Abstract
PNPase is a major exoribonuclease that plays an important role in the degradation, processing, and polyadenylation of RNA in prokaryotes and organelles. This phosphorolytic processive enzyme uses inorganic phosphate and nucleotide diphosphate for degradation and polymerization activities, respectively. Its structure and activities are similar to the archaeal exosome complex. The human PNPase was recently localized to the intermembrane space (IMS) of the mitochondria, and is, therefore, most likely not directly involved in RNA metabolism, unlike in bacteria and other organelles. In this work, the degradation, polymerization, and RNA-binding properties of the human PNPase were analyzed and compared to its bacterial and organellar counterparts. Phosphorolytic activity was displayed at lower optimum concentrations of inorganic phosphate. Also, the RNA-binding properties to ribohomopolymers varied significantly from those of its bacterial and organellar enzymes. The purified enzyme did not preferentially bind RNA harboring a poly(A) tail at the 3′ end, compared to a molecule lacking this tail. Several site-directed mutations at conserved amino acid positions either eliminated or modified degradation/polymerization activity in different manners than observed for the Escherichia coli PNPase and the archaeal and human exosomes. In light of these results, a possible function of the human PNPase in the mitochondrial IMS is discussed.
Keywords: polyadenylation, RNA degradation, PNPase, human, phosphorolysis
INTRODUCTION
Polynucleotide phosphorylase (PNPase) (EC 2.7.7.8) was the first enzyme to be identified that catalyzes the formation of polynucleotides from ribonucleotides. However, unlike RNA polymerases, PNPase neither requires a template nor can transcribe one. When a mixture of ribonucleotide diphosphates (NDPs) serves as a substrate for the polymerization reaction, the ensuing polymerization reaction forms a random copolymer.
PNPase catalyzes both processive 3′ → 5′ phosphorolysis and 5′ → 3′ polymerization of RNA (Littauer and Soreq 1982; Grunberg-Manago 1999; Littauer and Grunberg-Manago 1999). In Escherichia coli, PNPase is mostly active in 3′ → 5′ phosphorolysis during RNA degradation and 3′ end processing, but recently a substantial degree of activity in the polymerization of heteropolymeric tails has also been reported (Mohanty and Kushner 2000; Mohanty et al. 2004). Moreover, PNPase is suggested to be the major polyadenylating enzyme in spinach chloroplasts, cyanobacteria, and Gram-positive bacteria (Yehudai-Resheff et al. 2001; Rott et al. 2003; Slomovic et al. 2006). PNPase was also reported to be a global regulator of virulence and persistence in Salmonella enterica (Clements et al. 2002). The crystallographic structure analysis of PNPase from the bacteria, Streptomyces antibioticus, revealed a homotrimeric, circular-shaped complex (Symmons et al. 2000, 2002). Part of the PNPase population in E. coli is associated with the endoribonuclease, RNase E, an RNA helicase, enolase, and possibly other proteins in a high-molecular weight complex termed the degradosome (Marcaida et al. 2006). However, PNPase in the chloroplast was revealed to be a homotrimeric complex without the association of other proteins (Baginsky et al. 2001). It is also possible that two homotrimeric complexes are associated together in spinach chloroplasts, as PNPase elutes from a size-exclusion column at about 600 kDa (Baginsky et al. 2001).
The amino acid sequences of PNPases from bacteria, as well as from the nuclear genomes of plants and mammals, display a high level of identity and feature similar structures composed of five motifs (Yehudai-Resheff et al. 2003; Leszczyniecka et al. 2004). There are two core domains that display different degrees of homology with the E. coli phosphorylase RNase PH, an α-helical domain between the two core domains, and two adjacent RNA-binding domains called KH and S1. These RNA-binding domains were characterized in other RNA-binding proteins. The domain structure of the bacterial and organellar PNPases is very well conserved in the exosomes of archaea and eukaryotic cells (Buttner et al. 2006; Houseley et al. 2006; Lin-Chao et al. 2007). Indeed, the archaeal exosome is a phosphorolytic complex that is very similar to PNPase and, in systems in which it exists, is responsible for the degradation and polymerization of RNA, resembling the PNPase found in cyanobacteria (Portnoy et al. 2005; Portnoy and Schuster 2006). The core structure of the eukaryotic exosome, which is responsible for the 3′ → 5′ exonucleolytic degradation of RNA in the nucleus and cytoplasm is also very similar to the PNPase and the archaeal exosome. While the human complex contains a phosphorolytic activity site, it could not be detected in the yeast exosome (Liu et al. 2006; Dziembowski et al. 2007). The E. coli and chloroplast PNPases display high binding affinity to poly(A); the responsible site is located in the S1 domain (Yehudai-Resheff et al. 2003). This phenomenon is important for its function in the polyadenylation-stimulated degradation pathway, in which the addition of poly(A) or poly(A)-rich sequences induces rapid exonucleolytic degradation (Lisitsky et al. 1997; Lisitsky and Schuster 1999). The enzymatic activity is significantly enhanced by inorganic phosphate at concentrations of 10–20 mM, and is sensitive to secondary structures in the RNA substrate (Yehudai-Resheff et al. 2001, 2003).
The human PNPase was identified in an overlapping pathway screen to discover genes displaying coordinated expression as a consequence of terminal differentiation and senescence of melanoma cells (Leszczyniecka et al. 2002). Unlike chloroplast and plant mitochondria, in which PNPase is located in the stroma and matrix, the human PNPase is mostly or exclusively located in the mitochondrial intermembrane space (IMS) (Chen et al. 2006; Rainey et al. 2006). In addition, PNPase was purified as an interacting protein with TCL1, a nonenzymatic oncoprotein promoting B- and T-cell malignancies and apoptotic stimuli caused PNPase mobilization following cytochrome c release to the cytoplasm (Chen et al. 2006; French et al. 2007). Considering its IMS location, it most likely does not play a major or direct role in mitochondrial RNA metabolism, contrary to its role in bacteria, chloroplast, and plant mitochondria (Slomovic et al. 2006).
There is no obvious RNA substrate for the human PNPase in the IMS of the mitochondria, and its function and possible RNA target are still unknown. As the first step to reveal the biological function of this enzyme in human cells, we considered biochemical characterization in comparison to the homologous bacterial and chloroplast enzymes. To this end, the RNA phosphorolysis, binding, and complex formation of the human PNPase were analyzed. We found that when expressed in E. coli and purified the human PNPase is active in both polymerization and degradation and, like the bacterial enzyme, forms a trimeric homopolymer. However, the human PNPase differs from the bacterial and chloroplast enzymes in its optimal inorganic phosphate concentration for degradation activity as well as its RNA-binding properties. Several site-directed mutations that eliminated the phosphorolytic activity of the E. coli enzyme or the archaeal and human exosomes have different effects on the human PNPase.
RESULTS
Human PNPase has a similar structure to that of the bacterial enzyme
The human PNPase belongs to an evolutionarily conserved enzyme family whose members were characterized in bacteria and organelles (Yehudai-Resheff et al. 2003; Leszczyniecka et al. 2004). This enzyme contains an N-terminal transit peptide of 37 amino acids that guides the protein into the mitochondrial IMS and is, thereafter, removed by cleavage (Chen et al. 2006; Rainey et al. 2006). Similar to its bacterial and organellar counterparts, the protein is composed of two core domains homologous to the bacterial phosphorylase/exoribonuclease RNase PH (Fig. 1). The phosphorylase activity site was located at the second core domain in the bacteria, Streptomyces antibioticus (Symmons et al. 2000) as well as in the homologous archaeal exosome protein Rrp41 (Lorentzen et al. 2005). In spinach chloroplasts, in addition to the activity of the second core domain, the first core domain is active in the degradation of polyadenylated RNA while the situation in E. coli is still unknown (Yehudai-Resheff et al. 2003). The first core domain is followed by an α-helical domain and the second core domain, by KH and S1 domains, which were shown in the bacterial and chloroplast enzymes to be involved in RNA binding (Fig. 1; Jarrige et al. 2002; Symmons et al. 2002; Yehudai-Resheff et al. 2003; Stickney et al. 2005). The S1 and KH domain motifs characterize many other RNA-binding proteins (Worrall and Luisi 2007).
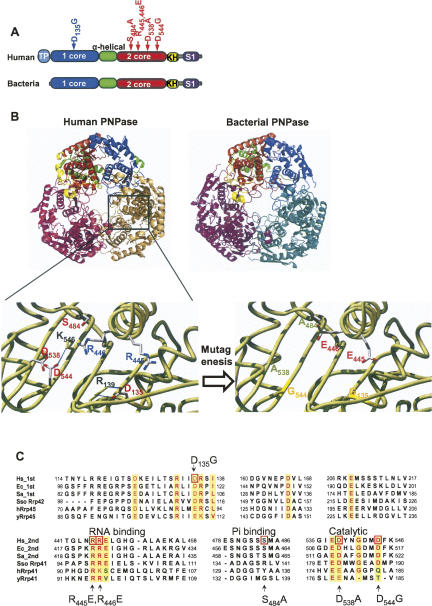
FIGURE 1.
