Abstract
The Simian picornavirus type 9 (SPV9) 5′-untranslated region (5′ UTR) has been predicted to contain an internal ribosomal entry site (IRES) with structural elements that resemble domains of hepacivirus/pestivirus (HP) IRESs. In vitro reconstitution of initiation confirmed that this 5′ UTR contains an IRES and revealed that it has both functional similarities and differences compared to HP IRESs. Like HP IRESs, the SPV9 IRES bound directly to 40S subunits and eukaryotic initiation factor (eIF) 3, depended on the conserved domain IIId for ribosomal binding and consequently for function, and additionally required eIF2/initiator tRNA to yield 48S complexes that formed elongation-competent 80S ribosomes in the presence of eIF5, eIF5B, and 60S subunits. Toeprinting analysis revealed that eIF1A stabilized 48S complexes, whereas eIF1 induced conformational changes in the 40S subunit, likely corresponding to partial opening of the entry latch of the mRNA-binding channel, that were exacerbated by eIF3 and suppressed by eIF1A. The SPV9 IRES differed from HP IRESs in that its function was enhanced by eIF4A/eIF4F when the IRES was adjacent to the wild-type coding sequence, but was less affected by these factors or by a dominant negative eIF4A mutant when potentially less structured coding sequences were present. Exceptionally, this IRES promoted binding of initiator tRNA to the initiation codon in the P site of 40S subunits independently of eIF2. Although these 40S/IRES/tRNA complexes could not form active 80S ribosomes, this constitutes a second difference between the SPV9 and HP IRESs. eIF1 destabilized the eIF2-independent ribosomal binding of initiator tRNA.
Keywords: IRES, eIF1, eIF1A, eIF4A, mRNA translation, ribosome
INTRODUCTION
The canonical translation initiation mechanism requires at least 11 eukaryotic initiation factors (eIFs) and separated 40S and 60S ribosomal subunits (Pestova et al. 2007). The latter are generated by dissociation of ribosomal post-termination complexes in a multistep process mediated by eIF1, eIF1A, eIF3, and its loosely associated eiF3j subunit (Pisarev et al. 2007a). eIFs 1, 1A, and 3 enhance association of eIF2•GTP•initiator tRNA (Met-tRNAi Met) to free 40S subunits. Binding of the resulting 43S preinitiation complex to mRNA near its 5′-terminal m7G “cap” is mediated by eIF4F, which consists of the eIF4E cap-binding subunit, the eIF4A ATP-dependent RNA helicase, and eIF4G (which binds eIFs 4E, 4A, and 3). 43S complexes require eIF1 and eIF1A to be able to scan to the initiation codon, which is recognized by the Met-tRNAi Met anticodon. Their base-pairing triggers subunit joining, during which eIF5 induces hydrolysis of eIF2-bound GTP and eIF5B mediates displacement of factors from the 48S complex and joining of a 60S subunit to yield an 80S ribosome.
As well as promoting scanning, eIF1 ensures the fidelity of initiation codon selection and links establishment of codon–anticodon base-pairing with eIF2-mediated GTP hydrolysis by preventing premature hydrolysis of eIF2-bound GTP and Pi release (Pestova et al. 1998a; Pestova and Kolupaeva 2002; Lomakin et al. 2003; Unbehaun et al. 2004; Algire et al. 2005; Maag et al. 2005; Lomakin et al. 2006; Cheung et al. 2007). A model that integrates these roles proposes that eIF1 and eIF1A act by switching the conformation of preinitiation complexes (Pestova and Hellen 2000; Pestova and Kolupaeva 2002; Unbehaun et al. 2004; Maag et al. 2005, 2006): eIF1A and particularly eIF1 are required for them to adopt an “open” scanning-competent conformation in which eIF1 is positioned so that it also represses premature activation of eIF2 by eIF5, and codon-anticodon base-pairing induces the complex to adopt a “closed” conformation characterized by tightening of the interaction of eIF1A with the 40S subunit and dissociation of eIF1, which activates hydrolysis of eIF2-bound GTP and Pi release.
A few eukaryotic mRNAs are translated as a result of internal ribosomal entry, a cap-independent mode of initiation that remains active when eIF4F's cap-binding activity is impaired. Many internal ribosomal entry sites (IRESs) remain poorly defined, particularly in cellular mRNAs, but three groups of viral IRESs have been characterized in detail and serve as models for this mode of initiation. These IRESs bind directly to canonical components of the translation apparatus, and initiation on all of them requires fewer eIFs than the canonical initiation mechanism, but they differ fundamentally in other respects. The ∼200-nucleotide (nt)-long dicistrovirus intergenic region (IGR) IRESs, such as that of Cricket paralysis virus, bind directly to 40S subunits and initiate translation without the involvement of eIFs (Wilson et al. 2000; Pestova et al. 2004). The ∼350-nt-long Hepacivirus (hepatitis C virus [HCV]) and Pestivirus (e.g., classical swine fever virus [CSFV]) (HP) IRESs bind directly to 40S subunits as well as eIF3, and require only them and eIF2•GTP•Met-tRNAi Met to form 48S complexes that can complete the remaining stages in initiation (Pestova et al. 1998b). Initiation on the ∼450-nt-long encephalomyocarditis virus (EMCV) IRES (which epitomizes a major group of picornavirus IRESs) requires all eIFs, except eIFs 4B and 4E, and involves binding of eIF4G/eIF4A to the IRES, followed by ATP-dependent recruitment of the 43S complex to the initiation codon (Pestova et al. 1996a,b). eIF1 ensures selection of the correct initiation codon on this IRES, whereas it reduces the efficiency of initiation on the CSFV IRESs (Pestova et al. 1998a).
HP IRESs consist of the ∼100-nt-long domain II, which enhances 48S complex formation and subunit joining, domain III, which contains a pseudoknot and several branched hairpins that together bind eIF3 and the 40S subunit, and a sequence between the pseudoknot and the initiation codon that in CSFV is unstructured and in HCV forms the hairpin domain IV (Reynolds et al. 1995; Honda et al. 1996; Pestova et al. 1998b; Sizova et al. 1998; Kolupaeva et al. 2000a,b; Kieft et al. 2001; Pisarev et al. 2005; Locker et al. 2007). The coding sequences adjacent to HCV and CSFV IRESs are unrelated but both unstructured, which contributes to initiation efficiency (Reynolds et al. 1995; Fletcher et al. 2002).
HP IRESs occur in two distinct genera of Flaviviridae, but putative HP-like IRESs have also recently been identified in several picornaviruses, of which those in porcine teschovirus type 1 (PTV1), porcine enterovirus 8 (PEV8), and Simian picornavirus type 1 (SPV1) have been experimentally confirmed (Pisarev et al. 2004, 2005; Chard et al. 2006a,b; Hellen and de Breyne 2007; Kapoor et al. 2007). The sporadic distribution of these elements in genera of Picornaviridae (which are not related to Flaviviridae) and in some but not all genera of Flaviviridae suggests that they have been exchanged between viruses by recombination. Despite their overall similarity, these elements are distinct and differ from HP IRESs in several respects, including the structures of the pseudoknot and the apical region of domain III, the size of domain II, and the sequence and likely structure of the proximal coding sequence. The putative Simian picornavirus type 9 (SPV9) IRES appears to be one of the most divergent in this group, and we therefore investigated whether its divergent elements lead to mechanistic differences in initiation on it from a strictly HP-like mechanism. The SPV9 IRES resembled HP IRESs in that eIF3 and 40S subunits bound directly to sites on it that were, despite sequence and structural differences, analogous to their binding sites on HP IRESs, and in that the addition of eIF2•GTP•Met-tRNAi Met led to formation of 48S complexes. However, the SPV9 IRES differed from HP IRESs in being partially dependent on the eIF4 group of factors in a manner that was obviated by mutation of sequences downstream from the initiation codon that were designed to reduce potential secondary structure. Moreover, unlike any IRES that has been examined to date, the SPV9 IRES promoted specific eIF2-independent binding of Met-tRNAi Met to the initiation codon in the peptidyl (P) site of the 40S subunit, although the resulting complexes were unable to form active 80S ribosomes. Toeprinting analysis also revealed distinct conformational changes that occurred upon binding of initiation factors to the SPV9 IRES/40S subunit complex.
RESULTS
The SPV9 5′ UTR contains an IRES
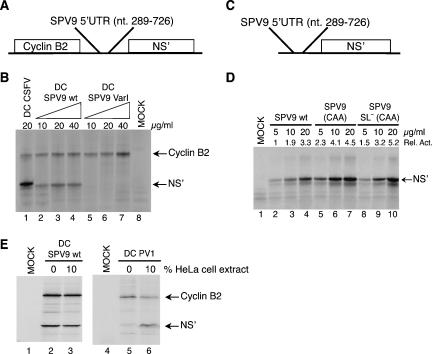
The 3′ half of the SPV9 5′ UTR resembles HP IRESs (Fig. 1A; Hellen and de Breyne 2007), suggesting that it too might be an IRES. To test this, SPV9 nucleotides 289–726 were inserted into DC SPV9 (wild type [wt]), between the cyclin B2 cistron and a downstream cistron encoding a truncated influenza NS protein (NS′) (Fig. 2A). SPV9 nucleotides 289–726 contain putative IRES domains II, III, and IV (Fig. 1A) and nucleotides 1–48 of the coding region, which were included because the corresponding HCV and CSFV sequences enhance IRES function by virtue of their unstructured nature (Reynolds et al. 1995; Fletcher et al. 2002). Uncapped DC SPV9 (wt) mRNA translated in rabbit reticulocyte lysate (RRL) yielded similar amounts of NS′ and cyclin B2 whereas translation of dicistronic CSFV mRNA yielded about as much cyclin B2 as DC SPV9 (wt) mRNA but a substantially larger amount of NS′ (e.g., Fig. 2B, lanes 1–4), indicating that the SPV9 IRES is less active than the CSFV IRES (Fig. 2B, lanes 1,3). We note that the NS′ open reading frame in DC CSFV is 18 nt longer than in DC SPV9 (wt) and consequently yields a larger NS′ protein. Translation of some picornavirus mRNAs in RRL is weak, aberrant, or both, but can be enhanced or corrected by addition of cytoplasmic extracts from permissive cells to the RRL (Dorner et al. 1984; Svitkin et al. 1988). Whereas addition of a cytoplasmic HeLa cell extract to the RRL enhanced poliovirus IRES function (Fig. 2E, lanes 5,6), it did not increase the activity of the SPV9 IRES nor did it alter the fidelity of SPV9 IRES-mediated translation of NS′ (Fig. 2E, lanes 2,3). SPV9 nucleotides 289–726 therefore comprise an IRES, and RRL contains adequate amounts of all factors required for its activity.
