cRT-PCR applications, oligoadenylated ND3 transcripts accumulated while the poly(A) fraction was barely detectable (Figs. 3A, center, 5A). Moreover, this oligoadenylation activity endured even when PNPase alone or PNPase and mtPAP were concurrently silenced.
In human mitochondria, along with stable mature 3′ end polyadenylation, truncated, adenylated molecules, considered to be degradation intermediates, are present as well (Slomovic et al. 2005). In this work, such nonabundant, internally adenylated COX1 transcripts were detected in both PNPase and mtPAP-silenced cells. Although the tails of molecules isolated from the PNPase knockdown cells were approximately of the same average length as those purified from control cells, the average tail length of truncated molecules found in cells treated with mtPAP siRNA was significantly lower: 12 adenosines compared to 27 in control cells. This suggests that internal polyadenylation may resemble that which occurs at the mature 3′ end; first, an oligo(A) tail is synthesized, followed by elongation of the tail to produce a poly(A) extension. Therefore, in both cases, during mtPAP silencing, oligo(A) tails endure while poly(A) tails do not. Hence, we propose that a second mitochondrial PAP, yet to be identified, is responsible for the oligoadenylation of mt-mRNA. We must, however, note the possibility that residual mtPAP could be responsible for the oligoadenylation activity, although this would be unlikely, as RT-PCR showed extremely efficient mtPAP knockdown and double transfections spanning 72 h were applied.
Together, the results of this work disclosed that human PNPase, although localized to the mitochondrial IMS, is extensively involved in the modulation of processing and polyadenylation of mitochondria mRNAs, most likely by indirect means. Revealing a possible additional mtPAP, an RNA substrate of PNPase in the IMS, and the biological function(s) of PNPase activity in the IMS and/or when released to the cytoplasm are the next challenging tasks in studies exploring human PNPase.
MATERIALS AND METHODS
Cells
HeLa cells were grown as a monolayer at 37°C, 5% CO2 in a DMEM medium (Sigma) supplemented with 10% fetal calf serum, 2 mM l-glutamine, and penicillin–streptomycin.
Construction of stable RNAi cell lines
Two different target locations within the PNPase mRNA, schematically termed “E” and “G,” were chosen using the siMAX RNAi design tool (MWG) (http://www.mwgbiotech.com/html/s_synthetic_acids/s_sirna_design.shtml). Oligonucleotides referring to the E and G target locations (all oligonucleotide sequences are available upon request) were annealed and ligated into pSUPER.retro.puro, an H1 promoter-driven RNAi retroviral vector (Oligoengine). Stable transfections of pSUPER.retro.puro vectors containing the corresponding oligonucleotides (10 μg), or a control vector lacking an oligomer insert were performed using DreamFect transfection agent (OZ Biosciences) in 10 mL of optimum GlutaMAX-1 medium (Invitrogen) according to the manufacturer's instructions. Six hours following the transfection, the medium was replaced with DMEM medium, and after an additional 18 h, puromycin (1 μg/mL) was added as a means of selection. For a negative control, a cell line harboring an empty vector (EV) was produced. To establish PNPase-silenced clonal stable cell lines, a number of colonies from the E and G transfections, expressing the shRNA sequences targeting the PNPase mRNA, were isolated and grown separately. After 2 weeks of selection, the PNPase shRNA-mediated silencing was assessed for each clone using both RNA and protein-blot assays, resulting in two retained E lines, E1 and E3, and one G line, G3, with substantial PNPase knockdown.
Transient siRNA application
For transient RNAi, targeting either PNPase or the human mitochondrial poly(A)-polymerase (mtPAP), 21-mer siRNA duplexes were designed. The PNPase–siRNA duplex was designed to target the PNPase transcript at location “E” described above for the stable RNAi treatment. The mtPAP duplex was planned according to previous work (Tomecki et al. 2004).
Cells were transfected with 400 pmol of siRNA duplex on 60-mm plates using lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions. A control mock transfection lacking siRNA was also performed. After 24 h, the cells were detached with trypsin (Biological Industries) and replated on 10-cm plates, followed, 24 h later, by a second transfection with 800 pmol of siRNA duplex. A total of 72 h after the initial transfection, RNA was harvested with Tri-Reagent (Sigma).
