Abstract
A recent crystal structure of the N15 α/β-T cell receptor (TCR) in complex with an Fab derived from the H57 Cβ-specific monoclonal antibody (mAb) shows the mAb fragment interacting with the elongated FG loop of the Cβ domain. This loop creates one side wall of a cavity within the TCR Ti-α/β constant region module (CαCβ) while the CD and EF loops of the Cα domain form another wall. The cavity size is sufficient to accommodate a single nonglycosylated Ig domain such as the CD3ε ectodomain. By using specific mAbs to mouse TCR-β (H57) and CD3ε (2C11) subunits, we herein provide evidence that only one of the two CD3ε chains within the TCR complex is located in close proximity to the TCR Cβ FG loop, in support of the above notion. Moreover, analysis of T cells isolated from transgenic mice expressing both human and mouse CD3ε genes shows that the heterologous human CD3ε component can replace the mouse CD3ε at this site. The location of one CD3ε subunit within the rigid constant domain module has implications for the mechanism of signal transduction throughout T cell development.
Each TCR consists of a clonotypic TCR heterodimer (Ti-α/β or Ti-γ/δ subunits) in complex with the invariant CD3 chains (γ, δ, ε, and ζ). The disulfide-linked heterodimer represents the peptide–MHC ligand binding unit, thereby determining the ligand specificity of an individual T cell. In contrast, the CD3 polypeptides which are in noncovalent association with a given Ti heterodimer, mediate TCR-base signal transduction (for review see references 1–5). Although CD3-γ and -δ are present in only one copy each (6–8), it appears that two copies of CD3ε and ζ exist per TCR complex (9–11). The signaling function of the CD3 components involves a conserved motif, termed an immunoreceptor tyrosine-based activation motif (ITAM) present in one to three copies in the cytoplasmic domain of each CD3 subunit (12, 13). The various CD3 subunits exhibit different interactions with intracellular signaling factors and induce distinct patterns of cellular protein tyrosine phosphorylation upon activation (14–19). How peptide–MHC ligand binding to a Ti-α/β or Ti-γ/δ heterodimer subsequently initiates signaling via the CD3 molecules is currently unknown.
Aside from their role in signal transduction, the CD3 subunits are also required for cell surface expression of the TCR heterodimers on mature T lymphocytes (20, 21), as well as for pre-T cell receptor function on immature CD4−CD8− double negative (DN)1 thymocytes (22, 23). Thus, without CD3ε or -ζ subunit expression there is a marked decrease or absence of TCR molecules on the cell surface as shown by in vitro analysis (20, 21, 24). In addition, in genetically engineered mouse strains in which these CD3 components are deleted by homologous recombination, there is a developmental blockade of thymocytes at the DN stage (25–29). The CD3δ subunit, in contrast to CD3ε and -ζ chains, is required for TCR expression only at a later stage of thymic development. The absence of CD3δ in a knockout mouse specifically blocks the thymic selection processes mediating the transition from double positive (DP) (CD4+CD8+) to single positive (CD4+CD8− or CD4−CD8+) thymocytes (30).
Although the overall stoichiometry of the TCR complex is commonly given as TCR-α/β–CD3γ/δ/ε2/ζ2, there is no direct structural evidence to support this subunit composition. Recently, a three-dimensional structure of the N15 vesicular stomatitis virus–specific/H-2Kb-restricted α/β-TCR (31) in complex with an Fab fragment from the H57 anti–mouse Cβ-specific mAb (32) provided a clue with which to infer new details about the association between the TCR-α/β heterodimer and CD3ε (33). We identified a cavity within the TCR-α/β C module formed by the Cβ FG loop, partially exposed Cβ domain strands, and conserved glycans from both Cα and Cβ domains that can accommodate a single Ig-like domain. Based on size and charge considerations, we suggested that the cavity probably represents the CD3ε binding site. To determine whether there is a physical proximity between the Cβ FG loop and the CD3ε chain, we performed a set of competition assays between the H57 and the CD3ε-specific 2C11 mAbs (34). The results of these experiments provide evidence that one of the two CD3ε subunits in a TCR complex is physically adjacent to the TCR-β constant region FG loop.
Materials and Methods
Transgenic Mice.
Transgenic mice expressing the human CD3ε gene (transgenic [tg] 600; reference 35) were provided by Dr. Cox Terhorst (Beth Israel Medical Center, Boston, MA) and are further referred to as hCD3εtg. This transgenic mouse strain contains 10–12 copies of the human CD3ε transgene in a hemizygous mouse. Since T cell development is blocked in the homozygous mice of this strain, we used hCD3εtg heterozygous mice for these studies. A littermate that does not contain the human CD3ε transgene was used as a control (wild-type, WT).
Molecular Modeling of the N15 TCR-H57 Fab Complex.
The molecular modeling was produced using the N15 TCR and H57 Fab complex crystal structure 3D coordinates (PDB code 1NFD; 33). The plot of the protein structure was created using the programs MOLSCRIPT (36) and RASTER3D (37).
Flow Cytometric Analysis.
The following mAbs were used: R-PE-labeled anti–mouse CD3ε (500A2; PharMingen, San Diego, CA), PE-labeled anti–mouse CD4 (H129.19; GIBCO BRL, Gaithersburg, MD), and Red613-labeled anti–mouse CD8 (53-6.7; GIBCO BRL). The mAbs hamster anti–mouse TCR-β (H57-597; reference 32), hamster anti–mouse CD3ε (2C11-145; reference 34), and mouse anti–mouse Vβ8.1,2,3 (F23.1; reference 38) were purified and labeled with FITC using the FluoReporter® FITC protein-labeling kit (Molecular Probes, Eugene, OR). Generation and purification of 2C11 and H57 Fab fragments were performed by using the ImmunoPure Fab Preparation Kit (Pierce, Rockford, IL). The purity of the Fab fragments was confirmed by SDS-PAGE, which demonstrated no residual intact IgG. Splenocytes from nontransgenic mice (WT) or mice heterozygous for human CD3ε (hCD3εtg) were triple stained as follows: cells were incubated with PE-labeled anti-CD4, Red613-labeled anti-CD8, and FITC-H57 or FITC-2C11 for 30 min at 4°C in PBS containing 2% FCS. For mAb competition assays, splenocytes were first incubated with unlabeled H57 or 2C11 mAbs at 10-fold molar excess of the FITC-labeled mAb for 30 min at 4°C, washed with PBS + 2% FCS, and triple stained as explained above. Flow cytometric analysis was performed on a FACScan (Becton Dickinson, San Jose, CA). Samples were gated on live cells based on forward and side scatter parameters. Data (10,000 events per sample) were collected in list mode using FACScan Research software and analyzed using LYSYS II software (Becton Dickinson). The mean fluorescence intensity (mFI) of the FITC-labeled H57 or FITC-labeled 2C11 labeled T cells (CD4+ or CD8+ splenocytes) was measured and the FITC mFI of non-T cell splenocytes (CD4− or CD8−; background) was deducted from this value in each sample.