Human and bacterial PNPases and the mutated versions used in this work. (A) The domain structures of the bacterial and human PNPases are presented schematically. The boxes represent the different domains as indicated, and the relative locations of the site-directed mutated amino acids are given. TP indicates the mitochondrial intermembrane transit peptide. (B) The resolved structure and predicted model of PNPase enzymes from Streptoomyces antibioticus (bacteria) and human (human) are shown. The trimeric doughnut-like shape is presented in an orientation enabling easy observation of the central channel. Each monomer is colored differently, and each domain is colored in one monomer of the enzyme, as shown in the scheme presented at the top. The homology-based modeling was performed using the Phyre Server at http://www.sbg.bio.ic.ac.uk/∼3dpssm/. The complex was built by applying the crystal symmetry of the structure using the PyMOL program. A closeup view of the locations of the modified amino acids and their particular locations is presented below. (C) Alignment of the amino acid sequences of the conserved parts of human (Hs), E. coli (Ec) and S. antibioticus (Sa) PNPases, as well as the hyperthermophilic archaea Sulfolobus solfataricus (Sso), human (hRrp45 and hRrp41), and yeast (yRrp45 and yRrp41) exosomes proteins. The first and second signs indicate the first and second core RNase PH-like domains for PNPase, respectively. The corresponding protein names are indicated for the exosomes. The yellow background shows the mostly conserved amino acids, which are colored red, and those mutated in this work are marked by red squares.
In order to observe the putative structure of the human PNPase, we used the coordinates of the homologous enzyme from the bacteria, S. antibioticus, that was deciphered by crystallization and X-ray diffraction analysis (Symmons et al. 2000). Figure 1B presents the predicted trimeric structure compared to the bacterial enzyme. The human PNPase exhibits a very similar structure to the bacterial enzyme, having a doughnut-shaped trimer with a central channel that is of the correct dimensions to accommodate a single-stranded RNA molecule (Symmons et al. 2000). The circular structure is composed of the first and second core domains, while the KH and S1 RNA-binding domains are located at the top, adjacent to the gate entrance into the circle (Symmons et al. 2000; Lorentzen and Conti 2005; Lorentzen et al. 2005; Buttner et al. 2006). In Figure 1B, in which the trimer structure is presented as viewed from above, the KH domain (in yellow) is shown, located above the circular structure.
To achieve a detailed analysis of the degradation, polymerization, and RNA-binding activities of the human PNPase, the protein was expressed in bacteria and purified to apparent homogeneity (Fig. 2A). In addition, several amino acids that are highly conserved when aligning the sequences of known PNPases (Fig. 1C), as well as the corresponding subunits of the archaeal, human, and yeast exosomes, which were predicted to be involved in the formation of the phosphorolysis site or the trimeric complex, based on the structural analysis, were modified by site-directed mutagenesis (Fig. 1B,C; Symmons et al. 2000; Lorentzen and Conti 2005; Lorentzen et al. 2005). The corresponding proteins were expressed in bacteria and purified also (Fig. 2A). Several additional conserved residues were modified as well; however, the corresponding proteins were not obtained in a soluble form when expressed in bacteria.
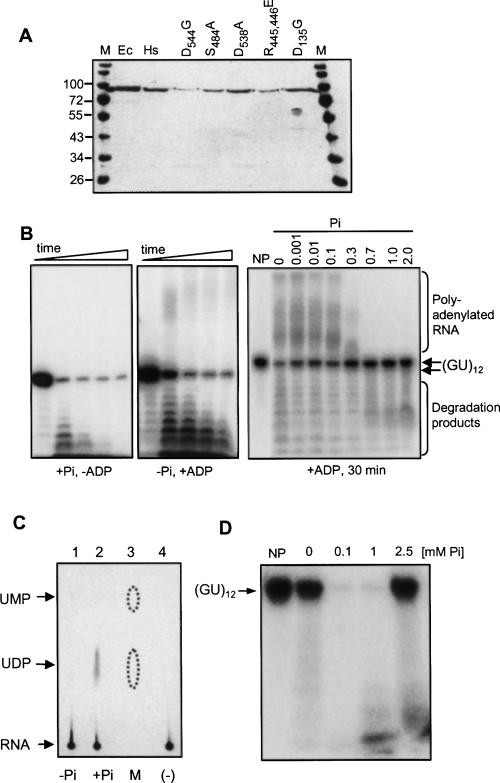
FIGURE 2.
Polymerization and degradation activities of human PNPase. (A) Coomassie-stained polyacrylamide gel profile of the recombinant proteins following expression in E. coli. Proteins were purified by nitrilotriacetic acid (NTA) agarose affinity chromatography, anion-exchange (MonoQ), and heparin affinity chromatography steps, and loaded onto a 10% SDS-PAGE gel. Molecular mass markers (in kDa) are shown at the left and right (M). Ec, E. coli PNPase; Hs, human PNPase. In other lanes, the purified proteins of the corresponding mutants as labeled by the modified amino acid residue are shown. (B) Human PNPase was incubated with [32P]-RNA (GU)12 oligonucleotide in the presence of either Pi (0.1 mM) or ADP (2.5 mM), as indicated in the figure, in order to promote degradation or polyadenylation activity, respectively. Samples were removed at 0, 5, 15, 30, and 60 min, and the RNA was analyzed by denaturing PAGE and autoradiography. The input RNA, as well as polymerized and partially degraded products, are indicated. In the right panel, incubation was for 30 min in the present of 2.5 mM ADP and Pi at the concentration indicated on the top. NP-no protein. (C) Thin-layer chromatography (TLC) analysis of the degradation products. Human PNPase was incubated with uniformly [32P] UTP-labeled RNA corresponding to part of the human mitochondrial transcript COX1. Following the incubation, the reaction products were spotted onto a polyethyleneimine TLC plate, which was developed with 0.9 M GnCl, dried, and autoradiographed. The PNPase activity was assayed either in the absence (lane 1) or presence of 0.1 mM Pi (lane 2). In lane 3, UMP and UDP were analyzed as markers on the same TLC plate and visualized by fluorescence quenching. The dotted lines mark the boundaries of the fluorescence pattern. In lane 4, a control reaction in which the RNA was incubated without the addition of protein, was analyzed. (D) Human PNPase activity in response to Pi concentrations. [32P]-labeled RNA oligonucleotide (GU)12 was incubated for 15 min with human PNPase at different Pi concentrations, as indicated. CaCl2 (50 μM) was added to the reaction mixture in order to precipitate the Pi already present in the sample. Following incubation, the RNA was analyzed by denaturing PAGE and autoradiography. No enzyme was added to the reaction in the lanes labeled (–).
Phosphorolytic activity of the human PNPase
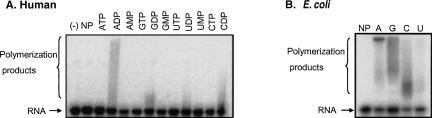
As a phosphorylase, PNPase incorporates Pi and ADP in degradation and polymerization processes, respectively (Littauer and Grunberg-Manago 1999). When a 24-nucleotide (nt) RNA molecule was incubated with the enzyme in the presence of Pi and without the addition of ADP, only the degradation of the RNA was observed (Fig. 2B, left panel). However, when the same substrate was incubated with the enzyme in the presence of ADP but without Pi, both degradation and polymerization activities were detected (Fig. 2B, middle panel). When both ADP and Pi were present at the reaction mixture, the direction of activity, either polyadenylation or degradation, was dependent on their relative concentrations (Fig. 2B, left panel). As expected of a phosphorylase, the byproduct of the degradation of [32P]UTP-labeled RNA was identified as UDP and the reaction was significantly stimulated by Pi (Fig. 2C). Optimal degradation activity was obtained at the relatively low concentration of 0.1 mM (Fig. 2D). This optimal concentration differs significantly from that of bacterial and chloroplast PNPases, in which the degradation reaction was stimulated at much higher Pi concentrations of 10–20 mM (Littauer and Grunberg-Manago 1999; Yehudai-Resheff et al. 2001; Jarrige et al. 2002). Indeed, at a Pi concentration of 10 mM, the degradation activity of the human PNPase was severely inhibited (Fig. 2C; data not shown). The low activity observed here without the addition of Pi was eliminated when CaCl2 was included in the reaction mixture in order to quench traces of Pi that were present in the reaction (cf Fig. 2B,C, which were performed without the addition of CaCl2, and Fig. 2D, in which 50 μM CaCl2 was added). This result suggested that the human PNPase can respond to much lower concentrations of Pi, changing its mode of activity between degradation and polymerization, compared to bacterial and chloroplast enzymes. The specificity of the enzyme for the polymerization reaction is, like that of the E. coli PNPase, high for ADP, with much less activity when incubated with other NDPs. No activity was observed with ATP nor the other NTPs, as well as mono phosphate nucleotides (Fig. 3).
