Abstract
Most mitochondrial mRNAs in kinetoplastid protozoa require post-transcriptional RNA editing that inserts and deletes uridylates, a process that is catalyzed by multiprotein editosomes. KREPA6 is the smallest of six editosome proteins that have predicted oligonucleotide-binding (OB) folds. Inactivation of KREPA6 expression results in disruption and ultimate loss of ∼20S editosomes and inhibition of procyclic form cell growth. Gel shift studies show that recombinant KREPA6 binds RNA, but not DNA, with a preference for oligo-(U) whether on the 3′ end of gRNA or as a (UU)12 homopolymer. Thus, KREPA6 is essential for the structural integrity and presence of ∼20S editosomes and for cell viability. It functions in RNA binding perhaps primarily through the gRNA 3′ oligo(U) tail. The significance of these findings to key steps in editing is discussed.
Keywords: Trypanosoma brucei, trypanosomatids, RNA editing, editosome, OB-fold, RNA-binding protein
INTRODUCTION
The protozoan parasite Trypanosoma brucei is responsible for African sleeping sickness in humans and nagana in cattle. RNA editing in T. brucei and related kinetoplastids is a unique, essential, and extensive form of post-transcriptional RNA processing in which uridylates (Us) are deleted and inserted in the majority of the mitochondrial pre-mRNAs as specified by guide RNAs (gRNAs). The edited mRNAs encode protein components of respiratory chain complexes I, III, IV, and V, as well as RPS12 and MURF2, a protein of unknown function (for reviews, see Madison-Antenucci et al. 2002; Simpson et al. 2004; Stuart et al. 2005). It functions in a stage-specific fashion resulting in differentially expressed proteins, perhaps to accommodate the alternate energy requirements within the insect (procyclic forms, PF) and animal hosts (bloodstream forms, BF) (Benne et al. 1986; Feagin et al. 1987, 1988; Koslowsky et al. 1990).
RNA editing is carried out by 20S editosomes, likely in coordination with other multiprotein complexes. The editosomes contain the catalysts that cleave the pre-mRNA, add or delete the Us, and ligate the processed products (Rusche et al. 1997; Madison-Antenucci et al. 1998; Panigrahi et al. 2001a). They can accurately edit single sites of model pre-RNAs in vitro (Seiwert and Stuart 1994; Kable et al. 1996; Igo Jr. et al. 2000, 2002). The current model of RNA editing by 20S editosomes envisions a series of dynamic interactions between the pre-mRNA and its cognate gRNA and between these RNAs and the RNA-binding proteins and catalysts within the editosomes (Madison-Antenucci et al. 2002; Stuart and Panigrahi 2002; Stuart et al. 2005 and references therein). Editing generally proceeds in the 3′ to 5′ direction and commences with a gRNA selection step via complementary base-pairing between the 5′ “anchor” region of each gRNA and the 8–10-base pair (bp) region of mRNA sequence adjacent to the block to be edited, followed by specific endonucleolytic cleavage of the pre-mRNA at an unpaired nucleotide immediately upstream of the anchor duplex. As guided by base complementarity within the central informational region of the gRNA, U removal or addition at the 3′ end of the pre-mRNA 5′ cleavage fragment is then catalyzed by a U-specific 3′-exoribonuclease (exoUase) or by a terminal uridylyl transferase (TUTase), respectively, followed by joining of the processed 5′ fragment with the 3′ mRNA fragment by RNA ligases. Multiple cycles of these catalytic steps are required for complete editing of the region specified by a single gRNA, and most mRNAs require sequential use of multiple gRNAs. This complex process must entail numerous protein–RNA and RNA–RNA interactions, the identification of which is still in its infancy (Sacharidou et al. 2006; Salavati et al. 2006; Yu and Koslowsky 2006).
Consistent with the expected complexity of the interactions between 20S editosomes and their RNA editing substrates, the sequences of all 20S editosome proteins have predicted catalytic and/or RNA interaction motifs (Schnaufer et al. 2003; Worthey et al. 2003; Stuart et al. 2005), many of which have been experimentally confirmed. The kinetoplastid RNA editing endonuclease1 (KREN1) that cleaves deletion sites, KREN2 that cleaves insertion sites, and the KREN3 editing endonuclease that specifically cleaves the COII insertion site have RNase III, U1-like zinc finger-like, and double-stranded RNA-binding (dsRBM) motifs (Carnes et al. 2005, 2007; Trotter et al. 2005). Similarly, the U-deletion editing exoUases KREX1 and KREX2 and the U-insertion editing TUTase KRET2 have 3′–5′ exonuclease and poly(A) polymerase domains, respectively (Aphasizhev et al. 2003; Ernst et al. 2003; Kang et al. 2005; N.L. Ernst, unpubl.). The editing RNA Ligases KREL1 and KREL2 also have motifs characteristic of their activities (McManus et al. 2001; Schnaufer et al. 2001; Rusché et al. 2001). All six members of the KREPA family of 20S editosome proteins have oligonucleotide-binding (OB) fold motifs (Schnaufer et al. 2003; Panigrahi et al. 2006; Salavati et al. 2006), and the KREPB4–8 family members have U1 zinc finger-like motifs (Worthey et al. 2003). The 20S editosomes have a common set of proteins that are physically and functionally arranged in subcomplexes that can catalyze the U removal and ligation steps of deletion editing or the U addition and ligation steps of insertion editing (Schnaufer et al. 2003). However, the KREN1, KREN2, and KREN3 endonucleases are in different 20S editosomes along with this common set of proteins but each with one or two specific proteins (Panigrahi et al. 2006). Thus, endonuclease activity appears associated with different 20S editosomes, each of which can catalyze the subsequent U deletion or U insertion and ligation steps of editing.
While components of 20S editosomes that account for the six key catalytic steps of editing have been identified, the functions of many other editosomal proteins have not been determined but must include RNA-binding and protein interaction. Some of the six KREPA family members have been shown to function in RNA-binding and/or protein interaction (Schnaufer et al. 2003; Brecht et al. 2005; Salavati et al. 2006). In addition to the aforementioned OB-fold motifs, KREPA1, KREPA2, and KREPA3 also contain two zinc-finger motifs (Panigrahi et al. 2001b). As shown in other organisms, OB-fold and zinc-finger domains function in nucleic acid or protein binding (Matthews and Sunde 2002; Theobald et al. 2003; Bochkarev and Bochkareva 2004). The OB-fold of KREPA4 resembles that of the S1 ribosomal protein, while the others more closely resemble that of the Escherichia coli DNA single-strand binding protein (SSB) (Schnaufer et al. 2003; Worthey et al. 2003; Salavati et al. 2006). KREPA1 and KREPA2 proteins interact with KREL2 and KREL1, respectively, and, in the case of KREPA1, enhance catalysis (Schnaufer et al. 2003). These proteins were suggested to provide the OB-fold in trans that may function in substrate binding (Schnaufer et al. 2003). KREPA3 binds duplex RNA or DNA in a zinc-dependent fashion (Brecht et al. 2005), while KREPA4 specifically binds the 3′ oligo(U) tail of gRNA (Salavati et al. 2006). Knockdown of expression of each of these proteins inhibits cell growth and, except perhaps for KREPA3, results in partial or full disruption of 20S editosomes (Drozdz et al. 2002; Huang et al. 2002; O'Hearn et al. 2003; Brecht et al. 2005; Salavati et al. 2006). Thus, KREPA family members have critical functions in RNA editing associated with RNA substrate binding and 20S complex integrity.
We examined KREPA6, the smallest (18 kDa) KREPA family member that is present among different 20S editosomes (Panigrahi et al. 2006), which interacts with several other editosome proteins (Schnaufer et al. 2003; A. Schnaufer, unpubl.) and the loss of which affects complex structure and U-deletion and U-insertion endonuclease activities in vitro (Law et al. 2007). Thus, while it is a critical member of 20S editosomes, its specific roles in RNA editing are unknown. We confirm here that KREPA6 is critical for normal growth of PF Trypanosome brucei and the structural integrity of 20S editosomes. Importantly, we also show further that loss of KREPA6 strongly affects RNA editing in vivo and that the protein binds RNA but not DNA in vitro, displaying high cooperativity, a similar affinity for pre-mRNA and gRNA, and a weaker affinity for their partially duplexed form. Also, KREPA6 preferentially binds to the gRNA 3′ oligo(U) tail. Thus, KREPA6 is a RNA-binding component of the 20S editosome that is essential for its structural integrity.
