Abstract
Small interfering RNAs (siRNAs) and microRNAs (miRNAs) guide catalytic sequence-specific cleavage of fully or nearly fully complementary target mRNAs or control translation and/or stability of many mRNAs that share 6–8 nucleotides (nt) of complementarity to the siRNA and miRNA 5′ end. siRNA- and miRNA-containing ribonucleoprotein silencing complexes are assembled from double-stranded 21- to 23-nt RNase III processing intermediates that carry 5′ phosphates and 2-nt overhangs with free 3′ hydroxyl groups. Despite the structural symmetry of a duplex siRNA, the nucleotide sequence asymmetry can generate a bias for preferred loading of one of the two duplex-forming strands into the RNA-induced silencing complex (RISC). Here we show that the 5′-phosphorylation status of the siRNA strands also acts as an important determinant for strand selection. 5′-O-methylated siRNA duplexes refractory to 5′ phosphorylation were examined for their biases in siRNA strand selection. Asymmetric, single methylation of siRNA duplexes reduced the occupancy of the silencing complex by the methylated strand with concomitant elimination of its off-targeting signature and enhanced off-targeting signature of the phosphorylated strand. Methylation of both siRNA strands reduced but did not completely abolish RNA silencing, without affecting strand selection relative to that of the unmodified siRNA. We conclude that asymmetric 5′ modification of siRNA duplexes can be useful for controlling targeting specificity.
Keywords: RNA interference, RNAi, off-target effects, gene silencing, siRNA, RISC
INTRODUCTION
Duplexes of 21-nucleotide (nt) small interfering RNAs trigger RNA interference (RNAi) in mammalian cells and are widely used for functional genetic studies or screens in cultured cells (for reviews, see Dorsett and Tuschl 2004; Echeverri and Perrimon 2006; Fuchs and Boutros 2006; Root et al. 2006; Krausz 2007). siRNA duplexes are designed to mimic the RNase III processing intermediates of naturally expressed dsRNAs, such as miRNAs, to effectively enter the RNAi pathway (for reviews, see Bartel 2004; Meister and Tuschl 2004; Filipowicz et al. 2005; Tomari and Zamore 2005). Naturally processed siRNAs or miRNAs carry 5′ phosphates and 3′-hydroxyl groups and have symmetric 2-nt 3′ overhangs (Elbashir et al. 2001; Lau et al. 2001). Synthetic siRNA duplexes with 5′-hydroxyl ends are rapidly phosphorylated inside cells by the cellular kinase Clp1 (Weitzer and Martinez 2007). Some classes of small RNAs are additionally 2′-O-methylated at their 3′ ends, depending on the species (Ebhardt et al. 2005; Yu et al. 2005; Vagin et al. 2006; Horwich et al. 2007; Kirino and Mourelatos 2007; Ohara et al. 2007; Pelisson et al. 2007; Saito et al. 2007). Mammalian miRNAs or siRNAs are not methylated, but the germline-specifically expressed piRNAs are 3′-end modified (Kirino and Mourelatos 2007; Ohara et al. 2007).
One strand of the siRNA duplex or miRNA/miRNA* molecule is assembled into an effector complex or RISC, while the other strand is degraded during the assembly process (Hutvágner and Zamore 2002; Martinez et al. 2002a). The effector complex contains at its heart an Ago/PIWI protein member (Hammond et al. 2001; Martinez et al. 2002a). Ago/PIWI proteins contain a conserved Piwi-Argonaute-Zwille (PAZ) and PIWI domain (for reviews, see Carmell et al. 2002; Peters and Meister 2007). The PAZ domain, which is also present in Dicer, specifically binds the characteristic 2-nt 3′ overhangs of RNase-III-processed dsRNAs (Song et al. 2003; Yan et al. 2003; Lingel et al. 2004; Ma et al. 2004). The PIWI domain contains a RNA 5′-phosphate binding (MID domain) and a RNase H domain (Parker et al. 2004, 2005; Song et al. 2004; Ma et al. 2005; Rivas et al. 2005; Yuan et al. 2005, 2006; Song and Joshua-Tor 2006). The MID domain anchors the 5′ end of the guide small RNAs (Ma et al. 2005; Parker et al. 2005; Rivas et al. 2005), and presumably also plays a role during RISC-loading by receiving and binding the guide strand 5′ phosphate (Nykänen et al. 2001).
Protein factors critically involved in siRNA or miRNA silencing complex assembly were first identified in Drosophila melanogaster. Duplex siRNAs are recognized by the heterodimer of RNase III Dcr-2 and the dsRNA-binding-domain protein R2D2, both of which are critical for formation of the Ago2-containing RISC (Liu et al. 2003, 2006). miRNA maturation in D. melanogaster is catalyzed by a heterodimeric complex of RNase III Dcr-1 and the dsRNA-binding-domain protein Loquacious/R3D1 (Förstemann et al. 2005; Jiang et al. 2005; Saito et al. 2005). R2D2 preferably binds the thermodynamically more stable end of the siRNA duplex and thereby directs strand selection (Tomari et al. 2004b). The assembly of RISC is ATP dependent, at least to a certain degree (for reviews, see Filipowicz 2005; Preall and Sontheimer 2005). In mammalian systems, Dicer, the dsRNA-binding proteins TARBP2 and/or PACT, and an Ago protein appear to form the RISC-loading complex (Chendrimada et al. 2005; Gregory et al. 2005; Haase et al. 2005; Maniataki and Mourelatos 2005; Lee et al. 2006).
Two pathways are known for the transition of the duplex siRNAs or miRNA/miRNA* processing intermediate into a single-stranded RNA-containing effector complex (Matranga et al. 2005; Rand et al. 2005; Leuschner et al. 2006). The first pathway requires near-perfect base-pairing of the small RNA strands and depends on the RNase H activity intrinsic to a subset of the siRNA-binding Ago proteins (Liu et al. 2004; Meister et al. 2004; Parker et al. 2004, 2005; Rand et al. 2004; Song et al. 2004; Ma et al. 2005; Miyoshi et al. 2005; Rivas et al. 2005; Yuan et al. 2005). RNase H active Ago proteins are able to receive the duplex siRNAs and guide the cleavage of the nonretained siRNA strand (often referred to as passenger, nonguide, or sense siRNA) (Matranga et al. 2005; Rand et al. 2005; Leuschner et al. 2006). Upon release of the cleavage products, the retained guide (or anti-sense) siRNA is able to recognize complementary or partially complementary mRNA targets. The second RISC loading pathway is used, when duplex siRNA or miRNA/miRNA* duplexes either encounter a RNase-H-deficient Ago protein member or when the duplexes are imperfectly paired across the center and cleavage site (like most miRNA/miRNA* duplexes), thereby preventing RNase H cleavage (Matranga et al. 2005). Presumably, a RNA helicase activity residing or transiently associating with the RISC-loading complex catalyzes the second RISC loading process (Tomari et al. 2004a; Meister et al. 2005; Robb and Rana 2007).
The duplex-initiated RISC assembly process appears to be bypassed if high concentrations of single-stranded siRNAs are added to cell lysates or transfected into cells (Martinez et al. 2002a). The role and the requirement for a 5′ phosphate in reconstituting RISC and its activity, however, remained somewhat controversial (Liu et al. 2004; Song et al. 2004; Ma et al. 2005).
The specificity of small-RNA-guided mRNA degradation was examined in detail including mRNA array analysis (Jackson et al. 2003; Lin et al. 2005; Birmingham et al. 2006; Jackson et al. 2006b). These studies revealed “off-targeting” activities of siRNAs that could not be separated from the “on-targeting” activity by simply decreasing the siRNA concentration. Some of the off-targets contained sequence segments of extensive complementarity to the siRNA, but many other off-targets showed only partial complementarity within their 3′-untranslated region (UTR) to the siRNAs, notably at the 5′ end of the siRNA guide strand. The latter observation was reminiscent of miRNA “seed” sequence (comprising positions 1–8) mediated target mRNA regulation (Lai 2002; Lewis et al. 2003; Stark et al. 2003; Rajewsky and Socci 2004; Lim et al. 2005; Linsley et al. 2007). Off-target signatures can be identified for both the sense and the anti-sense siRNA strands, although strand biases during the assembly of RISC affect the targeting efficiencies of the two strands (Khvorova et al. 2003; Schwarz et al. 2003). In selecting siRNA sequences one takes now into consideration the differential thermodynamic stability of the siRNA ends to favor the incorporation of the target mRNA complementary guide siRNA (for review, see Pei and Tuschl 2006). Other strategies proposed to control off-targeting activities included the introduction of 2′-O-methyl-ribose residues into the seed sequences of the siRNAs, which reduces off-targeting without detectable drop in on-targeting (Jackson et al. 2006a).