Proliferation assay
Cells from each line (25 × 103) were plated in 10-cm2 wells, detached, and recounted after 96 h. Fold-increase values were calculated by dividing the initial and final cell counts for each line.
RNA purification, random-primed RT-PCR, and Northern blots
Total RNA was purified using Tri-Reagent (Sigma) or Invisorb kit (Invitek), according to manufacturer's instructions. Random-primed RT was performed on 3 μg of total RNA using StrataScript 5.0 Multi-Temp Reverse Transcriptase (Stratagene) with 500 ng of random primers (Amersham Biosciences) according to manufacturer's instructions and normalized with β-actin. For PCR reactions, duration was 95°C, 1 min; 58°C, 1 min; and 72°C, 1 min; followed by a final 72°C 10-min extension. RNA-blot analysis was performed as described previously (Meierhoff et al. 2003; Slomovic et al. 2005). The hybridization probe was a 520-bp DNA fragment of the PNPase cDNA, amplified from random-primer generated cDNA and 32P-labeled by random priming. For COXI, a 32P-labeled DNA oligo was applied as a probe. RNA amounts were normalized according to the β-actin transcript using a riboprobe generated as previously described (Slomovic et al. 2005).
Western blot analysis
Cells were harvested in SDS-denaturing lysis buffer (1% SDS, 10% glycerol, and 0.1 M DTT), syringed with a 21-gauge needle, and cleared by centrifugation (12,000 RPM at 4°C for 15 min). Proteins were analyzed by 10% SDS-PAGE gel, transferred to a nitrocellulose membrane, which was then blotted with a PNPase chicken polyclonal antibody (kindly obtained from P.B. Fisher, Columbia University, New York City), and normalized with an actin rabbit polyclonal antibody (SC Biotechnology).
Oligo(dT)-primed RT-PCR isolation of truncated polyadenylated COXI RNA
First-strand cDNA, generated from total RNA with a T9-adapter oligo (Slomovic et al. 2005), was used as a template for PCR primed with the adapter oligo and a gene-specific forward primer, generally termed F1 (Fig. 4B). Next, a second round of PCR with a nested forward primer, F2, and the adapter oligo was applied and followed by T/A cloning to pGEM-T Easy vector (Promega). Then, competent cells transformed with the vector population were then screened for specificity with two PCR reactions, F2 + adapter and F3 (a second nested primer) + adapter. Positive colonies were sequenced (Macrogen).
cRT-PCR sequencing and cRT-PCR labeling assays
Circular reverse transcription PCR (cRT-PCR) labeling and sequencing were used to determine both 5′ and 3′ extremities of various mtRNA as described (Fig. 2A; Gagliardi et al. 2004; Perrin et al. 2004) Briefly, 5 μg of total RNA was circularized with T4 RNA ligase (Fermentas), followed by phenol–chloroform extraction and ethanol precipitation. cDNA was synthesized using a gene-specific reverse primer, generally termed R1, and StrataScript 5.0 (Stratagene) and used as a template for a PCR reaction, primed with a nested reverse primer, R2, and a forward primer, F1. The region amplified contained the junction of the 5′ and 3′ extremities. A second round of PCR with the R2 primer and a nested forward primer, F2, was applied. Products were either T/A-cloned and sequenced (cRT-PCR sequencing) or used to template a third round of PCR (cRT-PCR labeling), but with one of the two primers’ 5′ end labeled with 32P, using polynucleotide kinase and [γ-32P]ATP. PCR amplification consisted of 30 cycles for the first and second reactions and 6 cycles for the third (cRT-PCR labeling). Duration was 95°C, 1 min; 58°C, 1 min; 72°C, 1 min; followed by a final 72°C 10-min extension. cRT-PCR labeling products were resolved by 10% denaturing PAGE and autoradiography (nt sequences of all primers are available upon request).
Analysis of mitochondrial protein synthesis
Procedure applied as described previously (Chrzanowska-Lightowlers et al. 2004). Briefly, ∼ 5 × 106 cells of each cell line (wt, EV, and PNPase-silenced cells) were labeled with a [35S]methionine–[35S]cysteine mix (50 μCi/mL) for 25 min or 120 min in met/cys-free DMEM-glucose medium (Sigma) containing 100 μg/mL emetine (Sigma) and 10% dialyzed FBS. Samples (30 μg of proteins) were resolved by 10% SDS-PAGE and autoradiography.