RESULTS
KREPA6 is important for growth and in vivo RNA editing
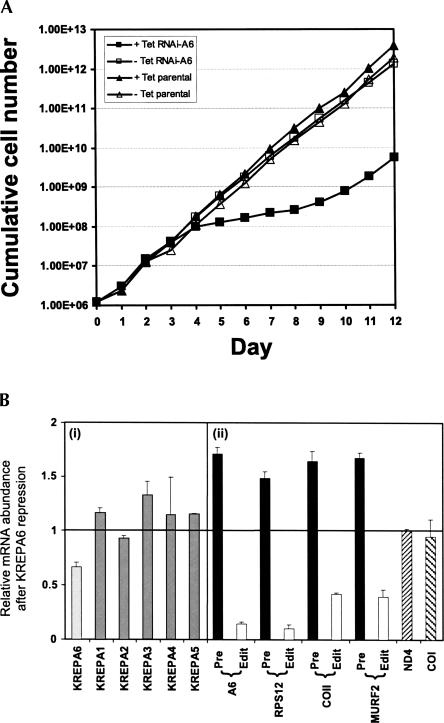
Conditional repression of KREPA6 expression by RNAi in PF T. brucei inhibited cell growth and accumulation of edited RNAs (Fig. 1). Induction of dsRNA that spans the entire KREPA6 coding sequence, using the two opposing promoter system of Wang et al. (2000), resulted in about a 40% knockdown of KREPA6 mRNA after 4 d compared to uninduced cells (Fig. 1B). Growth inhibition became apparent after 4 d, with cells exhibiting reduced motility, and persisted for another 5 d (Fig. 1A). During this period, most cells had reduced motility, and formation of cell debris was observed, mostly at days 6–8, suggesting cell death (data not shown). Growth resumed after ∼9 d and was accompanied by the reappearance of KREPA6 protein based on Western analysis (data not shown), suggesting loss of KREPA6 repression as sometimes observed (for example, see O'Hearn et al. 2003; Luo et al. 2004). Quantitative real-time RT-PCR (qPCR) analysis indicated that the level of KREPA6 mRNA was reduced substantially (Fig. 1B, light gray bar), whereas the levels of the KREPA1–5 mRNAs, which encode related editosome proteins, were unchanged or slightly increased (Fig. 1B, gray bars), indicating that knockdown was specific for KREPA6 mRNA. Western analysis of fractions of glycerol gradient purified total cell lysates from KREPA6 RNAi cells with anti-KREPA6 polyclonal antibodies revealed that KREPA6 protein was dramatically reduced after 2 d of RNAi knockdown and essentially undetectable after 4 d, while the level of a control mitochondrial protein, 2-OGDCp, a component of the 2-oxoglutarate dehydrogenase complex, was essentially unaffected (Fig. 2). The apparent greater reduction of KREPA6 protein compared to the mRNA may reflect the use of qPCR primers to KREPA6 3′ UTR lying outside the region targeted by RNAi, or an effect of dsRNA expression on translation of the KREPA6 mRNA rather than its stability. Overall, the RNAi knockdown dramatically reduced the cellular level of KREPA6 protein and proliferation of PF T. brucei cells.
FIGURE 1.
KREPA6 repression by RNAi inhibits cell growth and reduces RNA editing in PF cells. (A) Growth of RNAi-A6 cells in the absence (uninduced, open squares) or presence of tetracycline (tet) (induced, solid squares) was monitored for 12 d. For comparison, growth of uninduced (open triangles) and induced (solid triangles) 29.13 parental strain cells was also determined. The cumulative cell number was calculated by multiplying the cell densities by the dilution factor. (B) Quantitative real-time PCR analysis of total RNA from RNAi-A6 cells in which KREPA6 was either expressed or repressed for 4 d. The relative abundance of each RNA is in comparison to when KREPA6 was expressed, with the line at 1 indicating no change, and bars above or below this line indicating an increase or decrease, respectively. The levels of 18S rRNA were used for normalization. The error bars indicate the standard deviation of three replicates. Relative mRNA levels of (i) the KREPA editosome proteins (KREPA6, light gray bar; KREPA1 to KREPA5, dark gray bars), (ii) selected mitochondrial pre-edited (black bars) and edited (white bars) mRNAs, and never-edited mRNAs (slashed bars).
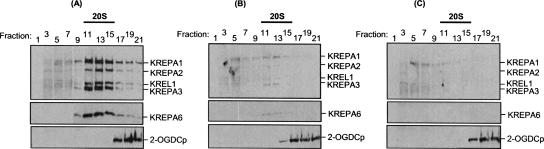
FIGURE 2.
KREPA6 repression leads to loss of 20S editosomes. RNAi-A6 cell lines were grown in the (A) absence or presence of tet for (B) 2 d or (C) 4 d. Equal numbers of cells were lysed and fractionated on glycerol gradients. Odd-numbered fractions were analyzed by Western blot with a mixture of monoclonal antibodies specific for editosome proteins KREPA1, KREPA2, KREL1, and KREPA3, and a polyclonal antibody to KREPA6. Blots were also probed with a monoclonal antibody recognizing a component (2-OGDCp) of the mitochondrial 2-oxoglutarate dehydrogenase complex as a control.
KREPA6 repression resulted in a 90% reduction of edited A6 and RPS12 mRNAs and 60% reduction of edited COII and MURF2 mRNAs after 4 d of RNAi induction as determined by qPCR (Fig. 1B, white bars). In contrast, the relative levels of the corresponding pre-edited mRNAs (Fig. 1B, black bars) were increased. In addition, the levels of ND4 and COI mitochondrial mRNAs (Fig. 1B, slashed bars) that do not undergo editing were essentially unaffected by the KREPA6 knockdown, similar to the nuclearly encoded KREPA1–5 mRNAs. Hence, KREPA6 repression resulted in loss of KREPA6 mRNA and protein, and RNA editing in vivo.
KREPA6 is essential for the integrity of 20S editosomes
Repression of KREPA6 expression by RNAi caused the loss of 20S editosomes (Fig. 2A–C, upper panels). RNAi-A6 PF cells were induced for 2 or 4 d, and whole cell lysates were fractionated on glycerol gradients and examined by Western analysis using monoclonal antibodies specific for the KREPA1, KREPA2, KREPA3, and KREL1 editosome components. Uninduced cells were analyzed in parallel. Each of these editosome proteins was dramatically reduced with virtually no detectable signal remaining in the 20S region (fractions 11–15) after 4 d of KREPA6 repression, closely paralleling the loss of KREPA6 protein (Fig. 2C). Approximately 90% of the strongest Western signal, corresponding to KREPA1 within the 20S region, was eliminated by day 2 of RNAi (Fig. 2B). A small amount of Western signal was detected in fractions 5–9, especially for KREPA1, possibly indicating a slight accumulation or retention of editosome subcomplexes as has been seen previously with knockdown of mRNAs for other editosome components (Huang et al. 2002; O'Hearn et al. 2003; Wang et al. 2003; Salavati et al. 2006; Babbarwal et al. 2007). This suggests that KREPA6 is critical to overall editosome integrity and perhaps assembly since the KREPA1, KREPA2, and KREPA3 mRNA levels, and presumably their translation and mitochondrial import, remained virtually unchanged at day 4 of RNAi induction (see Fig. 1B). The absence of these proteins probably reflects their degradation as a result of perturbed 20S editosome structural integrity or assembly.
