Abstract
Protective immunity to infection by many intracellular pathogens requires recognition by cytotoxic T lymphocytes (CTLs) of antigens presented on major histocompatibility complex (MHC) class I molecules. To be presented for recognition by pathogen-specific CTLs, these antigens must gain access to the host cell class I processing pathway. In the case of intracellular bacterial pathogens, the majority of bacterial proteins are retained within the bacterial membrane and therefore remain inaccessible to the host cell for antigen processing. We have isolated a CTL clone from a C57BL/6 mouse infected with the intracellular gram-positive bacterium Listeria monocytogenes (LM) and have identified the source of the antigen. Using a genomic expression library, we determined that the clone recognizes an antigenic N-formyl peptide presented by the nonpolymorphic murine MHC class Ib molecule, H2-M3. Several lengths of this peptide were able to sensitize cells for lysis by this CTL clone. The source of this antigenic peptide is a 23–amino acid polypeptide encoded at the start of a polycistronic region. Analysis of mRNA secondary structure of this region suggests that this polypeptide may be a leader peptide encoded by a transcriptional attenuator.
Keywords: Listeria, H2-M3, cytotoxic T lymphocyte, antigen, leader peptide
Intracellular pathogens, including viruses and bacteria such as Mycobacterium tuberculosis, Salmonella typhimurium, and Listeria monocytogenes (LM),1 are major causes of infectious diseases. As they grow and replicate within permissive host cells, they are protected from the humoral immune response (1, 2). However, infected cells can be recognized and destroyed by CD8+ CTLs (3). This population of effector cells prevents the replication and spread of the pathogen and allows for clearance by other immune effectors, such as antibodies or nonpermissive phagocytic immune cells.
CD8+ CTLs recognize antigens that are produced by the infecting organism and then processed by the host cell and presented by MHC class I molecules. Pathogen-derived proteins able to access the cytosol are degraded into peptides by proteasomes and transported via the transporter associated with antigen processing into the endoplasmic reticulum, where they associate with nascent class I molecules. In viral infections, host cell ribosomes are used for protein production; therefore, viral proteins are directly exposed to the class I processing pathway. However, bacterial proteins are produced within the bacterial membrane and are sequestered from the host cell cytosol. Thus, the bacterial products available for presentation by MHC class I molecules are confined to those that are secreted or shed by the bacteria (4). To understand which antigens are able to elicit CTL-mediated immune responses to intracellular bacteria, it is necessary to identify those molecules that enter the host cell cytosol and, therefore, the MHC class I processing pathway.
The MHC class I molecules are divided into two subgroups: the polymorphic class Ia molecules, which present peptides of eight or nine amino acids in length, and the nonpolymorphic class Ib molecules, which present peptides as well as nonpeptide ligands (5). Two of these MHC class Ib molecules are thought to have evolved to present hydrophobic antigens derived from intracellular bacterial pathogens (6, 7). Human CD1, for example, is known to bind and present several glycolipid moieties derived from mycobacterial cell wall components (8, 9). The murine class Ib molecule, H2-M3, has a high affinity for peptides that contain an N-formyl methionine as the NH2-terminal amino acid (10, 11). In prokaryotes, the initial methionine of each protein is formylated. In eukaryotes, only proteins from mitochondria and chloroplasts initiate with N-formyl methionine. Therefore, H2-M3 preferentially presents antigens derived from intracellular bacteria (12).
The gram-positive bacterium LM is an intracellular pathogen that infects hepatocytes, endothelial cells, and permissive (nonbacteriocidal) macrophages. Upon infection of the host cell, secretion by LM of the hemolysin listeriolysin O (LLO) results in lysis of the phagosomal membrane and entry of the bacteria into the host cell cytosol (13). This cytosolic localization facilitates entry of antigenic molecules released from the bacteria into the host cell class I processing pathway (14). Thus, LM is well suited for the study of MHC class I–restricted presentation of antigens derived from intracellular bacterial pathogens (15).
CD8+ CTLs are necessary for protective immunity against infection with LM (16, 17). Adoptive transfer of spleen cells from mice immunized with a sublethal dose of LM confers protection to naive animals (18–20). In some cases, immunity can be transferred between mice of disparate MHC haplotypes, suggesting antigen presentation by nonpolymorphic MHC molecules (21, 22). Subsequent studies have demonstrated that H2-M3 is able to present LM antigens to CTL clones isolated from LM-infected mice (6, 23). To date, two LM-derived peptides presented by H2-M3 have been identified. One is a pentapeptide that was identified by tandem mass spectrometry of LM bacterial culture supernatant preparations (24). It has the sequence f-MIVIL, and the protein from which it is derived has not yet been identified. The sequence of the second peptide was elucidated by screening an LM genomic expression library for production of antigenic activity (25). Genomic inserts coding for activity were sequenced, and a peptide of six or seven amino acids in length with the sequence f-MIGWII(A) was identified as the active peptide. A novel membrane protein, called LemA, was identified as the source of the MIGWII(A) peptide.
Since antigenic peptides presented by MHC class I molecules are derived generally from cytosolic proteins, it would be expected that bacterial products that efficiently gain access to the host cell cytosol would be preferentially presented (4). LM resides in the host cell cytosol, and indeed, all of the identified peptides from LM that are presented by MHC class Ia molecules are products of secreted proteins. Four epitopes presented on the murine class Ia H-2Kd molecule have been identified. They are derived from LLO (26), p60 (27, 28), and a metalloprotease (29), all of which are proteins secreted by LM. The protein source of the f-MIVIL peptide is unknown; however, it is present in bacterial culture supernatants, indicating that it is secreted or shed by the bacteria (23, 24). LemA is believed to be a bacterial membrane protein with an Nout–Cin topology; thus, the H2-M3–presented epitope is on the outside surface of the bacterial membrane and is exposed to the host cell cytosol. Identification of additional bacterial antigens presented by MHC class I molecules and the proteins from which they are derived will establish whether exposure to the host cell cytosol is a common feature shared by these antigens. In addition, characterization of the nature and source of antigenic peptides presented by nonpolymorphic MHC class Ib molecules may enable the development of vaccine strategies that circumvent the problem of MHC restriction encountered in outbred populations.