Results
A TCR Constant Domain Module Cavity as a Putative CD3ε Binding Site.
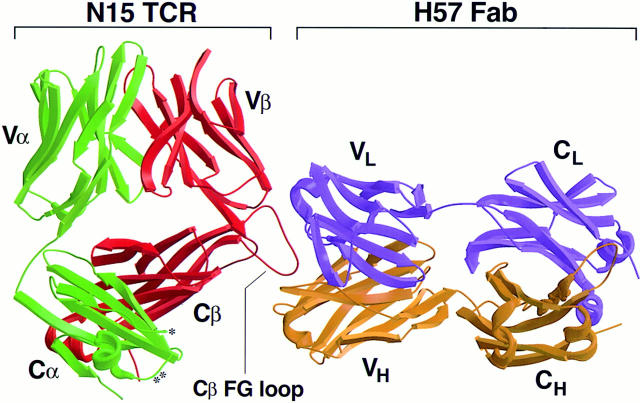
The three-dimensional structure of a complex between the N15 α/β-TCR and the anti–TCR-β chain mAb H57 Fab fragment resolved crystallographically to 2.8Å has revealed several interesting features about TCR structure relative to antibody structure. As shown in Fig. 1, there is an obvious difference between the ligand binding surface of the V domain modules of the two receptors (VαVβ versus VhVl), with a relatively flat antigen binding surface in the case of the TCR, versus a concave surface in the case of the Fab. The flatness of the N15 antigen binding surface and the concavity of the H57 Fab surface are complementary to the surface of their respective ligands, Kb and Cβ. Perhaps the most striking difference between these immunoreceptors is the symmetry of the Fab molecule compared with asymmetry of the TCR molecule. This TCR asymmetry results from the unusual arrangement of the Cα and Cβ domains relative to one another. Moreover, as shown in Fig. 1, the H57 Fab binds to the 12-residue Cβ FG loop conserved among TCR Cβ domains from multiple species (39). This loop architecture is uniquely rigidified by a hydrophobic minicore and an internal hydrogen bonding network. In addition, we have noted that this loop creates one side wall of a cavity, while the CD and EF loops of the Cα domain form the other side. The floor of the cave, which measures ∼25Å in depth, ∼20Å in height, and ∼25Å in width, is presumably formed by the plasma membrane on the T cell surface. The murine CD3ε subunit, which is predicted to have an Ig fold (40), is nonglycosylated unlike CD3γ and CD3δ, and could readily fit into a cavity of this size.
Figure 1.
Model of the N15 α/β-TCR heterodimer complexed with the H57 anti-TCR Cβ-specific mAb Fab fragment. Based on x ray coordinates and molecular modeling as described in Materials and Methods, a structure of the complex is shown. The TCR-α chain is in green, the TCR-β chain is in red, the H57 Fab heavy chain is in gold, and H57 Fab light chain is in purple. The Cβ FG loop, which is the epitope recognized by the mAb H57, is indicated. This loop creates one side wall of a cavity (whose center is marked by the Cβ label) within the TCR Ti α/β constant region while the CD (*) and EF (**) loops of the Cα domain form the other side. The cavity size is sufficient to accommodate a single nonglycosylated Ig domain such as the CD3ε chain.
Nonreciprocal Cross-blocking of 2C11 and H57 mAbs.
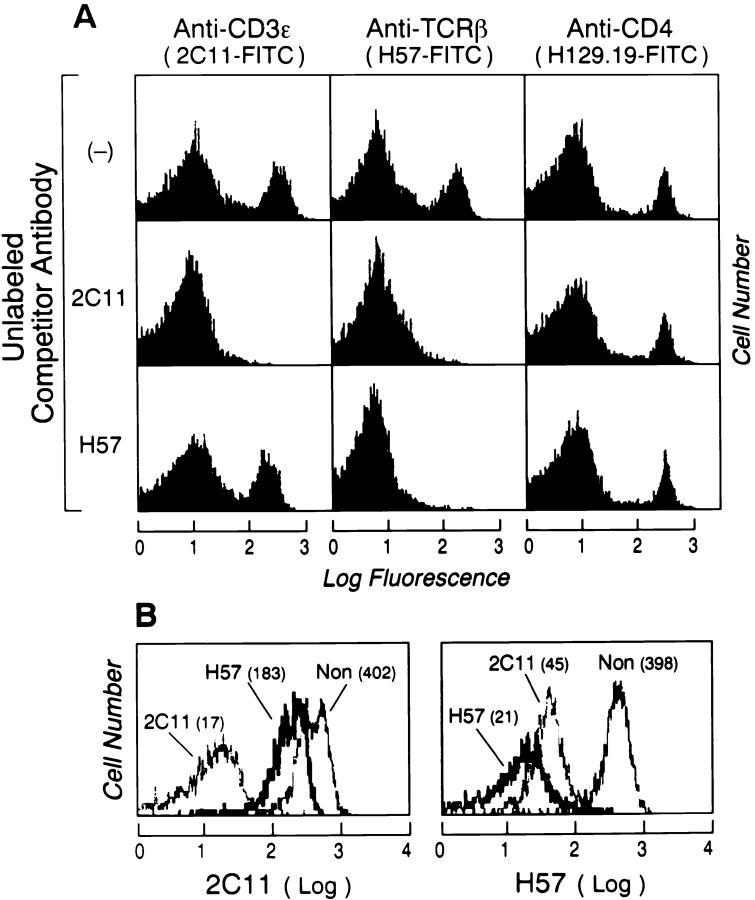
To investigate our hypothesis that a CD3ε subunit might occupy this cavity within the C module and hence lie proximal to the Cβ FG loop, a set of competition assays between the H57 and 2C11 mAbs were performed using direct immunofluorescence analysis by FACS®. 2C11 was previously shown to specifically bind to the mouse CD3ε subunit (34). As shown in Fig. 2 A, unlabeled competitor 2C11 or H57 was added before the addition of FITC-2C11, FITC-H57, or FITC-labeled anti-CD4, and log fluorescence was determined by quantitative FACS® analysis. As expected, unlabeled 2C11 blocks the subsequent binding of FITC-2C11 and, likewise, unlabeled H57 blocks the binding of FITC-H57. In contrast, neither unlabeled 2C11 nor H57 alters the binding of the unrelated FITC-H129.19. More importantly, note that under these conditions the anti-CD3ε mAb completely blocks the binding of FITC-H57 mAb, whereas H57 only partially reduces the mFI of FITC-2C11 binding to CD3ε on mouse splenic T cells. A second anti-CD3ε mAb, 500A2, also completely blocked FITC-H57 binding and, as with FITC-2C11, the binding of PE-500A2 was reduced by 50% when T cells were preincubated with unlabeled H57 (data not shown). Moreover, similar results were obtained by using 2C11 Fab or H57 Fab fragments for inhibition analysis. That the findings are not secondary to “general” steric blockade of the TCR by any mAb or Fab fragment binding to the complex is evident from the inability of Vβ-specific antibodies to block FITC-H57 binding on TCR transgenic T lymphocytes (data not shown).