ATP depletion and quantification
For ATP depletion assays, cells were pre-incubated for 2.5 h in growth medium with 5 mM azide and 10 mM deoxyglucose. In order to assess ATP levels either after ATP depletion or RNAi-mediated PNPase silencing, the ATP bioluminescence assay kit CLS II (Roche Molecular Biochemicals) was used. Briefly, 6 × 105 cells, washed twice in PBS, were resuspended in boiling lysis buffer (100 mM Tris, 4 mM EDTA, pH 7.7) and centrifuged at 1000g. Appropriate sample volumes were then transferred from the supernatant to bioluminescence tubes, and luciferase was added at a volume ratio of 1:1 to each sample immediately before measurement with a TRI-CARB 2100TR luminometer. Normalization was performed according to the protein concentrations of the supernatant that were determined using the BioRad protein assay reagent.
Mitochondrial membrane potential (Δψ)
To measure and compare Δψ, 1.5 × 106 cells from each line in Hara buffer (30 mM HEPES, pH 7.1, 75 mM sucrose, 20 mM glucose, 5 mM KH2PO4, 40 mM KCl, 0.5 mM EDTA, 1 M MgCl2) were perforated on ice with digitonin at 20 μM for 10 min. BSA was then added to a final concentration of 0.3% followed by succinate at 5 mM. Tetramethylrhodamine methyl ester (TMRM) was added to a final concentration of 5 μM, and fluorescence was measured by a Cary Eclipse fluorescence spectrophotometer (Varian) until mitochondrial saturation was reached (minimum fluorescence due to TMRM quenching). At this point, the uncoupler, carbonylcyanide m-chlorophenylhydrazone (CCCP), was added, which caused collapse of the membrane potential resulting in maximum fluorescence from which the minimum value (at mitochondrial saturation) was subtracted in order to obtain the Δψ (Scaduto and Grotyohann 1999; Duan et al. 2003).
ACKNOWLEDGMENTS
We thank Paul B. Fisher, Piotr Stepien, and Rafal Tomecki for the human PNPase antibodies and assistance in mtPAP RNAi targeting. We also thank Z.M. Chrzanowska-Lightowlers and Anne Chomyn for help with the protein labeling protocol and Dominique Gagliardi and Hagar Katzir on instruction for cRT-PCR and membrane potential measurement procedures, respectively. This work was supported by grants from the Israel Science Foundation (ISF), the United States–Israel Binational Science Foundation (BSF), and the United States–Israel Binational Agricultural Research and Development Fund (BARD).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.697308.
REFERENCES
- Bollenbach, T.J., Schuster, G., Stern, D.B. Cooperation of endo- and exoribonucleases in chloroplast mRNA turnover. Prog. Nucleic Acid Res. Mol. Biol. 2004;78:305–337. doi: 10.1016/S0079-6603(04)78008-3. [DOI] [PubMed] [Google Scholar]
- Buttner, K., Wenig, K., Hopfner, K.P. The exosome: A macromolecular cage for controlled RNA degradation. Mol. Microbiol. 2006;61:1372–1379. doi: 10.1111/j.1365-2958.2006.05331.x. [DOI] [PubMed] [Google Scholar]
- Chen, H.W., Rainey, R.N., Balatoni, C.E., Dawson, D.W., Troke, J.J., Wasiak, S., Hong, J.S., McBride, H.M., Koehler, C.M., Teitell, M.A., et al. Mammalian polynucleotide phosphorylase is an intermembrane space RNase that maintains mitochondrial homeostasis. Mol. Cell. Biol. 2006;26:8475–8487. doi: 10.1128/MCB.01002-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrzanowska-Lightowlers, Z.M., Temperley, R.J., Smith, P.M., Seneca, S.H., Lightowlers, R.N. Functional polypeptides can be synthesized from human mitochondrial transcripts lacking termination codons. Biochem. J. 2004;377:725–731. doi: 10.1042/BJ20031556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condon, C. RNA processing and degradation in Bacillus subtilis . Microbiol. Mol. Biol. Rev. 2003;67:157–174. doi: 10.1128/MMBR.67.2.157-174.