In this study, we describe an LM-derived N-formyl peptide presented by H2-M3. Using a genomic LM expression library, we determined the sequence of this peptide. Several lengths of this peptide are recognized by an H2-M3– restricted CTL clone, with the hexameric peptide having activity at concentrations in the femtomolar range. In addition, we have determined that this peptide is derived from a 23–amino acid leader polypeptide. This polypeptide is encoded at the start of a polycistronic mRNA region and may be a by-product of transcriptional attenuation.
Materials and Methods
Mice and Cell Lines.
C57BL/6, BALB/c, and DBA/2 mice were purchased from The Jackson Laboratory (Bar Harbor, ME). Cell lines were cultured at 37°C in 7% CO2 in IMDM (Sigma Chemical Co., St. Louis, MO) supplemented with 5 mM Hepes, 2 mM glutamine, 1 mM hydroxypyruvate, 50 mM 2-mercaptoethanol, nonessential amino acids, 100 IU/ml penicillin, 100 mg/ml streptomycin, 50 mg/ml gentamycin, and 10% fetal bovine serum (cIMDM; Intergen Co., Purchase, NY). Serum-free IMDM (sfIMDM) is identical to cIMDM in formulation, except BSA (Sigma Chemical Co.) 0.5% (wt/vol) was substituted for fetal bovine serum as a source of protein. BMC2.3 is a C57BL/ 6-derived H-2b macrophage–like cell line obtained from Dr. Kenneth Rock (Harvard University, Boston, MA). EL4 is a C57BL/6-derived H-2b thymoma cell line. P815 is a DBA/2-derived H-2d mastocytoma cell line. M12.C3 is a BALB/c-derived H-2d lymphoma cell line. MC.12 is an H-2d macrophage–like cell line. SVCAS2F6 is a fibroblast cell line that expresses a mutant (null) allele of H2-M3, and 13S2 is the SVCAS2F6 line that has been transfected with a chimeric MHC class I molecule containing the cytoplasmic, transmembrane, and α3 domains from H-2Ld and the α1 and α2 domains from wild-type H2-M3 (30). L9.6 is a C57BL/6 × BALB/c F1–derived CD8+ CTL clone that recognizes the murein hydrolase p60 epitope 217-225 presented by H-2Kd and obtained from Dr. Eric Pamer (Yale University, New Haven, CT). Recombinant murine GM-CSF (rmuGM-CSF) was produced from cultured Chinese hamster ovary (CHO) cells transfected with the murine GM-CSF gene (gift of Dr. Steven Dow, National Jewish Medical and Research Center, Denver, CO). Approximately 2–5 × 106 CHO/GM-CSF cells were cultured in 100 ml cIMDM in 75 cm2 tissue culture flasks for 4–5 d at 37°C in 7% CO2. Culture supernatant was harvested and used as a source of rmuGM-CSF for culture of primary bone marrow macrophages. Primary bone marrow macrophages were grown in six-well tissue culture plates in 5 ml cIMDM/well supplemented with 8% rmuGM-CSF/CHO cell culture supernatant.
Peptide Synthesis.
Peptides were synthesized using standard solid phase Fmoc chemistry on a peptide synthesizer (model 433A; Applied Biosystems, Foster City, CA). Protecting groups were removed from resin-bound amino acid residues using hydrogen fluoride. Fmoc amino acids were purchased from AnaSpec, Inc. (San Jose, CA), and N-formyl methionine was purchased from Bachem Bioscience Inc. (King of Prussia, PA). Peptides were purified by reverse-phase HPLC on a Luna 10μ C18(2) 250 × 2.12 mm column (Phenomenex, Inc., Torrance, CA) using an HPLC pump (model 510; Waters Associates, Milford, MA). Integrity of the peptides was confirmed using a Voyager reverse-phase matrix-assisted laser desorption ionization time-of-flight mass spectrometer (PerSeptive Biosystems, Framingham, MA). Peptides were dissolved in 100% DMSO, aliquoted, and stored at −20°C. Stock solutions were diluted in PBS as needed.
CTL Clones and Assays.
CTL clones were generated by immunizing 8–10-wk-old female C57BL/6 mice i.p. with 104 CFU LM (serotype 1/2a). Spleen and lymph node cells were harvested on day 8 after infection. Erythrocytes were lysed with Tris-buffered ammonium chloride (Sigma Chemical Co.), and spleen and lymph node cells were cultured in flat-bottomed 24-well plates in 2 ml cIMDM/well. Spleen and lymph node cells, 2 × 105/well, were restimulated with 2 × 104 irradiated (18,000 rads) LM-infected MC.12 or BMC2.3 cells/well in the presence of 4 × 106 irradiated (3,000 rads) C57BL/6 spleen cells/well. The MC12 and BMC2.3 cell lines were infected with LM at a multiplicity of infection (MOI) of 10:1 for 3 h before establishing the restimulation culture. Responder cells were cloned by limiting dilution in flat-bottomed 96-well plates after 1 wk in culture. CTL clones were restimulated every 7–8 d in 24-well plates and cultured in cIMDM supplemented with 4% supernatant from concanavalin A–stimulated rat spleen cells.
Target cells were labeled with [51Cr]NaCr2O4 for 1–1.5 h. Target cells that were to be infected with LM were cultured in antibiotic-free cIMDM for a minimum of 72 h before assay. LM from a log phase culture were washed three times in PBS, then added to target cells at an MOI of 10:1. Gentamycin sulfate (Sigma Chemical Co.) at a final concentration of 10 μg/ml was added 30 min after infection to kill extracellular bacteria. LM culture supernatants and peptides were incubated with target cells for 1–2 h at 37°C before addition of effectors. Peptide titration curves for N-formyl and nonformylated peptides were performed in sfIMDM to eliminate peptide degradation by serum proteases.
CTL assays were performed in 96-well round-bottomed plates. Target cells were plated at 104 cells/well in 100 μl cIMDM, and CTLs were added at the indicated E/T ratio in 100 μl cIMDM. Plates were centrifuged for 3 min at 800 g, then incubated for 4 h at 37°C in 7% CO2. Plates were centrifuged for 3 min at 1,500 g, and 100 μl of medium/well was collected and counted for 1 min on an automatic γ counter. All assays were performed in triplicate. Spontaneous release was determined from wells containing target cells in cIMDM with no effector cells. Total release was determined from wells containing target cells in cIMDM plus 1% Triton-X 100 (Sigma Chemical Co.) without effector cells. Percent specific release was calculated as follows: [(cpm released in the presence of effector cells − cpm of nonspecific release)/(cpm of total release − cpm of nonspecific release)] × 100.