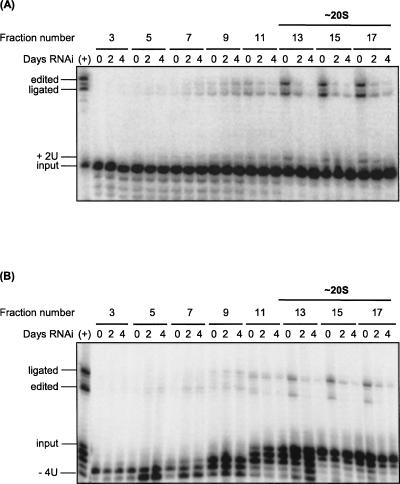
Repression of KREPA6 also resulted in loss of in vitro RNA editing activities (Fig. 3). Glycerol gradient fractions of crude mitochondrial lysates of RNAi-A6 cells in which KREPA6 was expressed (uninduced) or repressed for 2 and 4 d were analyzed by pre-cleaved insertion and deletion editing assays. These assays bypass the first step of editing, the endonucleolytic cleavage of the mRNA, and thus efficiently measure the subsequent TUTase and ligase activities associated with insertion editing (Fig. 3A), and exoUase and ligase activities associated with deletion editing (Fig. 3B), respectively. KREPA6 repression resulted in loss of TUTase, exoUase, and ligase activities in vitro in the 20S region of the gradient, consistent with the substantial loss of 20S editosome proteins (Fig. 3, fractions 13–17). Residual TUTase, exoUase, and ligase activities were detected in these fractions, and a small increase in these activities was observed in fractions 7–9 after RNAi induction, suggesting that some complete editosomes remained and some functional subcomplexes were generated upon KREPA6 loss, consistent with the results from the Western analysis (see Fig. 2). Taken together, these data demonstrate that KREPA6 is essential for the structural and functional integrity of 20S editosomes.
FIGURE 3.
KREPA6 repression results in a substantial reduction of RNA editing activities in vitro. Crude mitochondrial extracts of RNAi-A6 cells induced with tet for the indicated number of days were fractionated on glycerol gradients. Odd-numbered gradient fractions were assayed for (A) pre-cleaved insertion and (B) pre-cleaved deletion editing activities in vitro. The radiolabeled input RNAs, input RNA with two Us added (+2U), or four Us removed (−4U), ligated products of unprocessed 5′ and 3′ input RNAs (ligated), and edited products are indicated. Positive control reactions (+) were performed using a fraction of ∼20S editosomes that contains peak editing activity. The region corresponding to 20S editosomes is indicated, based on Western blot analyses of the gradients with antibodies specific for known editosome proteins as in Figure 2 (data not shown).
KREPA6 binds to pre-edited mRNA and gRNA cooperatively and with similar affinities in vitro
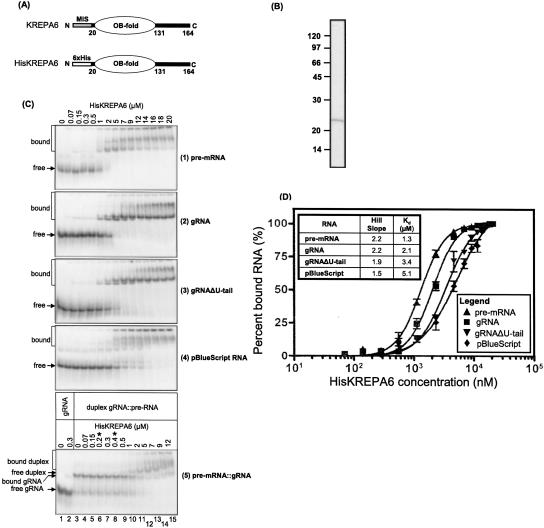
Since the presence of an OB-fold motif in KREPA6 (Fig. 4A; Schnaufer et al. 2003; Worthey et al. 2003) suggested that it might function in substrate RNA binding during editing, its RNA-binding ability was assessed by electrophoretic mobility-shift assay (EMSA). Recombinant His-tagged KREPA6 that was expressed in E. coli (HisKREPA6) and purified to near homogeneity (Fig. 4B) was incubated with 5′ end-labeled synthetic RNAs of comparable lengths representing pre-edited ATPase subunit 6 mRNA (A6 pre-mRNA), its cognate gRNA gA6[14] (gRNA), gA6[14] minus the oligo(U) tail (gRNAΔU-tail), annealed pre-mRNA and gRNA (pre-mRNA∷gRNA), and a pBluescript RNA (pBS), and resolved on native gels, as described in Materials and Methods. HisKREPA6 bound these RNAs in a protein concentration-dependent manner, resulting in three shifted bands (Fig. 4C) and accumulation of radioactive signal in the wells of the gel. The majority of the shifted bands occurred at higher concentrations of HisKREPA6, as did material that did not leave the well, especially in the case of pre-mRNA and pBS. The bands were particularly distinct with the gRNAs and pre-mRNA, suggesting relatively stable or well-defined protein–RNA complexes. This was less so with partial duplex pre-mRNA∷gRNA and pBS. Essentially all of each RNA was bound at ∼7 μM HisKREP6, except pBS RNA, which was completely bound at ∼20 μM HisKREPA6.
FIGURE 4.
Recombinant KREPA6 binds both gRNA and pre-mRNA cooperatively with similar affinities. (A) Schematic representation of native pre-processed KREPA6 and the recombinant TEV cleavable N-terminally histidine-tagged version, HisKREPA6, used in this study, highlighting the single central OB-fold domain. The mitochondrial import signal (MIS) of KREPA6 was replaced with a hexa-histidine tag. (B) Coomassie-stained SDS-PAGE gel of affinity-purified HisKREPA6 protein used in EMSA. Due to the affinity tag, it migrates at ∼22 kDa. (C) EMSA gel analysis of HisKREPA6 binding to (1) ATPase 6 pre-mRNA (pre-mRNA), (2) its cognate gRNA, gA6[14] with a 3′-oligo(U) tail (gRNA), (3) gA6[14] without a 3′-oligo(U) tail (gRNAΔU-tail), (4) control pBluescript RNA, and (5) partial duplexes of A6 pre-mRNA and gA6[14] (pre-mRNA∷gRNA). [32P]- radiolabeled RNA was incubated with various concentrations of HisKREPA6 and resolved on native gels. The unbound radiolabeled RNA (free) and HisKREPA6 bound RNA complexes (bound) are indicated. Note that the range of HisKREPA6 concentrations used in partial duplex RNA binding (panel 5) is different (0–12 μM) from the other panels (0–20 μM), and has two additional concentrations (asterisks). (D) HisKREPA6/RNA-binding curves indicating the relative amount of RNA bound at different HisKREPA6 concentrations. In each case, the amount of RNA bound includes all of the shifted bands in the gel including the signals in the wells (C) and is the average of three replicates. Binding data were fit as described in Materials and Methods. Quantitation of the amount of HisKREPA6 bound to duplex pre-mRNA∷gRNA could not be done accurately due to the overlap of gel shifts between HisKREPA6 bound to gRNA or pre-mRNA∷gRNA.
The complex and partially overlapping pattern of gel-shifted bands did not permit a rigorous determination of apparent equilibrium dissociation constants (K D app) for each individual HisKREPA6/RNA complex. Thus, an isotherm of the overall binding of HisKREPA6 with each RNA, referring to all retarded bands including those in the wells of the gel, was approximated from the best statistical fit of each binding curve to various equilibrium binding models (Fig. 4D). Each resulting best fit conformed to the Hill equation with coefficients greater than 1, indicating cooperative binding of HisKREPA6 to RNA (Fig. 4D, inset), and each estimated overall K D app was in the low micromolar range, indicating a moderate affinity of HisKREPA6 for each RNA tested. F-test analysis of the overall K D app values and the Hill plots indicated that the differences between each RNA species were statistically significant (P-values < 0.0001) (data not shown). A6 pre-mRNA, gRNA, and gRNAΔU-tail RNAs each had significantly higher Hill coefficients and lower K D app values for HisKREPA6 binding compared to pBS RNA, indicating both higher cooperativity and affinity of HisKREPA6 binding for these editing-related RNAs compared with pBS. The complex gel-shift patterns and binding models together suggest that either there are multiple HisKREPA6 editing RNA-binding interactions that influence each other, or concentration dependent HisKREPA6 oligomerization (which could be influenced by RNA binding), or both. Absence of the oligo(U) tail (i.e., gRNAΔU tail) significantly reduced both cooperativity and affinity, indicating that this tail contributes to the binding energy. The overall K D app values for all RNA species may be underestimated since it is likely that <100% of the recombinant protein in the preparation was active. Determination of the K D app for the HisKREPA6/duplex pre-mRNA∷gRNA complex (Fig. 4C, panel 5) could not be performed accurately due to the overlap of this complex with HisKREPA6/gRNA (Fig. 4C, lane 2, bound gRNA) and duplexed pre-RNA∷gRNA without protein (Fig. 4C, lane 3, free duplex), and therefore was not included here.