Figure 2.
The anti-CD3ε mAb (2C11) blocks the binding of the anti– TCR-β mAb (H57). (A) Splenocytes (106 per sample) were incubated either with no antibody or with 10 μg/ml unlabeled mAbs 2C11 or H57 for 30 min, then washed and reincubated with 1 μg/ml of the FITC-labeled mAbs 2C11 or H57 or anti–mouse CD4 mAb. After washing, the cells were analyzed on a FACScan as described in Materials and Methods. Each unlabeled mAb blocks the identical FITC-labeled antibody. The unlabeled 2C11 blocks FITC-H57 (center histogram), whereas H57 only partially blocks the binding of FITC-2C11 to CD3ε (lower left histogram). Neither 2C11 nor H57 block the FITC-labeled anti-CD4 mAb (right column). (B) The same competition experiment was performed as described above but after the preincubation with the unlabeled competitor mAb the splenocytes were triple-stained with FITC-H57 or FITC-2C11 and both PE–anti-CD4 and Red613–anti-CD8. Histograms show the FITC-H57 or FITC-2C11 staining of T cells (after gating for CD4+ and CD8+ cells) in the presence or absence (Non) of the unlabeled competitor mAbs. The mFI of each curve is indicated in parenthesis. Only CD4+ data is shown.
Only One of the Two CD3ε Subunits Is Proximal to the Cβ FG Loop.
To quantify the mFI reduction in FITC-2C11 resulting from unlabeled H57 preincubation in individual CD4 and CD8 T cell subsets, we performed the analogous competition experiments by three-color analysis, in addition using PE-labeled anti–mouse CD4 and Red613-labeled anti–mouse CD8. We collected data comparable to that shown in Fig. 2 A but on individual splenic CD4+ and CD8+ T lymphocytes. As shown in Fig. 2 B (left) for CD4+ T cells, although unlabeled 2C11 completely blocked the binding of FITC-2C11 to the mouse T cells, the unlabeled H57 mAb reduced the binding of the FITC-2C11 by ∼50% (mFI, from 402 to 183). On the other hand, unlabeled 2C11 almost completely blocked the binding of FITC-H57 (mFI, from 398 to 45; Fig. 2 B, right). Identical results were obtained when CD8+ T cells subset were examined. Three conclusions may be drawn from these data. First, one CD3ε subunit is close to the H57 mAb binding site on the Cβ FG loop. Second, given that there are two CD3ε components per TCR (10, 11) and the H57 mAb can only block 50% of the FITC-2C11 binding, the second CD3ε subunit must exist at a distance from the Cβ FG loop. Third, this postulated TCR subunit arrangement is found in both CD4+ and CD8+ T cell subpopulations.
The Human CD3ε Subunit as well as the Mouse CD3ε Subunit Associates with the CαCβ Module.
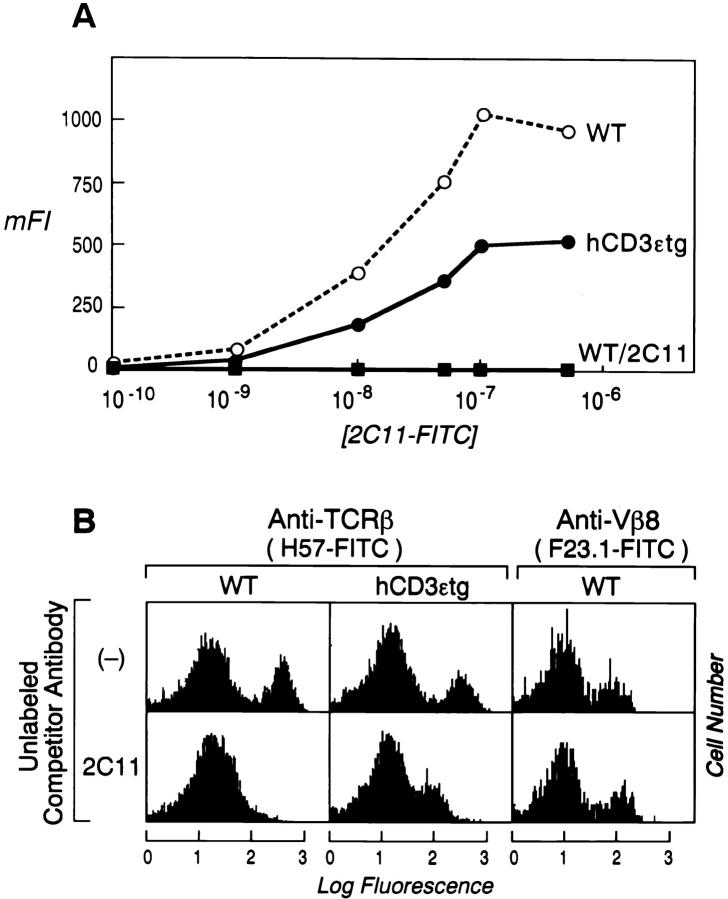
To test whether replacement of the murine CD3ε chain with the human CD3ε subunit might alter the binding affinity of the H57 mAb to the Cβ FG epitope, we used a well-characterized transgenic mouse strain (tg 600) engineered to express the human CD3ε component (reference 35; here referred to as hCD3εtg). Splenocytes from littermates that do not express the human CD3ε (WT), as well as a heterozygous hCD3εtg mouse, were stained with directly labeled FITC-2C11 mAb at concentrations ranging from 5 × 10−6–10−10 M. As shown in Fig. 3 A, the hCD3εtg mouse expressed only ∼50% of the normal cell surface level of mouse CD3ε. However, 2C11 binding affinity to the mouse CD3ε subunit is not altered by the presence of the human CD3ε component (K d ∼10−8 M). The capacity of the unlabeled 2C11 mAb to block binding of FITC-H57 to heterozygous hCD3εtg T cells was also examined. As shown in Fig. 3 B, the same amount of unlabeled 2C11 that blocks FITC-H57 binding to the WT mouse T cells only partially blocks the FITC-H57 binding to hCD3εtg T cells. This suggests that the human CD3ε subunit can replace the mouse CD3ε subunit in agreement with the finding of others (10, 41), and that it can occupy the cavity within the mouse TCR CβCα module. Fig. 3 B also demonstrates that the incubation of the mouse splenocytes with the 2C11 mAb does not block the binding of H57 to the TCR-β chain by downmodulation of the TCR complex from the T cell surface (42, 43). Thus, unlabeled 2C11 preincubation does not change the binding of the anti Vβ8 mAb, FITC-F23.1, to T cells.