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher, M.P. Degradation of RNA in bacteria: Comparison of mRNA and stable RNA. Nucleic Acids Res. 2006;34:659–666. doi: 10.1093/nar/gkj472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher, M.P., Li, Z. Exoribonucleases and their multiple roles in RNA metabolism. Prog. Nucleic Acid Res. Mol. Biol. 2001;66:67–105. doi: 10.1016/s0079-6603(00)66027-0. [DOI] [PubMed] [Google Scholar]
- Dreyfus, M., Regnier, P. The poly(A) tail of mRNAs: Bodyguard in eukaryotes, scavenger in bacteria. Cell. 2002;111:611–613. doi: 10.1016/s0092-8674(02)01137-6. [DOI] [PubMed] [Google Scholar]
- Duan, S., Hajek, P., Lin, C., Shin, S.K., Attardi, G., Chomyn, A. Mitochondrial outer membrane permeability change and hypersensitivity to digitonin early in staurosporine-induced apoptosis. J. Biol. Chem. 2003;278:1346–1353. doi: 10.1074/jbc.M209269200. [DOI] [PubMed] [Google Scholar]
- Dubin, D.T., Montoya, J., Timko, K.D., Attardi, G. Sequence analysis and precise mapping of the 3′ ends of HeLa cell mitochondrial ribosomal RNAs. J. Mol. Biol. 1982;157:1–19. doi: 10.1016/0022-2836(82)90510-1. [DOI] [PubMed] [Google Scholar]
- Fernandez-Silva, P., Enriquez, J.A., Montoya, J. Replication and transcription of mammalian mitochondrial DNA. Exp. Physiol. 2003;88:41–56. doi: 10.1113/eph8802514. [DOI] [PubMed] [Google Scholar]
- French, S.W., Dawson, D.W., Chen, H.W., Rainey, R.N., Sievers, S.A., Balatoni, C.E., Wong, L., Troke, J.J., Nguyen, M.T., Koehler, C.M., et al. The TCL1 oncoprotein binds the RNase PH domains of the PNPase exoribonuclease without affecting its RNA degrading activity. Cancer Lett. 2007;248:198–210. doi: 10.1016/j.canlet.2006.07.006. [DOI] [PubMed] [Google Scholar]
- Gagliardi, D., Stepien, P.P., Temperley, R.J., Lightowlers, R.N., Chrzanowska-Lightowlers, Z.M. Messenger RNA stability in mitochondria: Different means to an end. Trends Genet. 2004;20:260–267. doi: 10.1016/j.tig.2004.04.006. [DOI] [PubMed] [Google Scholar]
- Gaines, G., Rossi, C., Attardi, G. Markedly different ATP requirements for rRNA synthesis and mtDNA light strand transcription versus mRNA synthesis in isolated human mitochondria. J. Biol. Chem. 1987;262:1907–1915. [PubMed] [Google Scholar]
- Grunberg-Manago, M. Messenger RNA stability and its role in control of gene expression in bacteria and phages. Annu. Rev. Genet. 1999;33:193–227. doi: 10.1146/annurev.genet.33.1.193. [DOI] [PubMed] [Google Scholar]
- Hayakawa, H., Sekiguchi, M. Human polynucleotide phosphorylase protein in response to oxidative stress. Biochemistry. 2006;45:6749–6755. doi: 10.1021/bi052585l. [DOI] [PubMed] [Google Scholar]
- Houseley, J., LaCava, J., Tollervey, D. RNA-quality control by the exosome. Nat. Rev. Mol. Cell Biol. 2006;7:529–539. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- Kudla, J., Hayes, R., Gruissem, W. Polyadenylation accelerates degradation of chloroplast mRNA. EMBO J. 1996;15:7137–7146. [PMC free article] [PubMed] [Google Scholar]
- Kushner, S.R. mRNA decay in Escherichia coli comes of age. J. Bacteriol. 2002;184:4658–4665. doi: 10.1128/JB.184.17.4658-4665.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushner, S.R. mRNA decay in prokaryotes and eukaryotes: Different approaches to a similar problem. IUBMB Life. 2004;56:585–594. doi: 10.1080/15216540400022441. [DOI] [PubMed] [Google Scholar]