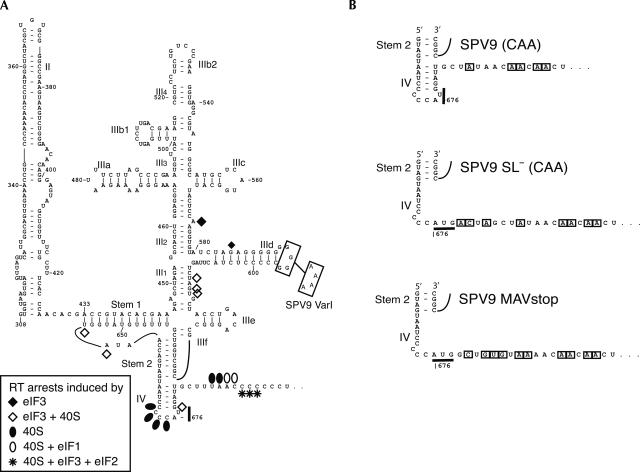
FIGURE 1.
(A) Model of the secondary and tertiary structure of the Simian picornavirus type 9 IRES (nucleotides 308–697) (Hellen and de Breyne 2007). The nomenclature of domains is based on proposals for the HCV IRES (Honda et al. 1996). The initiation codon AUG676 and the substitutions GGGG593–596AAAA in SPV9 (Var1) are indicated. Sites at which primer extension was arrested or enhanced by binding of eIF3, of 40S subunits, of 40S subunits with eIF3 or eIF1, and of the 48S complex to the IRES are indicated by symbols (shown at the lower left). (B) Sequences flanking the initiation codon of SPV9 (CAA), SPV9 SL− (CAA), and SPV9 MAVstop variants, in which substitutions relative to the wild-type coding sequence are boxed.
FIGURE 2.
Effect of cis-acting elements on SPV9 IRES function. (A) Schematic representation of dicistronic mRNAs containing cyclin B2 and influenza NS′ cistrons separated by either CSFV nucleotides 1–442, SPV9 (wt), or SPV9 (Var1). (C) Schematic representation of monocistronic mRNAs containing the NS′ cistron preceded by SPV9 (wt), SPV9 (CAA), or SPV9 SL− (CAA). (B,D) Translation of dicistronic (B) and monocistronic mRNAs (D) at the indicated concentrations in RRL containing [35S]methionine. Products were analyzed by gel electrophoresis and autoradiography. SPV9 IRES-mediated initiation (D) was quantified using a Molecular Dynamics PhosphoImager relative to translation of SPV9 (wt) mRNA (5 μg/mL) (lane 2), which was defined as 1. (E) The influence of a cytoplasmic S10 extract added to RRL at the indicated proportions on translation of DC SPV9 (wt) mRNA (lanes 2,3) and DC PV1 mRNA (lanes 5,6) was quantified using a Molecular Dynamics PhosphorImager after separation of translation products by gel electrophoresis. Lanes labeled “MOCK” correspond to RRL incubated without exogenous mRNA.
The G-rich loop of domain IIId is a feature of all HP-like IRESs (Pisarev et al. 2005). It is a primary determinant of ribosomal binding, and mutation of it led to loss of HCV and CSFV IRES function (Kolupaeva et al. 2000b; Kieft et al. 2001). G → A substitution of SPV9 IIId loop residues GGGG593–596 (Fig. 1A) in DC SPV9-Var1 also abrogated SPV9 IRES function (Fig. 2B, lanes 5–7).
The SPV9 initiation codon, AUG676, is sequestered in a predicted stem–loop that constitutes domain IV (ΔG = −3.4kcal/mol), which is adjacent to a coding region that arrests reverse transcriptase (RT) in toeprinting assays and it is thus also presumably involved in secondary structure formation (see below). To characterize the influence of these two potentially structured elements on IRES function, monocistronic mRNAs were made that contained the wt IRES and the adjacent wt coding region (SPV9 [wt]), the wt IRES and an adjacent coding region containing CAA repeats (designed to destabilize potential secondary structure formed by the wild-type C-rich region) (SPV9 [CAA]), or a mutated IRES, in which the predicted domain IV is destabilized, and the adjacent coding region was substituted by CAA repeats (SPV9 SL− [CAA]) (Fig. 1B). Translation of these uncapped mRNAs in RRL showed that SPV9 IRES function was enhanced up to twofold by mutation of the coding region downstream from the IRES, whereas additional mutations that disrupted the predicted domain IV did not result in further translational activation (Fig. 2D, lanes 2–4,5–7, 8–10).
The SPV9 IRES binds directly and stably to 40S ribosomal subunits
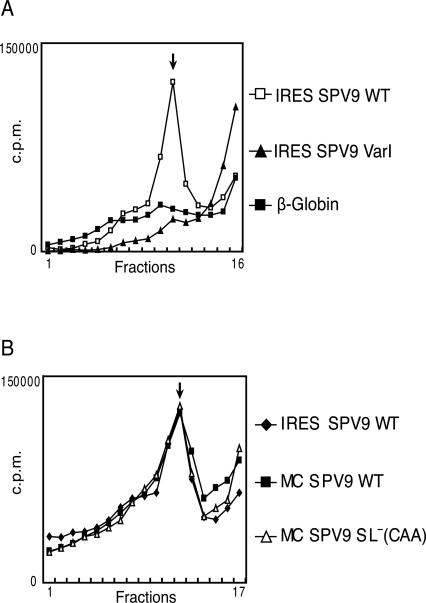
HP IRESs bind directly to 40S subunits, forming stable binary complexes that can be resolved by sucrose density gradient centrifugation (Pestova et al. 1998b; Pestova and Hellen 1999). The SPV9 IRES also formed stable binary complexes with 40S subunits, and their assembly was abrogated by the GGGG593–596AAAA mutation (Fig. 3A), indicating that the domain IIId loop acts at the same step in initiation on SPV9 as on CSFV and HCV IRESs (Kolupaeva et al. 2000b; Kieft et al. 2001). Binding of the SPV9 IRES to 40S subunits in this assay was unaffected by the presence of a downstream cistron (compare IRES SPV9 wt, which corresponds to SPV9 nucleotides 289–726, with the monocistronic MC SPV9 wt mRNA) nor was it affected by mutations downstream from the initiation codon that enhanced translation (compare MC SPV wt and MC SPV9 SL− [CAA] mRNAs; Fig. 3B). Thus, as in HP IRESs, the coding sequence does not act as a primary determinant of the IRES–40S subunit interaction.
FIGURE 3.
Factor-independent binding of 40S subunits to the SPV9 IRES. Ribosomal complexes were assembled by incubating (A) [32P]UTP-labeled SPV9 (wt), SPV9 (Var1), or β-globin mRNAs or (B) [32P]UTP-labeled SPV9 (wt), MC SPV9 (wt), or MC SPV9 SL− (CAA) mRNAs (as indicated) with 40S subunits. Ribosomal complexes were analyzed by centrifugation through a 10%–30% sucrose density gradient. Sedimentation was from right to left. The position of binary IRES–40S subunit complexes is indicated. Fractions from upper parts of the gradient have been omitted for greater clarity.
In toeprinting assays, association of 40S subunits with SPV9 (CAA) or SPV9 SL− (CAA) mRNAs induced RT stops at positions +12–13 relative to A (+1) of the initiation codon AUG676 and enhanced stops at −4–−1 (Fig. 1A, filled circles; Figs. 4, 6, see below), confirming that the IRES binds 40S subunits independently of Met-tRNAi Met and eIFs and indicating that AUG676 is placed in or near the ribosomal P site. The corresponding toeprint on SPV9 (wt) mRNA was substantially obscured by the background stops caused by the proximal C-rich region of the coding sequence (Fig. 4F).
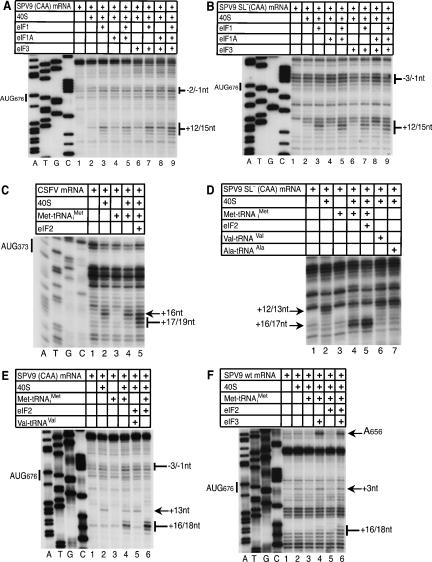
FIGURE 4.
eIF2-independent formation of 48S initiation complexes. MC SPV9 (CAA) (A,E), MC SPV9 SL− (CAA) (B,D), DC CSFV (C), and SPV9 (wt) mRNA (F) were incubated with 40S subunits, eIF2, eIF3, Met-tRNAi Met, Ala-tRNAAla, and Val-tRNAVal as indicated above each panel. Reaction conditions are described in Materials and Methods. Extension by AMV-RT of a primer annealed to the NS′ cistron was arrested at sites indicated on the right (A,B,D–F) or left (D) as positions relative to the A (position +1) of the SPV9 initiation codon (A,B,D,E) or the CSFV initiation codon. Reference lanes A, T, G, and C depict (A,B,E,F) SPV9 (CAA) and (C) CSFV sequences. For clarity, only a portion of each gel is shown.