Figure 3.
Human CD3ε can replace the mouse CD3ε within the rigid TCR-α/β constant domain module. (A) Splenocytes from a nontransgenic mouse (WT) or a mouse heterozygous for human CD3ε (hCD3εtg) were triple stained with the indicated molar concentration of FITC-2C11 as well as with PE–anti-CD4 and Red613–anti-CD8. The mFI of the FITC-2C11 labeled T cells was calculated as explained in Materials and Methods. Note that although the hCD3εtg T cells express only half of the WT CD3ε, the 2C11 binding affinity to the mouse CD3ε is not altered by the presence of the human CD3ε subunit. 2C11 preincubation completely blocks FITC-2C11 binding in WT mice. (B) Splenocytes from WT or hCD3εtg mice were preincubated with no antibody or with the unlabeled 2C11, washed and then reincubated with the FITC-labeled mAb H57 or the anti-Vβ8 (F23.1) as explained in the legend of Fig. 1. Although the total copy number of the mouse CD3ε subunits in hCD3εtg T cells is half of that in WT animals, 2C11 mAb only partially blocks the FITC-H57 binding in the hCD3εtg mouse (lower center histogram). Note that preincubation of the T cells with 2C11 had no effect on the TCR expression level since the anti-Vβ8 mAb binds to a subset of T cells expressing Vβ8 as well with or without 2C11 competition (right).
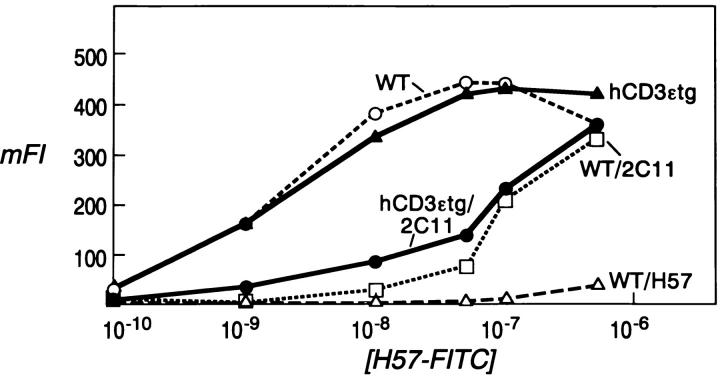
The Binding of the H57 mAb to the Cβ FG Loop Is Unaffected by Human Versus Mouse Origin of the CD3ε Component.
In contrast to the results comparing mouse CD3ε in WT and hCD3εtg mice where the level of 2C11 expression is ∼50% on the latter (Fig. 3 A), H57 reactivity is equivalent in both (Fig. 4).This implies that the affinity of H57 mAb to the Cβ FG loop is not altered by the presence of the human CD3ε component. Had human CD3ε altered the affinity of H57, then the curves of mFI at different molar concentrations of FITC-H57 would have shown a difference in the two mouse strains. Unlabeled 2C11 competitor mAb shifts the binding curve by ∼2 logs, at concentrations of FITC-H57 below 10−6 M. At the highest concentration of FITC-H57 (10−6 M), the labeled antibody binds similarly in the presence or absence of unlabeled competitor 2C11 mAb. These finding are consistent with the notion that the epitopes defined by H57 and 2C11 mAbs are distinct but nevertheless partially overlapping.
Figure 4.
Human CD3ε does not alter the affinity of H57 for its epitope. Splenocytes from WT or hCD3εtg mice were triple stained with the indicated molar concentration of FITC-H57 and with the labeled anti-CD4 and anti-CD8 with or without preincubation with an unlabeled competitor mAb (designated after the named mouse strain). The mFI of FITC-H57 binding to the T cells (CD4+ or CD8+ cells) was calculated for each sample as explained in Materials and Methods. Results are given for CD4+ T cells and identical data was observed for CD8+ T cells.
Discussion
The crystal structure of the complex between the N15 TCR and an Fab fragment of the anti-Cβ mAb H57 led us to the hypothesis that one CD3ε may physically associate with the H57 mAb epitope on Cβ (33). In this paper we demonstrate that one of the two CD3ε subunits of the TCR lies adjacent to the Cβ FG loop.
The overall shape of the TCR C domain module is remarkably asymmetric (Fig. 1). The Cβ domain bends more acutely towards the Vβ domain compared with the angle formed between the Cα and Vα domains. Cβ also has an unusually long and well-structured FG loop (the H57 mAb epitope) projecting down from the Vβ-Cβ interface in all TCRs studied to date (33, 44, 45). Although half of the Cβ domain's ABED sheet is surface exposed, it does not make contact with the Cα domain. As described in detail by Wang et al. (33), this asymmetry creates a cave-like structure or cavity below the β chain sufficient in size to accommodate a single Ig domain. The partially exposed ABED β sheet of the Cβ domain forms an extensive ceiling. The CD and EF loops of the Cα domain, along with the glycans attached to CαN185, CαN121, and CβN186 form one side wall, and the FG loop of Cβ and the glycan emanating from CβN236 form the other side wall of the cavity. The glycans project outward and do not obstruct the cavity. The floor of the cave is presumably formed by the plasma membrane on the T cell surface (33). It is noteworthy that the accessible surface at the α/β-TCR cavity contains multiple basic residues.