FIGURE 3.
Of the NDPs, human PNPase displays a preference for ADP during polymerization activity. [32P]-labeled RNA oligonucleotide (GU)12 was incubated with human (A) or E. coli (B) proteins and 2.5 mM of each of the corresponding nucleotides, as indicated at the top of the figure. For the E. coli enzyme (B), only the NDPs nucleotides were assayed. Samples were withdrawn at 10 min, and the RNA analyzed by denaturing PAGE and autoradiography. The input RNA and polymerized products are shown on the left. The input RNA (lane [–]) as well as a control reaction in which no protein was added (lane NP) were also analyzed.
Human PNPase degrades polyadenylated and nonpolyadenylated RNA at similar rates
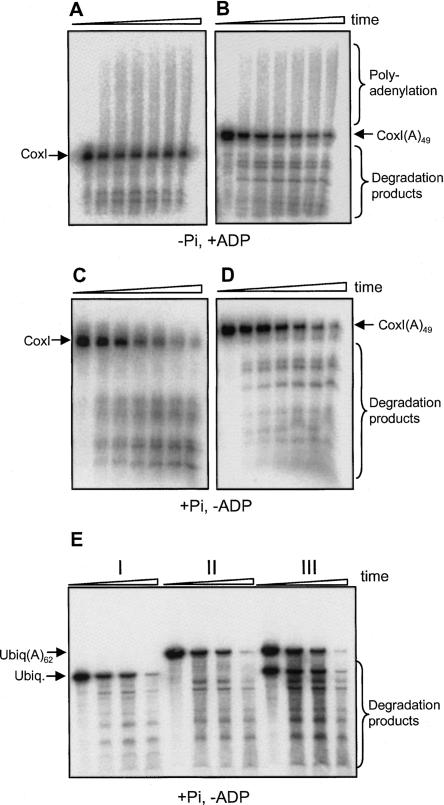
The PNPases of bacteria and chloroplasts preferentially degrade and polyadenylate RNA molecules containing a poly(A) tail at the 3′ end. This preferential activity on polyadenylated transcripts has been attributed to the high binding affinity to poly(A) tracks located at the S1 domain in the spinach PNPase (Lisitsky et al. 1997; Lisitsky and Schuster 1999; Yehudai-Resheff et al. 2003). To analyze whether the human PNPase shares this preferential activity, RNA related to a region of the mitochondrial COX1 gene, either with or without the addition of a 49 nt poly(A) tail, was incubated with the human PNPase in either optimal polymerization or degradation conditions. The results revealed no differences in either the polymerization or the degradation activities (Fig. 4). Similar results were obtained with polyadenylated or nonpolyadenylated RNA substrates of varying lengths, derived from additional genes (Fig. 4E; data not shown). Moreover, when polyadenylated and nonpolyadenylated transcripts related to the ubiquitin mRNA were mixed together and assayed for degradation, the same rate was observed for both substrates, indicating no competition of the polyadenylated RNA (Fig. 4E). In contrast, competition between polyadenylated and nonpolyadenylated transcripts, resulting with the stabilization of the nonpolyadenylated one, has been described previously for the E. coli and spinach chloroplast PNPases (Lisitsky et al. 1997; Lisitsky and Schuster 1999; Yehudai-Resheff et al. 2003). Therefore, we concluded that, unlike bacteria and chloroplast PNPases, the human enzyme does not degrade polyadenylated RNA preferentially. This conclusion was strengthened by the observation described in the continuation of the manuscript that human PNPase does not display high binding affinity to poly(A). Our results differ from a previous analysis of this protein in which preferential degradation of polyadenylated RNA was detected (French et al. 2007).
FIGURE 4.
Similar activities of human PNPase on nonpolyadenylated and polyadenylated substrates. [32P] RNA corresponding to part of the mitochondrial gene COX1, either without (A,C) or with the addition of 49 adenosins at the 3′ end (B and D) was incubated with the protein and 0.1 mM Pi (C,D) or 2.5 mM ADP (A,B). In panel E, degradation assay was performed to non-polyadenylated ubiquitin transcript (I), the same transcript with the addition of 64 adenosines (II), and the two transcripts mixed together (III). Samples were withdrawn at 0, 15, 30, and 120 min, and the RNA analyzed by denaturing PAGE and autoradiography.
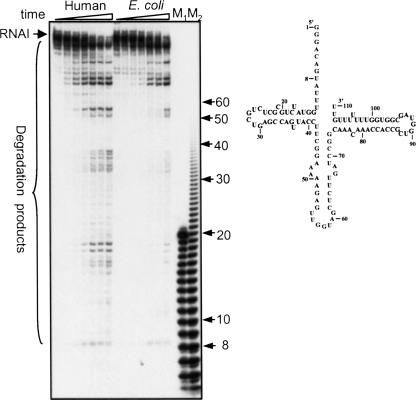
In order to determine the degradation efficiency that the human PNPase displays when incubated with RNA molecules containing extensive structural features, we analyzed the RNAI of E. coli. This RNA, which oversees the plasmid replication in the bacterial cell, is characterized by a tRNA-like structure, shown to hamper degradation by bacterial PNPases, to a certain degree (Kaberdin et al. 1996). When the human or E. coli PNPases were incubated at the same concentration with the RNAI transcript, it was successfully degraded, resulting in a very similar pattern of degradation products (Fig. 5). We concluded that once titrated to conditions for optimal degradation activity in terms of the concentrations of Pi and ADP, both the human and E. coli PNPases similarly degrade structured RNA molecules.
FIGURE 5.
Similar degradation activities of human and E. coli PNPases. [32P] RNA corresponding to the E. coli RNAI (111 nt) was incubated with the human or E. coli PNPases (100 ng each). Pi was added to a final concentration of 0.1 or 10 mM for the human and E. coli enzymes, respectively. Samples were collected at 0, 5, 15, 30, 60, 90, and 120 min, and the RNA was analyzed by denaturing PAGE and autoradiography. RNA markers were fractionated in the lanes marked M1 and M2. The length in nucleotides is shown on the right. The predicted secondary structure of RNAI is shown to the right.
Human PNPase displays different RNA-binding properties compared to bacterial and organellar PNPases
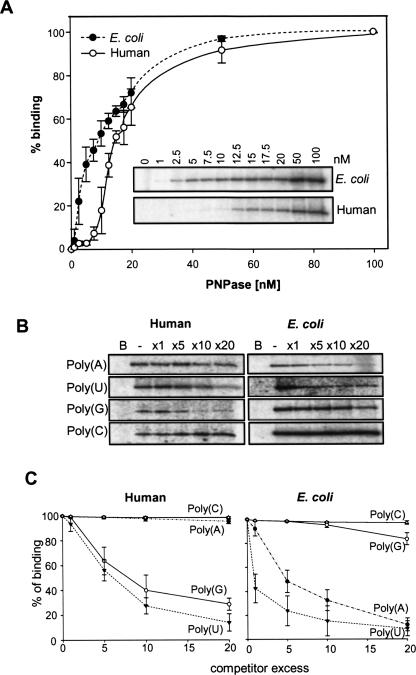
Bacterial and spinach PNPase proteins were previously analyzed and displayed high binding affinity to poly(A) and poly(U), in correlation with this enzyme's necessity to compete for polyadenylated RNA (Lisitsky et al. 1997; Lisitsky and Schuster 1999). In order to determine the binding curve, RNA-binding of the purified protein was analyzed in a UV-cross-linking assay using increasing amounts of PNPase. As shown in Figure 6A, while the E. coli enzyme displayed a saturation binding curve with a Kd of 11 nM, in agreement with previous reports (Bermudez-Cruz et al. 2002; Schubert et al. 2004; Stickney et al. 2005; Amblar et al. 2007), an apparent sigmoid curve, typical of cooperative binding, was observed for the human enzyme. The Kd observed for the human PNPase was ∼16 nM.
FIGURE 6.