FIGURE 6.
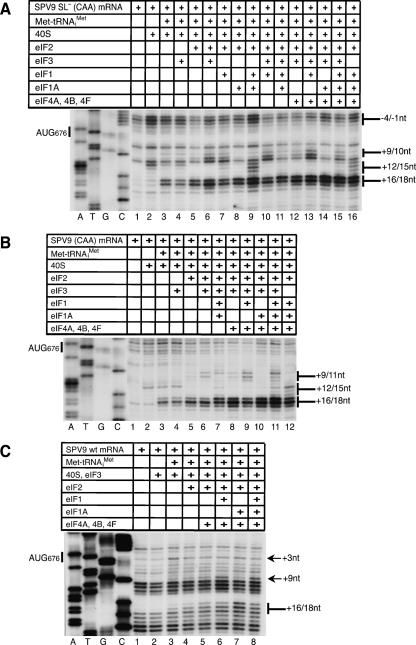
The influence of initiation factors and proximal coding sequences on 48S complex formation on the SPV9 IRES. (A) MC SPV9 SL− (CAA), (B) MC SPV9 (CAA), and (C) DC SPV9 (wt) mRNA were incubated with 40S subunits, eIF2, eIF3, eIF1, eIF1A, eIF4A, eIF4B, eIF4F, and Met-tRNAi Met as indicated, in standard conditions. Extension by AMV-RT of a primer annealed to the NS′ cistron was arrested at sites indicated on the right as positions relative to the A (position +1) of the initiation codon. Reference lanes A, T, G, and C depict the SPV9 (CAA) sequence.
Binding of 40S subunits was influenced by eIFs 1, 1A, and 3. eIF1A and eIF3 individually, and more strongly together, enhanced the +12–13 toeprints without changing their pattern. However, the toeprint pattern was changed by eIF1, either alone or in combination with eIFs 1A and 3. Thus, eIF1 particularly enhanced the +13 toeprint and led to the appearance of additional stops at +14–15 (Fig. 1A, open circles; Fig. 4A,B, lanes 3,5,7,9), which suggests that binding of eIF1 induces conformational changes either in the 40S subunit itself or in the IRES/40S subunit complex.
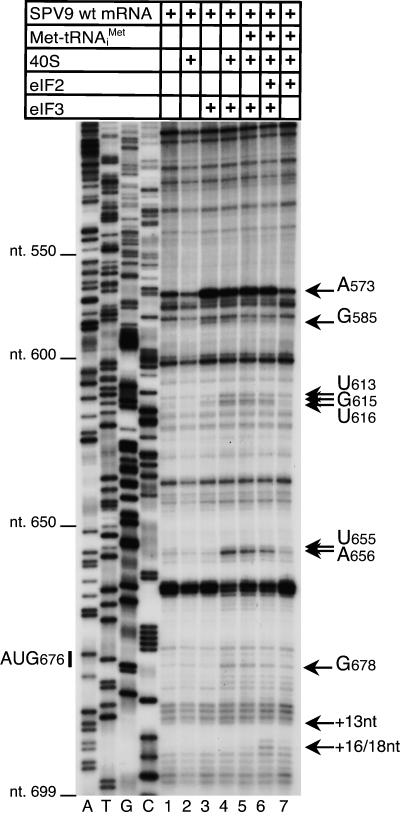
Complexes assembled on wt or mutated SPV9 mRNA and that contained both eIF3 and 40S subunits also yielded additional toeprints of different intensities at U613, GU615–6, UA655–6, and G678 (Fig. 1A, open diamonds; Fig. 5, lanes 4–6; data not shown) that were either the consequence of conformational changes in the IRES or represented stable new contacts of these nucleotides (which are outside the mRNA-binding cleft of the 40S subunit) with eIF3, the 40S subunit, or both.
FIGURE 5.
Influence of eIF3 on the IRES–40S subunit complex. DC SPV9 (wt) mRNA was incubated with 40S subunits, eIF2, eIF3, and Met-tRNAi Met as indicated, in standard conditions. Extension by AMV-RT of a primer annealed to the NS′ cistron was arrested at sites indicated on the right either as nucleotides in the SPV9 5′ UTR or as positions relative to the A (position +1) of the initiation codon. Reference lanes A, T, G, and C depict the SPV9 (CAA) sequence.
eIF3 binds specifically to the SPV9 IRES
A characteristic of HP IRESs is that they bind eIF3 specifically. The wt SPV9 IRES also bound eIF3 in the absence of 40S subunits and other eIFs, strongly enhancing RT stops at A573 and less strongly at G585 (Fig. 5, lanes 1,3). The position of the A573 stop (Fig. 1A, filled diamond) is analogous to stops at bulged A residues in helix III2 induced by eIF3 bound to bovine viral diarrhea virus (BVDV), CSFV, and HCV IRESs (Pestova et al. 1998b; Sizova et al. 1998; Pestova and Hellen 1999).
Factor-independent and eIF2-mediated attachment of Met-tRNAi Met to ribosomal complexes assembled on the SPV9 IRES
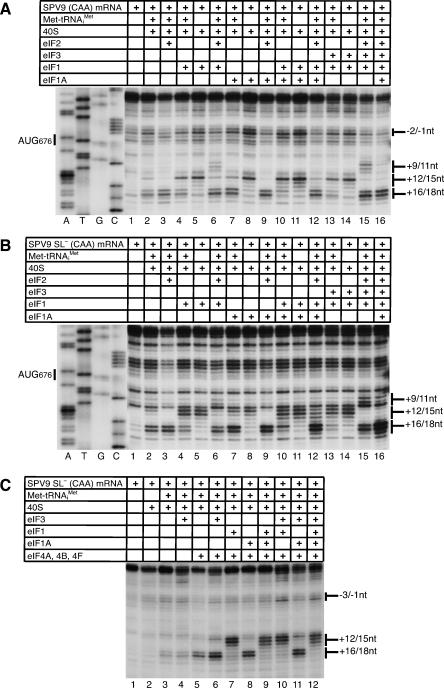
40S subunits attach directly to HP-like IRESs such as the CSFV IRES, and subsequent binding of eIF2•GTP•Met-tRNAi Met leads to efficient 48S complex formation, marked by a shift forward of toeprints that correspond to the leading edge of the 40S subunit (Pestova et al. 1998b). In contrast, assembly of 40S subunits and eIF2•GTP•Met-tRNAi Met to form 48S complexes on SPV9 wt mRNA yielded only very weak toeprints at the +16–+17 positions that are characteristic of 48S complexes (Fig. 4F, lane 5). Any potential stop at +18 was obscured by endogenous stops induced by the proximal coding region of this mRNA. Translation in RRL showed that initiation on the wt SPV9 IRES is less efficient than on the CSFV IRES, but it is not possible to judge accurately the efficiency of 48S complex formation by toeprinting because toeprints on the wt SPV9 IRES are partially obscured/masked by endogenous overlapping stops on this mRNA. However, these stops were eliminated by the mutations in the coding region of SPV9 (CAA) and SPV9 SL− (CAA) mRNAs, and it was therefore apparent that these mutated IRESs supported relatively efficient assembly of 48S complexes in the presence of 40S subunits and eIF2•GTP•Met-tRNAi Met ternary complexes, yielding prominent +16–18 toeprints (Fig. 4D, lane 5; Fig. 4E, lane 6).
Surprisingly, 48S complex formation on SPV9 (CAA) and SPV9 SL− (CAA) mRNAs was reduced only twofold on omission of eIF2•GTP (Fig. 4D, lanes 4,5; Fig. 4E, lanes 4,6). These characteristic +16–18 toeprints were not induced in the absence of 40S subunits or when Met-tRNAi Met was replaced by Val-tRNAVal or Ala-tRNAAla (Fig. 4D, lanes 3,4,6,7; Fig. 4E, lanes 3–5). Met-tRNAi Met therefore binds specifically and stably to AUG676 of the SPV9 IRES in ribosomal complexes in the absence of eIF2 on mutant mRNAs in which the coding sequence downstream from the IRES is predicted to be unstructured, and the IRES, 40S subunits, and Met-tRNAi Met are thus sufficient to form 48S complexes on such mRNAs. Formation of [35S-Met]tRNA/SPV9 RNA/40S subunit complexes could not be monitored by sucrose density gradient centrifugation (data not shown), which is consistent with the previously reported instability of binding of initiator tRNA to 40S subunits in the absence of eIF2 (Unbehaun et al. 2004). Stable binding of Met-tRNAi Met to IRES/40S subunit complexes in the absence of eIF2•GTP and other factors is specific to SPV9 and was not observed on the CSFV IRES (Fig. 4C, lanes 2,4,5; Pestova et al. 1998b). The influence of eIF2 on 48S complex formation on SPV9 wt mRNA could not be evaluated reliably because this process was not readily monitored by toeprinting even in the presence of eIF2 (Fig. 4F, lanes 3,5). Although the stops characteristic of 48S complexes were partially obscured on SPV9 wt mRNA, a low level of eIF2-independent ribosomal binding of Met-tRNAi Met was apparent in reactions containing eIF3 (Fig. 4F, lanes 3,4; Fig. 6C, lanes 2,3).
The SPV9 5′ UTR contains 19 AUG triplets upstream of AUG676, including eight in the nucleotides 289–726 fragment used in these experiments, but assembly of 48S complexes was not detected on any of them in in vitro reconstituted initiation reactions in the presence of any combination of factors (Fig. 5; data not shown).