Culture of LM and E. coli Containing the LM Genomic Library.
The virulent LM strain used in these studies was EGD serotype 1/2a. LM were cultured in brain heart infusion broth (BHI; Difco Laboratories Inc., Detroit, MI) at 37°C on a rotary shaker at 200 rpm. The LM genomic expression library was the gift of L.L. Lenz and M.J. Bevan (University of Washington, Seattle, WA), and its construction has been described previously (25). Individual LM genomic clones in E. coli DH10β were grown in 0.2 ml of BHI supplemented with 100 μg/ml spectinomycin in 96-well flat-bottomed plates at 37°C for 24 h. Bacteria were lysed by three rounds of freezing at −80°C and thawing at 37°C. Bacterial lysate was added at 10% to individual wells containing 51Cr-labeled BMC2.3 for 1 h, then CTLs were added at an E/T ratio of 5:1. Target cells were assayed for specific lysis as described. All positive transformants identified were grown in 3 ml BHI for 24 h and retested to confirm the presence of antigen. It was found during these experiments that incubation of target cells with bacterial lysates substantially increased the spontaneous release. Supernatants from positive cultures contained levels of antigenic activity comparable with bacterial lysates, but were less toxic to the target cells. After the initial round of screening, transformed bacteria were no longer subjected to freeze-thawing, and culture supernatants were tested directly in 51Cr release assays.
DNA Sequencing and PCR Screening.
Restriction mapping of plasmid DNA from positive transformants was performed using restriction endonucleases from Promega Corp. (Madison, WI) according to the manufacturer's instructions. LM genomic DNA was sequenced initially using the M13 forward primer 5′-GTAAAACGACGGCCAG and the M13 reverse primer 5′-CAGGAAACAGCTATGAC. Sequencing data thus generated were used to design additional sets of internal sequencing primers. All sequencing was performed at the National Jewish Medical and Research Center core sequencing facility by dye terminator cycle sequencing on a DNA sequencer (ABI Prism 377; Perkin-Elmer Corp., Norwalk, CT).
Sequencing data were used to design PCR primer pairs that would produce overlapping DNA fragments from the genomic LM DNA insert. These PCR products were ligated directly into the pCR 2.1 cloning vector (Invitrogen Corp., Carlsbad, CA) according to the manufacturer's instructions and transformed into DH5α E. coli. Transformants were grown in 200 μl BHI with 50 μg/ml ampicillin (Sigma Chemical Co.) at 37°C for 24 h, and supernatants were screened for antigenic activity in a standard 51Cr release assay as described for the LM genomic expression library. The PCR primer pair 5′-AAGCAGAGAAAAAATGTC and 5′-CAAACGTCATTCCGGTATT amplifies a 1.64-kb insert with antigenic activity from genomic clones H6.1 and E11.4. PCR primers 5′-ACGATAGTCACAAACCGTG and 5′-ACACAATGAAAACACCCC produce a 270-bp fragment, and primers 5′-TTTAATGATTGTAACGTTA and 5′-AAATTACATACATCCCCA produce a 330-bp fragment, that both encode antigenic activity and overlap by 60 bp.
Results and Discussion
CTL Clone C10.4 Recognizes a Listeria Peptide Presented by H2-M3.
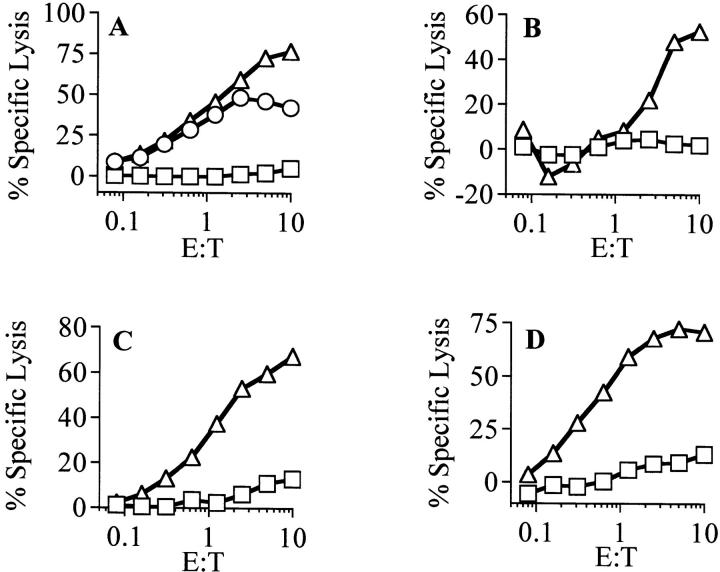
Previous studies have shown that CTLs derived from mice immunized with a sublethal dose of LM are able to lyse target cells infected with LM and, in some cases, target cells exposed to supernatant from LM cultures (3, 23). To generate LM-specific CTL clones restricted by nonpolymorphic MHC class Ib molecules, spleen and lymph node cells from LM-infected C57BL/6 (H-2b) mice were restimulated in vitro using an allogeneic (H-2d) macrophage–like cell line infected with LM. Cell lines derived from spleen and lymph node cells were tested for their ability to selectively lyse LM-infected target cells in a standard 51Cr release cytotoxicity assay. Cell lines able to lyse LM-infected allogeneic macrophage–like target cells were identified and cloned by limiting dilution. The CTL clone C10.4, isolated from the lymph nodes of an LM-infected mouse, was selected because of its ability to specifically lyse both syngeneic and allogeneic macrophage–like cell lines infected in vitro with LM. In addition, C10.4 was found to lyse the syngeneic macrophage–like cell line BMC2.3 exposed to LM culture supernatant, but not BMC2.3 incubated with uninoculated culture medium (Fig. 1 A). Using LM culture supernatant as a source of antigen, we established that C10.4 was able to lyse target cells of diverse MHC haplotypes. Target cells from H-2b, H-2d, and H-2q mice incubated with LM culture supernatant were lysed by C10.4 (Fig. 1, A–D). This lack of MHC class Ia restriction suggested two possibilities: that antigen presentation was MHC class Ia permissive or that presentation was MHC class Ib restricted.