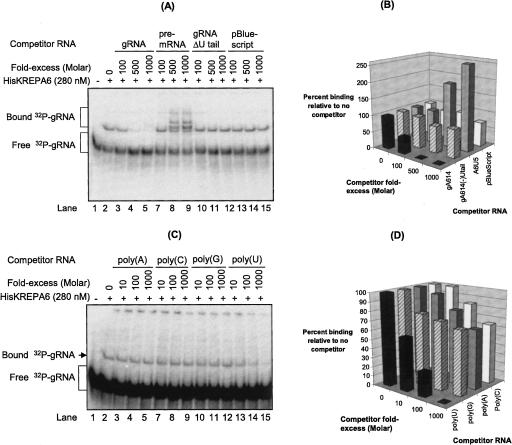
Recombinant KREPA6 protein preferentially binds to gRNA with a 3′ oligo(U) tail
Gel-shift competition assays revealed a general correlation between the affinity of HisKREPA6 for the gRNA based from overall K D app measured as above (see Fig. 4D) and binding specificities associated with the 3′ oligo(U) tail (Fig. 5A,B). A 500–1000-fold molar excess of unlabeled gRNA essentially abolished the gel-shifted radiolabeled gRNA band while similar excesses of unlabeled gRNAΔU-tail or pBS resulted in a reduction of <15% (Fig. 5A,B). Incubation with pre-mRNA resulted in additional bands that include the partial gRNA:mRNA duplex (see Fig. 4E) and, presumably, additional RNA/protein complexes, based on sensitivity of the higher-shifted bands to protease digestion (data not shown). The observed competition is consistent with the higher K D app values of HisKREPA6 for gRNA-ΔU tail and pBS relative to gRNA (see Fig. 4C, bottom panel). Thus, the oligo(U) tail may be an important element for the binding of HisKREPA6 to the gRNA, either acting as a direct binding target and/or by stabilizing a structure favorable for binding. This was paralleled by complementary gel-shift competition assays with homopolymeric poly(A), poly(G), poly(C), and poly(U) RNAs of heterogeneous lengths (10–70 nucleotides [nt]) (Fig. 5C,D). Poly(U) was found to be the most effective competitor of HisKREPA6 binding to gA6[14] gRNA, essentially eliminating the gel shift at 1000-fold molar excess, while the other homopolymers reduced it by ∼40% (Fig. 5C,D).
FIGURE 5.
Recombinant KREPA6 preferentially binds poly(U) and gRNA with an oligo(U) tail. (A) EMSA gel analysis of HisKREPA6 binding to radiolabeled gA6[14] gRNA with an oligo(U) tail in the absence or presence of excess unlabeled competitor RNAs. (Lanes 8,9) The multiple bands observed in the presence of excess unlabeled pre-mRNA are due to partially duplexed gRNA∷pre-mRNA and/or HisKREPA6 binding to the duplex RNA. (B) Quantitation of the amount of gRNA binding in A, indicating the percent of gRNA bound in the presence of competitor RNAs relative to their absence. (A, lane 2) The amount of HisKREPA6 bound to gRNA in the absence of competitor was set to 100%. The competitor RNAs were gRNA with a U tail (black bars), gRNAΔU tail (striped bars), pre-mRNA (gray bars), and pBlueScript control RNA (white bars). The apparent increase in HisKREPA6 gRNA binding in the presence of excess pre-mRNA >100% is due at least in part to formation of partial gRNA∷pre-mRNA duplexes. (C) EMSA analysis of HisKREPA6 binding to radiolabeled gRNA with a U tail in the absence or presence of excess unlabeled homopolymeric competitor RNAs. Molar excess was based on nucleotide gram weight estimate because of the heterogeneous polymer sizes (10–70 nt) of the RNAs. (D) Quantitation of the amount of gRNA binding in C, indicating the percent of gRNA bound in the presence of competitor RNAs relative to their absence. (C, lane 2) As in B, the amount of HisKREPA6 bound to gRNA in the absence of competitor was set to 100%. The competitors were poly(U) (black bars), poly(G) (striped bars), poly(A) (gray bars), and poly(C) (white bars).
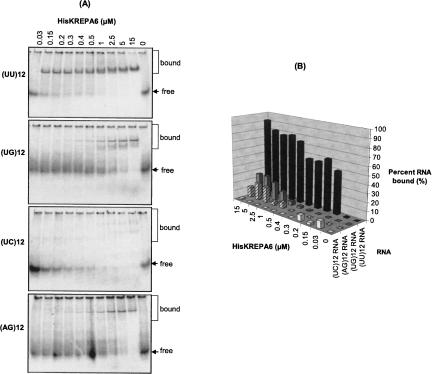
Recombinant KREPA6 protein is a poly(U) RNA-binding protein
HisKREPA6 also had a preference for oligo(U) in gel-shift assays using synthetic 24-mer (UU)12, (UG)12, (UC)12, and (AG)12 RNAs (Fig. 6). When limiting concentrations (0.5–2 nM) of 5′-end-labeled RNA were incubated at room temperature with increasing excess amounts of purified HisKREPA6 (30 nM–15 μM), only (UU)12 was bound by HisKREPA6 with high affinity (Fig. 6A,B). HisKREPA6 bound more than 50% of (UU)12 at ∼200 nM protein, indicating a K D app around this value. Synthetic (AG)12 and (UG)12 resulted in gel-shifted bands at higher HisKREPA6 concentrations although even at the highest concentration of HisKREPA6 (∼15 μM) less than 50% of the input RNA was shifted. Essentially no (UC)12 RNA was bound by HisKREPA6. The RNA degradation observed at high concentrations of purified HisKREPA6 is likely due to residual nucleases in the preparation. HisKREPA6 also did not shift two 24-mer deoxyribonucleotide-analogs, d(UU)12 and d(AG)12, to any significant extent (data not shown), indicating the specificity of HisKREPA6 for RNA.
FIGURE 6.
Recombinant KREPA6 has a higher affinity for poly(U) RNA. (A) EMSA analysis of HisKREPA6 binding to synthetic 24-mer RNAs. (B) Quantitation of the RNA binding in A indicating the percent of RNA bound with increasing HisKREPA6 concentrations. The RNA ligands were 5′-end-labeled (UC)12 (white bars), (AG)12 (striped bars), (UG)12 (gray bars), and (UU)12 (black bars). Owing to the apparent RNA degradation in lanes containing high concentrations of HisKREPA6, the amount of radioactive free RNA in the absence of HisKREPA6 (lane 11 of each gel) was used to estimate percent binding as detailed in the Materials and Methods. Also because of this RNA degradation, apparent K D estimations could not be accurately determined.
DISCUSSION
We conclude from the results reported here that KREPA6 has a critical role in the structural integrity of ∼20S editosomes and that it also appears to function in RNA binding. Repression by RNAi of KREPA6 expression resulted in essentially complete loss of ∼20S editosomes with little accumulation of free editosome proteins or subcomplexes. Not surprisingly, this resulted in the loss of both editing and the viability of PF T. brucei, which are reliant on this process. Recombinant KREPA6 was found by gel retardation studies to have a moderate affinity for RNA in general and a greater affinity for oligo(U) but no affinity for DNA. These results indicate that KREPA6 is an integral component of ∼20S editosomes and suggest that it functions in RNA binding that may entail binding of the 3′ oligo(U) tail of gRNAs.
Effects of KREPA6 repression on 20S editosome structure and cell growth
The extensive loss of 20S editosomes upon knockdown of KREPA6 expression (Fig. 2) indicates that it is essential to editosome structural integrity. A recent study reported a similar finding (Law et al. 2007). The concomitant loss of other editosome proteins also indicates that integration of these proteins into the editosome is essential for their stability since limiting amounts of KREPA6 appears to compromise the assembly and/or the stability of the 20S editosomes, leading to rapid turnover of editosome proteins. These structural effects appear more severe relative to similar inactivation of expression of editosome proteins KREPA1 or KREPA3 by RNAi in PF T. brucei, which caused only slight changes in editosome sedimentation (Drozdz et al. 2002; O'Hearn et al. 2003; Brecht et al. 2005). However, the results reported here are similar to the consequences of inactivation of expression of KREPA2, KREPA4, and KREPB4 in PFs (Huang et al. 2002; Salavati et al. 2006; Babbarwal et al. 2007) and KREPB5 (TbMP44) in BFs (Wang et al. 2003). KREPA6 has been shown to directly interact with KREPA1 and to partially associate with the heterotrimeric insertion and deletion subcomplexes of editosomes (Schnaufer et al. 2003) as well as to directly interact with various other editosome proteins (A. Schnaufer, unpubl.). Therefore, KREPA6 appears to have a central role in establishing and/or maintaining key protein–protein interactions within 20S editosomes, as hypothesized previously (Stuart et al. 2005). KREPA4, KREPB4, and KREPB5 are of similarly critical importance, while KREPA1 and KREPA2 appear to be less central to overall editosome stability, given the formation of stable subcomplexes upon their inactivation. Nevertheless all these noncatalytic proteins, and perhaps others in the editosome, are critical for 20S editosome assembly and/or stability.