Influence of eIF3 on 48S complex formation on wild-type and mutant SPV9 mRNAs
Inclusion of eIF3 in reaction mixtures containing 40S subunits, Met-tRNAi Met, and the wt SPV9 IRES led to the appearance of weak +16–17 toeprints that are characteristic of 48S complexes (Fig. 4F, lanes 3,4). eIF3 enhanced 48S complex formation on SPV9 (wt) mRNA to a greater extent when eIF2•GTP was also present (Fig. 4F, lanes 5,6; Fig. 5, lanes 6,7). Although eIF3 did not enhance 48S complex formation on SPV9 (CAA) or SPV9 SL− (CAA) mRNAs in reaction mixtures that included Met-tRNAi Met but not eIF2•GTP (Fig. 6A,B, lanes 3,4), it enhanced 48S complex formation on SPV9 (CAA) and particularly on SPV9 SL− (CAA) mRNA in the presence of eIF2•GTP (Fig. 6A,B, lanes 5,6). These results suggest that eIF3 may induce the resulting complex to adopt a conformation that stabilizes binding of Met-tRNAi Met, particularly if it is a constituent of eIF2•GTP•Met-tRNAi Met ternary complexes. Importantly, the presence of eIF3 in reaction mixtures containing 40S subunits and eIF2•GTP•Met-tRNAi Met led to the appearance of additional toeprints at +9–11 positions (Fig. 6A,B, lanes 5,6), which could also be a manifestation of conformational changes in ribosomal complexes induced by eIF3. As noted above, inclusion of eIF3 with 40S subunits bound to the SPV9 IRES without other eIFs yielded additional toeprints (Fig. 5, lanes 2–4) that are consistent with induced conformational changes in these ribosomal complexes.
The influence of eIF1 and eIF1A on 48S complex formation
eIF1 and eIF1A play key functions in scanning and AUG recognition in the canonical initiation mechanism but destabilize 48S complexes on the CSFV IRES (Pestova et al. 1998a; Pestova and Kolupaeva 2002). The ability of SPV9 (CAA) and SPV9 SL− (CAA) mRNAs to form 48S complexes in the presence of only 40S subunits and Met-tRNAi Met permitted the effects of eIF1 and eIF1A on ribosomal complexes to be characterized in the presence of any combination of factors. eIF1A stabilized 48S complexes assembled on SPV9 SL− (CAA) and SPV9 (CAA) mRNAs with Met-tRNAi Met in the absence of eIF2•GTP (Fig. 7A,B, lanes 2,7) whereas eIF1 destabilized such complexes (Fig. 7A,B, lanes 2,4), even when the stabilizing eIF1A was present (Fig. 7A,B, lanes 2,7,10). The destabilizing effect of eIF1 on these complexes was exacerbated slightly by eIF3 (Fig. 7A,B, lanes 4,13). In all cases, the presence of eIF1 resulted in strong decreases in the intensity of +16–18 toeprints that are characteristic for 48S complexes, and simultaneous increases in the intensity of +12–15 toeprints that are characteristic of binary IRES/40S subunit complexes assembled in the absence of Met-tRNAi Met.
FIGURE 7.
Effect of eIF1 and eIF1A on 48S complex formation. MC SPV9 (CAA) (A) and MC SPV9 SL− (CAA) (B,C) mRNAs were incubated with 40S subunits, eIF2, eIF3, eIF1, eIF1A, eIF4A, eIF4B, eIF4F, and Met-tRNAi Met as indicated, in standard conditions. Extension by AMV-RT of a primer annealed to the NS′ cistron was arrested at sites indicated on the right as positions relative to the A (position +1) of the initiation codon. Reference lanes A, T, G, and C depict the SPV9 (CAA) sequence.
The presence of eIF2•GTP strongly protected 48S complexes against destabilization by eIF1 in assembly reactions containing 40S subunits, Met-tRNAi Met, and SPV9 SL− (CAA) or SPV9 (CAA) mRNAs, both in the presence and the absence of eIF1A (Fig. 7A,B, cf. lanes 2,4,6 and 7,10,12). Partial destabilization of 48S complexes by eIF1 was nevertheless apparent in these reactions by the enhanced prominence of toeprints at +12–15, particularly at +12. The presence of eIF1 in reactions that contained eIF2•GTP•Met-tRNAi Met also led to the appearance of additional toeprints at +9–10 (Fig. 6A, lanes 5,7,10,13,15) or at +9–11 positions (Fig. 6B, lanes 9,11), which were also observed in the presence of eIF3 (Fig. 6A,B, lanes 5,6), suggesting that eIF3 and eIF1 might induce similar conformational changes in ribosomal complexes. In some cases when eIF1 and eIF3 were both present, +9–11 toeprints were more prominent than when eIF1 and eIF3 were present individually (Fig. 6A, lanes 6,7,10; Fig. 7A,B, lanes 6,15). Although toeprints were partially obscured by strong endogenous RT stops on the C-rich structure-prone coding sequence of wt SPV9 mRNA, it was apparent that eIF1 and eIF3 induced similar changes in ribosomal complexes assembled on wt SPV9 mRNA, particularly at position +9, and acted synergistically (Fig. 6C). eIF2•GTP and Met-tRNAi Met were required for the eIF1/eIF3-induced +9–11 toeprints to appear (cf. Fig. 7A,B, lanes 4–6 and lanes 13–15; Fig. 6A, lanes 4–6) whereas eIF3 was not required if eIF1 was present (Fig. 6A, lanes 7,10; Fig. 7A,B, lanes 3,6,15).
eIF1A suppressed the appearance of the +9–11 toeprints when eIF3 (Fig. 6A, lanes 12,14; Fig. 6B, lanes 8,10) and eIF1 (e.g., Fig. 6A, lanes 7,9; Fig. 7A, lanes 6,12) were present individually on SPV9 SL− (CAA) or SPV9 (CAA) mRNAs. The influence of eIF1A was significantly less when eIF1 and eIF3 were present together (Fig. 6A, lanes 10,11,13,15; Fig. 6B, lanes 9,11).
eIF4A, 4B, and 4F enhance translation initiation on the SPV9 IRES
Inclusion of eIFs 4A, 4B, and 4F in reaction mixture substantially enhanced 48S complex formation on SPV9 (CAA) mRNA in the presence of eIFs 2 and 3 (Fig. 6B, cf. lanes 6 and 8) or in the presence of eIFs 2, 3, 1, and 1A (Fig. 6B, cf. lanes 7 and 11). In identical conditions, the stimulatory effect of eIFs 4A, 4B, and 4F on 48S complex formation on SPV9 SL− (CAA) mRNA was lower (Fig. 6A, cf. lanes 6,12 and 11,15). The requirement for eIFs 4A, 4B, and 4F therefore decreased in parallel with the decrease in the degree of potential secondary structure downstream from the pseudoknot. Interestingly, the stimulatory effect of eIFs 4A, 4B, and 4F on SPV9 SL− (CAA) mRNA became very pronounced when eIF2 was omitted from the reaction mixture (Fig. 7C, cf. lanes 3,5 and 4,6), which implies that, in the absence of eIF2, attachment of Met-tRNAi Met may be more sensitive to the secondary structure of the IRES in the vicinity of the initiation codon. eIFs 4A, 4B, and 4F neither protected 48S complexes assembled in the absence of eIF2 from the destabilizing action of eIF1 (Fig. 7C, lanes 7,9,10,12) nor, in the presence of eIF2•GTP•Met-tRNAi Met, influenced the appearance of +9–11 toeprints that are induced in the presence of eIF1 and/or eIF3 (Fig. 6A, lanes 6,8,10,13).
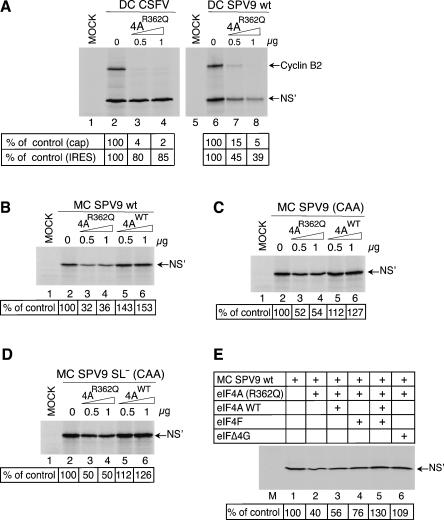
Initiation on mRNAs that require eIF4F (or eIF4A/eIF4G) for ribosomal recruitment is inhibited by dominant negative eIF4AR362Q mutant (Pause et al. 1994) whereas initiation by mechanisms that are eIF4A-independent (such as on the CSFV IRES) is not (Pestova et al. 1998b). The differential stimulatory effect of eIFs 4A, 4B, 4F on 48S complex formation on SPV9 mRNAs described above suggests that translation on these mRNAs might be sensitive to eIF4AR362Q mutant and that any potential sensitivity might depend on the secondary structure in these mRNAs around the initiation codon. Consistent with previous reports (Pestova et al. 1998b), and in contrast to end-dependent translation of cyclin B2 (Fig. 8A, lanes 2–4, 6–8), CSFV IRES-mediated translation of the second cistron of bicistronic mRNA was 85% resistant to inhibition by eIF4AR362Q whereas translation of the second cistron driven by the SPV9 IRES was only ∼40% resistant to inhibition (Fig. 8A, lanes 2–4, 6–8). The level of resistance of translation of monocistronic SPV9 mRNA to inhibition by eIF4AR362Q was very similar to that of the second cistron in bicistronic SPV9 mRNA (Fig. 8B, lanes 2,4), and therefore to avoid complications due to competition for initiation components between IRES-mediated and end-dependent modes of initiation that might arise when using dicistronic mRNAs, the effects of eIF4AR362Q on translation of variant SPV9 mRNAs with differing sequences downstream from the initiation codon were assessed using monocistronic mRNAs. SPV9 wt mRNA was slightly, but reproducibly, more sensitive to inhibition than the mutant RNAs in which domain IV and/or the adjacent structure-prone coding region were mutated (Fig. 8B–D, cf. lanes 2–4). These results are consistent with the results of in vitro reconstitution and confirm that, although eIF4F is not an essential factor for initiation on the SPV9 IRES, it nevertheless enhances 48S complex formation on this IRES, particularly when wild type rather than mutated sequences are present immediately downstream from AUG676. Inhibition of translation by eIF4AR362Q is due to sequestration of eIF4G into inactive eIFG/eIF4A complexes (Pause et al. 1994). Consistently, the relatively modest inhibition of SPV9 translation by eIF4AR362Q was overcome by addition of eIF4F or eIFΔ4G but not of wt eIF4A to RRL (Fig. 8E): eIF4A is one of the most abundant initiation factors, so that addition of eIFΔ4G to RRL restores levels of the eIF4G/eIF4A complex. Consistently, supplementation of RRL with wt eIF4A modestly enhanced translation of SPV9 wt mRNA and had a smaller stimulatory effect on translation of SPV9 (CAA) and SPV9 SL− (CAA) mRNAs (Fig. 8B–D, lanes 2,5,6). Taken together, these observations suggest that eIF4F may enhance productive ribosomal attachment by contributing to accommodation of mRNA flanking the SPV9 initiation codon in the mRNA-binding channel of the 40S subunit.