- Kwak, J.E., Wickens, M. A family of poly(U) polymerases. RNA. 2007;13:860–867. doi: 10.1261/rna.514007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leszczyniecka, M., Kang, D.C., Sarkar, D., Su, Z.Z., Holmes, M., Valerie, K., Fisher, P.B. Identification and cloning of human polynucleotide phosphorylase, hPNPase old-35, in the context of terminal differentiation and cellular senescence. Proc. Natl. Acad. Sci. 2002;99:16636–16641. doi: 10.1073/pnas.252643699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisitsky, I., Klaff, P., Schuster, G. Addition of poly(A)-rich sequences to endonucleolytic cleavage sites in the degradation of spinach chloroplast mRNA. Proc. Natl. Acad. Sci. 1996;93:13398–13403. doi: 10.1073/pnas.93.23.13398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littauer, U.Z., Grunberg-Manago, M. Polynucleotide phosphorylase. In: Creighton T.E., editor. The encyclopedia of molecular biology. Wiley; New York: 1999. pp. 1911–1918. [Google Scholar]
- Meierhoff, K., Felder, S., Nakamura, T., Bechtold, N., Schuster, G. HCF152, an Arabidopsis RNA binding pentatricopeptide repeat protein involved in the processing of chloroplast psbB-psbT-psbH-petB-petD RNAs. Plant Cell. 2003;15:1480–1495. doi: 10.1105/tpc.010397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty, B.K., Kushner, S.R. Polynucleotide phosphorylase functions both as a 3′ to 5′ exonuclease and a poly(A) polymerase in Escherichia coli . Proc. Natl. Acad. Sci. 2000;97:11966–11971. doi: 10.1073/pnas.220295997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty, B.K., Maples, V.F., Kushner, S.R. The Sm-like protein Hfq regulates polyadenylation dependent mRNA decay in Escherichia coli . Mol. Microbiol. 2004;54:905–920. doi: 10.1111/j.1365-2958.2004.04337.x. [DOI] [PubMed] [Google Scholar]
- Montoya, J., Ojala, D., Attardi, G. Distinctive features of the 5′-terminal sequences of the human mitochondrial mRNAs. Nature. 1981;290:465–470. doi: 10.1038/290465a0. [DOI] [PubMed] [Google Scholar]
- Nagaike, T., Suzuki, T., Katoh, T., Ueda, T. Human mitochondrial mRNAs are stabilized with polyadenylation regulated by mitochondria-specific poly(A) polymerase and polynucleotide phosphorylase. J. Biol. Chem. 2005;280:19721–19727. doi: 10.1074/jbc.M500804200. [DOI] [PubMed] [Google Scholar]
- Ojala, D., Montoya, J., Attardi, G. tRNA punctuation model of RNA processing in human mitochondria. Nature. 1981;290:470–474. doi: 10.1038/290470a0. [DOI] [PubMed] [Google Scholar]
- Perrin, R., Lange, H., Grienenberger, J.M., Gagliardi, D. AtmtPNPase is required for multiple aspects of the 18S rRNA metabolism in Arabidopsis thaliana mitochondria. Nucleic Acids Res. 2004;32:5174–5182. doi: 10.1093/nar/gkh852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piwowarski, J., Grzechnik, P., Dziembowski, A., Dmochowska, A., Minczuk, M., Stepien, P.P. Human polynucleotide phosphorylase, hPNPase, is localized in mitochondria. J. Mol. Biol. 2003;329:853–857. doi: 10.1016/s0022-2836(03)00528-x. [DOI] [PubMed] [Google Scholar]
- Portnoy, V., Palnizky, F., Yehudai-Resheff, S., Glaser, F., Schuster, G. Analysis of the human polynucleotide-phosphorylase (PNPase) reveals differences in RNA binding and response to phosphate compared to its bacterial and chloroplast counterparts. RNA. 2008 doi: 10.1261/rna.698108. (this issue). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainey, R.N., Glavin, J.D., Chen, H.W., French, S.W., Teitell, M.A., Koehler, C.M. A new function in translocation for the mitochondrial i-AAA protease Yme1: Import of polynucleotide phosphorylase into the intermembrane space. Mol. Cell. Biol. 2006;26:8488–8497. doi: 10.1128/MCB.01006-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regnier, P., Arraiano, C.M. Degradation of mRNA in bacteria: Emergence of ubiquitous features. Bioessays. 