RNA-binding properties of human and E. coli PNPases. (A) Human PNPase displays a sigmoidal curve which suggests cooperative binding. Human and E. coli PNPases (10 ng each) were analyzed for RNA-binding by a UV light cross-linking assay using increasing amounts of PNPase, as indicated in the figure inset. The radioactive signals obtained in three independent experiments (error bars are represented) were quantified and plotted in order to determine the shape of the binding curve and the Kd observed. The black line with circled points represents the human PNPase and the dashed line with filled points represents the E. coli PNPase. The Kd observed for E.coli and human PNPases were 11 and 16 nM, respectively. (B) High affinity binding of human PNPase to poly(U) and poly(G). Human or E. coli PNPases were incubated with [32P]-RNA corresponding to (GU)12, and a homoribopolymer at a molar excess, as shown in the figure. Following UV cross-linking and ribonuclease digestion, the label transfer from RNA to protein was analyzed by SDS-PAGE and autoradiography. In the lane labeled B, no protein was added to the assay as a control. (C) Quantization of the competition assays. The UV light cross-linking competition assays were performed as described for (B), and the intensity of the UV light cross-linking band without competitor was defined as 100%. Data shown are the average of at least three experiments.
Next, we examined whether the human PNPase, similar to other PNPases analyzed to date, binds poly(A) with higher affinity than poly(G) and poly(C). To this end, competition UV-cross-linking assays, in which increasing amounts of the corresponding ribohomopolymer compete with [32P]-(GU)12 RNA for binding of the protein, were carried out. The efficiency by which the ribohomopolymer competes for protein binding reflects its affinity. Surprisingly, while, as previously reported, the E. coli enzyme displayed high binding affinities to poly(U) and poly(A) (Lisitsky and Schuster 1999), the human PNPase bound poly(U) and poly(G) with high affinity and displayed very low binding affinity to poly(A) (Fig. 6B,C). The low affinity to poly(A) sequences is in line with the observation that, unlike the case of the E. coli and chloroplast PNPases, the human enzyme displayed no preferential activity for polyadenylated RNA (Fig. 4).
Unlike the spinach chloroplast PNPase, the human PNPase does not bind the E. coli enzyme to form a heterotrimeric complex when expressed in E. coli
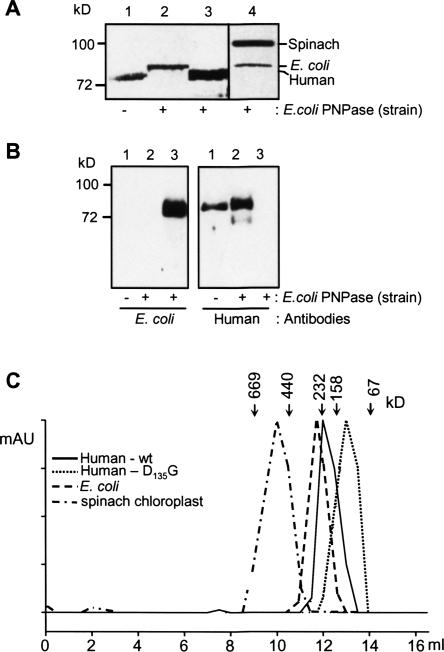
When expressing the recombinant spinach PNPase protein in E. coli, we previously found that a heterotrimeric functional complex, consisting of both spinach and E. coli subunits, was formed (Yehudai-Resheff et al. 2003). In this case, the presence of bacterial subunits in the trimer interfered with the determination of the activities and RNA-binding properties of the recombinant spinach PNPase. We then, therefore, expressed the spinach protein in an E. coli strain in which the endogenous PNPase gene was interrupted by the genomic insertion of DNA and therefore, the bacterial protein was not expressed (Lopez et al. 1999). In the present work, since we were concerned that a similar phenomenon could hamper characterization of the human PNPase, we initially expressed the protein in the E. coli strain in which PNPase is not expressed. In order to examine whether, similar to the spinach PNPase, the human enzyme can form a heterotrimeric complex composed of E. coli and human PNPase subunits, the human PNPase was produced in nonmutated E. coli strain expressing the intrinsic protein. The recombinant his6 tagged human PNPase was purified by affinity chromatography and analyzed on SDS-PAGE in order to see if an associated E. coli PNPase coeluted with it. As shown in Figure 7, the result revealed that no bacterial PNPase copurified with the his6 tagged human PNPase, indicating that a heterotrimeric PNPase complex was not obtained (Fig. 7A, lane 3). However, when spinach chloroplast PNPase was expressed in this strain, the E. coli polypeptide copurified with the recombinant spinach protein (Fig. 7A, lane 4). In order to verify that even a minute amount of E. coli PNPase did not bind the human polypeptide, we analyzed the purified recombinant proteins with specific antibodies. No signal was obtained with antibodies specific to the E. coli PNPase when probing the purified human PNPase (Fig. 7B, left panel, lane 2). We concluded that, unlike the spinach chloroplast PNPase, human polypeptides cannot form a heterotrimer with the E. coli protein.
FIGURE 7.
Unlike the spinach chloroplast PNPase, the human protein does not form a complex with its E. coli counterpart. (A) Human (lanes 1 and 3), E. coli (lane 2), or spinach chloroplast (lane 4) PNPases were expressed in E. coli strain in which the expression of the endogenous PNPase was knocked-out (lane 1), or in E. coli strain containing the endogenous PNPase (lanes 2, 3, and 4). The expressed proteins were purified as described in Materials and Methods and analyzed on SDS-PAGE followed by silver staining. The identities of the different PNPases are indicated on the right. The E. coli endogenous PNPase was copurified with the spinach protein (lane 4) but not with the human one (lane 3). (B) Recombinant human PNPase was expressed and purified in E. coli strains either lacking (lane 1) or containing (lane 2) endogenous PNPase. In lane 3, a purified recombinant E. coli PNPase was loaded. The proteins were analyzed by SDS-PAGE and immunoblotted to a nitrocellulose membrane that was incubated with antibodies against the E. coli PNPase or the human PNPase, as indicated in the figure. (C) Recombinant human PNPase forms a high molecular weight complex of ∼300 kDa. Recombinant proteins (∼5 μg) were fractionated on a Superdex 200 size-exclusion column. The elution profile of molecular-weight markers: Thyroglobulin 665 kDa, Feritin 440 kDa, Catalase 232 kDa, Aldolase 158 kDa, and Albumin 67 kDa, fractionated on the same column, is indicated at the top.
Recombinant human PNPase forms a homotrimeric complex; formation is impeded by a D135G mutation located at the first core
Next, we analyzed whether human PNPase, like the E. coli enzyme, forms a homotrimer of ∼240 kDa (Amblar et al. 2007). Alternatively, the spinach chloroplast enzyme is characterized as a 580-kDa complex that is possibly comprised of two homotrimers (Symmons et al. 2000; Baginsky et al. 2001). Analysis of the complex formation of the various enzymes by fractionation on a size-exclusion column revealed that the human enzyme migrated at a molecular weight of 230 kDa, similar to E. coli and unlike the spinach chloroplast homolog (Fig. 7C). The same size was measured previously, upon fractionation on a blue-native gel (Chen et al. 2006). A site-directed mutant, in which one amino acid located in the first core domain was changed (D135G), was characterized by the inability to form the homotrimer complex, and the protein was eluted from the column in a monomeric form of ∼90 kDa (Fig. 7C).
Changing conserved amino acids resulted in the altering of phosphorolytic and RNA-binding activities
In order to determine which amino acids are important for the phosphorolytic and RNA-binding activities of PNPase, several residues that are evolutionarily conserved between bacterial and organellar PNPases, as well as the archaeal, yeast, and human exosomes, were modified via site-directed mutagenesis (Fig. 1). The corresponding modified proteins were then analyzed for activity and RNA-binding (Fig. 8). Although other conserved amino acids, such as G509D and G513R, were modified as well, the corresponding proteins could not be obtained when expressed in bacteria in a soluble form and could therefore not be analyzed.
FIGURE 8.
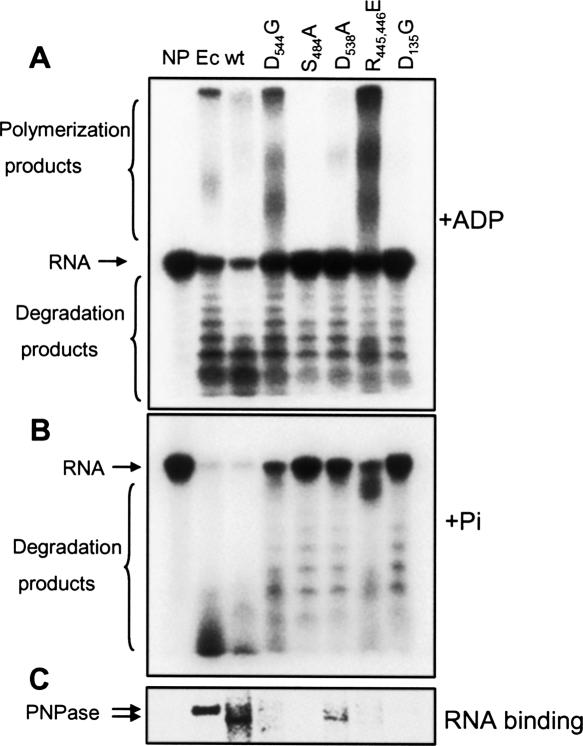
Polymerization, degradation, and RNA-binding activities of the different mutants. (A,B) [32P]-labeled RNA oligonucleotide (GU)12 was incubated with wild-type (wt) PNPase or site-directed mutants as indicated. Reactions contained 2.5 mM of ADP and incubated for 10 min in (A) and 0.1 mM Pi incubating for 15 min in (B). RNA was purified and analyzed by denaturing PAGE and autoradiography. The input RNA, as well as the polymerized and degradation products, are indicated on the left. A control reaction in which no protein was added is shown in lane NP. (C) Wild-type (wt) human PNPase and its site-directed mutants (10 ng each) were analyzed for RNA-binding by the UV light cross-linking assay using [32P] RNA corresponding to the (GU)12. Following UV cross-linking and ribonuclease digestion, the label transfer from RNA to protein was analyzed by SDS-PAGE and autoradiography. In the lane labeled NP, no protein was added to the assay as a control. Ec, purified E. coli PNPase.