FIGURE 8.
The influence of proximal coding sequences on the sensitivity of SPV9 IRES-mediated translation to inhibition by trans-dominant eIF4AR362Q. (A) RRL was preincubated alone (lanes 2,6) or with the indicated amounts (μg) of eIF4AR362Q mutant (lanes 3,4,7,8) for 5 min at 30°C and then incubated for 55 min at 30°C without exogenous mRNA (lanes 1,5), with DC CSFV mRNA (lanes 2–4) or DC SPV9 (wt) mRNA (lanes 6–8). (B–D) RRL was pre-incubated alone (lanes 1,2) or with the indicated amounts (μg) of eIF4Awt (lanes 3,4) or eIF4AR362Q mutant (lanes 5,6) for 5 min at 30°C and then incubated for 55 min at 30°C with (A) DC CSFV, (B) DC SPV9 (wt), (B) MC SPV9 (wt), (C) MC SPV9 (CAA), (D) MC SPV9 SL− (CAA) mRNAs (10 μg/mL), or (B–D, lane 1) without exogenous mRNA. (E) MC SPV9 (wt) mRNA (10 μg/mL) was incubated in pretreated RRL without (lane 1) or with 1 μg eIF4AR362Q mutant (lanes 2–6) and with 1 μg of eIF4Awt, eIF4F, or eIFΔ4G as indicated for 55 min at 30°C. (Lane M) Incubation of RRL without exogenous mRNA or factors for 55 min at 30°C. Translation products were visualized by autoradiography after electrophoresis on NuPAGE 4%–12% Bis-Tris-Gel. SPV9 IRES-mediated initiation was quantified using a Molecular Dynamics PhosphorImager relative to translation in the absence of exogenous eIF4A, which was defined as 100%.
eIF2•GTP•Met-tRNAi Met and eIF3 are necessary and sufficient to form active 48S complexes on the SPV9 IRES
In vitro reconstituted translation reactions were extended to include the remaining major step in initiation (subunit joining) and several cycles of elongation to confirm the activities of 48S complexes assembled in the presence of different combinations of factors. The wt SPV9 coding sequence (Met–Asp–Cys–Phe) was altered to Met–Ala–Val–Stop, yielding SPV9 MAVstop mRNA (Fig. 9C), to circumvent potential complications in toeprinting assays due to the strong RT stops that occur downstream from the initiation codon on the wt SPV9 IRES, and to permit codon-dependent ribosomal elongation steps to be monitored in toeprinting assays done using available pure aminoacyl-tRNAs.
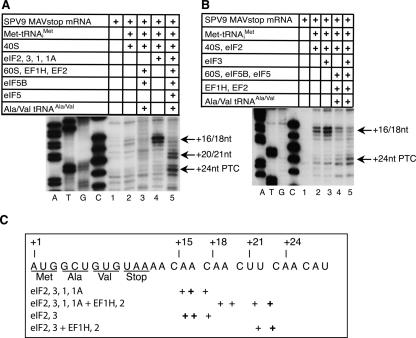
FIGURE 9.
Factor requirements for assembly of 48S complexes that can form elongation-competent ribosomes. (A,B) MC SPV9 MAVstop mRNA was incubated for 10 min at 37°C with 40S subunits, eIFs 1, 1A, 2 and 3, and Met-tRNAi Met as indicated to assemble 48S complexes. 80S ribosomes were formed by incubating these 48S complexes with eIF5, eIF5B, and 60S subunits for 10 min at 37°C, and were programmed to complete two cycles of codon-dependent elongation by incubation with eEF1H, eEF2, Ala-tRNAAla, and Val-tRNAVal for 10 min at 37°C. A primer annealed to the NS′ cistron and extended with AMV-RT was arrested at sites indicated on the right as positions relative to the A (position +1) of the AUG codon. Reference lanes A, T, G, and C depict (panel A) SPV9 (CAA) and (panel B) the SPV9 MAVstop sequences. PTC refers to the ribosomal pretermination complex, which contains peptidyl-tRNA in the P site and the stop codon in the A site. (C) Summary of the positions of ribosomal complexes assembled on SPV9 MAVstop mRNA in the presence of initiation and elongation factors as indicated. The positions of toeprints relative to nucleotide +1 of the AUG codon in the P site before translocation are indicated schematically below the SPV9 MAVstop sequence.
48S complexes formed efficiently on SPV9 MAVstop mRNA in the presence of eIFs 2, 3, 1, and 1A (Fig. 9A, lane 4) or in the presence of eIFs 2 and 3 (Fig. 9B, lane 3), yielding characteristic toeprints at positions +16–18. In the presence of eIF2 alone, 48S complex formation was about two times lower (Fig. 9B, lane 2). Less efficient 48S complex formation also occurred in the presence of only Met-tRNAi Met (Fig. 9A, lane 2).
Upon addition of eIFs 5 and 5B, 60S subunits, elongation factors eEF1H and eEF2, Ala-tRNAAla and Val-tRNAVal, 48S complexes obtained in the presence of eIFs 2, 3, 1, and 1A or in the presence of eIFs 2 and 3 underwent two consecutive elongation steps, very strongly reducing the intensity of +16–18 toeprints and leading to the appearance of +20–21 and +24 toeprints (Fig. 9A,B, lane 5). eIF3 was required for formation of 48S complexes that could efficiently undergo downstream translation events (Fig. 9B, lanes 4,5). 48S complexes assembled in the absence of initiation factors were not competent to undergo elongation and, moreover, were destabilized by addition of eIF5B, 60S subunits, eEF1H, and eEF2 (Fig. 9A, lanes 2,3).
DISCUSSION
The SPV9 RNA genome contains one of a group of IRESs that resembles HP IRESs (Hellen and de Breyne 2007). Here, we report reconstitution of the entire initiation process on it in vitro from individual purified components to yield 80S ribosomes that can undergo cycles of programmed elongation. Characterization of the initiation mechanism revealed aspects that resemble initiation on HP IRESs and others that are unique to SPV9. This analysis also yielded evidence for conformational changes induced in ribosomal initiation complexes by eIF3, eIF1, and eIF1A that suggest insights into their mechanism of action during canonical end-dependent initiation.
The mechanism of initiation on the SPV9 IRES
Although there are differences between the mechanisms of initiation on SPV9 and HP IRESs that may be accounted for by their structural differences, the similarities are notable: like HP IRESs (Pestova et al. 1998b; Pestova and Hellen 1999), the SPV9 IRES binds 40S subunits directly such that the initiation codon is placed in or near the P site. eIF3 also binds directly to the apical half of domain III, and eIF2•GTP•Met-tRNAi Met and eIF3 are required for assembly of 48S complexes that can form elongation-competent 80S ribosomes. These properties are comparable to aspects of initiation on the HP-like PTV1 IRES, which also binds eIF3 and 40S subunits and requires only a 40S subunit and eIF2•GTP•Met-tRNAi Met to form a 48S complex that is stabilized by eIF3 (Pisarev et al. 2004). Notably, the PTV IRES may be as much as 100 nt shorter than the SPV9 IRES, and IRES domain II and IIIb/IIIc in it are predicted to be shorter than in the SPV9 IRES (Pisarev et al. 2004; Chard et al. 2006b; Hellen and de Breyne 2007). Although 40S subunits bound directly to SPV9 and PTV1 IRESs to form complexes that can be detected by sucrose density gradient centrifugation, such binary complexes do not yield toeprints in the pseudoknot that are characteristic of HP IRESs (Pestova et al. 1998b; Pestova and Hellen 1999; Pisarev et al. 2004). However, such toeprints did appear in this region when the SPV9 IRES was bound to a 40S subunit in the presence of eIF3 (Fig. 5), which suggests that eIF3 might induce conformational changes that result in establishing/tightening of specific contacts between the SPV9 IRES and the 40S subunit that are characteristic for HP IRESs. The fundamental similarity in the mode of binding of these IRESs to 40S subunits is underscored by the observation that the G-rich loop of domain IIId is a primary determinant of ribosomal binding to the SPV9 IRES and consequently for its activity, just as for HCV and CSFV IRESs (Kolupaeva et al. 2000b; Kieft et al. 2001). Binding of eIF3 to these IRESs yields toeprints at identical positions (Figs. 1, 5; Pestova et al. 1998b; Sizova et al. 1998; Pestova and Hellen 1999), but the major sequence differences in this region suggest that structural (rather than sequence) determinants likely play a major role in recognition of the IRES by eIF3.