RNA binding of KREPA6
Another important characteristic of KREPA6 uncovered in this study, in addition to its role in protein interaction and editosome structural stability, is its ability to bind RNA with an apparent preference for oligo(U) RNA. This RNA-binding ability is consistent with its predicted OB-fold structure (Schnaufer et al. 2003), a domain associated with nucleic acid recognition/binding in various proteins (Theobald et al. 2003; Bochkarev and Bochkareva 2004). The six KREPA family members all have predicted OB-fold domains near their C termini, and we previously suggested that KREPA1 and KREPA2 may provide this substrate recognition/binding function in trans to the catalytic proteins with which they directly interact within the editosome (Schnaufer et al. 2003). RNA binding would also be expected to function in associating pre-mRNA and gRNA with the editosome and in relocating these RNAs during the multiple coordinated steps of editing.
KREPA6 exhibited a similarly complex pattern of binding to various synthetic RNAs, generating multiple but discrete, concentration-dependent gel-shifted products, including material that did not enter the gel (Fig. 4C). Hence, KREPA6 may have either multiple or alternate binding sites on the RNAs, bind multiple RNA ligands simultaneously, or have oligomeric conformations that influence binding. The strong positive cooperativity of KREPA6 binding to RNA (Fig. 4D) is consistent with these possibilities. Understanding the functional significance of this behavior by KREPA6 will require determining the stoichiometry of KREPA6 within the editosome.
Remarkably, although KREPA6 binds all of the RNAs tested with similar, moderate affinities (range of overall K D app's = 1.3–5.1 μM), a preference of KREPA6 for oligo(U) binding is clearly evident from the competition and direct binding studies (Figs. 5, 6). Therefore, the kinetics of substrate binding, which was not addressed in our studies, may significantly contribute to the relative specificity observed. The preference of KREPA6 for oligo(U), at least for recombinant protein in vitro, implies interaction with the gRNA 3′ oligo(U) tail, although interaction with stretches of U to be deleted or that are inserted at specific sites during editing cannot be excluded. For example, the presence of the oligo(U) tail might result in more stable binding of KREPA6 to gRNA (i.e., result in a reduced OFF rate constant, k OFF). This interaction may significantly contribute to the binding of gRNA to the editosome complex, perhaps analogous to the kinetic contribution of the mRNA 3′-poly(A) tail to ribosome binding during translation in eukaryotes via the poly(A)-binding protein (Tarun Jr. and Sachs 1995; Sachs and Buratowski 1997). Detailed kinetic studies will be necessary to uncover the underlying mechanism for the observed specificity.
Despite the preference of the KREPA6 protein for oligo(U), its moderate affinity binding of RNA, essentially regardless of sequence, may reflect a general RNA affinity that is important to the process of RNA editing. Hence, KREPA6 may utilize both its general RNA-binding properties and its higher affinity and relative preference for oligo(U) for distinct but functionally related purposes. The former characteristic could provide a low-affinity interaction platform for gRNA and/or pre-mRNA during editing. On the other hand, since it is inconceivable that each of the more than 1200 different gRNAs are uniquely recognized by the 20S editosome, preferential recognition of a common element such as the gRNA oligo(U) tail via KREPA6 could facilitate positioning of the anchor duplex (which is another common structural element in RNA editing) and editing sites with the respective RNA-binding and catalytic domains of the editosome proteins. Furthermore, such potential functions can be integrated with those of other members of the KREPA family or other RNA-binding components of the editosome. For example, like KREPA6, KREPA4 also binds gRNA with a preference for the oligo(U) tail and is essential for editing and for editosome structural integrity, although it appears less stably associated with the deletion and insertion subcomplexes than KREPA6 (Salavati et al. 2006). Therefore, it is conceivable that KREPA6 and KREPA4 might be involved in a “hand-off” process via their respective OB-fold domains to coordinate the forward progression of RNA editing steps. This subunit switching is a recurring and unifying theme that has emerged from mechanistic studies in DNA replication, repair, and recombination (Stauffer and Chazin 2004). In these transactions, the OB-fold domains of various DNA-binding proteins act cooperatively in handing off the DNA substrate, either by direct competition for binding sites or allosteric structural rearrangements to facilitate dynamic restructuring of DNA processing complexes in the course of DNA processing. KREPA6 and KREPA4, probably along with other predicted OB-fold proteins in the editosome, may therefore play analogous roles within editosome complexes during RNA editing. Such RNA-binding functions of the KREPA-family proteins must be further integrated with the predicted RNA-binding motifs of other editosome proteins, such as zinc fingers, dsRBMs, and Pumilio motifs. Clearly, both structural information from protein/RNA complexes and detailed studies of the dynamic events that occur during RNA editing will be needed to validate our hypothesis.
MATERIALS AND METHODS
Plasmids
An RNAi construct for inducible knockdown of KREPA6 expression was prepared in the pZJM vector, which harbors opposing tetracycline (tet)-inducible T7 promoters flanking the cloning site (Wang et al. 2000; Morris et al. 2001). A 563-bp XhoI/HindIII fragment encompassing the entire KREPA6 open reading frame plus 48 and 17 nt of the 5′ and 3′ UTR, respectively, was generated by PCR amplification of PF T. brucei genomic DNA (strain 29.13) using 5′-GACCTCGAGAAGGGGGTTCGTATTCTGTT-3′ as the forward primer and 5′-GCCAAGCTTGCGGAAACACAAGGGCATTA-3′ as the reverse primer that contain the relevant restriction sites (underlined italics). The PCR product was initially cloned into pGEM-T Easy (Promega) then released by digestion with HindIII and XhoI and subcloned into pZJM that was digested with the same enzymes, thus generating RNAi plasmid pZJMA6–1. The HisKREPA6 expression plasmid pProEX-HisKREPA6–1 was the kind gift of Junpeng Deng and Wim Hol (J. Deng and W. Hol, unpubl.). It expresses a TEV-cleavable N-terminally His-tagged KREPA6 without the mitochondrial targeting signal from a pProEX HTa vector (Invitrogen).
Transfection, RNAi induction and growth
PF T. brucei strain 29–13 cells (Wirtz et al. 1999) were transfected as described previously (Schnaufer et al. 2001), using 15 μg of pZJMA6–1 plasmid DNA that was linearized by digestion with NotI. The resultant stable cell lines were selected by growth in SDM-79 medium containing 10% FBS and supplemented with 15 μg/mL G418, 25 μg/mL hygromycin, and 2.5 μg/mL phleomycin, and designated RNAi-A6 PF. Correct integration into the 18S rRNA locus was confirmed by PCR using vector-specific and genome-specific primers. RNAi was induced in half of the culture after growth in the same medium to a density of 2.0 × 107 cells/mL, dilution to 1.2 × 106 cells/mL, and addition of 1 μg/mL tet. The induced and uninduced cultures were maintained between 1.2 × 106 and 2.0 × 107 cells/mL, and cell density was monitored daily using a particle counter (Beckman).