Figure 1.
Clone C10.4 recognizes LM antigen present in bacterial culture supernatant in an MHC class Ia–unrestricted manner. (A) The macrophage-like cell line BMC2.3 (H-2b) presents antigen to CTL clone C10.4 when infected in vitro with live washed LM (open circles), MOI 10:1. BMC2.3 are also lysed when incubated with supernatant from LM cultures (open triangles). Target cells from mice with different MHC haplotypes were incubated with 48-h LM culture supernatant and then assayed for lysis by C10.4. Primary bone marrow macrophages from (B) DBA/1 (H-2q) were lysed in the presence of LM culture supernatant but not when incubated with uninoculated culture medium (open squares). The H-2d B cell lymphoma cell line M12C3 (C) and the H-2d mastocytoma cell line P815 (D) were lysed only in the presence of LM culture supernatant.
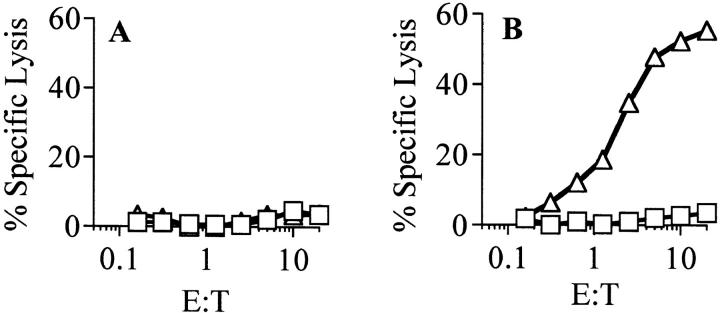
The murine MHC class Ib molecule H2-M3 has been shown to present LM-derived peptides containing an N-formyl methionine residue to CD8+ CTLs (23). Although H2-M3 is highly conserved among most mouse strains, a null allele from Mus musculus castaneus, which is unable to present antigen for recognition by H2-M3–restricted CTLs, has been described (31). A fibroblast cell line derived from the B10.CAS2 mouse, which carries this null allele, is unable to present antigens to CTLs restricted by H2-M3. The ability of this cell line to present H2-M3–restricted antigens is restored by transfection with a chimeric MHC class I molecule consisting of the α1 and α2 domains of wild-type H2-M3 and the α3 transmembrane and cytoplasmic domains of H-2Ld (30). To determine whether H2-M3 was the presenting molecule, both transfected and untransfected B10.CAS2 fibroblasts were incubated with LM culture supernatant. The C10.4 CTL clone was able to kill B10.CAS2 cells transfected with the chimeric H2-M3wt gene, but did not kill the untransfected cells (Fig. 2, A and B). Thus, the H2-M3 molecule presents the antigen from the LM culture supernatant to C10.4. Further biochemical analysis, including protease digestion of LM culture supernatant, indicated that the antigen presented by H2-M3 was proteinaceous (data not shown).
Figure 2.
Antigen presentation to C10.4 is restricted by H2-M3. The ability of C10.4 to lyse fibroblasts expressing mutant H2-M3cas2 or H2-M3wt in the presence of LM culture supernatant was tested. (A) Untransfected B10.CAS2 fibroblasts expressing the mutant H2-M3cas2 MHC class I molecule were incubated with LM culture supernatant (open triangles) or uninoculated culture medium (open squares) as a negative control. The fibroblast target cells were then assayed for lysis by C10.4. (B) B10.CAS2 fibroblasts transfected with the H2-M3wt MHC class I molecule were incubated with LM culture supernatant or uninoculated culture medium as in A, then assayed for lysis by C10.4.
Two LM-derived N-formyl peptides presented by H2-M3 to LM-specific CTL clones have been described (24, 25). Neither of these peptides was able to sensitize cells for lysis by C10.4. These peptides were also examined for their ability to competitively inhibit the antigenic activity present in LM culture. Both peptides had a half-maximal inhibitory concentration (IC50) of ∼10 nM when mixed with LM culture supernatant and tested for the ability to sensitize EL4 target cells for lysis by C10.4 (data not shown). Thus, the N-formyl LM peptides and the antigen recognized by C10.4 were presented by the same MHC class I molecule.
Isolation of Antigenic Activity Using an LM Genomic Library.
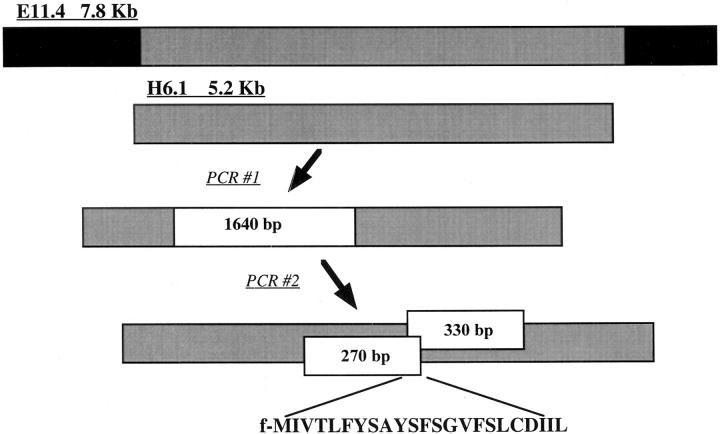
The antigen recognized by C10.4 was identified by screening an LM genomic expression library for antigenic activity. Construction of this library has been described previously (25). The library consists of approximately six LM genome equivalents cloned as 5–8-kb inserts into the plasmid expression vector pAT29 and transformed into DH10β E. coli. The library was screened by testing supernatant from individual E. coli transformants for the ability to sensitize BMC2.3 target cells for lysis by C10.4. Of 2,400 genomic clones tested, two positives were selected for further analysis. Plasmid DNA from these positive transformants was isolated and subjected to digestion with restriction endonucleases. Restriction maps of these two clones suggested they were overlapping (data not shown). These two LM DNA inserts were sequenced, and the sequence data were compared for homology. We found that the entire insert designated H6.1 was contained within the sequence of the insert designated E11.4 (Fig. 3).