Changing the aspartic acid residue at position 544, which is involved in the formation of the catalytic site, to glycine (D544G), significantly reduced degradation activity but surprisingly enhanced polymerization. In contrast, in the E. coli PNPase, the equivalent change, (D492G), resulted in the loss of phosphorolytic activity but also abolished polyadenylation activities by 90% (Table 1; Jarrige et al. 2002). Similar results of enhancing polymerization and significantly inhibiting degradation activities were obtained when arginines, 445 and 446, involved in RNA binding at the catalytic site, were converted to glutamic acid (R445,446E). This observation is interesting, since modifying the corresponding conserved arginines in the E. coli PNPase, (R398,399D), the archaeal exosome, (R98,99E of Rrp41), and human exosome, (R94V,K95A of Rrp41), resulted in the complete inhibition of phosphorolytic activity for E. coli PNPase and the human exosome, as well as the degradation activity for the archaeal complex (Table 1; Jarrige et al. 2002; Lorentzen et al. 2005; Liu et al. 2006). Changing serine 484 and aspartic acid 538 to alanine (S484G and D538A) practically eliminated both the polyadenylation and degradation activities of the human PNPase (Fig. 8). When the corresponding residues in the human and archaeal exosomes, involved in the formation of the Pi binding and the catalytic active sites, were modified, the result was elimination of activity, as well (Table 1).
TABLE 1.
Changes in polymerization and degradation activities of site-directed mutants compared with the corresponding mutations in E. coli PNPase (Jarrige et al. 2002), the S. solfataricus (archaea) exosome (Buttner et al. 2005; Lorentzen and Conti 2005; Lorentzen et al. 2005), and human exosome (Liu et al. 2006) core proteins
Interestingly, changing aspartic acid 135, which is located in the first core domain, to glycine, (D135G), a mutation shown above to cause the inability to form the homotrimer complex (Fig. 7), resulted in the almost complete inhibition of degradation and polyadenylation activities. The corresponding mutation in the E. coli PNPase, (D96G), resulted with almost complete loss of the degradation activity but only slightly reduced the polymerization activity (Table 1; Jarrige et al. 2002).
The RNA-binding property of the modified proteins, as disclosed by the UV-cross-linking assay, was drastically reduced compared to the wild-type and the E. coli proteins even in the D544G and R445,446E mutants, in which the polymerization activity was significantly enhanced (Fig. 8C). Together, the mutational analyses revealed both similarities and differences between the activities of the mutated PNPase, compared with parallel mutations in the E. coli enzyme and the archaeal and human exosomes (Table 1).
Domain swapping of the S1 domain between the E. coli and human PNPases, conferred high poly(A) binding affinity to the human enzyme
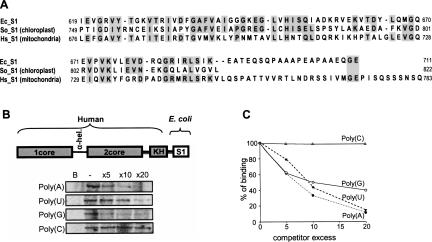
Previously, we have found that the S1 domain of the spinach chloroplast PNPase is responsible for high binding affinity to poly(A) (Yehudai-Resheff et al. 2003). This phenomenon is important for the activity of this enzyme in bacteria, chloroplast, and plant mitochondria in the polyadenylation stimulated RNA degradation pathway (Lisitsky et al. 1997; Lisitsky and Schuster 1999; Yehudai-Resheff et al. 2003). The human PNPase is located in the mitochondria IMS and does not seems to be directly involved in this pathway, and accordingly, displayed low binding affinity to poly(A) (Fig. 6). Alignment of the amino acids sequence of the S1 domain of the E. coli, spinach chloroplast, and human PNPase revealed significant amounts of conserved amino acids but not drastic differences that could possibly account for the diverse binding properties (Fig. 9A). Nevertheless, when the S1 domain of the human enzyme was replaced with that of the E. coli PNPase, high binding affinity to poly(A) was observed, indicating that the S1 domain is responsible for this phenomenon (Fig. 9B). Interestingly, the high binding affinity to poly(G), a characteristic of human but not E. coli PNPase, was still observed in the human PNPase harboring the E. coli S1 domain (Fig. 9B). This result suggests that the high affinity to poly(G) characteristic of the human PNPase is not located at the S1 domain.
FIGURE 9.
Changing the S1 domain of human PNPase to that of the E. coli enzyme resulted with high binding affinity to poly(A). (A) Alignment of the amino acid sequences of the S1 domain of E. coli (Ec_S1), spinach chloroplast (So_S1), and human (Hs_S1) PNPases. Conserved amino acids are indicated with a gray background. (B) High-affinity binding of the chimera protein to poly(U), poly(A), and poly(G). The chimera protein, as schematically presented, was incubated with [32P]-RNA corresponding to (GU)12, and a homoribopolymer at a molar excess, as shown in the figure. Following UV cross-linking and ribonuclease digestion, the label transfer from RNA to protein was analyzed by SDS-PAGE and autoradiography. In the lane labeled B, no protein was added to the assay as a control. (C) Quantization of the competition assays. The UV light cross-linking competition assays were performed as described for (B), and the intensity of the UV light cross-linking band without competitor was defined as 100%.
DISCUSSION
In this study, the phosphorolytic activity and RNA-binding properties of the human PNPase were studied and compared to bacterial and organellar PNPases. In addition, conserved amino acids in PNPases and the archaea and human exosomes, which were shown to be located at the activity sites, were mutated, and the modified proteins analyzed for activity and RNA-binding properties. The human PNPase was found to be active in the phosphorolysis of RNA, resembling its bacterial and organellar homologs, as previously described (Hayakawa and Sekiguchi 2006; Sarkar and Fisher 2006; French et al. 2007). Differences in the Pi concentrations for optimal activity and RNA-binding properties, between the human PNPase and other PNPases, were detected as well.
The characterization of the human PNPase is of great interest because of its distinct location in the human cell in comparison to plant organellar PNPases. While in plant organelles PNPase is located in the mitochondrial matrix and the chloroplast stroma, the human PNPase is located in the IMS of the mitochondrion, where no RNA has yet been observed. This unique location is probably reflected by diverse functions acquired in the evolution of plant and animal PNPases. In bacteria, PNPase is directly involved in the degradation, processing, and nontemplate polymerization of RNA (Kushner 2004; Deutscher 2006). Indeed, in cyanobacteria, PNPase is exclusively responsible for the polyadenylation activity of the RNA degradation pathway, and deletion of the PNPase gene is lethal (Rott et al. 2003). Similarly, in hyperthermophilic and methanogenic archaea, the archaeal exosome, which is very similar to PNPase, is responsible for both polyadenylation and degradation of RNA (Portnoy et al. 2005; Portnoy and Schuster 2006). PNPase also plays a direct role in the degradation and processing of organellar transcripts in the mitochondria and chloroplast of plants (Walter et al. 2002; Perrin et al. 2004a,b). However, no PNPase exists in yeast; possibly a general case for all fungi (Dziembowski et al. 2003). Interestingly, it was recently observed that the yeast exosome exhibits no phosphorolytic activity, implying that with neither PNPase nor a phosphorolytic exosome, no RNA phosphorolytic activity occurs in yeast (Liu et al. 2006; Dziembowski et al. 2007).