Here we study the role of the 5′-terminal phosphate during RISC assembly from duplex and single-stranded siRNAs using 5′-O-methyl-modified siRNAs. We show that the 5′-phosphorylation status within a duplex siRNA is an important determinant of strand incorporation into RISC, and we demonstrate that selective 5′-O-methylation can be used to control strand-specific off-targeting activity. The phosphorylation status of single-stranded siRNAs has little impact on the nonnatural RISC assembly and the subsequent activity of RISC.
RESULTS
5′ phosphates are required for reconstitution of RISC from double-stranded but not single-stranded siRNAs
To revisit the requirements for 5′ phosphates described for reconstituting RISC in D. melanogaster (Nykänen et al. 2001) or human cell lysates (Martinez et al. 2002a; Liu et al. 2004; Song et al. 2004; Ma et al. 2005), we prepared single- and double-stranded siRNAs with uridine and thymidine 5′-end modifications (Fig. 1A). 5′-O-methyl-thymidine is currently the only nucleotide readily available for solid-phase synthesis to render the ribose 5′ ends of siRNAs refractory to phosphorylation in cell lysates (Nykänen et al. 2001). HeLa cells and lysates contain hClp1 kinase, which rapidly phosphorylates 5′-hydroxyl termini of dsRNA or dsDNA as well as single-stranded RNA (Martinez et al. 2002a; Weitzer and Martinez 2007). To control for the concomitant introduction of a 5-methyl group with 2′-deoxythymidine incorporation into RNA, we also prepared siRNAs with 5′-hydroxyl-2′-deoxythymidine, 5′-hydroxyl-uridine, and 5′-phosphorylated uridine-containing siRNAs.
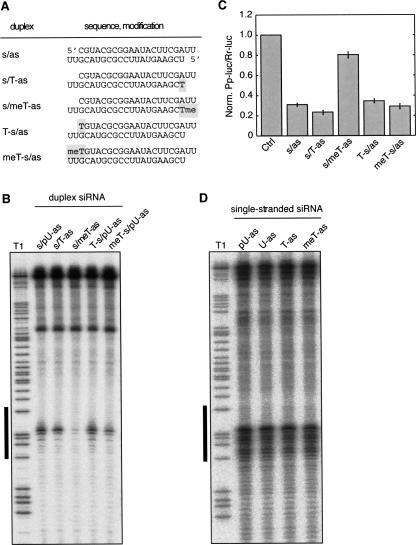
FIGURE 1.
5′ phosphates are required for reconstitution of RISC from double-stranded but not single-stranded siRNAs. (A) Schematic presentation of the luciferase duplex siRNAs used in B and C. (B) HeLa S100 extract was incubated with the siRNAs shown in A. After preincubation, a 32P-cap-labeled RNA substrate was added, and the cleaved RNA fragments were analyzed on a denaturing sequencing gel. T1 refers to partial nuclease T1 digestion of the target RNA. The black line to the left indicates the segment of the target RNA covered by the used siRNAs. (C) The effect of duplexes 1–5 and a control siRNA duplex on inhibition of the firefly luciferase (Pp-luc) expression relative to Renilla luciferase (Rr-luc) in a dual-luciferase assay. The ratios of the signals of Pp-luc/Rr-luc for duplexes 1–5 were normalized to that of the control siRNA. The plotted data were averaged from three independent experiments ±SD. (D) FLAG/HA-tagged Ago2 was transiently transfected into HEK 293 cells. Tagged proteins were immunoprecipitated from the lysates using anti-FLAG beads, and RISC activity was reconstituted by adding single-stranded siRNA against the luciferase mRNA either with a 5′ phosphate (lane 1), without a 5′ phosphate (lane 2), with a 5′-hydroxyl-2′-dT (lane 3), or with a 5′-methoxy-2′-dT (lane 4). The beads were subsequently incubated with a 32P-cap-labeled RNA substrate and analyzed by denaturing RNA-PAGE followed by phospho-imaging. T1 refers to partial nuclease T1 digestion of the luciferase target RNA. p indicates 5′ phosphate; Me, 5′-O-methyl group.
HeLa S100 cell lysates were incubated with double-stranded siRNA derivatives followed by addition of 5′ 32P-labeled complementary target mRNA segments. Irrespective of the modification of the sense (passenger) strand, 5′-hydroxyl- or 5′-phosphate-modified anti-sense strands mediate target RNA cleavage. In contrast, 5′-O-methylated anti-sense siRNA showed substantially reduced activity (Fig. 1B). The siRNA duplexes were cognate to firefly luciferase (Pp-luc) mRNA, and they were cotransfected with plasmids encoding the Pp-luc target and sea pansy control luciferase (Rr-luc) genes into HeLa cells. Consistent with the biochemical results, only the duplex with 5′-O-methyl-modified anti-sense strand showed reduced silencing activity (Fig. 1C). Together, these observations were pointing to a role of the 5′ phosphate of the anti-sense strand during RISC loading or RISC activity.
Two possibilities can be envisioned responsible for the reduced silencing activity of 5′-O-methylated anti-sense strand duplex siRNAs: (1) loading of the anti-sense strand into RISC was compromised, and/or (2) the anti-sense strand-loaded RISC had reduced activity because of conformational restraints imposed by an unoccupied Ago2 5′-phosphate binding pocket (Liu et al. 2004; Ma et al. 2005; Parker et al. 2005; Rivas et al. 2005). We therefore tested if we were able to load RISC using the single-stranded anti-sense siRNAs. Because single-stranded siRNAs are more susceptible to nucleases present in cell lysates than duplex siRNAs, we immunopurified FLAG/HA-affinity-tagged Ago2 protein complexes from HEK 293 cell lysates, and subsequently incubated them with single-stranded siRNAs and target RNA substrate. Surprisingly, the single-stranded siRNAs reconstituted RISC activity irrespective of their 5′ modification status (Fig. 1D).
These data suggest that the 5′ phosphate plays an important role during the process of RISC loading, that a 5′-phosphate-sensing mechanism can be bypassed using single-stranded siRNAs, and that the Ago2 5′-phosphate binding pocket does not need to be occupied to mediate target mRNA cleavage.
Asymmetric 5′-O-methylation of duplex siRNAs directs strand selection during RISC formation
The loss of silencing activity of duplex siRNAs in which only the anti-sense strand was 5′-O-methyl-modified could be due either to preferential loading of the sense strand into RISC under these conditions or to a defective recognition of the siRNA duplex by some RNAi machinery protein at a stage prior to RISC assembly. To monitor the asymmetry of siRNA strand incorporation and target RNA cleavage, we synthesized two pairs of siRNA duplexes that were predicted to be symmetrically and asymmetrically incorporated into RISC based on the differences in thermodynamic stability at their duplex termini (Khvorova et al. 2003; Schwarz et al. 2003).
We first characterized biochemically the symmetric siRNA duplex (Fig. 2A) by incubating it in lysates from HEK 293 cells transiently transfected with FLAG/HA-affinity-tagged Ago2. HEK 293 cells were chosen because they are efficiently transfected at large scale with FLAG/HA-Ago2 expression plasmids. The siRNAs that coimmunoprecipitated with FLAG/HA-Ago2 were analyzed by Northern blotting using probes complementary to either the anti-sense or the sense strand. Signals for the anti-sense strand were detected when the siRNA duplex contained unmodified or 5′-O-methyl sense strand but not when the anti-sense strand was 5′-O-methylated (Fig. 2B). Signals for the sense strand were detected when the siRNA duplex contained unmodified or 5′-O-methyl anti-sense strand but not when the sense strand was 5′-O-methylated (Fig. 2B). We then confirmed that symmetrically or asymmetrically loaded FLAG/HA-Ago2 immunoprecipitates cleaved siRNA-complementary 32P-cap-labeled target RNAs (Tuschl et al. 1999) as expected from their strand-loading ratios determined by Northern blotting (Fig. 2C).