Figure 3.
Strategy for cloning DNA fragments with antigenic activity from an LM genomic library. Using sequence data from LM genomic DNA inserts E11.4 and H6.1, PCR primers were designed to produce overlapping fragments of 1.5–2.0 kb. A 1.64-kb fragment was found with activity. PCR primers were designed to produce overlapping fragments contained within the 1.64-kb sequence. These PCR products were designed to be of 200–400 bp in length. Two fragments with a 60-bp overlap were found with activity. Sequence information from this 60-bp fragment was used to identify a 23–amino acid polypeptide containing an N-formyl methionine with antigenic activity.
Using these sequence data, PCR primers were designed to amplify DNA fragments of 1.5–2.0 kb from the LM inserts. These DNA fragments were designed to overlap each other by 100–200 bp and to cover the entire sequence of both LM inserts. It was found that when these PCR products were ligated directly into the pCR2.1 cloning vector and transformed into DH5α E. coli, some of them produced antigenic activity that could be recovered from bacterial culture supernatant (data not shown). These culture supernatants were screened to identify PCR products encoding antigenic activity. The sequences of PCR products encoding activity were then used to produce a second set of PCR products 200–400 bp in length that overlapped by 50–80 bp. In this way, two DNA fragments with antigenic activity and overlapping by 60 bp were isolated from the H6.1 and E11.4 inserts (Fig. 3).
Identification of an Antigenic N-formyl Peptide.
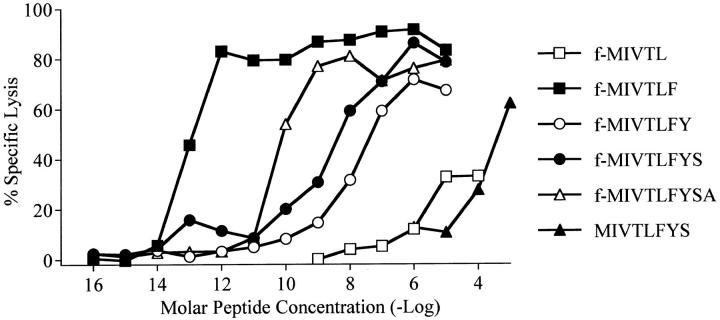
Based on the data presented above and the fact that the presenting molecule is H2-M3, we expected that the antigenic peptide would contain an N-formyl methionine residue. The 60-bp insert was examined in all six reading frames for possible ATG or GTG translation start sites. One open reading frame (ORF) containing an ATG start site followed by several hydrophobic residues was identified. The full-length sequence of this ORF encodes a 23–amino acid polypeptide we have termed AttM. Since H2-M3 has a strong preference for hydrophobic peptides containing an N-formyl methionine (11, 32, 33), this sequence was a likely candidate to encode the antigenic peptide recognized by C10.4. The heptameric peptide encoded by the first 21 bp of this sequence was synthesized and found to contain antigenic activity at micromolar peptide concentrations (Fig. 4).
Figure 4.
Several lengths of an N-formyl peptide target cells for lysis by C10.4. Peptides beginning with the NH2-terminal methionine of the 23– amino acid polypeptide identified from the genomic LM library were synthesized and tested for antigenic activity. C10.4 cells were assayed for the ability to lyse EL4 target cells coated with peptide at an E/T of 5:1. Target cells were incubated for 1–2 h at 37°C in sfIMDM to prevent digestion of peptide by serum-derived proteases. N-formyl peptides of five (open squares), six (filled squares), seven (open circles), eight (filled circles), and nine (open triangles) amino acids in length were found to have activity. Peptides were titrated and tested for activity. The 6-mer retained activity at lower peptide concentrations than the other N-formyl peptides tested. A nonformylated 8-mer peptide (filled triangles) required a 105 higher concentration for half-maximal lysis compared with the N-formyl 8-mer peptide.
H2-M3 is able to present peptides of different lengths for recognition by the same T cell clone (11, 25). Therefore, the 5-, 6-, 7-, 8-, and 9-mer versions of the peptide were synthesized and assayed for their ability to sensitize EL4 cells for lysis by C10.4. The 6-mer was found to have the greatest activity, with half-maximal lysis of EL4 occurring at concentrations of ∼80 fM (Fig. 4). Higher concentrations of the longer peptides were required to sensitize EL4 cells for lysis by C10.4. Eliminating the sixth residue, phenylalanine, almost completely abrogated recognition by C10.4. Previous studies have shown that H2-M3 is able to bind N-formyl peptides of five amino acids or shorter with high affinity (34), suggesting that the phenylalanine at position 6 of this peptide is crucial for TCR interaction with the H2-M3–peptide complex.
When target cells were incubated with the 8- or 9-mer peptides in the presence of fetal bovine serum, lower concentrations of peptide were required for half-maximal lysis, suggesting that the bovine serum contained peptidase activity that cleaved the COOH-terminal end of the peptides. The crystal structure of H2-M3 elucidated by Wang et al. (33) demonstrates that the COOH-terminal residues of bound peptides extend beyond the peptide binding groove and suggests that COOH-terminal trimming of peptides could occur after binding of the peptide to H2-M3. Elimination of the N-formyl moiety from peptides presented by H2-M3 results in a 102–104-fold increase in the peptide concentration needed for half-maximal lysis by CTLs specific for the N-formyl peptide (25, 35). Compared with the N-formyl 8-mer peptide, ∼105-fold more of the nonformylated 8-mer peptide was required for half-maximal cell lysis by C10.4 (Fig. 4). Therefore, the ability of the peptide to bind H2-M3 is highly dependent on the N-formyl moiety.
AttM is Encoded at the Transcriptional Start Site of a Polycistronic Region.