2000;22:235–244. doi: 10.1002/(SICI)1521-1878(200003)22:3<235::AID-BIES5>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Rott, R., Zipor, G., Portnoy, V., Liveanu, V., Schuster, G. RNA polyadenylation and degradation in cyanobacteria are similar to the chloroplast but different from Escherichia coli . J. Biol. Chem. 2003;278:15771–15777. doi: 10.1074/jbc.M211571200. [DOI] [PubMed] [Google Scholar]
- Ryan, C.M., Read, L.K. UTP-dependent turnover of Trypanosoma brucei mitochondrial mRNA requires UTP polymerization and involves the RET1 TUTase. RNA. 2005;11:763–773. doi: 10.1261/rna.7248605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar, D., Fisher, P.B. Polynucleotide phosphorylase: An evolutionary conserved gene with an expanding repertoire of functions. Pharmacol. Ther. 2006;112:243–263. doi: 10.1016/j.pharmthera.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Sarkar, D., Park, E.S., Emdad, L., Randolph, A., Valerie, K., Fisher, P.B. Defining the domains of human polynucleotide phosphorylase (hPNPaseOLD-35) mediating cellular senescence. Mol. Cell. Biol. 2005;25:7333–7343. doi: 10.1128/MCB.25.16.7333-7343.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaduto R.C., Jr, Grotyohann, L.W. Measurement of mitochondrial membrane potential using fluorescent rhodamine derivatives. Biophys. J. 1999;76:469–477. doi: 10.1016/S0006-3495(99)77214-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slomovic, S., Laufer, D., Geiger, D., Schuster, G. Polyadenylation and degradation of human mitochondrial RNA: The prokaryotic past leaves its mark. Mol. Cell. Biol. 2005;25:6427–6435. doi: 10.1128/MCB.25.15.6427-6435.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slomovic, S., Portnoy, V., Liveanu, V., Schuster, G. RNA polyadenylation in prokaryotes and organelles: Different tails tell different tales. Crit. Rev. Plant Sci. 2006;25:65–77. [Google Scholar]
- Temperley, R.J., Seneca, S.H., Tonska, K., Bartnik, E., Bindoff, L.A., Lightowlers, R.N., Chrzanowska-Lightowlers, Z.M. Investigation of a pathogenic mtDNA microdeletion reveals a translation-dependent deadenylation decay pathway in human mitochondria. Hum. Mol. Genet. 2003;12:2341–2348. doi: 10.1093/hmg/ddg238. [DOI] [PubMed] [Google Scholar]
- Tomecki, R., Dmochowska, A., Gewartowski, K., Dziembowski, A., Stepien, P.P. Identification of a novel human nuclear-encoded mitochondrial poly(A) polymerase. Nucleic Acids Res. 2004;32:6001–6014. doi: 10.1093/nar/gkh923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter, M., Kilian, J., Kudla, J. PNPase activity determines the efficiency of mRNA 3′-end processing, the degradation of tRNA and the extent of polyadenylation in chloroplasts. EMBO J. 2002;21:6905–6914. doi: 10.1093/emboj/cdf686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yehudai-Resheff, S., Hirsh, M., Schuster, G. Polynucleotide phosphorylase functions as both an exonuclease and a poly(A) polymerase in spinach chloroplasts. Mol. Cell. Biol. 2001;21:5408–5416. doi: 10.1128/MCB.21.16.5408-5416.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yehudai-Resheff, S., Zimmer, S.L., Komine, Y., Stern, D.B. Integration of chloroplast nucleic acid metabolism into the phosphate deprivation response in Chlamydomonas reinhardtii . Plant Cell. 2007;19:1023–1038. doi: 10.1105/tpc.106.045427. [DOI] [PMC free article] [PubMed] [Google Scholar]