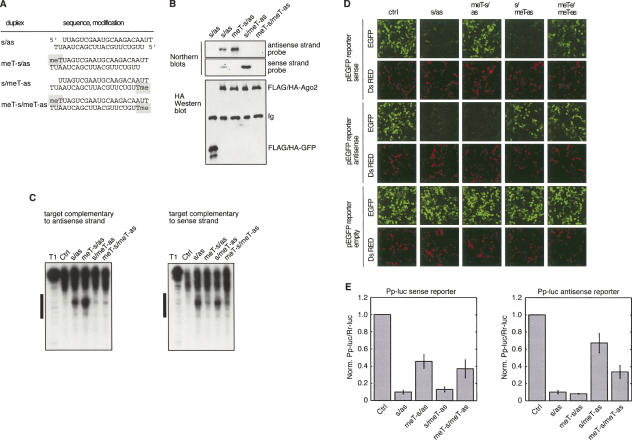
FIGURE 2.
siRNA 5′-O-methylation inhibits RISC loading and RISC activity. (A) Schematic presentation of the symmetric RISC loading siRNA duplexes used in B, C, D, and E. (B) FLAG/HA-Ago2 and FLAG/HA-EGFP were transiently transfected into HEK 293 cells. Cell lysates were preincubated with siRNA duplexes allowing for RISC loading. RISCs were immunoprecipitated using anti-FLAG antibodies, and the precipitated proteins were analyzed using anti-HA antibodies (lower panel). Ig indicates the heavy chain of the immunoglobulin. The bound siRNA strands were examined by Northern blotting (upper panel). (C) HeLa cell extracts were preincubated with the indicated siRNA duplexes allowing for RISC loading. Control (Ctrl) refers to luciferase siRNA duplex. 32P-cap-labeled substrates either complementary to the sense strand or the anti-sense strand were subsequently added and the cleaved RNA products were analyzed by 4% denaturing RNA PAGE. T1 indicates digestion of the substrate using nuclease T1. The bar to the left of the image indicates the sequence of the substrate RNA, which is complementary to the used siRNAs. (D) 5′-O-methylated siRNAs inhibits RNAi in living cells. Plasmids encoding EGFP, EGFP fused to a complementary target site for the anti-sense strand or EGFP fused to a complementary target site for the sense strand were cotransfected with control (ctrl), luciferase siRNAs, or the indicated siRNA duplex. A plasmid encoding the DS Red gene was cotransfected and served as a transfection control. (E) Plasmids containing either a Pp-luciferase gene fused to a complementary target site for the anti-sense strand or a complementary target site for the sense strand of the siRNA were cotransfected with Rr-luciferase and the indicated siRNAs. GFP siRNA was utilized as control (ctrl) siRNA. The Pp-luc/Rr-luc ratios were normalized to that of the control siRNAs.
To measure the cell-based silencing activities from the assembly of the anti-sense and the sense siRNA strand into RISC, we introduced anti-sense and sense-complementary sequence segments into the 3′ UTR of EGFP as well as Pp-luc reporters. The results were consistent with our biochemical observations in HEK 293 lysates (Fig. 2D,E). Symmetric 5′-O-methylation of both siRNA strands lead to an overall reduced gene silencing activity in the luciferase reporter assay, without changing the symmetry of the residual cleavage activity when compared with the unmodified siRNA duplex. In contrast, a single asymmetric modification did not alter the activity attributable to the unmodified siRNA strand. Together, these observations indicate that the RISC assembly of symmetrical siRNA duplexes can be influenced using asymmetric 5′-O-methylation, whereby the methylation of one strand directs incorporation of the complementary strand into RISC.
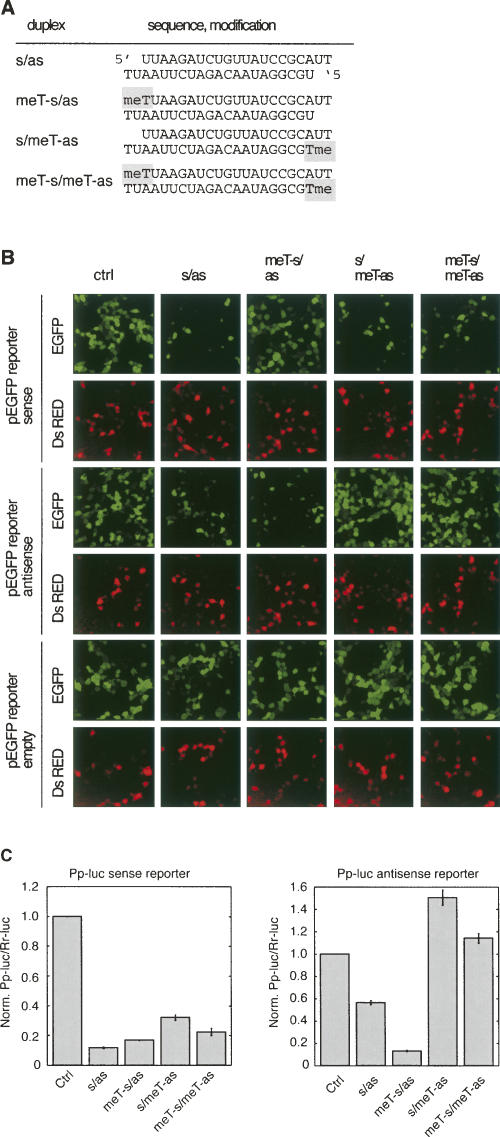
We next evaluated whether strand selection of a thermodynamically asymmetrical siRNA duplex could be controlled by 5′-O-methylation (Fig. 3A). The activities of the modified and unmodified siRNA duplexes were determined using the EGFP and luciferase reporter assay described above (Fig. 3B,C). The unmodified siRNA duplex preferentially repressed the target complementary to the sense strand, as expected from the design of the siRNA. The 5′-O-methylation of the anti-sense siRNA reduced its cleavage activity about twofold. Reciprocally, 5′-O-methylation of the sense siRNA strand reduced its cleavage activity twofold, while the anti-sense strand cleavage activity was significantly increased. Similar to the observations made for the symmetric siRNA duplex above, double 5′-O-methylation weakened the silencing activity of both the anti-sense and sense strands. These observations indicate that 5′-O-methylation of siRNA strands can influence siRNA strand incorporation into RISC, even in the context of a thermodynamically asymmetric siRNA.
FIGURE 3.
siRNA 5′-O-methylation inhibits RISC loading and RISC activity. (A) Schematic presentation of the asymmetric RISC loading siRNA duplexes used in B and C. (B) The same experiments and controls described in Figure 2D were carried out using the asymmetrically RISC loading siRNAs duplex as well as complementary EGFP target constructs. (C) The same experiments and controls described in Figure 2E were performed using the asymmetrical RISC loading siRNAs.
Strand-specific 5′-O-methylation also controls siRNA off-targeting activity
Off-target effects are, like on-target effects, strictly sequence-specific and caused either by near-perfect complementarity between the central region of the siRNA and its targets or by seed-sequence complementarity between the siRNA and the target 3′ UTR (Jackson et al. 2003, 2006b; Birmingham et al. 2006). To assess if strand selection from asymmetrically 5′-O-methylated siRNA duplexes could be used for controlling siRNA off-targeting activity, we determined the gene expression profiles of HeLa cells transfected with 5′-O-methylated or unmodified siRNA duplexes. The symmetrical siRNA duplex, whose both sense and anti-sense strands are incorporated into RISC, was selected for the analysis (Fig. 2A).
The choice of cell line and transfection reagents was critical for identifying siRNA-strand-specific off-targets. We first tested Lipofectamine 2000 Transfection Reagent (Invitrogen) in HEK 293 cells, but variations in gene expression, caused presumably by mild toxicity of the formulated transfection reagent, made it impossible to detect siRNA-sequence-dependent off-target signatures. Next, we tested Lipofectamine RNAiMAX transfection reagent (Invitrogen) in HEK 293 cells. Though the siRNA and mock transfection yielded reproducible and stable expression profiles, we did not detect any off-targeting signature using either Affymetrix or Agilent mRNA microarrays (data not shown). Finally, we examined HeLa cells transfected with Lipofectamine RNAiMAX using Affymetrix microarrays, and we were able to identify the expected off-targeting effects. Using the same approach, we also detected the previously reported off-targets of a siRNA duplex targeting the PIK3CB gene (PIK3CB-6340) (Jackson et al. 2006b), indicating that our Affymetrix array platform was sufficiently sensitive for off-target analysis in this cell type.
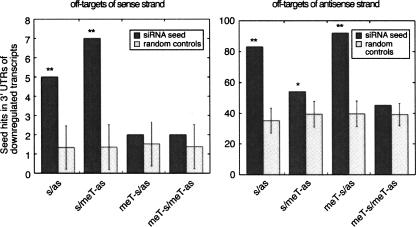
The off-target effects were quantified by analyzing the frequency of seed-complementary sites (where the seed was defined as nucleotides 1–7, 2–8, or 1–8 of the siRNA) for real and randomized siRNAs within the 3′ UTRs of mRNAs that were down-regulated 1 d after siRNA transfection (Fig. 4). Consistent with our biochemical analysis, which indicated that both strands of our unmodified siRNA duplex were loaded into RISCs, we found seed-complementary site enrichment for both siRNA strands. 5′-O-methylation of the sense strand increased the off-targeting activity of the anti-sense strand, while decreasing its own off-targeting activity, and vice versa. Note that the sense strand has intrinsically fewer off-targets compared with the anti-sense strand. This is because the sense strand contains a CG dinucleotide in its seed sequence and CG-containing motifs are underrepresented in the genome compared to other dinucleotides. The random controls for the sense strand, being selected to have a similar number of seed-complementary sites in the entire set of 3′ UTRs as the sense strand, also have low numbers of seed matches in the set of down-regulated UTRs. This leads to a higher variance in the expected number of seed-complementary sites for the sense compared with the anti-sense strand. Nonetheless, the seed enrichment for the sense strand is statistically significant in the case of the s/as and s/meT-as constructs. These analyses thus indicate that chemical modification can limit the off-targeting activity to only one strand of the siRNA duplex.