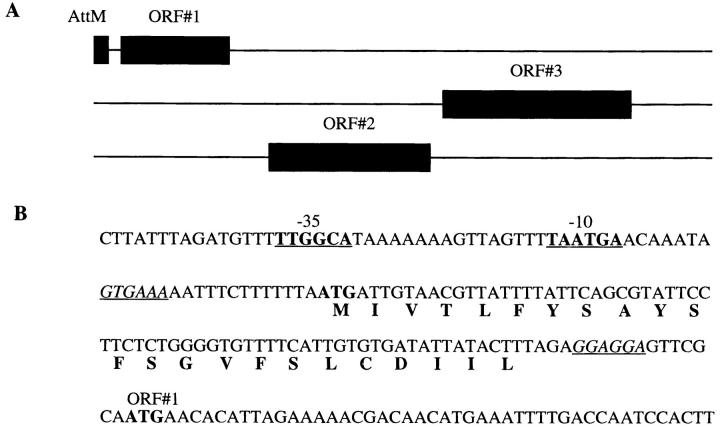
To better understand the nature of the AttM polypeptide, we examined the DNA sequence data obtained from the H6.1/E11.4 LM genomic inserts. We found that the region immediately upstream of the attM translation start site contained a consensus sequence for an RNA polymerase binding site (−35), a Pribnow box (−10), and a ribosome binding site (Fig. 5). Examination of the sequence data downstream from the polypeptide coding region revealed a polycistronic region containing at least three ORFs. ORF 1 contains a ribosome binding site consensus sequence immediately 5′ to the translation start site (Fig. 5), as do ORF 2 and ORF 3 (GenBank accession number AF039207). The presence of these sites is consistent with the AttM polypeptide being a transcription start site of a polycistronic operon.
Figure 5.
The 23–amino acid antigenic polypeptide (AttM) is encoded at the transcription start site of a polycistronic region. (A) The DNA sequence encoding the 23–amino acid polypeptide containing the antigenic peptide sequence is immediately upstream of a polycistronic region containing at least three ORFs. (B) The DNA sequence immediately 5′ of the AttM polypeptide translation start site contains an RNA polymerase binding site consensus sequence (−35, underlined bold), a Pribnow box (−10, underlined bold), and a ribosome binding site consensus sequence (underlined italic). Immediately after the AttM stop codon is a ribosome binding site consensus sequence for the first ORF of the polycistronic region (underlined italic). The ATG translation start sites for the AttM polypeptide and for ORF 1 as well as the single-letter amino acid translation of the AttM polypeptide are shown (bold).
The amino acid sequence of the three ORFs as well as the AttM polypeptide were examined for sequence homology with proteins of known sequence using the basic local alignment sequence tool (BLAST) at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov). No significant homology was found for either the AttM polypeptide or for ORF 1. ORF 2 was found to have significant sequence homology (P <10−12) with members of the XylS/AraC family of transcriptional regulatory proteins (36, 37). Members of this protein family possess a COOH-terminal region which contains a DNA binding motif that is highly conserved. The NH2-terminal portion is not conserved and is presumed to bind effector molecules that determine specificity. ORF 3 had significant homology (P <10−10) with several bacterial NADH-dependent dehydrogenases. In fact, a region homologous to ORFs 2 and 3 and with similar orientation and spacing is present in the yisR and the yisS genes of Bacillus subtilis (GenBank accession number Y09476 [38]). Therefore, these ORFs may be part of a functional operon conserved in other gram-positive bacteria.
AttM Is a Candidate for a Leader Peptide Encoded by a Transcriptional Attenuator.
The role of transcriptional attenuators in the regulation of the expression of genes involved in the biosynthesis of amino acids, nucleotides, and metabolic enzymes has been well documented for prokaryotes, eukaryotes, and viruses (39–41). A common feature of these transcriptional attenuators is the formation of stable secondary mRNA structure at the transcriptional start site of the operon. Formation of this secondary structure prevents ribosome binding and thus translation of the first ORF of the operon. The formation of secondary mRNA structure can be prevented if the ribosome stalls during translation of the attenuator region of the operon, allowing the binding of ribosomes to the downstream ORFs and translation. In all of the transcriptional attenuators studied to date, the attenuator leader region encodes a short polypeptide. This polypeptide appears to be a by-product of the translation of the attenuator leader region that occurs during regulation of the operon. Although no biological function has been described for any of these polypeptides, an attenuator leader peptide could serve as a source of antigenic peptide during the host immune response to intracellular bacterial infection.
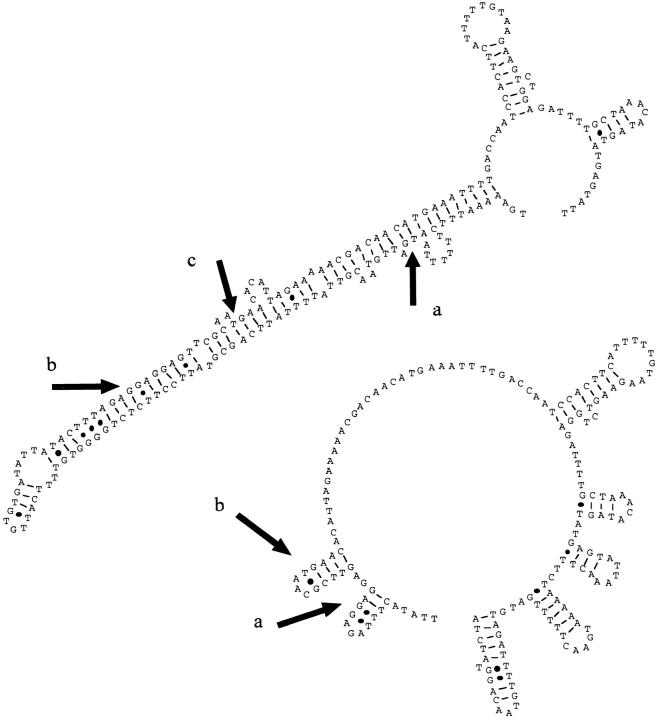
The length of the AttM polypeptide, its location at the beginning of a polycistronic region, and the presence of consensus sequences for the binding of RNA polymerase and ribosomes immediately 5′ of the AttM translation start site suggested that it may be a leader peptide for a transcriptional attenuator. We used computer analysis to examine the 300-bp region of DNA that included the attM gene and the 5′ section of ORF 1 for formation of secondary structure (42–44). When the region encoding the AttM polypeptide was included, a large stable hairpin structure was predicted that would include the ribosome binding site for ORF 1. Formation of such a structure would prevent ribosome binding and translation (Fig. 6, top). The exclusion of the nucleotides encoding AttM, as would occur if ribosome stalling were to take place in this region, eliminates the large hairpin structure and frees the Shine-Dalgarno sequence of ORF 1 for ribosome binding and initiation of translation (Fig. 6, bottom).