As described above, the human PNPase was recently found to be located in the IMS of mitochondria and is, therefore, excluded from the matrix where the mitochondrial transcripts are present (Chen et al. 2006; Rainey et al. 2006). This observation suggests that it is not directly involved in the metabolism of mitochondrial RNA, contrary to the case of plants. On the other hand, silencing the expression of PNPase in human cells was found to significantly affect, perhaps indirectly, the amount and length of stable poly(A) tails which characterized the 3′ ends of mitochondrial transcripts (Slomovic and Schuster 2008). Therefore, and since there is not an obvious known RNA substrate in the IMS, it was of great interest to analyze the phosphorolytic and RNA-binding properties of the human PNPase in comparison to the bacterial and plant PNPases that are directly involved in RNA metabolism.
Differences between human and previously characterized PNPases
The RNA degradation activity of human PNPase has been previously shown for purified recombinant or in vitro translated protein (Leszczyniecka et al. 2002; Sarkar et al. 2005; French et al. 2007). The results of the present work disclosed that the human PNPase exhibits phosphorolytic activity that could efficiently polymerize or degrade all of the RNA molecules that were examined. There are two major differences between the human and the thus far characterized bacterial and organellar PNPases. The first difference is that while the bacterial and plant organellar enzymes disclosed a high binding affinity to poly(A), the human PNPase binds poly(A) with very low affinity (Fig. 6). The high affinity poly(A)-binding of bacterial and plant organellar PNPases is required for activity because of their direct involvement in the polyadenylation-stimulated RNA degradation pathway. Due to its binding preference to polyadenylated RNA, following the polyadenylation of RNA by poly(A)-polymerase or PNPase, PNPase binds and degrades the polyadenylated molecule. Thus, polyadenylated RNA molecules compete with nonpolyadenylated molecules for PNPase binding (or additional exoribonucleases) in bacteria and plant organelles (Schuster et al. 1999; Regnier and Arraiano 2000; Dreyfus and Regnier 2002; Slomovic et al. 2006). Since, in human mitochondria PNPase is mostly or exclusively not located in the matrix, it is most likely not directly involved in a polyadenylation-stimulated RNA degradation pathway, and therefore high binding affinity to poly(A) is not required. All characterized PNPases bind the ribohomopolymer, poly(U), with high affinity. However, while bacterial and chloroplast PNPases exhibited low binding affinity to poly(G) (Lisitsky and Schuster 1999; Yehudai-Resheff et al. 2003), the human enzyme binds this ribohomopolymer with high affinity. Whether or not this is related to its biological function is yet to be revealed.
The second difference is the optimal concentration of Pi for degradation activity. While the E. coli and chloroplast PNPases display optimal activity at Pi concentrations exceeding 10 mM, the human PNPase optimal activity was found at a concentration of 0.1 mM and is actually inhibited at higher concentrations. While the Pi concentration is believed to be 10–20 mM in bacteria and the organellar matrixes, its concentration in the mitochondrial IMS in human cells is unknown. It could be suggested that either the human enzyme acquired optimal activity at a low Pi concentration for its function within the IMS, or that other PNPases were modified during evolution to function at higher Pi concentrations in bacteria and within the organellar matrix.
There are two possibilities to explain the IMS location of this enzyme: There may be an RNA substrate in the IMS that is subjected to the phosphorolytic activity of PNPase which plays an important function in the fine tuning of the NDP/Pi concentrations inside the mitochondria, which may also influence the NTP concentrations. In human cells in which PNPase was silenced by RNAi, significant effects upon the length and amount of poly(A) tails, which characterize mitochondrial transcripts, were detected, together with changes in ATP concentration (Slomovic and Schuster 2008). Furthermore, proper ATP concentrations are essential for efficient polyadenylation and transcription of mitochondrial RNA (Dubin et al. 1982).
Another possibility is that, similar to endonuclease G, PNPase does not have a designated function within the IMS; rather, it is stored in this compartment, released during apoptosis, and functions in the phosphorolysis of a substrate found in the cytoplasm (Chen et al. 2006). Further studies concerning the biological and molecular properties of the human PNPase will explore these hypotheses and reveal the biological function of this enzyme.
MATERIALS AND METHODS
Production of recombinant PNPase and its mutated versions
The corresponding DNA sequence of the mature protein (without the transit peptide) was amplified by PCR using the primers GGTACCCGAGCTGTGGCCGTGGACTTA and GTCGACTCACTGAGAATTAGATGATGACTGTGA and the PNPase cDNA, kindly obtained from P.B. Fisher (Columbia University). The Acc65I and SalI restriction sites that were added are bold and underlined. The different point mutations in the full-length (wild-type [wt]) protein were introduced using the site-directed mutagenesis kit (Stratagene). To create the chimera protein in which the S1 domain of human PNPase was replaced by that from the E. coli enzyme, a BamHI site was introduced between KH and S1 domains using a site-directed mutagenesis kit (Stratagene). The E. coli S1 domain, to which the same restriction site was introduced, was cloned using this site and the XhoI restriction site located in the pet20b vector.
For expression in E. coli, the PCR products were inserted into the Pet 20b vector with the addition of a His6 tag at the C terminus. The proteins were expressed in the BL-21 DE3 pLysS or the strain ENS134-3 lacking the endogenous PNPase and containing the T7 RNA polymerase [BL21(DE3)(lacZ:Tn10 malPp534:PT7lacZ-Arg5)(pnp:Tn5)] (Lopez et al. 1999), kindly obtained from Marc Dreyfus (Ecole Normale Supérieure). The proteins were purified according to the manufacturer's protocol using a NTA-agarose affinity column with additional purification steps using MonoQ and Heparin columns (Pharmacia). The wt and mutant D91G proteins were purified additionally by fractionation on the size-exclusion column Superdex 200. All of the proteins were purified to the stage of one Coomassie-stained band on SDS-PAGE (Fig. 1), and no contaminating ribonuclease activity was detected. In order to examine possible RNA content of the PNPase preparation, RNA was extracted from a purified fraction containing 50 μg of PNPase and analyzed by gel fractionation and silver staining. Less than 1 ng of RNA was detected (data not shown). In addition, incubation of the enzyme with Pi for 30 min (phosphorolysis) or extensive RNase A treatment did not reveal any differences. Therefore, the purified PNPase contains only minor traces, if any, of RNA. Specific antibodies for the E. coli and human PNPases were kindly obtained from Agamemnon J. Carpousis (Centre National de la Recherche Scientifique) and Paul B. Fisher (Columbia University), respectively.
Synthetic RNAs and antibodies
cDNAs encoding the 100 nt of the 3′ end of the human mitochondrial transcript COX1 with or without the addition of 49 adenosines were described (Slomovic et al. 2005). Part of the cDNA encoding 267 nt of the ubb (ubiquitin), with or without the addition of 62 adenosines was used to generate the corresponding transcript. These cDNAs were cloned to the pGEM vector, and the RNAs were transcribed using T7 RNA polymerase and radioactively labeled with [32P]UTP (Portnoy and Schuster 2006). The full-length transcription products were purified from 5% denaturing polyacrylamide gels. The plasmid for the transcription of the E. coli RNAI was described previously (Portnoy and Schuster 2006). The (GU)12 oligonucleotide was radioactively labeled at the 5′ end using [32P] γ-ATP and polynucleotide kinase.
Polyadenylation and degradation assays
Polyadenylation and degradation activities of the recombinant proteins were assayed as previously described (Yehudai-Resheff et al. 2003). [32P]-RNA was incubated with the corresponding proteins in buffer E (20 mM HEPES, pH 7.9, 60 mM KCl, 12.5 mM MgCl2, 0.1 mM EDTA, 2 mM DTT, and 17% glycerol) at 37°C with the addition of Pi. When polyadenylation was assayed, ADP or the corresponding nucleotide was added and the Pi omitted. Following incubation, the RNA was isolated and analyzed by denaturing PAGE and autoradiography. For a thin-layer chromatography analysis of the degradation products, an aliquot of each sample was spotted on a polyethyleneimine thin layer chromatography plate, which was then developed with 0.9 M GnCl, dried, and exposed to autoradiography (Yehudai-Resheff et al. 2003). Nucleosides di-, and three phosphates (5 μg of each) were separated on the same plate and visualized by fluorescence quenching.
UV light cross-linking assay
UV light cross-linking of proteins to radiolabeled RNA was performed as previously described (Lisitsky et al. 1997). The proteins were mixed with [32P]-RNA in a buffer containing 10 mM HEPES-NaOH, pH 7.9, 30 mM KCl, 6 mM MgCl2, 0.05 mM EDTA, 2 mM DTT, and 8% glycerol and cross-linked immediately at a UV light cross-linker that was set at 1.8 J (Hoefer). This was followed by digestion of the RNA with 10 μg of RNase A and 30 units of RNase T1 at 37°C for 1 h. The proteins were then fractionated by SDS-PAGE and analyzed by autoradiography. For the competition assay, the protein was first mixed with the ribohomopolymers and then the radiolabeled RNA was added.