FIGURE 4.
siRNA off-target analysis of the symmetric RISC loading siRNA duplexes. Enrichment of “seed”-complementary sequences for sense (left panel) and anti-sense (right panel) strands relative to random controls in the 3′ UTRs of transcripts that are down-regulated upon siRNA transfection (see Materials and Methods). Dark gray and light gray bars show the number of occurrences of seed-complementary sites for the siRNAs used in the study and random controls, respectively. Double and single stars indicate enrichments that are significant at 0.01 and 0.05 level.
Comparison of the effects of 5′-O-methylation and duplex-destabilizing mutations on strand selection
The differential thermodynamic stability of siRNA duplex termini impacts siRNA strand selection (Khvorova et al. 2003; Schwarz et al. 2003). Thermodynamic biases can be introduced by varying the G/C content of the termini of the siRNA duplex or by placing destabilizing, non-Watson–Crick base pairs (mismatches) at one of the termini. We therefore wanted to compare the strand bias introduced by 5′-O-methylation with that introduced by mismatches using the asymmetrical siRNA duplex described above, whose sense strand is preferentially incorporated into RISC.
We placed mismatches in the G/C-rich terminus by altering the sequence of the anti-sense siRNA from positions 1 to 5 (Fig. 5A). According to the current model of strand selection, destabilizing the G/C-rich termini should lower the bias for incorporation of the sense strand of this siRNA duplex and should enhance the incorporation of the anti-sense strand into RISC. What we observed was that mismatches only minimally reduced the activity of the sense siRNA-containing RISC measured by the sense reporter, while 5′-O-methylation of the sense siRNA showed a more pronounced effect (Fig. 5B).
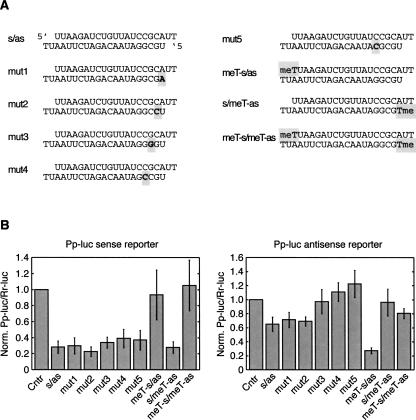
FIGURE 5.
Influence of thermodynamic stability vs. 5′-O-methyl modification. (A) Schematic presentation of the asymmetric RISC loading siRNA duplexes used in B. (B) Plasmids containing either a Pp-luciferase gene fused to a complementary target site for the anti-sense strand or a complementary target site for the sense strand of the siRNA were cotransfected with Rr-luciferase and the indicated siRNAs. GFP siRNA was utilized as control (Ctrl) siRNA.
Anti-sense siRNA strand incorporation was determined by the anti-sense reporter assay. Mismatches introduced by altering the sequence of the anti-sense siRNA trivially lead to mismatches between the anti-sense siRNA and its reporter, resulting in the lack of cleavage activity for mutants of positions 3–5 of the anti-sense siRNA. Mutations placed at position 1 or 2 of the anti-sense siRNA showed similar activity as the unmodified siRNA duplex, consistent with unaltered behavior of the sense reporter, indicating that mismatches at position 1 or 2 were insufficient to alter asymmetry of RISC assembly. In contrast, the 5′-O-methylation of the sense strand, led to a much more pronounced activity of the anti-sense siRNA. Together, these experiments showed that 5′-O-methylation of the 5′ end of siRNAs was more effective in changing strand preferences compared with alterations of thermodynamic stability induced by duplex-destabilizing mismatches.
DISCUSSION
Our analysis describes for the first time the consequences of strand-specific 5′-O-methyl-modification of siRNA duplexes on the assembly and activity of RISC. 5′-O-methylation of the terminal ribose blocked the phosphorylation of siRNAs by cellular kinases (Nykänen et al. 2001; Martinez et al. 2002b; Weitzer and Martinez 2007). The requirement for 5′ phosphates during the assembly of RISC was noted previously using symmetrically 5′-O-methylated siRNA duplexes in D. melanogaster embryo lysates (Nykänen et al. 2001), and similarly, we also found that double 5′-O-methyl modification reduced RISC assembly in human cells. Strikingly, the placement of a single 5′-O-methyl only reduced the incorporation of the methylated strand without impairing the incorporation of the unmodified siRNA strand. It even appeared that modification of the sense siRNA strand enhanced the incorporation of the unmodified anti-sense strand, and vice versa. The effect of the 5′-O-methyl modification was also strong enough to counteract the otherwise strong strand preference of an asymmetrically loading siRNA duplex. This observation emphasizes the importance of 5′-phosphate recognition during RISC assembly and its potential use for siRNA design and application.
The molecular events responsible for 5′ phosphate recognition during RISC assembly remain to be defined. Presumably the contacts are made while placing the 5′ phosphate of the guide siRNA strand into the 5′-phosphate binding pocket of Ago (Ma et al. 2005; Parker et al. 2005). The sensing of the 5′ phosphate also appears to take place in the context of a duplex siRNA or a partially unwound duplex siRNA, because artificial loading of RISC with single-stranded 5′-O-methyl-modified or unmodified siRNAs was possible and led to similar RISC-mediated cleavage activities. Interestingly, a bulky 5′-fluorescein modification coupled via a 5′-phosphodiester linkage to the guide strand of a siRNA duplex did not affect its silencing efficiency (Harborth et al. 2003). It will be interesting to explore the effects of other 5′-hydroxyl modifications, for example, bulky alkyl groups (e.g., tertiary butyl), on RISC assembly.
Although siRNA duplexes are widely used in research as reagents for gene silencing, sequence-specific off-target effects can be problematic. Off-target effects are typically caused by siRNA sense and anti-sense strands, and the strand-specific reduction of off-target effects using 5′-O-methyl modifications will help to improve siRNA specificity. However, 5′-O-methylation will only eliminate off-target effects caused by the 5′-O-methylated strand of the siRNA duplex. Moreover, this may also lead to an enhanced off-target effect of the unmodified strand as its loading into RISC may increase; it should nonetheless allow for lowering the dose of siRNAs needed in gene silencing experiments and will aid in controlling potential side effects caused by competition of siRNA with miRNA pathways (Grimm et al. 2006).
Because not all siRNAs that are cognate to a given target mRNA are equally effective, computational tools have been developed based on experimental data to increase the likelihood of selecting effective siRNAs (for review, see Pei and Tuschl 2006). Though these methods facilitate selection of functional siRNAs, they do not yet alleviate the need for experimental validation. 5′-O-methylation of the passenger strand adds an additional constraint and might be useful in the context of generating genome-wide siRNA libraries, where the validation of each single siRNA is technically not feasible. Depending on the assay, high-throughput RNAi screens are sensitive to seed-sequence-mediated off-targeting effects (Lin et al. 2005), and reducing the likelihood for contributions of the passenger siRNA is useful.
Although it has been proposed that thermodynamic asymmetry can be readily introduced into siRNAs using LNAs (Elmén et al. 2005), minimizing the degree of chemical modification needed to control asymmetry may be beneficial from the point of view of manufacturing or potential therapeutic use. It was also interesting to observe that the introduction of conventional mismatches for destabilizing one of the siRNA termini was less effective then the selective placement of a 5′-O-methyl group.
In summary, in this study we describe a novel approach of biasing siRNA strand selection from duplex siRNA based on 5′-phosphate sensing during RISC assembly.
MATERIALS AND METHODS
Oligonucleotide synthesis
siRNAs were chemically synthesized using RNA phosphoramidites (Pierce) on an Äkta Oligopilot 10 DNA/RNA synthesizer (GE Healthcare Life Sciences) at a 1 μmol scale. The synthesis, deprotection and precipitation were performed according to the manufacturer's protocol. 5′-O-methylated siRNAs were purchased from Dharmacon. The sequences of the siRNAs used in this study are as follows: luciferase siRNA anti-sense strand, 5′-UCGAAGUAUUCCGCGUACGUU, sense strand, 5′−CGUACGCGGAAUACUUCGAUU; GFP siRNA anti-sense strand, 5′-GGCAAGCUGACCCUGAAGUUT, sense strand, 5′-ACUUCAGGGUCAGCUUGCCUT; symmetrically RISC-loaded siRNA anti-sense strand, 5′-UUGUCUUGCAUUCGACUAAUT, sense strand, 5′-UUAGUCGAAUGCAAGACAAUT; and asymmetrically RISC-load siRNA anti-sense strand, 5′-UUAAGAUCUGUUAUCCGCAUT, sense strand, 5′-UGCGGAUAACAGAUCUUAAUT. For analysis of the importance of the 5′ phosphate, the 5′ uridine residues were substituted by 5′-O-methyl-2′-deoxythymidine or 2′-deoxythymidine as control.