The murine CD3ε subunit consists of 87 residues in the extracellular segment, and has Ig-like characteristics suggesting that this segment can readily fold into a small Ig domain (40). CD3ε is the only CD3 subunit with a nonglycosylated Ig-like ectodomain in the TCR complex of man and mouse (40). Moreover, the CD3ε subunit has twice as many acidic residues as basic ones and is therefore negatively charged (pI = 4.5, whereas the predicted pIs of the CD3γ and CD3δ extracellular domains are more basic, being 8.76 and 5.82, respectively). The charge complementation between the acidic residues of the CD3ε subunit and the basic cavity also argues in favor of the CD3ε subunit occupying this site. The initial hypothesis regarding the proximity of CD3ε and the Cβ FG loop was confirmed here by a set of competition assays in which the 2C11 and H57 mAbs, which bind to epitopes on CD3ε and TCR-β, respectively, were able to alter each other's binding capacity. We further showed that although there are two CD3ε subunits per TCR-β subunit, only one of the CD3ε components is in close proximity to the Cβ FG loop. Given that the γ/δ-TCR includes CD3 components (46) and is predicted to contain an equivalent insertion in the corresponding constant domain loop (47) analogous to the Cβ FG loop, we can reasonably predict that a cavity with a comparable arrangement for CD3ε exists in γ/δ T cells.
If the TCR C module cavity associates with one CD3ε, where is the second CD3ε located? In vitro translation studies have shown that when CD3ε is translated alone it tends to form disulfide-linked oligomers. However, using more physiological TCR-α/β–CD3 expression conditions, namely simultaneous cotranslation of all the TCR subunits, the cotranslation of CD3γ or CD3δ was sufficient to keep CD3ε in a monomeric state (48). Both CD3γ and CD3δ compete for binding to CD3ε (49). In addition, it was shown that the CD3δ/ε pair associates with TCR-α, whereas the CD3γ/ε pair associates with TCR-β. This association takes place upon formation of an intrachain disulfide bridge between TCR-α and -β (10). Additional studies have shown that CD3ε and TCR-β pair via their ectodomains, whereas the association of the other CD3 components with TCR-α and -β is largely mediated by interactions within their transmembrane regions. The proximity of TCR-β and CD3γ has been suggested by cross-linking experiments in humans (50) and mice (7). Based on these findings, it is likely that the CD3ε subunit that is physically associated with the Cβ FG loop is paired with CD3γ and that the second CD3ε, which pairs with the extracellular domain of the CD3δ subunit, associates via its transmembrane domain with the TCR-α (51). The ability of the CD3ε/γ dimer–specific mAb 7D6 (52) to partially inhibit FITC-H57 binding to T cells is also consistent with this view (data not shown).
An interesting feature of the potential interaction of CD3ε within the TCR C domain module relates to certain critical Cα residues that are preserved in pre-Tα (pTα), a 30-kD glycoprotein whose expression is restricted to early CD4−CD8− DN T lineage cells (53–55). The pre-TCR, a disulfide-linked heterodimer of a functionally rearranged TCR-β chain and the pTα chain, noncovalently associated with the CD3 components is expressed in DN immature thymocytes (56). During development, the DN to DP thymocyte transition is induced by an as yet unknown ligand that binds to the pre-TCR, resulting in expression of mature type α/β TCR heterodimers on DP thymocytes (57– 59). Although there is only 12% identity between pTα and Cα in humans and mice (60), the Cα residues involving important polar interactions with the Cβ domain are all conserved in the pTα sequence. Given that conserved structural elements account for close to half of the pTα–Cα amino acid sequence identities, it is very obvious that the heterodimer interface of pTα–Cβ will be extremely similar to that of Cα–Cβ. With this in mind and since genetic disruption of the CD3ε gene results in T cell development blockage at the DN stage, it is likely that the CD3ε subunit is accommodated in the pre-TCR module in a manner similar to its mode in the mature α/β-TCR.
One can imagine that certain mouse CD3ε residues contribute to the contacts made between H57 and the TCR. In this regard, the total buried molecular surface area at the N15 TCR-H57 Fab interaction is 1460A2 (720A2 for N15 and 740A2 for H57) (33), consistent with the range of values observed for other intact protein antigen–Fab interactions (61–63). However, one unusual feature of the N15– H57 interaction is the predominance of contacts made by the light chain (430A2 buried surface) rather than the heavy chain (310A2 buried surface) (33). This unusual characteristic led us to postulate that the mAb H57 heavy chain may interact primarily with TCR Cβ but in addition with some CD3ε residues. In agreement with that notion is the large rotational mobility (∼20°) of the Fab relative to the TCR Cβ FG loop observed in the two independent copies of the TCR–Fab complexes in the asymmetric unit of the crystal (33). Nevertheless, analysis of the hemizygous hCD3εtg mice revealed that although those T cells contain only half the surface copy number of the mouse CD3ε, H57 affinity is unaltered. This result suggests that the assembly of the TCR complex is not affected by the expression of the human CD3ε, and that the extracellular domain of the human CD3ε subunit can fit into the mouse TCR C domain module cavity (Fig. 3 B). Given that the extracellular domain of the mouse and human CD3ε share 53% identity in their amino acid sequence, it is possible that any putative CD3ε residues that interact with the H57 heavy chain are conserved among mouse and human CD3ε.
Although the precise functional role of the FG loop in Cβ is far from clear, it is likely to have an important impact on signaling and/or TCR assembly and structure. The unique FG loop is conserved among sequences of mouse, rat, human, and rabbit TCRs (39). Although absent in TCRs sequences from some other species, it is replaced by a potential glycosylated addition site whose glycan may serve a similar structural function (33). Undoubtedly this loop influences the mobility and disposition of Vβ relative to Cβ domains. Although the minihydrophobic patch of this loop as resolved from the crystal structure fixes the overall loop orientation relative to the Vβ and Cβ domains, local movements are permitted (33). How, if at all, peptide–MHC interactions might transfer information to the CD3 signaling subunits by affecting the FG Cβ loop remains to be tested experimentally.
Acknowledgments
We thank Drs. Linda K. Clayton and Raute Sunder-Plassmann for helpful comments on the manuscript and Dr. Cox Terhorst for generous provision of the human CD3ε transgenic mice.