Structure prediction and complex model building
Homology-based modeling of the three-dimensional structure of human PNPase was performed using the Phyre Server at http://www.sbg.bio.ic.ac.uk/∼3dpssm/. The visualization of the human PNPase trimer complex was performed by inserting the monomeric model described above into a pseudorhombohedral (H32) space group with dimensions similar to those of the Streptomyces antibioticus crystal structure (PDB code 1E3H) using PyMOL (Delano).
ACKNOWLEDGMENTS
We thank Paul B. Fisher and Piotr Stepien for the human PNPase cDNA and antibodies and Marc Dreyfus for the PNPase minus strain of E. coli. We thank Agamemnon J. Carpousis for E. coli PNPase antibodies. We also thank Shimyn Slomovic for critical reading and editing the manuscript and Rafal Tomecki for help with the RNA-binding experiments. This work was supported by grants from the Israel Science Foundation (ISF), the United States–Israel Binational Science Foundation (BSF), and the United States–Israel Binational Agricultural Research and Development Fund (BARD). S.Y.-R. was supported by the Ministry of Absorption fellowship.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.698108.
REFERENCES
- Amblar, M., Barbas, A., Gomez-Puertas, P., Arraiano, C.M. The role of the S1 domain in exoribonucleolytic activity: Substrate specificity and multimerization. RNA. 2007;13:317–327. doi: 10.1261/rna.220407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baginsky, S., Shteiman-Kotler, A., Liveanu, V., Yehudai-Resheff, S., Bellaoui, M., Settlage, R.E., Shabanowitz, J., Hunt, D.F., Schuster, G., Gruissem, W. Chloroplast PNPase exists as a homo-multimer enzyme complex that is distinct from the Escherichia coli degradosome. RNA. 2001;7:1464–1475. [PMC free article] [PubMed] [Google Scholar]
- Bermudez-Cruz, R.M., Garcia-Mena, J., Montanez, C. Polynucleotide phosphorylase binds to ssRNA with same affinity as to ssDNA. Biochimie. 2002;84:321–328. doi: 10.1016/s0300-9084(02)01385-8. [DOI] [PubMed] [Google Scholar]
- Buttner, K., Wenig, K., Hopfner, K.P. Structural framework for the mechanism of archaeal exosomes in RNA processing. Mol. Cell. 2005;20:461–471. doi: 10.1016/j.molcel.2005.10.018. [DOI] [PubMed] [Google Scholar]
- Buttner, K., Wenig, K., Hopfner, K.P. The exosome: A macromolecular cage for controlled RNA degradation. Mol. Microbiol. 2006;61:1372–1379. doi: 10.1111/j.1365-2958.2006.05331.x. [DOI] [PubMed] [Google Scholar]
- Chen, H.W., Rainey, R.N., Balatoni, C.E., Dawson, D.W., Troke, J.J., Wasiak, S., Hong, J.S., McBride, H.M., Koehler, C.M., Teitell, M.A., et al. Mammalian polynucleotide phosphorylase is an intermembrane space RNase that maintains mitochondrial homeostasis. Mol. Cell. Biol. 2006;26:8475–8487. doi: 10.1128/MCB.01002-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements, M.O., Eriksson, S., Thompson, A., Lucchini, S., Hinton, J.C., Normark, S., Rhen, M. Polynucleotide phosphorylase is a global regulator of virulence and persistency in Salmonella enterica . Proc. Natl. Acad. Sci. 2002;99:8784–8789. doi: 10.1073/pnas.132047099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher, M.P. Degradation of RNA in bacteria: Comparison of mRNA and stable RNA. Nucleic Acids Res. 2006;34:659–666. doi: 10.1093/nar/gkj472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfus, M., Regnier, P. The poly(A) tail of mRNAs: Bodyguard in eukaryotes, scavenger in bacteria. Cell. 2002;111:611–613. doi: 10.1016/s0092-8674(02)01137-6. [DOI] [PubMed] [Google Scholar]
- Dubin, D.T., Montoya, J., Timko, K.D., Attardi, G. Sequence analysis and precise mapping of the 3′ ends of HeLa cell mitochondrial ribosomal RNAs. J. Mol. Biol. 1982;157:1–19. doi: 10.1016/0022-2836(82)90510-1. [DOI] [PubMed] [Google Scholar]
- Dziembowski, A., Piwowarski, J., Hoser, R., Minczuk, M., Dmochowska, A., Siep, M., van der Spek, H., Grivell, L., Stepien, P.P. The yeast mitochondrial degradosome. Its composition, interplay between RNA helicase and RNase activities and the role in mitochondrial RNA metabolism. J. Biol. Chem. 2003;278:1603–1611. doi: 10.1074/jbc.M208287200. [DOI] [PubMed] [Google Scholar]
- Dziembowski, A., Lorentzen, E., Conti, E., Seraphin, B. A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat. Struct. Mol. Biol. 2007;14:15–22. doi: 10.1038/nsmb1184. [DOI] [PubMed] [Google Scholar]
- French, S.W., Dawson, D.W., Chen, H.W., Rainey, R.N., Sievers, S.A., Balatoni, C.E., Wong, L., Troke, J.J., Nguyen, M.T., Koehler, C.M., et al. The TCL1 oncoprotein binds the RNase PH domains of the PNPase exoribonuclease without affecting its RNA degrading activity. Cancer Lett. 2007;248:198–210. doi: 10.1016/j.canlet.2006.07.006. [DOI] [PubMed] [Google Scholar]
- Grunberg-Manago, M. Messenger RNA stability and its role in control of gene expression in bacteria and phages. Annu. Rev. Genet. 1999;33:193–227. doi: 10.1146/annurev.genet.33.1.193. [DOI] [PubMed] [Google Scholar]
- Hayakawa, H., Sekiguchi, M. Human polynucleotide phosphorylase protein in response to oxidative stress. Biochemistry. 2006;45:6749–6755. doi: 10.1021/bi052585l. [DOI] [PubMed] [Google Scholar]
- Houseley, J., LaCava, J., Tollervey, D. RNA-quality control by the exosome. Nat. Rev. Mol. Cell Biol. 2006;7:529–539. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- Jarrige, A., Brechemier-Baey, D., Mathy, N., Duche, O., Portier, C. Mutational analysis of polynucleotide phosphorylase from Escherichia coli . J. Mol. Biol. 2002;321:397–409. doi: 10.1016/s0022-2836(02)00645-9. [DOI] [PubMed] [Google Scholar]
- Kaberdin, V.R., Chao, Y.H., Lin-Chao, S. RNase E cleaves at multiple sites in bubble regions of RNA I stem–loops yielding products that dissociate differentially from the enzyme. J. Biol. Chem. 1996;271:13103–13109. doi: 10.1074/jbc.271.22.13103. [DOI] [PubMed] [Google Scholar]
- Kushner, S.R. mRNA decay in prokaryotes and eukaryotes: Different approaches to a similar problem. IUBMB Life. 2004;56:585–594. doi: 10.1080/15216540400022441. [DOI] [PubMed] [Google Scholar]
- Leszczyniecka, M., Kang, D.C., Sarkar, D., Su, Z.Z., Holmes, M., Valerie, K., Fisher, P.B. Identification and cloning of human polynucleotide phosphorylase, hPNPase old-35, in the context of terminal differentiation and cellular senescence. Proc. Natl. Acad. Sci. 2002;99:16636–16641. doi: 10.1073/pnas.252643699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leszczyniecka, M., DeSalle, R., Kang, D.C., Fisher, P.B. The origin of polynucleotide phosphorylase domains. Mol. Phylogenet. Evol. 2004;31:123–130. doi: 10.1016/j.ympev.2003.07.012. [DOI] [PubMed] [Google Scholar]
- Lin-Chao, S., Chiou, N.T., Schuster, G. The PNPase, exosome, and RNA helicases as the building components of evolutionarily conserved RNA degradation machines. J. Biomed. Sci. 2007;14:523–532. doi: 10.1007/s11373-007-9178-y. [DOI] [PubMed] [Google Scholar]
- Lisitsky, I., Schuster, G. Preferential degradation of polyadenylated and polyuridinylated RNAs by bacterial exoribonuclease polynucleotide phosphorylase (PNPase) Eur. J. Biochem. 1999;261:468–474. doi: 10.1046/j.1432-1327.1999.00285.x. [DOI] [PubMed] [Google Scholar]
- Lisitsky, I., Kotler, A., Schuster, G. The mechanism of preferential degradation of polyadenylated RNA in the chloroplast: The exoribonuclease 100RNP/PNPase displays high binding affinity for poly(A) sequence. J. Biol. Chem. 1997;272:17648–17653. doi: 10.1074/jbc.272.28.17648. [DOI] [PubMed] [Google Scholar]
- Littauer, U.Z., Grunberg-Manago, M. Polynucleotide phosphorylase. In: Creighton T.E., editor. The encyclopedia of molecular biology. Wiley; New York: 1999. pp. 1911–1918. [Google Scholar]