Despite these similarities, there are also significant differences between initiation on SPV9 and HP IRESs. Initiation on the SPV9 IRES with an adjacent wild-type coding sequence was less efficient than on the CSFV IRES (Fig. 2B). A significant difference between SPV9 and CSFV IRESs is that the initiation codon is sequestered in the predicted domain IV only in the former, and it is adjacent to a C-rich sequence in the coding region that is unlike the proximal A-rich coding region of CSFV (Fletcher et al. 2002). The observation that substitutions in the C-rich sequence enhanced SPV9 IRES activity might plausibly be accounted for by minimization of secondary structure in this region, in an analogous manner to impairment of IRES function by increasing secondary structure within the ribosomal loading site of HCV and CSFV IRESs (Honda et al. 1996; Fletcher et al. 2002) and, conversely, to enhanced ribosomal binding to and function of the BVDV IRES as a result of pseudorevertant substitutions that minimize secondary structure in this region (Myers et al. 2001). Initiation on SPV9 (wt) was moderately enhanced by eIFs 4A, 4B, and 4F, and, consistently, translation of SPV9 mRNA in RRL was also moderately inhibited by the negative trans-dominant eIF4AR362Q mutant in a manner that was overcome by eIF4F or eIFΔ4G. Although eIF4A/eIF4F are not essential for SPV9 IRES function, their ability to influence its activity constitutes a notable difference between the SPV9 IRES and HP IRESs, whose activity is not influenced by eIF4A (Pestova et al. 1998b; Fletcher et al. 2002; Bordeleau et al. 2006).
The mechanism by which eIF4A/eIF4F enhance 48S complex formation on the SPV9 IRES is not known, but the correlation between (a) the greater enhancement by these factors of 48S complex formation on and translation mediated by the wt IRES than on the (CAA) and SL− (CAA) variants, (b) the slightly stronger inhibition by eIF4(R362Q) of the function of the wt IRES than of these variant IRESs in in vitro translation, and (c) the fact that factor-independent binding of the SPV9 (CAA) and SPV9 SL− (CAA) variants but not the wt IRES to 40S subunits yielded toeprints downstream from the initiation codon together suggests that eIF4A/eIF4F promote accommodation of mRNA around the initiation codon in the mRNA-binding cleft of the 40S subunit.
Exceptionally, the SPV9 IRES (unlike the CSFV IRES) promoted binding of Met-tRNAi Met to the 40S subunit P site independently of eIF2 on mRNAs that had an unstructured proximal coding sequence. This ability was also evident on wild-type SPV9 IRES-containing mRNA in the presence of eIF3, which may induce conformational changes in the IRES–40S subunit complex. Although binding of Met-tRNAi Met in the P site in the absence of eIF2 did not lead to formation of elongation-competent 80S ribosomes, this observation reveals a second difference between SPV9 and HP IRESs. The basis for this activity of the SPV9 IRES may include induced conformational changes in the 40S subunit that differ subtly from those induced by HP IRESs. HCV IRES domain II enhances binding of the coding sequence in the mRNA-binding cleft of 40S subunits by inducing conformational changes, including rotation of the head relative to the body, opening of the entry latch of the mRNA-binding cleft, and closure of the cleft at the exit side (Kolupaeva et al. 2000a, Spahn et al. 2001). The larger size of domain II of the SPV9 IRES compared to HCV and CSFV-like IRESs (Lukavsky et al. 2003; Hellen and de Breyne 2007; Locker et al. 2007) may result in it making additional contacts with the 40S subunit resulting in additional conformational changes in the bound ribosome compared to those caused by binding of the HCV IRES (Spahn et al. 2001) which may account for the ability of the SPV9 IRES to promote eIF2-independent binding of Met-tRNAi Met to the P site.
We suggested that HP-like IRESs are modular elements that have exchanged between flaviviruses (which include HCV and CSFV) and some picornaviruses (such as SPV9) by recombination (Hellen and de Breyne 2007). Data presented here indicate that domain III of the SPV9 IRES functions like those of HCV and CSFV IRESs in binding independently to eIF3 and to 40S subunits, and suggest that other differences may be attributable to flanking sequences. Differences in downstream sequences can account for the influence of eIF4A/eIF4B/eIF4F on SPV9 IRES function, whereas the IRES' ability to promote binding of Met-tRNAi Met to the P site may be due to differences in domain II, although the latter possibility remains to be tested.
Factor-induced conformational changes in ribosomal 48S initiation complexes
The exceptional ability of the SPV9 IRES to promote binding of Met-tRNAi Met in the P site independently of eIFs permitted toeprinting to be used to characterize the influence of all factors on the stability and conformation of 48S complexes individually. eIF1A stabilized the 48S complex in the presence and in the absence of eIF2, as revealed by the increased prominence of characteristic +16–17 toeprints, whereas eIF1 destabilized 48S complexes formed without eIF2•GTP, yielding toeprints at +12–15 that are characteristic of binary 40S subunit/IRES complexes. Their appearance likely reflects ejection of Met-tRNAi Met from 48S complexes, which suggests that, despite the presence of a cognate P site codon, Met-tRNAi Met binds to 40S subunits in the absence of eIF2 with an orientation that is incompatible either with the presence of eIF1 or with the conformational changes that eIF1 induces in the 40S subunit. 48S complexes containing eIF2•GTP•Met-tRNAi Met resisted destabilization by eIF1. However, eIF1 induced a significant proportion of such 48S complexes to yield toeprints at +9–11 positions. These toeprints were also induced by eIF3 alone and, more strongly, with eIF1 but were suppressed by eIF1A. Their position is analogous to that of an aberrant set of toeprints (“complex III”) that we previously noted on assembly of 48S complexes by the canonical initiation mechanism (Battiste et al. 2000). These toeprints become prominent in the absence of eIF1A or when wild-type eIF1A is replaced by mutants containing substitutions that likely disrupt contacts with the 40S subunit (Battiste et al. 2000; Pestova and Hellen 2001; Pestova and Kolupaeva 2002; Unbehaun et al. 2004; Fekete et al. 2005; Lomakin et al. 2006). We suggested previously that 43S complexes can adopt different conformations depending on their associations with eIF1 and eIF1A, and therefore toeprints at the approximate +9 position could reflect the conformation of ribosomal complexes in which mRNA is incompletely fixed in the mRNA-binding cleft, thus permitting reverse transcriptase to advance beyond the leading edge of the 48S complex (Battiste et al. 2000; Pestova and Hellen 2000).
The factor-dependent changes in toeprints reported here are consistent with the current model for initiation codon selection, in which eIF1, in cooperation with eIF1A, promotes 43S complexes to adopt an “open” scanning-competent conformation that is maintained until scanning complexes encounter the initiation codon (Pestova et al. 1998a; Pestova and Hellen 2000). The conformational changes that lead 43S complexes to adopt this “open” conformation likely correspond to opening of the entry latch and exit channel of the mRNA-binding cleft of the 40S subunit (Passmore et al. 2007). Base-pairing between the initiation codon and the Met-tRNAi Met anticodon, which is accompanied by the establishment of specific interactions of the −3 and +4 “context” nucleotides of mRNA with eIF2α and 18S rRNA, respectively (Pisarev et al. 2005), leads to tightening of eIF1A's interaction with the 40S subunit and subsequent dissociation of eIF1, which switches this ribosomal complex to a “closed” conformation (Pestova and Hellen 2000; Unbehaun et al. 2004; Algire et al. 2005; Maag et al. 2005; Lomakin et al. 2006; Pisarev et al. 2006; Cheung et al. 2007; Pestova et al. 2007). Incomplete release of eIF1, potentially exacerbated by the absence of eIF1A or by mutations in eIF1A that lead to incomplete tightening of its interaction with the 40S subunit, would lead to the 40S subunit adopting a conformation in which the entry latch of the mRNA-binding channel remains partially open, thus leading to the appearance of the toeprints at the +9–11 positions. eIF1A likely binds in the ribosomal aminoacyl (A) site, like its prokaryotic homolog IF1 (Battiste et al. 2000; Carter et al. 2001; Pestova and Hellen 2001). We suggest that the changes induced by eIF1A in the small ribosomal subunit might resemble those induced by IF1 (Carter et al. 2001), and could thus also tighten the grip of the small subunit on mRNA. Data reported here suggest that the tighter interaction of eIF1A with the 40S subunit (Maag et al. 2006) would stabilize binding of mRNA in the mRNA-binding channel of 48S complexes until the subunit joining stage, when eIF1A is released with eIF5B (Fringer et al. 2007).
MATERIALS AND METHODS
Plasmids
Expression vectors for His6-tagged wild-type eIFs 1, 1A, 4A, 4B, 4GI737–1600 (“Δ4G”), 5, and 5B, for mutant eIF4AR362Q and transcription vectors for dicistronic CSFV IRES-containing mRNA (pXL CSFV 1–423 NS′, renamed DC CSFV here), β-globin mRNA (pBS−[β-globin]), and tRNAi Met (pUC Met-tRNA) have been described (Kolupaeva et al. 2007; Pisarev et al. 2007b). The tRNAAla and tRNAVal vectors pBR Ala-tRNA (GCU) and pUC57 Val-tRNA (GUG) were made by inserting appropriate DNA sequences into pBR322 and pUC57, respectively, between a T7 promoter and a BstN1 restriction site (Pestova and Hellen 2003; Pisarev et al. 2007a).
DNA corresponding to SPV9 nucleotides 289–726 (GenBank AY064717) flanked by 5′ Sal1 and 3′ BamH1 sites was synthesized (Epoch Biolabs) and sequenced after insertion into pBluescript II SK (Invitrogen) to yield pBS-SPV9 (wt) and into DC CSFV, replacing the CSFV IRES, to yield dicistronic DC SPV9 (wt). Analogous vectors pBS-SPV9 (Var1) and DC SPV9 (Var1) contained the mutation GGGG593–596AAAA. Monocistronic (MC) SPV9 (wt) was made by inserting a SalI/NotI fragment from DC SPV9 (wt) into pBS-SK. The sequence 5′-ATGGATTGCTTTAACCCCCCCTTCCCCCT-3′ downstream from the SPV9 initiation codon AUG676 (underlined) was modified to 5′-ATGGATTGCTATAACAACAACTTCAACAT-3′, 5′-ATG ACTAGCTATAACAACAACTTCAACAT-3′, or 5′-ATGGCTGTGTAAAACAACAACTTCAACAT-3′ (substitutions in italics) by PCR, yielding MC SPV9 (CAA), MC SPV9 SL− (CAA), and SPV9 (MAVstop), respectively. DNA corresponding to poliovirus type 1 (Mahoney) nucleotides 70–798 was inserted into DC CSFV, replacing the CSFV IRES, to yield dicistronic DC PV1.