This work was supported by National Institute of Health grants AI-19807 (to E.L. Reinherz) and AI-39098 (to H.-C. Chang).
Abbreviations used in this paper
- DN
double negative
- DP
double positive
- mFI
mean fluorescence intensity
- pTα
pre-Tα
- tg
transgenic
- WT
wild-type
References
- 1.Meuer SC, Acuto O, Hercend T, Schlossman SF, Reinherz EL. The human T-cell receptor. Annu Rev Immunol. 1984;2:23–50. doi: 10.1146/annurev.iy.02.040184.000323. [DOI] [PubMed] [Google Scholar]
- 2.Marrack P, Kappler J. The antigen-specific, major histocompatibility complex–restricted receptor on T cells. Adv Immunol. 1986;38:1–30. doi: 10.1016/s0065-2776(08)60005-x. [DOI] [PubMed] [Google Scholar]
- 3.Clevers H, Alarcon B, Wileman T, Terhorst C. The T cell receptor/CD3 complex: a dynamic protein ensemble. Annu Rev Immunol. 1988;6:629–662. doi: 10.1146/annurev.iy.06.040188.003213. [DOI] [PubMed] [Google Scholar]
- 4.Davis MM, Bjorkman PJ. T-cell antigen receptor genes and T-cell recognition. Nature. 1988;334:395–402. doi: 10.1038/334395a0. [DOI] [PubMed] [Google Scholar]
- 5.Ashwell JD, Klausner RD. Genetic and mutational analysis of the T-cell antigen receptor. Annu Rev Immunol. 1990;8:139–167. doi: 10.1146/annurev.iy.08.040190.001035. [DOI] [PubMed] [Google Scholar]
- 6.Alarcon B, Berkhout B, Breitmeyer J, Terhorst C. Assembly of the human T cell receptor–CD3 complex takes place in the endoplasmic reticulum and involves intermediary complexes between the CD3-γ -δ -ε core and single T cell receptor α and β chains. J Biol Chem. 1988;263:2953–2961. [PubMed] [Google Scholar]
- 7.Koning F, Maloy WL, Coligan JE. The implications of subunit interactions for the structure of the T cell receptor–CD3 complex. Eur J Immunol. 1990;20:299–305. doi: 10.1002/eji.1830200211. [DOI] [PubMed] [Google Scholar]
- 8.Manolios N, Letourneur F, Bonifacino JS, Klausner RD. Pairwise, cooperative and inhibitory interactions describe the assembly and probable structure of the T-cell antigen receptor. EMBO (Eur Mol Biol Organ) J. 1990;10:1643–1651. doi: 10.1002/j.1460-2075.1991.tb07687.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baniyash M, Garcia-Morales P, Bonifacino JS, Samelson LE, Klausner RD. Disulfide linkage of the ζ and η chains of the T cell receptor. J Biol Chem. 1989;263:9874–9878. [PubMed] [Google Scholar]
- 10.de la Hera A, Muller U, Olsson C, Isaaz S, Tunnacliffe A. Structure of the T cell antigen receptor (TCR): two CD3ε subunits in a functional TCR/CD3 complex. J Exp Med. 1991;173:7–17. doi: 10.1084/jem.173.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Punt JA, Roberts JL, Kearse KP, Singer A. Stoichiometry of the T cell antigen receptor (TCR) complex: each TCR/CD3 complex contains one TCR α, one TCR β, and two CD3ε chains. J Exp Med. 1994;180:587–593. doi: 10.1084/jem.180.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reth M. Antigen receptor tail clue. Nature. 1989;338:383–384. [PubMed] [Google Scholar]
- 13.Cambier JC. Antigen and Fc receptor signaling. The awesome power of the immunoreceptor tyrosine-based activation motif (ITAM) J Immunol. 1995;155:3281–3285. [PubMed] [Google Scholar]
- 14.Letourneur F, Klausner RD. Activation of T cells by a tyrosine kinase activation domain in the cytoplasmic tail of CD3ε. Science. 1992;255:79–82. doi: 10.1126/science.1532456. [DOI] [PubMed] [Google Scholar]
- 15.Ravichandran KS, Lee KK, Songyang Z, Cantley LC, Burn P, Burakoff SJ. Interaction of Shc with the ζ chain of the T cell receptor upon T cell activation. Science. 1993;262:902–905. doi: 10.1126/science.8235613. [DOI] [PubMed] [Google Scholar]
- 16.Exley M, Varticovski L, Peter M, Sancho J, Terhorst C. Association of phosphatidylinositol 3-kinase with a specific sequence of the T cell receptor ζ chain is dependent on T cell activation. J Biol Chem. 1994;269:15140–15146. [PubMed] [Google Scholar]
- 17.Isakov N, Wange RL, Burgess WH, Watts JD, Aebersold R, Samelson LE. ZAP-70 binding specificity to T cell receptor tyrosine-based activation motifs: the tandem SH2 domains of ZAP-70 bind distinct tyrosine-based activation motifs with varying affinity. J Exp Med. 1995;181:375–380. doi: 10.1084/jem.181.1.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Osman N, Turner H, Lucas S, Reif K, Cantrell DA. The protein interactions of the immunoglobulin receptor family tyrosine-based activation motifs present in the T cell receptor ζ subunits and the CD3 γ, δ and ε chains. Eur J Immunol. 1996;26:1063–1068. doi: 10.1002/eji.1830260516. [DOI] [PubMed] [Google Scholar]
- 19.Sunder-Plassmann R, Lialios F, Madsen M, Koyasu S, Reinherz EL. Functional analysis of immunoreceptor tyrosine-based activation motif (ITAM)–mediated signal transduction: the two YxxL segments within a single CD3ζ-ITAM are functionally distinct. Eur J Immunol. 1997;27:2001–2009. doi: 10.1002/eji.1830270826. [DOI] [PubMed] [Google Scholar]
- 20.Sussman JJ, Bonifacino JS, Lippincott-Schwartz J, Weissman AM, Saito T, Klausner RD, Ashwell JD. Failure to synthesize the T cell ζ chain: structure and function of a partial T cell receptor complex. Cell. 1988;52:85–95. doi: 10.1016/0092-8674(88)90533-8. [DOI] [PubMed] [Google Scholar]