Quantitative real-time PCR (qPCR)
Oligonucleotide primers for qPCR were designed using the Primer Express software (Applied Biosystems) and purchased from Invitrogen. Primers corresponding to sequences that are specific for KREPA6 mRNA, are outside the OB-fold motif, and are in its 3′ UTR, were used to distinguish it from other KREPA family members (Worthey et al. 2003). Primers specific for pre-edited and edited mRNAs (A6, RPS12, COII, MURF2), never-edited mRNAs (COI and ND4), and control 18S rRNA were previously reported (Carnes et al. 2005; Trotter et al. 2005). Briefly, total cellular RNA was extracted using Trizol essentially as described in the manufacturer's protocol (GIBCO BRL) from ∼1 × 108 KREPA6 RNAi PF cells in which RNAi was induced by 4 d of growth in 1 μg/mL tet and from non-induced cells. Ten micrograms of RNA was DNase-treated using the DNA-free kit (Ambion), following the manufacturer's instructions, and the quality of the resultant RNA was assessed using a Bioanalyzer (Agilent Technologies). cDNA was generated from intact total cellular RNA using random hexamer primers and the TaqMan Reverse Transcription Reagent Kit (Applied Biosystems) according to the manufacturer's instructions. Control reactions without reverse transcriptase were included. Amplification reactions (25 μL total) were set up in triplicate in a 96-well plate format, and each reaction contained 2.5 μL of appropriate dilutions of cDNA (typically between 1:7 and 1:50) and 5 μL each of 1.5 μM stock of the forward and reverse primer pairs, and 12.5 μL of SYBR Green PCR Master Mix (Applied Biosystems). qPCR reactions were run in the ABI Prism 700 Sequence Detection System (Applied Biosystems) under the following amplification conditions: 10 min at 95°C followed by 40 cycles of 15 sec at 95°C and 1 min at 60°C at each cycle. PCR efficiencies were calculated by linear regression using LinRegPCR (Ramakers et al. 2003), and amplification was analyzed using the Pfaffl method (Pfaffl 2001). Data were normalized to 18S rRNA, and relative changes in mRNA abundance after RNAi induction were expressed as fold-changes relative to uninduced control cells.
Glycerol gradient fractionations and Western analysis
RNAi-A6 cells were induced with 1 μg/mL tet for 0, 2, or 4 d (i.e., until prior to growth inhibition), and lysates were prepared from 1 × 108 total cells or from mitochondria isolated from 1.25 × 1010 cells of each induction as described previously (Harris et al. 1990), using lysis buffer with protease inhibitors [20 mM HEPES at pH 7.9, 10 mM Mg(OAc)2, 50 mM KCl, 1 mM EDTA, 1 μg/mL pepstain, 2 μg/mL leupeptin, and 1 mM pefabloc]. One milliliter of cleared lysates were layered onto an 11-mL 10%–30% glycerol gradient that contained 10 mM Tris-Cl (pH 7.2), 10 mM MgCl2, 100 mM KCl, 1 mM DTT, 1 μg/mL pepstatin, 2 μg/mL leupeptin, and 1 mM pefabloc. Total cell lysates were centrifuged for 9 h and mitochondrial extracts for 12 h both at 38,000 rpm in a SW40 rotor (Beckman) at 4°C. Half-milliliter fractions were collected from the top, aliquoted, and quick-frozen in liquid nitrogen and stored at −80°C for Western and in vitro RNA editing assays.
For Western analysis, 30 μL fractions were electrophoresed in pre-cast 10% Tris-HCl SDS-PAGE gels (Bio-Rad) and transferred to PVDF membrane as described previously (Trotter et al. 2005). The membranes were probed with monoclonal antibodies specific for KREPA1, KREPA2, KREPA3, and KREL1 (Panigrahi et al. 2001b), rabbit polyclonal antibodies specific for KREPA6 (Schnaufer et al. 2003), or a monoclonal antibody recognizing a protein, designated here as 2-OGDCp, of the mitochondrial 2-oxoglutarate dehydrogenase complex (A. Panigrahi, pers. comm.).
RNA synthesis and in vitro RNA editing
Synthetic 24-mer RNAs (UU12, UG12, UC12, and AG12) and DNAs (dAG12 and dUU12) were purchased and de-protected according to the manufacturer's protocol (Dharmacon). The A6 short/TAG.1 ATP synthase subunit 6 pre-mRNA and its cognate gA6[14] gRNA with or without the oligo(U) tail were transcribed in vitro from PCR products as described previously (Seiwert et al. 1996) and used in gel-shift analyses (Salavati et al. 2006). In vitro pre-cleaved insertion was assayed using 5′-labeled 5′-CL18 with 3′-CL13pp and gPCA6–2A RNAs, while pre-cleaved deletion used 5′-labeled U5-5′, U5-3′, and gA6[14]PC-del RNAs as previously described (Igo Jr. et al. 2000, 2002).
HisKREPA6 protein expression and purification
The pProEX-HisKREPA6–1 plasmid encoding the recombinant KREPA6 protein with a TEV cleavable N-terminal His6 tag was transformed and expressed in E. coli BL-21(DE3). Cells were grown to an OD600 of 0.5–0.7 at 37°C in LB broth containing 100 μg/mL ampicillin and induced with 1 mM isopropyl β-D-1-thiogalactopyranoside for 20 h at 18°C. The protein was purified as follows with all steps at 4°C. Two grams of fresh or quick-frozen, washed cells were resuspended in 6 mL of wash buffer A (50 mM Tris-HCl at pH 8.0) that contained Complete Mini-EDTA free Protease Inhibitor Cocktail (Roche). They were lysed by 30-min incubation at 4°C in 100 μg/mL lysozyme (Sigma), and the lysate was cleared by 30 min of centrifugation at 10,000g after adding NaCl to 1.0 M and sonicating to reduce viscosity. In order to remove nucleic acids, the cleared lysate was mixed with 5 mL of Q-Sepharose beads (Amersham Biosciences) that were pre-equilibrated in buffer A containing 1.0 M NaCl and incubated for 60 min with gentle shaking and poured into a glass column, and the flowthrough was collected. HisKREPA6 protein was purified from the flowthrough by binding to a 0.5 mL column of nickel-nitrilotriacetic acid (QIAGEN) equilibrated with the same buffer washing stepwise with 5 mL of buffer B (20 mM Tris-HCl at pH 8.0, 100 mM KCl, 10% glycerol) containing 2 and 20 mM imidazole, then eluting with 1.5 mL of buffer B containing 250 mM imidazole. Each step of HisKREPA6 purification was monitored by SDS-PAGE and Western analysis using rabbit anti-KREPA6 and anti-His antibodies. A single protein band with an apparent size of 22 kDa was seen in Western and stained gels. The purified protein was dialyzed against two changes of buffer C (10 mM Tris-HCl at pH 7.2, 100 mM KCl, 10% glycerol, and 1 mM DTT) and resulted in ∼1 mL of 4–5 mg/mL protein based on comparison of serial dilutions of the protein and known amounts of BSA as determined by Bradford analysis using the Coomassie Plus Kit (Pierce) and comparison of serial dilutions of the protein and known amounts of BSA in stained SDS-PAGE gels. Aliquots were stored at −80°C.
RNA-binding assays
Electrophoretic mobility shift assays (EMSA) were performed as previously described (Salavati et al. 2006) except as indicated. Briefly, for single-strand RNA binding, 2–8 fmol of [γ-32P] 5′-end-labeled RNA were incubated with increasing concentrations of purified HisKREPA6 protein in 20-μL reactions with RBB-1 buffer (20 mM Tris-HCl at pH 7.4, 150 mM KCl, 5 mM Mg2Cl, 1 mM DTT, 0.1 μg/μL BSA, 2 units/μL RNAsin) for 30 min at room temperature. Reactions were incubated, mixed with gel-loading dye (0.05% bromophenol blue, 0.05% xylene cyanol, 8% sucrose), and loaded directly on a native 10% TBE gel (Bio-Rad), pre-run at 110 V for 30 min with 0.5% TBE buffer at 4°C. The gel was electrophoresed for 1.5 h under pre-running conditions, fixed in 10% isopropanol plus 7% acetic acid for 30 min, and air-dried with cellophane sheets overnight. For the duplex RNA-binding assay, equimolar (250 nM) amounts of internally labeled [32P]UTP gRNA and unlabeled A6 short/TAG.1 pre-mRNA were annealed in 20 μL of 50 mM Tris (pH 7.5), 0.1 mM EDTA, 100 mM KCl, 3% glycerol, 2 mM MgCl2, and 1 μL of RNA Secure (Ambion) in a GeneAmp System 9700 PCR machine (Applied Biosystems) for 10 min at 60°C, cooled to 27°C at 2°/min, and held at this temperature for 20 h (D. Koslowsky, unpubl.). Annealing efficiency was determined by electrophoresis in pre-cast 10% TBE native gels (Bio-Rad). One-tenth (25 nM) of the annealed RNA was used in each reaction for gel-shift analyses as described above for single-strand RNA binding.