Figure 6.
The 23–amino acid AttM polypeptide may be the product of a putative transcriptional attenuator. (Top) The nucleotide sequence encoding the AttM polypeptide and the 5′ region of ORF 1 was examined for the ability to form secondary structure using a computer program designed to predict secondary structure from DNA sequences based on free energy of formation (MuFold [42–44]). Formation of a large duplex containing the translation start sites of the AttM polypeptide (a) and ORF 1 (c), as well as the ribosome binding site consensus sequence of ORF 1 (b), was predicted. (Bottom) Eliminating the region encoding the AttM polypeptide prevents formation of this secondary structure. This makes the ribosome binding site (a) and the ATG translation start site (b) of ORF 1 available for ribosome binding and initiation of protein translation.
Intracellular bacterial pathogens are adapted to survive in the host cell environment, either in the cytosol or in intracellular vacuoles. To enter the host cell MHC class I processing pathway, proteins derived from live intracellular bacteria have to be secreted by the bacteria or released from the bacterial cell surface (4). Indeed, the class I–presented peptides identified from LM thus far are all derived from the secreted proteins LLO, p60, and metalloprotease (45). The LemA-derived N-formyl peptide presented by H2-M3 is a membrane protein whose Nout–Cin topology exposes the antigenic epitope to the host cell cytosol (25). The AttM polypeptide characterized here does not appear either to be a membrane protein or to be specifically targeted for secretion by LM. However, antigenic activity can be recovered from culture medium in which LM are grown, indicating the AttM polypeptide is released by the bacteria. This release of antigenic peptide is biologically relevant, as antigen presentation occurs in cells infected with live virulent Listeria. How the AttM polypeptide escapes the bacterial cytoplasm is unknown, but the small size and the hydrophobicity of AttM may contribute to its release from the bacteria to the host cell cytosol. This would enhance entry of peptide to the MHC class I processing pathway of infected cells as well as protect the N-formyl group of the peptide from bacterial deformylases.
Our findings suggest that in addition to proteins specifically targeted for secretion and membrane surface proteins, other bacterial proteins may be accessible to the host cell MHC class I processing pathway and serve as sources of antigen. It is possible that other leader peptides serve as sources of T cell antigens presented by MHC class I or perhaps class II molecules. In addition, these leader peptides could be a source of the short N-formyl bacterial peptides that bind to chemotactic receptors found on monocytes and neutrophils (46–48). Identification of additional leader peptide–derived T cell antigens may establish that these polypeptides, which are by-products of transcriptional regulation, are a source of immunologically relevant molecules.
Acknowledgments
The authors wish to thank K. Rock for the BMC2.3 cell line, R.R. Rich and J.R. Rodgers for the SVCAS2F6 and 13S2 cell lines, E. Pamer for the L9.6 CTL clone, and S. Dow for the rmuGM-CSF/CHO cell line. We are especially grateful to A.L. Marrs for synthesis of N-formyl peptides. We thank T.A. Potter, R. Berg, S. Hayes, and E. Zahradka for critical review of the manuscript and for helpful discussions.
This work was supported in part by National Institutes of Health grants HD-26841, AI-35194, and AI-22295 (all to U.D. Staerz).
Abbreviations used in this paper
- BHI
brain heart infusion broth
- CHO
Chinese hamster ovary
- LLO
listeriolysin O
- LM
Listeria monocytogenes
- MOI
multiplicity of infection
- ORF
open reading frame
- rmu
recombinant murine
- sf
serum-free.
References
- 1.Unanue ER. Studies in listeriosis show the strong symbiosis between the innate cellular system and the T-cell response. Immunol Rev. 1997;158:11–25. doi: 10.1111/j.1600-065x.1997.tb00988.x. [DOI] [PubMed] [Google Scholar]
- 2.Lane FC, Unanue ER. Requirement of thymus (T) lymphocytes for resistance to listeriosis. J Exp Med. 1972;135:1104–1112. doi: 10.1084/jem.135.5.1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Libero G, Kaufmann SH. Antigen-specific Lyt-2+ cytolytic T lymphocytes from mice infected with the intracellular bacterium Listeria monocytogenes. . J Immunol. 1986;137:2688–2694. [PubMed] [Google Scholar]
- 4.Pamer EG. Cellular immunity to intracellular bacteria. Curr Opin Immunol. 1993;5:492–496. doi: 10.1016/0952-7915(93)90028-q. [DOI] [PubMed] [Google Scholar]
- 5.Stroynowski I, Lindahl KF. Antigen presentation by non-classical class I molecules. Curr Opin Immunol. 1994;6:38–44. doi: 10.1016/0952-7915(94)90031-0. [DOI] [PubMed] [Google Scholar]
- 6.Kurlander RJ, Shawar SM, Brown ML, Rich RR. Specialized role for a murine class I-b MHC molecule in prokaryotic host defenses. Science. 1992;257:678–679. doi: 10.1126/science.1496381. [DOI] [PubMed] [Google Scholar]
- 7.Maher JK, Kronenberg M. The role of CD1 molecules in immune responses to infection. Curr Opin Immunol. 1997;9:456–461. doi: 10.1016/s0952-7915(97)80095-7. [DOI] [PubMed] [Google Scholar]
- 8.Beckman EM, Porcelli SA, Morita CT, Behar SM, Furlong ST, Brenner MB. Recognition of a lipid antigen by CD1-restricted alpha beta+ T cells. Nature. 1994;372:691–694. doi: 10.1038/372691a0. [DOI] [PubMed] [Google Scholar]
- 9.Sieling PA, Chatterjee D, Porcelli SA, Prigozy TI, Mazzaccaro RJ, Soriano T, Bloom BR, Brenner MB, Kronenberg M, Brennan PJ, et al. CD1-restricted T cell recognition of microbial lipoglycan antigens. Science. 1995;269:227–230. doi: 10.1126/science.7542404. [DOI] [PubMed] [Google Scholar]