Transcription of mRNA
DC CSFV, DC PV1, DC SPV9 (wt), DC SPV9 (Var1), MC SPV9 (wt), MC SPV9 (CAA), MC SPV9 SL− (CAA) and MC SPV9 (MAVstop) linearized with EcoR1, pBS-SPV9 (wt) and pBS-SPV9 (Var1) linearized with SapI, pUC Met-tRNA, pBR Ala-tRNA and pUC57 Val-tRNA linearized with BstNI, and pBS−(β-globin) linearized with BamH1 were transcribed in vitro using T7 RNA polymerase, with or without [32P]UTP (111 TBq/mmol, MP Biomedicals), as described (Kolupaeva et al. 2007; Pisarev et al. 2007b).
In vitro translation assays
mRNAs were translated for 1 h at 30°C in rabbit reticulocyte lysate (RRL) (Promega) optimized for the SPV9 IRES by addition of 0.5 mM MgOAc and 70 mM KCl and supplemented with 20 μM complete amino acids (minus methionine), 0.5 mCi/mL [35S]methionine (43.5 TBq/mmol), and recombinant wild-type eIF4A, eIF4AR362Q, eIFΔ4G, or native eIF4F as indicated in the text. The dicistronic poliovirus DC PV1 mRNA was translated for 1 h at 30°C in RRL supplemented with 20 μM complete amino acids (minus methionine), 0.5 mCi/mL [35S]methionine (43.5 TBq/mmol), and amounts of cytoplasmic S10 extracts of HeLa cells (Pestova et al. 1991) as indicated in the text.
Translation products were analyzed by electrophoresis using NuPAGE 4%–12% Bis-Tris-Gel (Invitrogen). Gels were exposed to X-ray film overnight and quantified using a Molecular Dynamics PhosphoImager.
Purification of initiation factors, elongation factors, 40S and 60S ribosomal subunits
40S and 60S ribosomal subunits and eIF2, eIF3, eIF4F, eEF1H, and eEF2 were purified from RRL, and recombinant eIF1, eIF1A, eIF4A, eIF4B, eIFΔ4G, eIF5, and eIF5B were expressed in Escherichia coli and purified (Pisarev et al. 2007b). Their activities were then verified (Kolupaeva et al. 2007; Pisarev et al. 2007b).
Assembly and analysis of ribosomal initiation complexes
Met-tRNAi Met was prepared using in vitro transcribed tRNA (Pisarev et al. 2007b). Ribosomal preinitiation complexes were assembled by incubating 1 pmol mRNA for 10 min at 37°C in a 40 μL reaction volume that contained 1 mM DTT, 90 mM KCl, 20 mM Tris pH 7.5, 2.5 mM MgOAc, 40 U RNase OUT (Invitrogen), 1 mM ATP, 0.2 mM GTP, 0.25 mM spermidine, 5 pmol Met-tRNAi Met, 3 pmol 40S subunit and eIF2 (10 pmol), eIF3 (8.5 pmol), eIF1 (75 pmol), eIF1A (75 pmol), eIF4A (30 pmol), eIF4B (12 pmol), and eIF4F (3.5 pmol). Elongation assays were done as described (Pestova et al. 2004; Alkalaeva et al. 2006) using reaction mixtures that contained combinations of components as described in the text, and 60S subunits (6 pmol), eIF5 (45 pmol), eIF5B (3.5 pmol), eEF2 (35 pmol), eEF1H (35 pmol), Ala-tRNAAla (5 pmol), and Val-tRNAVal (5 pmol). Primer extension analysis was done by adding 1 μL MgOAC (320 mM), 4 μL deoxynucleoside triphosphates (5 mM dCTP, dGTP, dTTP; 1 mM dATP), 0.1 mCi [α-32P]-dATP, 1 μg oligonucleotide (NS′ primer: 5′-CTCGTTTGCGGACATGCC-3′), and 5 U avian myeloblastosis virus reverse transcriptase (AMV-RT, Promega) to reaction mixtures, followed by incubation for 40 min at 37°C. cDNAs were phenol-extracted, ethanol-precipitated, and analyzed by electrophoresis through 6% polyacrylamide sequencing gels. cDNAs were compared against SPV9 dideoxynucleotide sequence ladders.
Sucrose density gradient analysis
Binary ribosomal complexes were assembled by incubating 40S subunits (6 pmol) with 1 μg [32P]UTP-labeled SPV9 nucleotides 289–726 wild-type RNA, SPV9 nucleotides 289–726 Var1 mutant RNA, MC SPV9 (wt), MC SPV9 SL− (CAA), or full-length β-globin mRNA, as indicated, and analyzed by centrifugation through 10%–30% sucrose density gradients (Kolupaeva et al. 2007). The optical density of fractionated gradients was measured at 260 nm and association of [32P]-labeled mRNA with 40S subunits was assayed by scintillation counting.
ACKNOWLEDGMENT
This work was supported by NIH grant AI-51340.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.696508.
REFERENCES
- Algire, M.A., Maag, D., Lorsch, J.R. Pi release from eIF2, not GTP hydrolysis, is the step controlled by start-site selection during eukaryotic translation initiation. Mol. Cell. 2005;20:251–262. doi: 10.1016/j.molcel.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Alkalaeva, E.Z., Pisarev, A.V., Frolova, L.Y., Kisselev, L.L., Pestova, T.V. In vitro reconstitution of eukaryotic translation reveals cooperativity between release factors eRF1 and eRF3. Cell. 2006;125:1125–1136. doi: 10.1016/j.cell.2006.04.035. [DOI] [PubMed] [Google Scholar]
- Battiste, J.L., Pestova, T.V., Hellen, C.U.T., Wagner, G. The eIF1A solution structure reveals a large RNA-binding surface important for scanning function. Mol. Cell. 2000;5:109–119. doi: 10.1016/s1097-2765(00)80407-4. [DOI] [PubMed] [Google Scholar]
- Bordeleau, M.E., Mori, A., Oberer, M., Lindqvist, L., Chard, L.S., Higa, T., Belsham, G.J., Wagner, G., Tanaka, J., Pelletier, J. Functional characterization of IRESes by an inhibitor of the RNA helicase eIF4A. Nat. Chem. Biol. 2006;2:213–220. doi: 10.1038/nchembio776. [DOI] [PubMed] [Google Scholar]
- Carter, A.P., Clemons W.M., Jr, Brodersen, D.E., Morgan-Warren, R.J., Hartsch, T., Wimberly, B.T., Ramakrishnan, V. Crystal structure of an initiation factor bound to the 30S ribosomal subunit. Science. 2001;291:498–501. doi: 10.1126/science.1057766. [DOI] [PubMed] [Google Scholar]
- Chard, L.S., Bordeleau, M.E., Pelletier, J., Tanaka, J., Belsham, G.J. Hepatitis C virus-related internal ribosome entry sites are found in multiple genera of the family Picornaviridae . J. Gen. Virol. 2006a;87:927–936. doi: 10.1099/vir.0.81546-0. [DOI] [PubMed] [Google Scholar]
- Chard, L.S., Kaku, Y., Jones, B., Nayak, A., Belsham, G.J. Functional analyses of RNA structures shared between the internal ribosome entry sites of hepatitis C virus and the picornavirus porcine teschovirus 1 Talfan. J. Virol. 2006b;80:1271–1279. doi: 10.1128/JVI.80.3.1271-1279.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung, Y.N., Maag, D., Algire, M.A., Fekete, C.A., Mitchell, S.F., Takacs, J.E., Shirokikh, N., Pestova, T., Lorsch, J.R., Hinnebusch, A.G. Dissociation of eIF1 from the 40S ribosomal subunit is a key step in start codon selection in vivo. Genes & Dev. 2007;21:1217–1230. doi: 10.1101/gad.1528307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorner, A.J., Semler, B.L., Jackson, R.J., Hanecak, R., Duprey, E., Wimmer, E. In vitro translation of poliovirus RNA: Utilization of internal initiation sites in reticulocyte lysate. J. Virol. 1984;50:507–514. doi: 10.1128/jvi.50.2.507-514.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fekete, C.A., Applefield, D.J., Blakely, S.A., Shirokikh, N., Pestova, T., Lorsch, J.R., Hinnebusch, A.G. The eIF1A C-terminal domain promotes initiation complex assembly, scanning and AUG selection in vivo. EMBO J. 2005;19:3588–3601. doi: 10.1038/sj.emboj.7600821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher, S.P., Ali, I.K., Kaminski, A., Digard, P., Jackson, R.J. The influence of viral coding sequences on pestivirus IRES activity reveals further parallels with translation initiation in prokaryotes. RNA. 2002;8:1558–1571. [PMC free article] [PubMed] [Google Scholar]
- Fringer, J.M., Acker, M.G., Fekete, C.A., Lorsch, J.R., Dever, T.E. Coupled release of eukaryotic translation initiation factors 5B and 1A from 80S ribosomes following subunit joining. Mol. Cell. Biol. 2007;27:2384–2397. doi: 10.1128/MCB.02254-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellen, C.U., de Breyne, S. A distinct group of hepacivirus/pestivirus-like IRESs in members of diverse Picornavirus genera: Evidence for modular exchange of functional noncoding RNA elements by recombination. J. Virol. 2007;81:5850–5856. doi: 10.1128/JVI.02403-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda, M., Brown, E.A., Lemon, S.M. Stability of a stem–loop involving the initiator AUG controls the efficiency of internal initiation of translation on hepatitis C virus RNA. RNA. 1996;2:955–968. [PMC free article] [PubMed] [Google Scholar]
- Kapoor, A., Victoria, J., Simmonds, P., Wang, C., Shafer, R.W., Nims, R., Nielsen, O., Delwart, E. A highly divergent picornavirus in a marine mammal. J. Virol. 2007 doi: 10.1128/JVI.01240-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieft, J.S., Zhou, K., Jubin, R., Doudna, J.A. Mechanism of ribosome recruitment by hepatitis C IRES RNA. RNA. 2001;7:194–206. doi: 10.1017/s1355838201001790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolupaeva, V.G., Pestova, T.V., Hellen, C.U.T. An enzymatic footprinting analysis of the interaction of 40S ribosomal subunits with the internal ribosomal entry site of hepatitis C virus. J. Virol. 2000a;74:6242–6250. doi: 10.1128/jvi.74.14.6242-6250.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolupaeva, V.G., Pestova, T.V., Hellen, C.U.T. Ribosomal binding to the internal ribosomal entry site of classical swine fever virus. RNA. 2000b;6:1791–1807. doi: 10.1017/s1355838200000662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolupaeva, V.G., de Breyne, S., Pestova, T.V., Hellen, C.U.T. In vitro reconstitution and biochemical characterization of translation initiation by internal ribosomal entry. Methods Enzymol. 2007;430:409–439. doi: 10.1016/S0076-6879(07)30016-5. [DOI] [PubMed] [Google Scholar]