Plasmids
The mammalian expression plasmids for FLAG/HA-tagged Ago2 and GFP were previously described (Meister et al. 2004) and are available from www.addgene.org. Reporter plasmids for measuring RISC activity of anti-sense and sense siRNA strands were generated as follows: Complementary pairs of DNA oligonucleotides bearing the siRNA target sequence and flanking SacI and NaeI restriction sites were annealed, digested with SacI and NaeI, and cloned into the 3′ UTR of the reporter vectors, using the SacI and NaeI sites of the pMIR-REPORT plasmid (Ambion) or the SacI and SmaI sites of the pEGFP-C2 plasmid (Clontech), respectively. Prior to this procedure, the pEGFP-C2 plasmid was modified by the insertion of a stop codon into the BglII site. The oligos containing the siRNA target sequences were as follows:
Target complementary to symmetric RISC loading anti-sense siRNA strand, 5′-CGCTGAGCTCATCGCCACCTTGTTTAAGCCTTAGTCGAATGCAAGACAAATTAGACCTACGCACTCCAGGCCGGCTCGC and 5′-GCGAGCCGGCCTGGAGTGCGTAGGTCTAATTTGTCTTGCATTCGACTAAGGCTTAAACAAGGTGGCGATGAGCTCAGCG;
Target complementary to symmetric RISC loading sense siRNA strand, 5′-CGCTGAGCTCATCGCCACCTTGTTTAAGCCTTGTCTTGCATTCGACTAAATTAGACCTACGCACTCCAGGCCGGCTCGC and 5′-GCGAGCCGGCCTGGAGTGCGTAGGTCTAATTTAGTCGAATGCAAGACAAGGCTTAAACAAGGTGGCGATGAGCTCAGCG;
Target complementary to asymmetric RISC loading anti-sense siRNA strand, 5′-CGCTGAGCTCATCGCCACCTTGTTTAAGCCGCGGATAACAGATCTTAAAATTAGACCTACGCACTCCAGGCCGGCTCGC and 5′-GCGAGCCGGCCTGGAGTGCGTAGGTCTAATTTTAAGATCTGTTATCCGCGGCTTAAACAAGGTGGCGATGAGCTCAGCG; and
Target complementary to asymmetric RISC loading sense siRNA strand, 5′-CGCTGAGCTCATCGCCACCTTGTTTAAGCCTTTAAGATCTGTTATCCGCATTAGACCTACGCACTCCAGGCCGGCTCGC and 5′-GCGAGCCGGCCTGGAGTGCGTAGGTCTAATGCGGATAACAGATCTTAAAGGCTTAAACAAGGTGGCGATGAGCTCAGCG.
Plasmids for in vitro transcription of RNA cleavage substrates were generated by cloning of the annealed oligodeoxynucleotides into the SacI/SmaI sites of the pIVEX2.4d plasmid (Roche).
In vitro transcription of RISC cleavage substrates
DNA templates for in vitro transcription of RNA cleavage substrates were generated by linearization of the respective pIVEX2.4d plasmids using BamHI restriction enzyme digestion. The linearized plasmid was used for run-off in vitro transcription using T7 RNA polymerase (Fermentas) according to the manufacturer's protocol. The transcripts were purified by denaturing PAGE, visualized by UV-shadowing, excised, and eluted overnight in 0.3 M NaCl at 4°C. The eluted RNA was precipitated by addition of 3 volumes of ethanol and collected by centrifugation. The cleavage substrate complementary to the luciferase targeting siRNAs was 32P-cap-labeled (Martinez et al. 2002a), and the cleavage was assayed as described previously (Meister et al. 2004). The sense and anti-sense 32P-cap-labeled cleavage substrates were 188 nt and 195 nt, respectively.
Northern blotting
The oligodeoxynucleotide probes 5′-TTAGTCGAATGCAAGACAA, and 5′-TTGTCTTGCATTCGACTAA were used for detection of the symmetrically RISC-loading siRNAs. Probes were radioactively labeled with T4 polynucleotide kinase (New England Biolabs) using [γ-32P]-ATP. RNA samples were separated by 15% denaturing PAGE and transferred to a nylon membrane (Hybond-N+, Amersham) by semi-dry electroblotting. The membrane was then subjected to UV cross-linking using the auto-crosslink function on the Stratalinker (Stratagene) and subsequently baked for 1 h at 80°C. The membrane was incubated with 15 mL of prehybridization buffer (5× SSC/20 mM sodium phosphate buffer (pH 7.2)/7% SDS/1× Denhardt's solution/3 mg of sonicated salmon sperm DNA) in a hybridization oven for 1 h rotating at 50°C. The prehybridization solution was then replaced with 15 mL of hybridization buffer containing 3,000,000 cpm of 32P-radiolabeled DNA probe and incubated overnight at 50°C. The membrane was washed twice with 100 mL of wash solution I (5× SSC/1% SDS) at 50°C for 10 min followed by a single wash step with wash solution II (1× SSC/1% SDS) at 50°C. The membrane was wrapped in plastic wrap and exposed to a film for 2 d.
Tissue culture and transfections
HEK 293 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 unit/mL penicillin, and 100 μg/mL streptomycin at 37°C in a 5% CO2-containing atmosphere. The stably transfected HeLa S3 FLAG/HA-Ago2 cell line (Meister et al. 2004) was cultured under the same conditions with the addition of 0.5 mg/mL G418. HEK 293 cells were cotransfected with reporter or control plasmids and siRNAs using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol in a 24-well format 24 h after seeding. Transfected cells were analyzed for luciferase activity or GFP fluorescence 24 h after transfection. For immunoprecipitation experiments, HEK 293 cells were transfected using the calcium chloride method. Cells were plated to 40% confluency 3–4 h before transfection on a 15-cm dish. Twenty micrograms of plasmid DNA were diluted in 858 μL of water and 122 μL 2M CaCl2. One milliliter of 2× HEPES-buffered saline (274 mM NaCl, 1.5 mM Na2HPO4, 54.6 mM HEPES-KOH at pH 7.1) was added drop-wise under gentle agitation. The transfection solution was then sprinkled onto the cells.
Microscopy
HEK 293 cells were seeded on coverslips and were cotransfected with either empty pEGFP-C2 as control or pEGFP-C2 bearing the target complementary to siRNA strand-specific sequences (0.2 μg/well), pDsRedmonomer-C1 (Clontech; 0.1 μg/well) and siRNA (20 pmol/well). After 24 h, cells were fixed in PBS with 4% formaldehyde for 30 min at room temperature, washed twice with PBS, and mounted to slides using Vectashield mounting medium (Vector Laboratories). Images were recorded using a Leica TCS SP2 confocal laser microscope and a 20× immersion oil objective. For GFP and DsRed images, 10 z-sections of the cells were recorded and processed to average projections using the Leica confocal software.
Dual luciferase assays
For experiments described in Figure 1C, 7000 HEK 293 cells per well were transferred into 96-well plates the day before transfection. The cells were then cotransfected with 0.2 μg pGL2-control (Promega), 0.02 μg pRL-TK (Promega), and 3.75 pmol siRNA duplexes (final concentration 25 nM) with Lipofectamine 2000 (Invitrogen; 0.75 μL). Luciferase activities were measured 20 h after transfection using the Dual Luciferase Assay Kit (Promega) and a Bio-Tek Clarity luminometer. The ratios of the signals of firefly (Pp) luciferase to seapansy (Rr) luciferase were calculated and normalized by dividing by the ratio for control siRNA against FLJ30525.3. The plotted data were averaged from triplicates ±SD For Figures 2–5, HEK 293 cells were cultured in 24-well plates and each cotransfected with either empty pMIR-REPORT (Pp-luc) control plasmid or pMIR-REPORT bearing the target complementary to siRNA strand-specific sequences (0.2 μg/well), pRL-SV40 control vector (Rr-luc) (Promega) (0.1 μg/well), and siRNA (20 pmol/well). The cells were lysed and assayed 24 h post-transfection following the Dual-Luciferase Reporter Assay system (Promega) instructions. Samples were analyzed on a Mithras LB 940 Multimode Microplate Reader (Berthold Technologies). All samples were assayed in triplicates.