HisKREPA6-bound and unbound RNAs were visualized using a Molecular Dynamics PhosphorImager, and scaled-up reactions performed in triplicate were quantified using the ImageQuant software. The overall apparent equilibrium dissociation constant (overall K D app), i.e., the HisKREPA6 protein concentration at which half of the RNA was bound, was estimated from the sum of all shifted radiolabel in each lane, including that in the well (which may be aggregated material), since the RNA/protein stoichiometry in each band is uncertain. The data were analyzed using GraphPad Prism Software (GraphPad Software, Inc.) to determine an overall binding model for each RNA. The EC50 values for each RNA, defined as the concentration of HisKREPA6 evoking 50% RNA binding, were taken as overall K D app and were determined using the Hill equation with a floating Hill coefficient and had P-values <0.0001 by the F-test.
RNA-gel-shift assays with competitor RNAs were carried out as described above using a fixed amount (6 pmol) of HisKREPA6 protein. The radiolabeled RNA was mixed with various molar excess amounts of unlabeled competitor RNA in buffer prior to the addition of HisKREPA6 protein. The amount of competition was estimated as the ratio of bound RNA in the presence of cold competitor and bound RNA without competitor.
ACKNOWLEDGMENTS
We thank G. Cross and E. Wirtz for providing T. brucei strain 29.13, P. Englund for the pZJM vector, and members of the Stuart laboratory for insightful comments. Funding for this work was provided by NIH grant GM042188 to K.S., initial joint support by AI014102 to K.S., NIH grant GM077418 to W.H., and also by NIH F-32 Ruth L. Kirschstein National Research Service Awards for Individual Post-doctoral Fellows grant AI68303 to S.Z.T.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.763308.
REFERENCES
- Aphasizhev, R., Aphasizheva, I., Simpson, L. A tale of two TUTases. Proc. Natl. Acad. Sci. 2003;100:10617–10622. doi: 10.1073/pnas.1833120100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babbarwal, V.K., Fleck, M., Ernst, N.L., Schnaufer, A., Stuart, K.D. An essential role of KREPB4 in RNA editing and structural integrity of the editosome in Trypanosoma brucei . RNA. 2007;13:737–744. doi: 10.1261/rna.327707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benne, R., Van den Burg, J., Brakenhoff, J.P.J., Sloof, P., Van Boom, J.H., Tromp, M.C. Major transcript of the frameshifted coxII gene from trypanosome mitochondria contain four nucleotides that are not encoded in the DNA. Cell. 1986;46:819–826. doi: 10.1016/0092-8674(86)90063-2. [DOI] [PubMed] [Google Scholar]
- Bochkarev, A., Bochkareva, E. From RPA to BRCA2: Lessons from single-stranded DNA binding by the OB-fold. Curr. Opin. Struct. Biol. 2004;14:36–42. doi: 10.1016/j.sbi.2004.01.001. [DOI] [PubMed] [Google Scholar]
- Brecht, M., Niemann, M., Schlüter, E., Müller, U.F., Stuart, K., Göringer, H.U. TbMP42, a protein component of the RNA editing complex in African trypanosomes has endo-exoribonuclease activity. Mol. Cell. 2005;17:621–630. doi: 10.1016/j.molcel.2005.01.018. [DOI] [PubMed] [Google Scholar]
- Carnes, J., Trotter, J.R., Ernst, N.L., Steinberg, A.G., Stuart, K. An essential RNase III insertion editing endonuclease in Trypanosoma brucei . Proc. Natl. Acad. Sci. 2005;102:16614–16619. doi: 10.1073/pnas.0506133102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnes, J., Trotter, J.R., Peltan, A., Fleck, M., Stuart, K. RNA editing in Trypanosoma brucei requires three different editosomes. Mol. Cell. Biol. 2007 doi: 10.1128/MCB.01374-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drozdz, M., Palazzo, S.S., Salavati, R., O'Rear, J., Clayton, C., Stuart, K. TbMP81 is required for RNA editing in Trypanosoma brucei . EMBO J. 2002;21:1791–1799. doi: 10.1093/emboj/21.7.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst, N.L., Panicucci, B., Igo R.P., Jr, Panigrahi, A.K., Salavati, R., Stuart, K. TbMP57 is a 3′ terminal uridylyl transferase (TUTase) of the Trypanosoma brucei editosome. Mol. Cell. 2003;11:1525–1536. doi: 10.1016/s1097-2765(03)00185-0. [DOI] [PubMed] [Google Scholar]
- Feagin, J.E., Jasmer, D.P., Stuart, K. Developmentally regulated addition of nucleotides within apocytochrome b transcripts in Trypanosoma brucei . Cell. 1987;49:337–345. doi: 10.1016/0092-8674(87)90286-8. [DOI] [PubMed] [Google Scholar]
- Feagin, J.E., Abraham, J.M., Stuart, K. Extensive editing of the cytochrome c oxidase III transcript in Trypanosoma brucei . Cell. 1988;53:413–422. doi: 10.1016/0092-8674(88)90161-4. [DOI] [PubMed] [Google Scholar]
- Harris, M.E., Moore, D.R., Hajduk, S.L. Addition of uridines to edited RNAs in trypanosome mitochondria occurs independently of transcription. J. Biol. Chem. 1990;265:11368–11376. [PubMed] [Google Scholar]
- Huang, C.E., O'Hearn, S.F., Sollner-Webb, B. Assembly and function of the RNA editing complex in Trypanosoma brucei requires band III protein. Mol. Cell. Biol. 2002;22:3194–3203. doi: 10.1128/MCB.22.9.3194-3203.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igo R.P., Jr, Palazzo, S.S., Burgess, M.L.K., Panigrahi, A.K., Stuart, K. Uridylate addition and RNA ligation contribute to the specificity of kinteoplastid insertion RNA editing. Mol. Cell. Biol. 2000;20:8447–8457. doi: 10.1128/mcb.20.22.8447-8457.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igo R.P., Jr, Weston, D.S., Ernst, N.L., Panigrahi, A.K., Salavati, R., Stuart, K. Role of uridylate-specific exoribonuclease activity in Trypanosoma brucei RNA editing. Eukaryot. Cell. 2002;1:112–118. doi: 10.1128/EC.1.1.112-118.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kable, M.L., Seiwert, S.D., Heidmann, S., Stuart, K. RNA editing: A mechanism for gRNA-specified uridylate insertion into precursor mRNA. Science. 1996;273:1189–1195. doi: 10.1126/science.273.5279.1189. [DOI] [PubMed] [Google Scholar]
- Kang, X., Rogers, K., Gao, G., Falick, A.M., Zhou, S., Simpson, L. Reconstitution of uridine-deletion precleaved RNA editing with two recombinant enzymes. Proc. Natl. Acad. Sci. 2005;102:1017–1022. doi: 10.1073/pnas.0409275102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koslowsky, D.J., Bhat, G.J., Perrollaz, A.L., Feagin, J.E., Stuart, K. The MURF3 gene of T. brucei contains multiple domains of extensive editing and is homologous to a subunit of NADH dehydrogenase. Cell. 1990;62:901–911. doi: 10.1016/0092-8674(90)90265-g. [DOI] [PubMed] [Google Scholar]
- Law, J.A., O'Hearn, S., Sollner-Webb, B. In Trypanosoma brucei RNA editing, TbMP18 (band VII) is critical for editosome integrity and for both insertional and deletional cleavages. Mol. Cell. Biol. 2007;27:777–787. doi: 10.1128/MCB.01460-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, S., Rohloff, P., Cox, J., Uyemura, S.A., Docampo, R. Trypanosoma brucei plasma membrane-type Ca2+-ATPase 1 (TbPMC1) and 2 (TbPMC2) genes encode functional Ca2+-ATPases localized to the acidocalcisomes and plasma membrane, and essential for Ca2+ homeostasis and growth. J. Biol. Chem. 2004;279:14427–14439. doi: 10.1074/jbc.M309978200. [DOI] [PubMed] [Google Scholar]
- Madison-Antenucci, S., Sabatini, R.S., Pollard, V.W., Hajduk, S.L. Kinetoplastid RNA-editing-associated protein 1 (REAP-1): A novel editing complex protein with repetitive domains. EMBO J. 1998;17:6368–6376. doi: 10.1093/emboj/17.21.6368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madison-Antenucci, S., Grams, J., Hajduk, S.L. Editing machines: The complexities of trypanosome RNA editing. Cell. 2002;108:435–438. doi: 10.1016/s0092-8674(02)00653-0. [DOI] [PubMed] [Google Scholar]