- Locker, N., Easton, L.E., Lukavsky, P.J. HCV and CSFV IRES domain II mediate eIF2 release during 80S ribosome assembly. EMBO J. 2007;26:795–805. doi: 10.1038/sj.emboj.7601549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomakin, I.B., Kolupaeva, V.G., Marintchev, A., Wagner, G., Pestova, T.V. Position of eukaryotic initiation factor eIF1 on the 40S ribosomal subunit determined by directed hydroxyl radical probing. Genes & Dev. 2003;17:2786–2797. doi: 10.1101/gad.1141803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomakin, I.B., Shirokikh, N.E., Yusupov, M.M., Hellen, C.U.T., Pestova, T.V. The fidelity of translation initiation: Reciprocal activities of eIF1, IF3 and YciH. EMBO J. 2006;25:196–210. doi: 10.1038/sj.emboj.7600904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukavsky, P.J., Kim, I., Otto, G.A., Puglisi, J.D. Structure of HCV IRES domain II determined by NMR. Nat. Struct. Biol. 2003;10:1033–1038. doi: 10.1038/nsb1004. [DOI] [PubMed] [Google Scholar]
- Maag, D., Fekete, C.A., Gryczynski, Z., Lorsch, J.R. A conformational change in the eukaryotic translation preinitiation complex and release of eIF1 signal recognition of the start codon. Mol. Cell. 2005;17:265–275. doi: 10.1016/j.molcel.2004.11.051. [DOI] [PubMed] [Google Scholar]
- Maag, D., Algire, M.A., Lorsch, J.R. Communication between eukaryotic translation initiation factors 5 and 1A within the ribosomal preinitiation complex plays a role in start site selection. J. Mol. Biol. 2006;356:724–737. doi: 10.1016/j.jmb.2005.11.083. [DOI] [PubMed] [Google Scholar]
- Myers, T.M., Kolupaeva, V.G., Mendez, E., Baginski, S.G., Frolov, I., Hellen, C.U.T., Rice, C.M. Efficient translation initiation is required for replication of bovine viral diarrhea virus subgenomic replicons. J. Virol. 2001;75:4226–4238. doi: 10.1128/JVI.75.9.4226-4238.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passmore, L.A., Schmeing, T.M., Maag, D., Applefield, D.J., Acker, M.G., Algire, M.A., Lorsch, J.R., Ramakrishnan, V. The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Mol. Cell. 2007;26:41–50. doi: 10.1016/j.molcel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- Pause, A., Methot, N., Svitkin, Y., Merrick, W.C., Sonenberg, N. Dominant negative mutants of mammalian translation initiation factor eIF-4A define a critical role for eIF-4F in cap-dependent and cap-independent initiation of translation. EMBO J. 1994;13:1205–1215. doi: 10.1002/j.1460-2075.1994.tb06370.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova, T.V., Hellen, C.U.T. Internal initiation of translation of bovine viral diarrhea virus RNA. Virology. 1999;258:249–256. doi: 10.1006/viro.1999.9741. [DOI] [PubMed] [Google Scholar]
- Pestova, T.V., Hellen, C.U.T. The structure and function of initiation factors in eukaryotic protein synthesis. Cell. Mol. Life Sci. 2000;57:651–674. doi: 10.1007/PL00000726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova, T.V., Hellen, C.U.T. Functions of eukaryotic factors in initiation of translation. Cold Spring Harb. Symp. Quant. Biol. 2001;66:389–396. doi: 10.1101/sqb.2001.66.389. [DOI] [PubMed] [Google Scholar]
- Pestova, T.V., Hellen, C.U.T. Translation elongation after assembly of ribosomes on the Cricket paralysis virus internal ribosomal entry site without initiation factors or initiator tRNA. Genes & Dev. 2003;17:181–186. doi: 10.1101/gad.1040803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova, T.V., Kolupaeva, V.G. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes & Dev. 2002;16:2906–2922. doi: 10.1101/gad.1020902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova, T.V., Hellen, C.U.T., Wimmer, E. Translation of poliovirus RNA: Role of an essential cis-acting oligopyrimidine element within the 5′ nontranslated region and involvement of a cellular 57-kilodalton protein. J. Virol. 1991;65:6194–6204. doi: 10.1128/jvi.65.11.6194-6204.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova, T.V., Hellen, C.U.T., Shatsky, I.N. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol. Cell. Biol. 1996a;16:6859–6869. doi: 10.1128/mcb.16.12.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova, T.V., Shatsky, I.N., Hellen, C.U.T. Functional dissection of eukaryotic initiation factor 4F: The 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol. Cell. Biol. 1996b;16:6870–6878. doi: 10.1128/mcb.16.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova, T.V., Borukhov, S.I., Hellen, C.U.T. Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature. 1998a;394:854–859. doi: 10.1038/29703. [DOI] [PubMed] [Google Scholar]
- Pestova, T.V., Shatsky, I.N., Fletcher, S.P., Jackson, R.J., Hellen, C.U.T. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes & Dev. 1998b;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova, T.V., Lomakin, I.B., Hellen, C.U.T. Position of the CrPV IRES on the 40S subunit and factor dependence of IRES/80S ribosome assembly. EMBO Rep. 2004;5:906–913. doi: 10.1038/sj.embor.7400240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova, T.V., Lorsch, J.R., Hellen, C.U.T. The mechanism of translation initiation in eukaryotes. In: Mathews M.B., et al., editors. Translational control in biology and medicine. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2007. pp. 87–128. [Google Scholar]
- Pisarev, A.V., Chard, L.S., Kaku, Y., Johns, H.L., Shatsky, I.N., Belsham, G.J. Functional and structural similarities between the internal ribosome entry sites of hepatitis C virus and porcine teschovirus, a picornavirus. J. Virol. 2004;78:4487–4497. doi: 10.1128/JVI.78.9.4487-4497.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev, A.V., Shirokikh, N.E., Hellen, C.U.T. Translation initiation by factor-independent binding of eukaryotic ribosomes to internal ribosomal entry sites. C. R. Biol. 2005;328:589–605. doi: 10.1016/j.crvi.2005.02.004. [DOI] [PubMed] [Google Scholar]
- Pisarev, A.V., Kolupaeva, V.G., Pisareva, V.P., Merrick, W.C., Hellen, C.U.T., Pestova, T.V. Specific functional interactions of nucleotides at key −3 and +4 positions flanking the initiation codon with components of the mammalian 48S translation initiation complex. Genes & Dev. 2006;20:624–636. doi: 10.1101/gad.1397906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev, A.V., Hellen, C.U.T., Pestova, T.V. Recycling of eukaryotic post-termination ribosomal complexes. Cell. 2007a;131:286–299. doi: 10.1016/j.cell.2007.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev, A.V., Unbehaun, A., Hellen, C.U.T., Pestova, T.V. Assembly and analysis of eukaryotic translation initiation complexes. Methods Enzymol. 2007b;430:147–177. doi: 10.1016/S0076-6879(07)30007-4. [DOI] [PubMed] [Google Scholar]
- Reynolds, J.E., Kaminski, A., Kettinen, H.J., Grace, K., Clarke, B.E., Carroll, A.R., Rowlands, D.J., Jackson, R.J. Unique features of internal initiation of hepatitis C virus RNA translation. EMBO J. 1995;14:6010–6020. doi: 10.1002/j.1460-2075.1995.tb00289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sizova, D.V., Kolupaeva, V.G., Pestova, T.V., Shatsky, I.N., Hellen, C.U.T. Specific interaction of eukaryotic translation initiation factor 3 with the 5′ nontranslated regions of hepatitis C virus and classical swine fever virus RNAs. J. Virol. 1998;72:4775–4782. doi: 10.1128/jvi.72.6.4775-4782.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spahn, C.M., Kieft, J.S., Grassucci, R.A., Penczek, P.A., Zhou, K., Doudna, J.A., Frank, J. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40s ribosomal subunit. Science. 2001;291:1959–1962. doi: 10.1126/science.1058409. [DOI] [PubMed] [Google Scholar]
- Svitkin, Y.V., Pestova, T.V., Maslova, S.V., Agol, V.I. Point mutations modify the response of poliovirus RNA to a translation initiation factor: A comparison of neurovirulent and attenuated strains. Virology. 1988;166:394–404. doi: 10.1016/0042-6822(88)90510-7. [DOI] [PubMed] [Google Scholar]
- Unbehaun, A., Borukhov, S.I., Hellen, C.U.T., Pestova, T.V. Release of initiation factors from 48S complexes during ribosomal subunit joining and the link between establishment of codon-anticodon base-pairing and hydrolysis of eIF2-bound GTP. Genes & Dev. 2004;18:3078–3093. doi: 10.1101/gad.1255704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, J.E., Pestova, T.V., Hellen, C.U., Sarnow, P. Initiation of protein synthesis from the A site of the ribosome. Cell. 2000;102:511–520. doi: 10.1016/s0092-8674(00)00055-6. [DOI] [PubMed] [Google Scholar]