Western blotting, extract preparation, and immunoprecipitation
Western blotting was performed as previously described (Meister et al. 2004). For immunoprecipitation, HEK 293 cells transiently transfected with FLAG-/HA-tagged Ago2 and HeLa S3 cells stably transfected with FLAG/HA-tagged Ago2 were harvested from 15 cm plates 48 h post-transfection. The cells were washed with PBS (pH 7.4) and subjected to lysis with 700 μL of lysis buffer (150 mM KCl, 25 mM Tris-HCl at pH 7.4, 2 mM EDTA at pH 8.0, 1 mM NaF, 0.5 mM DTT, 0.05% NP40, 0.5 mM AEBSF) for 10 min at 4°C. The cells were subsequently scraped off of the plate and centrifuged at 17,200g for 10 min. The supernatant was incubated with 15 μL of anti-FLAG M2 agarose beads (Sigma), which were activated by washing once with 0.1 M glycine-HCl (pH 2.5) and equilibrated by washing with 1.5 M Tris-HCl (pH 8.8), for 3 h at 4°C with rotation. The beads were collected and washed three times with 300 mM NaCl/5 mM MgCl2/0.05% NP40/50 mM Tris-HCl (pH 7.5) and once with PBS (pH 7.5). To isolate the RISC-incorporated siRNA, the beads were then incubated in 300 μL of proteinase K solution consisted of 2× proteinase K buffer (300 mM NaCl/25 mM EDTA at pH 8.0/2% SDS/200 mM Tris-HCl at pH 7.5 and proteinase K at 1 mg/mL concentration) for 10 min at 37°C. The RNA was phenol/chloroform-extracted and ethanol precipitated.
RNA cleavage assays
In vitro transcribed cleavage substrates were 5′-cap labeled as described previously (Martinez et al. 2002a). In a typical RNA cleavage reaction, 100 nM of siRNA was incubated in a 15 μL reaction containing 50% HeLa S100 extract, 1 mM ATP, 0.2 mM GTP, 10 U/mL RNasin (Promega), 100 mM KCl, 1.5 mM MgCl2, 0.5 mM DTT, 10 mM HEPES-KOH (pH 7.9) at 30°C. After 30 min, 5 nM of the cap-labeled cleavage substrate was added and further incubated for 1.5 h at 30°C. The reactions were stopped by adding 200 μL of proteinase K buffer containing 1 mg/mL proteinase K. The RNA was subsequently isolated using phenol/chloroform extraction, and the cleavage products were analyzed by 8% denaturing RNA PAGE. The labeled RNA was detected by phosphoimaging and autoradiography.
Microarray mRNA expression analysis
HeLa cells were plated in a six-well plate with a volume of 2.5 mL at a density such that next day they are ∼75% confluenent for transfection. A transfection solution of 0.5 mL was utilized for transfection for the final siRNA duplex concentration of 50 nM using RNAiMAX (Invitrogen). Total RNA was extracted 24 h post-transfection with Trizol (Invitrogen) and purified using the RNeasy Mini Kit (Qiagen). Three micrograms of purified RNA was used to synthesize the first strand of cDNA using ArrayScript reverse transcriptase (Ambion, catalog no. 1791) and an oligo(dT) primer bearing a T7 promoter. The single-stranded cDNA was converted into dsDNA by DNA polymerase I in the presence of Escherichia coli RNase H and DNA ligase. After column purification, the dsDNA was served as a template for in vitro transcription in a reaction containing biotin-labeled UTP, unlabeled NTPs, and T7 RNA polymerase. The amplified, biotin-labeled, anti-sense RNA (aRNA) was purified, and quality was assessed using the Agilent 2100 Bioanalyzer and the RNA 6000 Nano kit. Twenty micrograms of labeled aRNA were fragmented, and 15 μg of the fragmented aRNA was hybridized to Affymetrix Human Genome U133 Plus 2.0 Array for 16 h at 45°C as described in the Affymetrix Technical Analysis Manual (Affymetrix). After hybridization, Gene Chips were stained with streptavidin-phycoerythrin, followed by an antibody solution (anti-streptavidin), and a second streptavidin-phycoerythrin solution, with all liquid handling performed by a GeneChip Fluidics Station 450. Gene Chips were then scanned with the Affymetrix GeneChip Scanner 3000. Agilent Whole Human Genome Oligonucleotide Microarray (catalog no. 4112F) analysis was performed by Cogenics.
Computational analyses of putative off-targets
We normalized the probe intensities for the five microarrays (transfection reagent only, symmetric RISC loading sense/anti-sense, MeO-sense/anti-sense, sense/MeO-anti-sense, MeO-sense/MeO-anti-sense siRNA duplexes) using the bioconductor (see http://www.bioconductor.org and Gentleman et al. 2004) and gcRMA software (Wu et al. 2004). To quantify off-target effects based on the frequency of seed-complementary sites in the 3′ UTRs, we selected, for each gene measured by the microarray, the transcript with median 3′-UTR length. This data set consisted of 14,997 transcripts. From all the probe sets corresponding unambiguously to a given gene, we selected the one that responded best (exhibited the highest variance) across a large number of experiments performed on the Affymetrix platform that we used. This probe set was used to monitor the per-transcript expression level across our experiments. From each experiment (sense/anti-sense versus mock transfection, MeO-sense/anti-sense versus mock transfection, sense/MeO-anti-sense versus mock transfection, MeO-sense/MeO-anti-sense versus mock transfection), we extracted the top 1%, i.e., 149, most down-regulated transcripts and computed the number of occurrences of matches to the 1–7, 2–8, 1–8 nucleotide positions of both the sense and the anti-sense strands in these set of transcripts. We compared these numbers with the number of occurrences expected for “random siRNAs,” which we calculated as follows. For each of the anti-sense and the sense strand, we determined the number of occurrences of seed-complementary sites in the entire set of 3′ UTRs. From all octameric sequences, we then selected the 5% (3277) whose reverse complements occurred with a frequency closest to that of siRNA-complementary sites in the entire set of 3′ UTRs. These served as “random siRNA controls.” We determined the number of occurrences of seed-complementary sites of these random siRNAs in the down-regulated set of 3′ UTRs, To correct for slight variations in this number that can be expected simply because the frequency of seed-complementary sites for real and random siRNAs in the 3′ UTRs overall are not identical, we adjusted the observed counts by a factor equal to the ratio of observed occurrences of the real siRNA-complementary sites to the random siRNA-complementary sites in the entire set of 3′ UTRs. We used the distribution of the values determined for random siRNAs to estimate the P-value of the number of occurrences of real siRNA-complementary sites.
ACKNOWLEDGMENTS
We thank S. Rottmüller for technical assistance; P. Linsley, A. Jackson, and J. Burchard for guidance on the technical aspects of microarray-based off-targeting analysis; C. Ender and An. Frohn for critically reading the manuscript; and S. Jentsch for support. We also acknowledge W. Zhang and C. Zhao for Affymetrix array profiling carried out in the Rockefeller University Genomics Resource Center. L.W. is recipient of a fellowship from the Boehringer-Ingelheim Funds, and D.G. is supported by the Swiss Institute of Bioinformatics. Y.P. was supported by the Ruth L. Kirschstein Fellowship from NIH/National Institute of General Medical Sciences. This work was also supported by the Max Planck Society, HHMI and NIH grant R01 GM068476-01.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.789808.