- Matthews, J.M., Sunde, M. Zinc fingers—Folds for many occasions. IUBMB Life. 2002;54:351–355. doi: 10.1080/15216540216035. [DOI] [PubMed] [Google Scholar]
- McManus, M.T., Shimamura, M., Grams, J., Hajduk, S.L. Identification of candidate mitochondrial RNA editing ligases from Trypanosoma brucei . RNA. 2001;7:167–175. doi: 10.1017/s1355838201002072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, J.C., Wang, Z., Drew, M.E., Paul, K.S., Englund, P.T. Inhibition of bloodstream form Trypanosoma brucei gene expression by RNA interference using the pZJM dual T7 vector. Mol. Biochem. Parasitol. 2001;117:111–113. doi: 10.1016/s0166-6851(01)00334-6. [DOI] [PubMed] [Google Scholar]
- O'Hearn, S., Huang, C.E., Hemann, M., Zhelonkina, A., Sollner-Webb, B. Trypanosoma brucei RNA editing complex: Band II is structurally critical and maintains band V ligase, which is nonessential. Mol. Cell. Biol. 2003;23:7909–7919. doi: 10.1128/MCB.23.21.7909-7919.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi, A.K., Gygi, S., Ernst, N., Igo R.P., Jr, Palazzo, S.S., Schnaufer, A., Weston, D., Carmean, N., Salavati, R., Aebersold, R., et al. Association of two novel proteins, TbMP52 and TbMP48, with the Trypanosoma brucei RNA editing complex. Mol. Cell. Biol. 2001a;21:380–389. doi: 10.1128/MCB.21.2.380-389.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi, A.K., Schnaufer, A., Carmean, N., Igo R.P., Jr, Gygi, S.P., Ernst, N.L., Palazzo, S.S., Weston, D.S., Aebersold, R., Salavati, R., et al. Four related proteins of the Trypanosoma brucei RNA editing complex. Mol. Cell. Biol. 2001b;21:6833–6840. doi: 10.1128/MCB.21.20.6833-6840.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi, A.K., Ernst, N.L., Domingo, G.J., Fleck, M., Salavati, R., Stuart, K.D. Compositionally and functionally distinct editosomes in Trypanosoma brucei . RNA. 2006;12:1038–1049. doi: 10.1261/rna.45506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakers, C., Ruijter, J.M., Deprez, R.H.L., Moorman, A.F.M. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 2003;339:62–66. doi: 10.1016/s0304-3940(02)01423-4. [DOI] [PubMed] [Google Scholar]
- Rusche, L.N., Cruz-Reyes, J., Piller, K.J., Sollner-Webb, B. Purification of a functional enzymatic editing complex from Trypanosoma brucei mitochondria. EMBO J. 1997;16:4069–4081. doi: 10.1093/emboj/16.13.4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusché, L.N., Huang, C.E., Piller, K.J., Hemann, M., Wirtz, E., Sollner-Webb, B. The two RNA ligases of the Trypanosoma brucei RNA editing complex: Cloning the essential Band IV gene and identifying the Band V gene. Mol. Cell. Biol. 2001;21:979–989. doi: 10.1128/MCB.21.4.979-989.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacharidou, A., Cifuentes-Rojas, C., Halbig, K., Hernandez, A., Dangott, L.J., De Nova-Ocampo, M., Cruz-Reyes, J. RNA editing complex interactions with a site for full-round U deletion in Trypanosoma brucei . RNA. 2006;12:1219–1228. doi: 10.1261/rna.2295706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs, A.B., Buratowski, S. Common themes in translational and transcriptional regulation. Trends Biochem. Sci. 1997;22:189–192. doi: 10.1016/s0968-0004(97)01051-7. [DOI] [PubMed] [Google Scholar]
- Salavati, R., Ernst, N.L., O'Rear, J., Gilliam, T., Tarun S., Jr, Stuart, K. KREPA4, an RNA binding protein essential for editosome integrity and survival of Trypanosoma brucei . RNA. 2006;12:819–831. doi: 10.1261/rna.2244106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnaufer, A., Panigrahi, A.K., Panicucci, B., Igo R.P., Jr, Salavati, R., Stuart, K. An RNA ligase essential for RNA editing and survival of the bloodstream form of Trypanosoma brucei . Science. 2001;291:2159–2162. doi: 10.1126/science.1058955. [DOI] [PubMed] [Google Scholar]
- Schnaufer, A., Ernst, N.L., O'Rear, J., Salavati, R., Stuart, K. Separate insertion and deletion sub-complexes of the Trypanosoma brucei RNA editing complex. Mol. Cell. 2003;12:307–319. doi: 10.1016/s1097-2765(03)00286-7. [DOI] [PubMed] [Google Scholar]
- Seiwert, S.D., Stuart, K. RNA editing: Transfer of genetic information from gRNA to precursor mRNA in vitro. Science. 1994;266:114–117. doi: 10.1126/science.7524149. [DOI] [PubMed] [Google Scholar]
- Seiwert, S.D., Heidmann, S., Stuart, K. Direct visualization of uridylate deletion in vitro suggests a mechanism for kinetoplastid RNA editing. Cell. 1996;84:831–841. doi: 10.1016/s0092-8674(00)81062-4. [DOI] [PubMed] [Google Scholar]
- Simpson, L., Aphasizhev, R., Gao, G., Kang, X. Mitochondrial proteins and complexes in Leishmania and Trypanosoma involved in U-insertion/deletion RNA editing. RNA. 2004;10:159–170. doi: 10.1261/rna.5170704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauffer, M.E., Chazin, W.J. Structural mechanisms of DNA replication, repair, and recombination. J. Biol. Chem. 2004;279:30915–30918. doi: 10.1074/jbc.R400015200. [DOI] [PubMed] [Google Scholar]
- Stuart, K., Panigrahi, A.K. RNA editing: Complexity and complications. Mol. Microbiol. 2002;45:591–596. doi: 10.1046/j.1365-2958.2002.03028.x. [DOI] [PubMed] [Google Scholar]
- Stuart, K.D., Schnaufer, A., Ernst, N.L., Panigrahi, A.K. Complex management: RNA editing in trypanosomes. Trends Biochem. Sci. 2005;30:97–105. doi: 10.1016/j.tibs.2004.12.006. [DOI] [PubMed] [Google Scholar]
- Tarun S.Z., Jr, Sachs, A.B. A common function for mRNA 5′ and 3′ ends in translation initiation in yeast. Genes & Dev. 1995;9:2997–3007. doi: 10.1101/gad.9.23.2997. [DOI] [PubMed] [Google Scholar]
- Theobald, D.L., Mitton-Fry, R.M., Wuttke, D.S. Nucleic acid recognition by OB-fold proteins. Annu. Rev. Biophys. Biomol. Struct. 2003;32:115–133. doi: 10.1146/annurev.biophys.32.110601.142506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotter, J.R., Ernst, N.L., Carnes, J., Panicucci, B., Stuart, K. A deletion site editing endonuclease in Trypanosoma brucei . Mol. Cell. 2005;20:403–412. doi: 10.1016/j.molcel.2005.09.016. [DOI] [PubMed] [Google Scholar]
- Wang, Z., Morris, J.C., Drew, M.E., Englund, P.T. Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J. Biol. Chem. 2000;275:40174–40179. doi: 10.1074/jbc.M008405200. [DOI] [PubMed] [Google Scholar]
- Wang, B., Ernst, N.L., Palazzo, S.S., Panigrahi, A.K., Salavati, R., Stuart, K. TbMP44 is essential for RNA editing and structural integrity of the editosome in Trypanosoma brucei . Eukaryot. Cell. 2003;2:578–587. doi: 10.1128/EC.2.3.578-587.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtz, E., Leal, S., Ochatt, C., Cross, G.A.M. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei . Mol. Biochem. Parasitol. 1999;99:89–101. doi: 10.1016/s0166-6851(99)00002-x. [DOI] [PubMed] [Google Scholar]
- Worthey, E.A., Schnaufer, A., Mian, I.S., Stuart, K., Salavati, R. Comparative analysis of editosome proteins in trypanosomatids. Nucleic Acids Res. 2003;31:6392–6408. doi: 10.1093/nar/gkg870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, L.E., Koslowsky, D.J. Interactions of mRNAs and gRNAs involved in trypanosome mitochondrial RNA editing: Structure probing of a gRNA bound to its cognate mRNA. RNA. 2006;12:1050–1060. doi: 10.1261/rna.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]