- 21.Hall C, Berkhout B, Alarco J, Sancho T, Wileman T, Terhorst C. Requirements for cell surface expression of the human TCR/CD3 complex in non-T cells. Int Immunol. 1991;3:359–368. doi: 10.1093/intimm/3.4.359. [DOI] [PubMed] [Google Scholar]
- 22.Shinkai Y, Alt FW. CD3ε-mediated signals rescue the development of CD4+CD8+ thymocytes in RAG-2−/−mice in the absence of TCR β chain expression. Int Immunol. 1994;6:995–1001. doi: 10.1093/intimm/6.7.995. [DOI] [PubMed] [Google Scholar]
- 23.Levelt CN, Mombaerts P, Iglesias A, Tonegawa S, Eichmann K. Restoration of early thymocyte differentiation in T cell receptor β-chain–deficient mutant mice by transmembrane signaling through CD3ε. Proc Natl Acad Sci USA. 1993;90:11401–11405. doi: 10.1073/pnas.90.23.11401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kappes DJ, Tonegawa S. Surface expression of alternative forms of the TCR/CD3 complex. Proc Natl Acad Sci USA. 1991;88:10619–10623. doi: 10.1073/pnas.88.23.10619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu C-P, Ueda R, She J, Sancho J, Wang B, Weddell G, Loring J, Kurahara C, Dudley EC, Hayday A, et al. Abnormal T cell development in CD3ζ−/−mutant mice and identification of a novel T cell population in the intestine. EMBO (Eur Mol Biol Organ) J. 1993;12:4863–4875. doi: 10.1002/j.1460-2075.1993.tb06176.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Love PE, Shores EW, Johnson MD, Tremblay ML, Lee EJ, Grinberg A, Huang SP, Singer A, Westphal H. T cell development in mice that lack the ζ chain of the T cell antigen receptor complex. Science. 1993;261:918–921. doi: 10.1126/science.7688481. [DOI] [PubMed] [Google Scholar]
- 27.Malissen M, Gillet A, Rocha B, Trucy J, Vivier E, Boyer C, Koentgen F, Brun N, Mazza G, Spanopoulou E, et al. T cell development in mice lacking the CD3ζ/η gene. EMBO (Eur Mol Biol Organ) J. 1993;12:4347–4355. doi: 10.1002/j.1460-2075.1993.tb06119.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohno H, Aoe T, Taki S, Kitamura D, Ishida Y, Rajewsky K, Saito T. Developmental and functional impairment of T cells in mice lacking CD3ζ chains. EMBO (Eur Mol Biol Organ) J. 1993;12:4357–4366. doi: 10.1002/j.1460-2075.1993.tb06120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malissen M, Gillet A, Ardouim L, Bouvier G, Trucy J, Ferrier P, Vivier E, Malissen B. Altered T cell development in mice with a targeted mutation of the CD3ε gene. EMBO (Eur Mol Biol Organ) J. 1995;14:4641–4653. doi: 10.1002/j.1460-2075.1995.tb00146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dave VP, Cao Z, Browne C, Alarcon B, Fernandez-Miguel G, Lafaille J, de la Hera A, Tonegawa S, Kappes DJ. CD3δ deficiency arrests development of the αβ but not the γδ T cell lineage. EMBO (Eur Mol Biol Organ) J. 1997;16:1360–1370. doi: 10.1093/emboj/16.6.1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shibata K-I, Imarai M, van Bleek GM, Joyce S, Nathenson SG. Vesicular stomatitis virus antigenic octapeptide N52-59 is anchored into the groove of the H-2Kbmolecule by the side chains of three amino acids and the main-chain atoms of the amino terminus. Proc Natl Acad Sci USA. 1992;89:3135–3139. doi: 10.1073/pnas.89.7.3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kubo RT, Born W, Kappler JW, Marrack P, Pigeon M. Characterization of a monoclonal antibody which detects all murine αβ T cell receptors. J Immunol. 1989;142:2736–2742. [Google Scholar]
- 33.Wang J, Lim K, Smolyar A, Teng M, Liu J, Tse AG, Liu J, Hussey RE, Chishti Y, Thomson CT, et al. Atomic structure of an αβ T cell receptor (TCR) heterodimer in complex with an anti-TCR Fab fragment derived from a mitogenic antibody. EMBO (Eur Mol Biol Organ) J. 1998;17:10–26. doi: 10.1093/emboj/17.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leo O, Foo M, Sachs DH, Samelson LE, Bluestone JA. Identification of a monoclonal antibody specific for a murine T3 polypeptide. Proc Natl Acad Sci USA. 1987;84:1374–1378. doi: 10.1073/pnas.84.5.1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang B, Biron C, She J, Higgins K, Sunshine M-J, Lacy E, Lonberg N, Terhorst C. A block in both early T lymphocyte and natural killer cell development in transgenic mice with high-copy numbers of the human CD3ε gene. Proc Natl Acad Sci USA. 1994;91:9402–9406. doi: 10.1073/pnas.91.20.9402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kraulis PJ. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structure. J Appl Cryst. 1991;24:946–950. [Google Scholar]
- 37.Merritt, E.A., and M.E.P. Murphy. 1994. Raster3D version 2.0.-A program for protorealistic molecular graphics. Acta Cryst. D50:869–873. [DOI] [PubMed]
- 38.Staerz UD, Rammensee HG, Benedotto JD, Bevan MJ. Characterization of murine monoclonal antibody specific for an allotype determinant on T cell antigen receptor. J Immunol. 1985;134:3994–4000. [PubMed] [Google Scholar]
- 39.Kabat, E.A., T.-T. Wu, H.M. Perry, K.S. Gottesman, and C. Foeller. 1991. Sequences of proteins of immunological interest. US Department of Health and Human Services, DHS, NIH, Bethesda, MD.