REFERENCES
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Birmingham, A., Anderson, E.M., Reynolds, A., Ilsley-Tyree, D., Leake, D., Fedorov, Y., Baskerville, S., Maksimova, E., Robinson, K., Karpilow, J., et al. 3′-UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat. Methods. 2006;3:199–204. doi: 10.1038/nmeth854. [DOI] [PubMed] [Google Scholar]
- Carmell, M.A., Xuan, Z., Zhang, M.Q., Hannon, G.J. The Argonaute family: Tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes & Dev. 2002;16:2733–2742. doi: 10.1101/gad.1026102. [DOI] [PubMed] [Google Scholar]
- Chendrimada, T.P., Gregory, R.I., Kumaraswamy, E., Norman, J., Cooch, N., Nishikura, K., Shiekhattar, R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett, Y., Tuschl, T. siRNAs: Applications in functional genomics and potential as therapeutics. Nat. Rev. Drug Discov. 2004;3:318–329. doi: 10.1038/nrd1345. [DOI] [PubMed] [Google Scholar]
- Ebhardt, H.A., Thi, E.P., Wang, M.B., Unrau, P.J. Extensive 3′ modification of plant small RNAs is modulated by helper component-proteinase expression. Proc. Natl. Acad. Sci. 2005;102:13398–13403. doi: 10.1073/pnas.0506597102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echeverri, C.J., Perrimon, N. High-throughput RNAi screening in cultured cells: A user's guide. Nat. Rev. Genet. 2006;7:373–384. doi: 10.1038/nrg1836. [DOI] [PubMed] [Google Scholar]
- Elbashir, S.M., Lendeckel, W., Tuschl, T. RNA interference is mediated by 21 and 22 nt RNAs. Genes & Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmén, J., Thonberg, H., Ljungberg, K., Frieden, M., Westergaard, M., Xu, Y., Wahren, B., Liang, Z., Ørum, H., Koch, T., et al. Locked nucleic acid (LNA) mediated improvements in siRNA stability and functionality. Nucleic Acids Res. 2005;33:439–447. doi: 10.1093/nar/gki193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz, W. RNAi: The nuts and bolts of the RISC machine. Cell. 2005;122:17–20. doi: 10.1016/j.cell.2005.06.023. [DOI] [PubMed] [Google Scholar]
- Filipowicz, W., Jaskiewicz, L., Kolb, F.A., Pillai, R.S. Post-transcriptional gene silencing by siRNAs and miRNAs. Curr. Opin. Struct. Biol. 2005;15:331–341. doi: 10.1016/j.sbi.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Förstemann, K., Tomari, Y., Du, T., Vagin, V.V., Denli, A.M., Bratu, D.P., Klattenhoff, C., Theurkauf, W.E., Zamore, P.D. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol. 2005;3:e236. doi: 10.1371/journal.pbio.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs, F., Boutros, M. Cellular phenotyping by RNAi. Brief Funct. Genomic Proteomic. 2006;5:52–56. doi: 10.1093/bfgp/ell007. [DOI] [PubMed] [Google Scholar]
- Gentleman, R.C., Carey, V.J., Bates, D.M., Bolstad, B., Dettling, M., Dudoit, S., Ellis, B., Gautier, L., Ge, Y., Gentry, J., et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory, R.I., Chendrimada, T.P., Cooch, N., Shiekhattar, R. Human RISC couples microRNA biogenesis and post-transcriptional gene silencing. Cell. 2005;123:631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- Grimm, D., Streetz, K.L., Jopling, C.L., Storm, T.A., Pandey, K., Davis, C.R., Marion, P., Salazar, F., Kay, M.A. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- Haase, A.D., Jaskiewicz, L., Zhang, H., Laine, S., Sack, R., Gatignol, A., Filipowicz, W. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 2005;6:961–967. doi: 10.1038/sj.embor.7400509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond, S.M., Boettcher, S., Caudy, A.A., Kobayashi, R., Hannon, G.J. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 2001;293:1146–1150. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- Harborth, J., Elbashir, S.M., Vandenburgh, K., Manninga, H., Scaringe, S.A., Weber, K., Tuschl, T. Sequence, chemical, and structural variation of small interfering RNAs and short hairpin RNAs and the effect on mammalian gene silencing. Antisense Nucleic Acid Drug Dev. 2003;13:83–105. doi: 10.1089/108729003321629638. [DOI] [PubMed] [Google Scholar]
- Horwich, M.D., Li, C., Matranga, C., Vagin, V., Farley, G., Wang, P., Zamore, P.D. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr. Biol. 2007;17:1265–1272. doi: 10.1016/j.cub.2007.06.030. [DOI] [PubMed] [Google Scholar]
- Hutvágner, G., Zamore, P.D. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- Jackson, A.L., Bartz, S.R., Schelter, J., Kobayashi, S.V., Burchard, J., Mao, M., Li, B., Cavet, G., Linsley, P.S. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- Jackson, A.L., Burchard, J., Leake, D., Reynolds, A., Schelter, J., Guo, J., Johnson, J.M., Lim, L., Karpilow, J., Nichols, K., et al. Position-specific chemical modification of siRNAs reduces “off-target” transcript silencing. RNA. 2006a;12:1197–1205. doi: 10.1261/rna.30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, A.L., Burchard, J., Schelter, J., Chau, B.N., Cleary, M., Lim, L., Linsley, P.S. Widespread siRNA “off-target” transcript silencing mediated by seed region sequence complementarity. RNA. 2006b;12:1179–1187. doi: 10.1261/rna.25706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, F., Ye, X., Liu, X., Fincher, L., McKearin, D., Liu, Q. Dicer-1 and R3D1-L catalyze microRNA maturation in Drosophila . Genes & Dev. 2005;19:1674–1679. doi: 10.1101/gad.1334005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova, A., Reynolds, A., Jayasena, S.D. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- Kirino, Y., Mourelatos, Z. Mouse Piwi-interacting RNAs are 2′-O-methylated at their 3′ termini. Nat. Struct. Mol. Biol. 2007;14:347–348. doi: 10.1038/nsmb1218. [DOI] [PubMed] [Google Scholar]
- Krausz, E. High-content siRNA screening. Mol. Biosyst. 2007;3:232–240. doi: 10.1039/b616187c. [DOI] [PubMed] [Google Scholar]
- Lai, E.C. Micro RNAs are complementary to 3′-UTR sequence motifs that mediate negative post-transcriptional regulation. Nat. Genet. 2002;30:363–364. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- Lau, N.C., Lim, L.P., Weinstein, E.G., Bartel, D.P. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans . Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- Lee, Y., Hur, I., Park, S.Y., Kim, Y.K., Suh, M.R., Kim, V.N. The role of PACT in the RNA silencing pathway. EMBO J. 2006;25:522–532. doi: 10.1038/sj.emboj.7600942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuschner, P.J., Ameres, S.L., Kueng, S., Martinez, J. Cleavage of the siRNA passenger strand during RISC assembly in human cells. EMBO Rep. 2006;7:314–320. doi: 10.1038/sj.embor.7400637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, B.P., Shih, I., Jones-Rhoades, M.W., Bartel, D.P., Burge, C.B. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Lim, L.P., Lau, N.C., Garrett-Engele, P., Grimson, A., Schelter, J.M., Castle, J., Bartel, D.P., Linsley, P.S., Johnson, J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Lin, X., Ruan, X., Anderson, M.G., McDowell, J.A., Kroeger, P.E., Fesik, S.W., Shen, Y. siRNA-mediated off-target gene silencing triggered by a 7 nt complementation. Nucleic Acids Res. 2005;33:4527–4535. doi: 10.1093/nar/gki762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingel, A., Simon, B., Izaurralde, E., Sattler, M. Nucleic acid 3′-end recognition by the Argonaute2 PAZ domain. Nat. Struct. Mol. Biol. 2004;11:576–577. doi: 10.1038/nsmb777. [DOI] [PubMed] [Google Scholar]
- Linsley, P.S., Schelter, J., Burchard, J., Kibukawa, M., Martin, M.M., Bartz, S.R., Johnson, J.M., Cummins, J.M., Raymond, C.K., Dai, H., et al. Transcripts targeted by the microRNA-16 family cooperatively regulate cell cycle progression. Mol. Cell. Biol. 2007;27:2240–2252. doi: 10.1128/MCB.02005-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Q., Rand, T.A., Kalidas, S., Du, F., Kim, H.E., Smith, D.P., Wang, X. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science. 2003;301:1921–1925. doi: 10.1126/science.1088710. [DOI] [PubMed] [Google Scholar]
- Liu, J., Carmell, M.A., Rivas, F.V., Marsden, C.G., Thomson, J.M., Song, J.J., Hammond, S.M., Joshua-Tor, L., Hannon, G.J. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- Liu, X., Jiang, F., Kalidas, S., Smith, D., Liu, Q. Dicer-2 and R2D2 coordinately bind siRNA to promote assembly of the siRISC complexes. RNA. 2006;12:1514–1520. doi: 10.1261/rna.101606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, J.B., Ye, K., Patel, D.J. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature. 2004;429:318–322. doi: 10.1038/nature02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, J.B., Yuan, Y.R., Meister, G., Pei, Y., Tuschl, T., Patel, D.J. Structural basis for 5′-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature. 2005;434:666–670. doi: 10.1038/nature03514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniataki, E., Mourelatos, Z. A human, ATP-independent, RISC assembly machine fueled by pre-miRNA. Genes & Dev. 2005;19:2979–2990. doi: 10.1101/gad.1384005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez, J., Patkaniowska, A., Urlaub, H., Lührmann, R., Tuschl, T. Single-stranded anti-sense siRNAs guide target RNA cleavage in RNAi. Cell. 2002a;110:563–574. doi: 10.1016/s0092-8674(02)00908-x. [DOI] [PubMed] [Google Scholar]