- Littauer, U.Z., Soreq, H. Polynucleotide phosphorylase. Academic Press; New York: 1982. [Google Scholar]
- Liu, Q., Greimann, J.C., Lima, C.D. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell. 2006;127:1223–1237. doi: 10.1016/j.cell.2006.10.037. [DOI] [PubMed] [Google Scholar]
- Lopez, P.J., Marchand, I., Joyce, S.A., Dreyfus, M. The C-terminal half of RNase E, which organizes the Escherichia coli degradosome, participates in mRNA degradation but not rRNA processing in vivo. Mol. Microbiol. 1999;33:188–199. doi: 10.1046/j.1365-2958.1999.01465.x. [DOI] [PubMed] [Google Scholar]
- Lorentzen, E., Conti, E. Structural basis of 3′ end RNA recognition and exoribonucleolytic cleavage by an exosome RNase PH core. Mol. Cell. 2005;20:473–481. doi: 10.1016/j.molcel.2005.10.020. [DOI] [PubMed] [Google Scholar]
- Lorentzen, E., Walter, P., Fribourg, S., Evguenieva-Hackenberg, E., Klug, G., Conti, E. The archaeal exosome core is a hexameric ring structure with three catalytic subunits. Nat. Struct. Mol. Biol. 2005;12:575–581. doi: 10.1038/nsmb952. [DOI] [PubMed] [Google Scholar]
- Marcaida, M.J., DePristo, M.A., Chandran, V., Carpousis, A.J., Luisi, B.F. The RNA degradosome: Life in the fast lane of adaptive molecular evolution. Trends Biochem. Sci. 2006;31:359–365. doi: 10.1016/j.tibs.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Mohanty, B.K., Kushner, S.R. Polynucleotide phosphorylase functions both as a 3′ to 5′ exonuclease and a poly(A) polymerase in Escherichia coli . Proc. Natl. Acad. Sci. 2000;97:11966–11971. doi: 10.1073/pnas.220295997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty, B.K., Maples, V.F., Kushner, S.R. The Sm-like protein Hfq regulates polyadenylation dependent mRNA decay in Escherichia coli . Mol. Microbiol. 2004;54:905–920. doi: 10.1111/j.1365-2958.2004.04337.x. [DOI] [PubMed] [Google Scholar]
- Perrin, R., Lange, H., Grienenberger, J.M., Gagliardi, D. AtmtPNPase is required for multiple aspects of the 18S rRNA metabolism in Arabidopsis thaliana mitochondria. Nucleic Acids Res. 2004a;32:5174–5182. doi: 10.1093/nar/gkh852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin, R., Meyer, E.H., Zaepfel, M., Kim, Y.J., Mache, R., Grienenberger, J.M., Gualberto, J.M., Gagliardi, D. Two exoribonucleases act sequentially to process mature 3′-ends of atp9 mRNAs in Arabidopsis mitochondria. J. Biol. Chem. 2004b;279:25440–25446. doi: 10.1074/jbc.M401182200. [DOI] [PubMed] [Google Scholar]
- Portnoy, V., Schuster, G. RNA polyadenylation and degradation in different Archaea; Roles of the exosome and RNase R. Nucleic Acids Res. 2006;34:5923–5931. doi: 10.1093/nar/gkl763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portnoy, V., Evguenieva-Hackenberg, E., Klein, F., Walter, P., Lorentzen, E., Klug, G., Schuster, G. RNA polyadenylation in Archaea: Not observed in Haloferax while the exosome polyadenylates RNA in Sulfolobus . EMBO Rep. 2005;6:1188–1193. doi: 10.1038/sj.embor.7400571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainey, R.N., Glavin, J.D., Chen, H.W., French, S.W., Teitell, M.A., Koehler, C.M. A new function in translocation for the mitochondrial i-AAA protease Yme1: Import of polynucleotide phosphorylase into the intermembrane space. Mol. Cell. Biol. 2006;26:8488–8497. doi: 10.1128/MCB.01006-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regnier, P., Arraiano, C.M. Degradation of mRNA in bacteria: Emergence of ubiquitous features. Bioessays. 2000;22:235–244. doi: 10.1002/(SICI)1521-1878(200003)22:3<235::AID-BIES5>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Rott, R., Zipor, G., Portnoy, V., Liveanu, V., Schuster, G. RNA polyadenylation and degradation in cyanobacteria are similar to the chloroplast but different from Escherichia coli . J. Biol. Chem. 2003;278:15771–15777. doi: 10.1074/jbc.M211571200. [DOI] [PubMed] [Google Scholar]
- Sarkar, D., Fisher, P.B. Polynucleotide phosphorylase: An evolutionary conserved gene with an expanding repertoire of functions. Pharmacol. Ther. 2006;112:243–263. doi: 10.1016/j.pharmthera.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Sarkar, D., Park, E.S., Emdad, L., Randolph, A., Valerie, K., Fisher, P.B. Defining the domains of human polynucleotide phosphorylase (hPNPaseOLD-35) mediating cellular senescence. Mol. Cell. Biol. 2005;25:7333–7343. doi: 10.1128/MCB.25.16.7333-7343.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert, M., Edge, R.E., Lario, P., Cook, M.A., Strynadka, N.C., Mackie, G.A., McIntosh, L.P. Structural characterization of the RNase E S1 domain and identification of its oligonucleotide-binding and dimerization interfaces. J. Mol. Biol. 2004;341:37–54. doi: 10.1016/j.jmb.2004.05.061. [DOI] [PubMed] [Google Scholar]
- Schuster, G., Lisitsky, I., Klaff, P. Update on chloroplast molecular biology: Polyadenylation and degradation of mRNA in the chloroplast. Plant Physiol. 1999;120:937–944. doi: 10.1104/pp.120.4.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slomovic, S., Schuster, G. Stable PNPase RNAi silencing: Its effect on the processing and adenylation of human mitochondrial RNA. RNA. 2008 doi: 10.1261/rna.697308. (this issue). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slomovic, S., Laufer, D., Geiger, D., Schuster, G. Polyadenylation and degradation of human mitochondrial RNA: The prokaryotic past leaves its mark. Mol. Cell. Biol. 2005;25:6427–6435. doi: 10.1128/MCB.25.15.6427-6435.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slomovic, S., Portnoy, V., Liveanu, V., Schuster, G. RNA polyadenylation in prokaryotes and organelles; Different tails tell different tales. Crit. Rev. Plant Sci. 2006;25:65–77. [Google Scholar]
- Stickney, L.M., Hankins, J.S., Miao, X., Mackie, G.A. Function of the conserved S1 and KH domains in polynucleotide phosphorylase. J. Bacteriol. 2005;187:7214–7221. doi: 10.1128/JB.187.21.7214-7221.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symmons, M.F., Jones, G.H., Luisi, B.F. A duplicated fold is the structural basis for polynucleotide phosphorylase catalytic activity, processivity, and regulation. Structure. 2000;8:1215–1226. doi: 10.1016/s0969-2126(00)00521-9. [DOI] [PubMed] [Google Scholar]
- Symmons, M.F., Williams, M.G., Luisi, B.F., Jones, G.H., Carpousis, A.J. Running rings around RNA: A superfamily of phosphate-dependent RNases. Trends Biochem. Sci. 2002;27:11–18. doi: 10.1016/s0968-0004(01)01999-5. [DOI] [PubMed] [Google Scholar]
- Walter, M., Kilian, J., Kudla, J. PNPase activity determines the efficiency of mRNA 3′-end processing, the degradation of tRNA and the extent of polyadenylation in chloroplasts. EMBO J. 2002;21:6905–6914. doi: 10.1093/emboj/cdf686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worrall, J.A., Luisi, B.F. Information available at cut rates: Structure and mechanism of ribonucleases. Curr. Opin. Struct. Biol. 2007;17:128–137. doi: 10.1016/j.sbi.2006.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yehudai-Resheff, S., Hirsh, M., Schuster, G. Polynucleotide phosphorylase functions as both an exonuclease and a poly(A) polymerase in spinach chloroplasts. Mol. Cell. Biol. 2001;21:5408–5416. doi: 10.1128/MCB.21.16.5408-5416.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yehudai-Resheff, S., Portnoy, V., Yogev, S., Adir, N., Schuster, G. Domain analysis of the chloroplast polynucleotide phosphorylase reveals discrete functions in RNA degradation, polyadenylation, and sequence homology with exosome proteins. Plant Cell. 2003;15:2003–2019. doi: 10.1105/tpc.013326. [DOI] [PMC free article] [PubMed] [Google Scholar]