- 40.Gold DP, Clevers H, Alarcon B, Dunlap S, Novotny J, Williams AF, Terhorst C. Evolutionary relationship between the T3 chains of the T cell receptor complex and the immunoglobulin supergene family. Proc Natl Acad Sci USA. 1987;84:7649–7653. doi: 10.1073/pnas.84.21.7649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Transy C, Moingeon P, Stebbins C, Reinherz EL. Deletion of the cytoplasmic region of the CD3ε subunit does not prevent assembly of functional T-cell receptor. Proc Natl Acad Sci USA. 1989;86:7108–7112. doi: 10.1073/pnas.86.18.7108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reinherz EL, Meuer S, Fitzgerald KA, Hussey RE, Levine H, Schlossman SF. Antigen recognition by human T lymphocytes is linked to surface expression of the T3 molecular complex. Cell. 1982;30:735–743. doi: 10.1016/0092-8674(82)90278-1. [DOI] [PubMed] [Google Scholar]
- 43.Meuer SC, Fitzgerald KA, Hussey RE, Hodgdon JC, Schlossman SF, Reinherz EL. Clonotypic structures involved in antigen-specific human T cell function: relationship to the T3 molecular complex. J Exp Med. 1983;157:705–719. doi: 10.1084/jem.157.2.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Christopher Garacia, K., M. Degano, R.L. Stanfield, A. Brunmark, M.R. Jackson, P.A. Peterson, L. Teyton, and I.A. Wilson. An αβ T cell receptor structure at 2.5Å and its orientation in the TCR-MHC complex. Science. 1996;274:209–219. [Google Scholar]
- 45.Garboczi DN, Ghosh P, Utz U, Fan Q-R, Biddison WE, Wiley DC. Structure of the complex between human T-cell receptor, viral peptide and HLA-A2. Nature. 1996;384:134–141. doi: 10.1038/384134a0. [DOI] [PubMed] [Google Scholar]
- 46.van Neerven J, Coligan JE, Koning F. Structural comparison of αβ and γδ T cell receptor–CD3 complexes reveals identical subunit interactions but distinct crosslinking patterns of T cell receptor chains. Eur J Immunol. 1990;20:2105–2111. doi: 10.1002/eji.1830200932. [DOI] [PubMed] [Google Scholar]
- 47.Bentley GA, Boulot G, Kajalainen K, Mariuzza RA. Crystal structure of the β chain of a T cell antigen receptor. Science. 1995;267:1984–1987. doi: 10.1126/science.7701320. [DOI] [PubMed] [Google Scholar]
- 48.Huppa JB, Ploegh HL. In vitro translation and assembly of a complete T cell receptor–CD3 complex. J Exp Med. 1997;186:393–403. doi: 10.1084/jem.186.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Geisler C. Failure to synthesize the CD3γ chain. Consequences for the T cell antigen receptor assembly, processing, and expression. J Immunol. 1992;148:2437–2445. [PubMed] [Google Scholar]
- 50.Brenner MB, Trowbridge IS, Strominger JL. Cross-linking of human T cell receptor proteins: association between the cell idiotype β subunit and the T3 glycoprotein heavy subunit. Cell. 1985;40:183–190. doi: 10.1016/0092-8674(85)90321-6. [DOI] [PubMed] [Google Scholar]
- 51.Manolios N, Kemp O, Li Z-G. The T cell antigen receptor α and β chains interact via distinct regions with CD3 chain. Eur J Immunol. 1994;24:84–92. doi: 10.1002/eji.1830240114. [DOI] [PubMed] [Google Scholar]
- 52.Coulie PG, Uyttenhove C, Wauters P, Manolios N, Klausner RD, Samelson LE, Van Snick J. Identification of a murine monoclonal antibody specific for an allotypic determinant on mouse CD3. Eur J Immunol. 1991;21:1703–1709. doi: 10.1002/eji.1830210718. [DOI] [PubMed] [Google Scholar]
- 53.Saint-Ruf C, Ungewiss K, Groettrup M, Bruno L, Fehling HJ, von Boehmer H. Analysis and expression of a cloned pre-T cell receptor gene. Science. 1994;266:1208–1212. [PubMed] [Google Scholar]
- 54.Ramiro AR, Trigueros C, Marquez C, San JL, Millan, Toribio ML. Regulation of pre-T cell receptor (pTα-TCRβ) gene expression during human thymic development. J Exp Med. 1996;184:519–530. doi: 10.1084/jem.184.2.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bruno L, Rocha B, Rolink A, von Boehmer H, Rodewald H-R. Intra- and extra-thymic expression of the pre-T cell receptor α gene. Eur J Immunol. 1995;25:1877–1882. doi: 10.1002/eji.1830250713. [DOI] [PubMed] [Google Scholar]
- 56.Wilson A, MacDonald HR. Expression of genes encoding the pre-TCR and CD3 complex during thymus development. Int Immunol. 1995;7:1659–1664. doi: 10.1093/intimm/7.10.1659. [DOI] [PubMed] [Google Scholar]
- 57.Shinkai Y, Koyasu S, Nakayama K, Murphy KM, Loh DY, Reinherz EL, Alt FW. Restoration of T cell development in RAG-2–deficient mice by functional TCR transgenes. Science. 1993;259:822–825. doi: 10.1126/science.8430336. [DOI] [PubMed] [Google Scholar]
- 58.Levelt CN, Wang B, Ehrfeld A, Terhorst C, Eichmann K. Regulation of T cell receptor (TCR)–β locus allelic exclusion and initiation of TCR-α locus rearrangement in immature thymocytes by signaling through the CD3 complex. Eur J Immunol. 1995;25:1257–1261. doi: 10.1002/eji.1830250519. [DOI] [PubMed] [Google Scholar]
- 59.Koyasu S, Clayton LK, Lerner A, Heiken H, Parkes A, Reinherz EL. Pre-TCR signaling components trigger transcriptional activation of a rearranged TCRα gene locus and silencing of the pTα locus: implication for intrathymic differentiation. Int Immunol. 1997;9:1475–1480. doi: 10.1093/intimm/9.10.1475. [DOI] [PubMed] [Google Scholar]
- 60.Del Porto P, Bruno L, Mattei M-G, von Boehmer H, Saint-Ruf C. Cloning and comparative analysis of the human pre-T-cell receptor α-chain gene. Proc Natl Acad Sci USA. 1995;92:12105–12109. doi: 10.1073/pnas.92.26.12105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stanfield RL, Takimoto-Kamimura M, Rini JM, Profy AT, Wilson IA. Major antigen-induced domain rearrangements in an antibody. Structure. 1993;1:83–93. doi: 10.1016/0969-2126(93)90024-b. [DOI] [PubMed] [Google Scholar]
- 62.Wilson IA, Stanfield RL. Antibody-antigen interactions. Curr Opin Sruct Biol. 1993;3:113–118. doi: 10.1016/0959-440x(94)90267-4. [DOI] [PubMed] [Google Scholar]
- 63.Padlan EA. Anatomy of the antibody molecule. Mol Immunol. 1994;31:169–217. doi: 10.1016/0161-5890(94)90001-9. [DOI] [PubMed] [Google Scholar]