- Martinez, L.A., Naguibneva, I., Lehrmann, H., Vervisch, A., Tchenio, T., Lozano, G., Harel-Bellan, A. Synthetic small inhibiting RNAs: efficient tools to inactivate oncogenic mutations and restore p53 pathways. Proc. Natl. Acad. Sci. 2002b;99:14849–14854. doi: 10.1073/pnas.222406899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matranga, C., Tomari, Y., Shin, C., Bartel, D.P., Zamore, P.D. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- Meister, G., Tuschl, T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- Meister, G., Landthaler, M., Patkaniowska, A., Dorsett, Y., Teng, G., Tuschl, T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Meister, G., Landthaler, M., Peters, L., Chen, P.Y., Urlaub, H., Luhrmann, R., Tuschl, T. Identification of novel Argonaute-associated proteins. Curr. Biol. 2005;15:2149–2155. doi: 10.1016/j.cub.2005.10.048. [DOI] [PubMed] [Google Scholar]
- Miyoshi, K., Tsukumo, H., Nagami, T., Siomi, H., Siomi, M.C. Slicer function of Drosophila Argonautes and its involvement in RISC formation. Genes & Dev. 2005;19:2837–2848. doi: 10.1101/gad.1370605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nykänen, A., Haley, B., Zamore, P.D. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell. 2001;107:309–321. doi: 10.1016/s0092-8674(01)00547-5. [DOI] [PubMed] [Google Scholar]
- Ohara, T., Sakaguchi, Y., Suzuki, T., Ueda, H., Miyauchi, K. The 3′ termini of mouse Piwi-interacting RNAs are 2′-O-methylated. Nat. Struct. Mol. Biol. 2007;14:349–350. doi: 10.1038/nsmb1220. [DOI] [PubMed] [Google Scholar]
- Parker, J.S., Roe, S.M., Barford, D. Crystal structure of a PIWI protein suggests mechanisms for siRNA recognition and slicer activity. EMBO J. 2004;23:4727–4737. doi: 10.1038/sj.emboj.7600488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, J.S., Roe, S.M., Barford, D. Structural insights into mRNA recognition from a PIWI domain-siRNA guide complex. Nature. 2005;434:663–666. doi: 10.1038/nature03462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei, Y., Tuschl, T. On the art of identifying effective and specific siRNAs. Na. Methods. 2006;3:670–676. doi: 10.1038/nmeth911. [DOI] [PubMed] [Google Scholar]
- Pelisson, A., Sarot, E., Payen-Groschene, G., Bucheton, A. A novel repeat-associated small interfering RNA-mediated silencing pathway downregulates complementary sense gypsy transcripts in somatic cells of the Drosophila ovary. J. Virol. 2007;81:1951–1960. doi: 10.1128/JVI.01980-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters, L., Meister, G. Argonaute proteins: Mediators of RNA silencing. Mol. Cell. 2007;26:611–623. doi: 10.1016/j.molcel.2007.05.001. [DOI] [PubMed] [Google Scholar]
- Preall, J.B., Sontheimer, E.J. RNAi: RISC gets loaded. Cell. 2005;123:543–545. doi: 10.1016/j.cell.2005.11.006. [DOI] [PubMed] [Google Scholar]
- Rajewsky, N., Socci, N.D. Computational identification of microRNA targets. Dev. Biol. 2004;267:529–535. doi: 10.1016/j.ydbio.2003.12.003. [DOI] [PubMed] [Google Scholar]
- Rand, T.A., Ginalski, K., Grishin, N.V., Wang, X. Biochemical identification of Argonaute 2 as the sole protein required for RNA-induced silencing complex activity. Proc. Natl. Acad. Sci. 2004;101:14385–14389. doi: 10.1073/pnas.0405913101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand, T.A., Petersen, S., Du, F., Wang, X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005;123:621–629. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- Rivas, F.V., Tolia, N.H., Song, J.J., Aragon, J.P., Liu, J., Hannon, G.J., Joshua-Tor, L. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat. Struct. Mol. Biol. 2005;12:340–349. doi: 10.1038/nsmb918. [DOI] [PubMed] [Google Scholar]
- Robb, G.B., Rana, T.M. RNA helicase A interacts with RISC in human cells and functions in RISC loading. Mol. Cell. 2007;26:523–537. doi: 10.1016/j.molcel.2007.04.016. [DOI] [PubMed] [Google Scholar]
- Root, D.E., Hacohen, N., Hahn, W.C., Lander, E.S., Sabatini, D.M. Genome-scale loss-of-function screening with a lentiviral RNAi library. Nat. Methods. 2006;3:715–719. doi: 10.1038/nmeth924. [DOI] [PubMed] [Google Scholar]
- Saito, K., Ishizuka, A., Siomi, H., Siomi, M.C. Processing of pre-microRNAs by the Dicer-1-Loquacious complex in Drosophila cells. PLoS Biol. 2005;3:e235. doi: 10.1371/journal.pbio.0030235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito, K., Sakaguchi, Y., Suzuki, T., Siomi, H., Siomi, M.C. Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylation of Piwi- interacting RNAs at their 3′ ends. Genes & Dev. 2007;21:1603–1608. doi: 10.1101/gad.1563607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz, D.S., Hutvágner, G., Du, T., Xu, Z., Aronin, N., Zamore, P.D. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- Song, J.J., Joshua-Tor, L. Argonaute and RNA—Getting into the groove. Curr. Opin. Struct. Biol. 2006;16:5–11. doi: 10.1016/j.sbi.2006.01.010. [DOI] [PubMed] [Google Scholar]
- Song, J.J., Liu, J., Tolia, N.H., Schneiderman, J., Smith, S.K., Martienssen, R.A., Hannon, G.J., Joshua-Tor, L. The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. Nat. Struct. Biol. 2003;10:1026–1032. doi: 10.1038/nsb1016. [DOI] [PubMed] [Google Scholar]
- Song, J.J., Smith, S.K., Hannon, G.J., Joshua-Tor, L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- Stark, A., Brennecke, J., Russell, R.B., Cohen, S.M. Identification of Drosophila microRNA targets. PLoS Biol. 2003;1:397–409. doi: 10.1371/journal.pbio.0000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomari, Y., Du, T., Haley, B., Schwarz, D.S., Bennett, R., Cook, H.A., Koppetsch, B.S., Theurkauf, W.E., Zamore, P.D. RISC assembly defects in the Drosophila RNAi mutant armitage. Cell. 2004a;116:831–841. doi: 10.1016/s0092-8674(04)00218-1. [DOI] [PubMed] [Google Scholar]
- Tomari, Y., Matranga, C., Haley, B., Martinez, N., Zamore, P.D. A protein sensor for siRNA asymmetry. Science. 2004b;306:1377–1380. doi: 10.1126/science.1102755. [DOI] [PubMed] [Google Scholar]
- Tomari, Y., Zamore, P.D. Perspective: Machines for RNAi. Genes & Dev. 2005;19:517–529. doi: 10.1101/gad.1284105. [DOI] [PubMed] [Google Scholar]
- Tuschl, T., Zamore, P.D., Lehmann, R., Bartel, D.P., Sharp, P.A. Targeted mRNA degradation by double-stranded RNA in vitro. Genes & Dev. 1999;13:3191–3197. doi: 10.1101/gad.13.24.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin, V.V., Sigova, A., Li, C., Seitz, H., Gvozdev, V., Zamore, P.D. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- Weitzer, S., Martinez, J. The human RNA kinase hClp1 is active on 3′ transfer RNA exons and short interfering RNAs. Nature. 2007;447:222–226. doi: 10.1038/nature05777. [DOI] [PubMed] [Google Scholar]
- Wu, Z., Irizarry, R.A., Gentleman, R., Martinez-Murillo, F., Spencer, F. A model-based background adjustment for oligonucleotide expression arrays. J. Am. Stat. Assoc. 2004;99:909–917. [Google Scholar]
- Yan, K.S., Yan, S., Farooq, A., Han, A., Zeng, L., Zhou, M.M. Structure and conserved RNA binding of the PAZ domain. Nature. 2003;426:468–474. doi: 10.1038/nature02129. [DOI] [PubMed] [Google Scholar]
- Yu, B., Yang, Z., Li, J., Minakhina, S., Yang, M., Padgett, R.W., Steward, R., Chen, X. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005;307:932–935. doi: 10.1126/science.1107130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, Y.R., Pei, Y., Ma, J.B., Kuryavyi, V., Zhadina, M., Meister, G., Chen, H.Y., Dauter, Z., Tuschl, T., Patel, D.J. Crystal structure of A. aeolicus Argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Mol. Cell. 2005;19:405–419. doi: 10.1016/j.molcel.2005.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, Y.R., Pei, Y., Chen, H.Y., Tuschl, T., Patel, D.J. A potential protein-RNA recognition event along the RISC-loading pathway from the structure of A. aeolicus Argonaute with externally bound siRNA. Structure. 2006;14:1557–1565. doi: 10.1016/j.str.2006.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]