- 10.Shawar SM, Vyas JM, Shen E, Rodgers JR, Rich RR. Differential amino-terminal anchors for peptide binding to H-2M3a or H-2Kb and H-2Db. J Immunol. 1993;151:201–210. [PubMed] [Google Scholar]
- 11.Shawar SM, Cook RG, Rodgers JR, Rich RR. Specialized functions of MHC class I molecules. I. An N-formyl peptide receptor is required for construction of the class I antigen Mta. J Exp Med. 1990;171:897–912. doi: 10.1084/jem.171.3.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shawar SM, Vyas JM, Rodgers JR, Cook RG, Rich RR. Specialized functions of major histocompatibility complex class I molecules. II. Hmt binds N-formylated peptides of mitochondrial and prokaryotic origin. J Exp Med. 1991;174:941–944. doi: 10.1084/jem.174.4.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tilney LG, Portnoy DA. Actin filaments and the growth, movement, and spread of the intracellular bacterial parasite, Listeria monocytogenes. . J Cell Biol. 1989;109:1597–1608. doi: 10.1083/jcb.109.4.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brunt LM, Portnoy DA, Unanue ER. Presentation of Listeria monocytogenesto CD8+ T cells requires secretion of hemolysin and intracellular bacterial growth. J Immunol. 1990;145:3540–3546. [PubMed] [Google Scholar]
- 15.Unanue ER. Why listeriosis? A perspective on cellular immunity to infection. Immunol Rev. 1997;158:5–9. doi: 10.1111/j.1600-065x.1997.tb00987.x. [DOI] [PubMed] [Google Scholar]
- 16.Harty JT, Schreiber RD, Bevan MJ. CD8 T cells can protect against an intracellular bacterium in an interferon gamma-independent fashion. Proc Natl Acad Sci USA. 1992;89:11612–11616. doi: 10.1073/pnas.89.23.11612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ladel CH, Flesch IE, Arnoldi J, Kaufmann SH. Studies with MHC-deficient knock-out mice reveal impact of both MHC I- and MHC II-dependent T cell responses on Listeria monocytogenesinfection [published erratum 1995. 154:4223] J Immunol. 1994;153:3116–3122. [PubMed] [Google Scholar]
- 18.Harty JT, Pamer EG. CD8 T lymphocytes specific for the secreted p60 antigen protect against Listeria monocytogenesinfection. J Immunol. 1995;154:4642–4650. [PubMed] [Google Scholar]
- 19.Harty JT, Bevan MJ. CD8+ T cells specific for a single nonamer epitope of Listeria monocytogenesare protective in vivo. J Exp Med. 1992;175:1531–1538. doi: 10.1084/jem.175.6.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bishop DK, Hinrichs DJ. Adoptive transfer of immunity to Listeria monocytogenes. The influence of in vitro stimulation on lymphocyte subset requirements. J Immunol. 1987;139:2005–2009. [PubMed] [Google Scholar]
- 21.Kaufmann SH, Rodewald HR, Hug E, De Libero G. Cloned Listeria monocytogenesspecific non-MHC-restricted Lyt-2+ T cells with cytolytic and protective activity. J Immunol. 1988;140:3173–3179. [PubMed] [Google Scholar]
- 22.Lukacs K, Kurlander RJ. MHC-unrestricted transfer of antilisterial immunity by freshly isolated immune CD8 spleen cells. J Immunol. 1989;143:3731–3736. [PubMed] [Google Scholar]
- 23.Pamer EG, Wang CR, Flaherty L, Lindahl KF, Bevan MJ. H-2M3 presents a Listeria monocytogenespeptide to cytotoxic T lymphocytes. Cell. 1992;70:215–223. doi: 10.1016/0092-8674(92)90097-v. [DOI] [PubMed] [Google Scholar]
- 24.Gulden PH, Fischer P, III, Sherman NE, Wang W, Engelhard VH, Shabanowitz J, Hunt DF, Pamer EG. A Listeria monocytogenespentapeptide is presented to cytolytic T lymphocytes by the H2-M3 MHC class Ib molecule. Immunity. 1996;5:73–79. doi: 10.1016/s1074-7613(00)80311-8. [DOI] [PubMed] [Google Scholar]
- 25.Lenz LL, Dere B, Bevan MJ. Identification of an H2-M3-restricted Listeriaepitope: implications for antigen presentation by M3. Immunity. 1996;5:63–72. doi: 10.1016/s1074-7613(00)80310-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pamer EG, Harty JT, Bevan MJ. Precise prediction of a dominant class I MHC-restricted epitope of Listeria monocytogenes. . Nature. 1991;353:852–855. doi: 10.1038/353852a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sijts AJ, Neisig A, Neefjes J, Pamer EG. Two Listeria monocytogenesCTL epitopes are processed from the same antigen with different efficiencies. J Immunol. 1996;156:683–692. [PubMed] [Google Scholar]
- 28.Pamer EG. Direct sequence identification and kinetic analysis of an MHC class I-restricted Listeria monocytogenesCTL epitope. J Immunol. 1994;152:686–694. [PubMed] [Google Scholar]
- 29.Busch DH, Bouwer HGA, Hinrichs D, Pamer EG. A nonamer peptide derived from Listeria monocytogenesmetalloprotease is presented to cytolytic T lymphocytes. Infect Immun. 1997;65:5326–5329. doi: 10.1128/iai.65.12.5326-5329.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vyas JM, Rich RR, Howell DD, Shawar SM, Rodgers JR. Availability of endogenous peptides limits expression of an M3a-Ld major histocompatibility complex class I chimera. J Exp Med. 1994;179:155–165. doi: 10.1084/jem.179.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Richards S, Bucan M, Brorson K, Kiefer MC, Hunt SW, Lehrach H, Lindahl KF. Genetic and molecular mapping of the Hmt region of mouse. EMBO (Eur Mol Biol Organ) J. 1989;8:3749–3757. doi: 10.1002/j.1460-2075.1989.tb08551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vyas JM, Shawar SM, Rodgers JR, Cook RG, Rich RR. Biochemical specificity of H-2M3a. Stereospecificity and space-filling requirements at position 1 maintain N-formyl peptide binding. J Immunol. 1992;149:3605–3611. [PubMed] [Google Scholar]
- 33.Wang CR, Castano AR, Peterson PA, Slaughter C, Lindahl KF, Deisenhofer J. Nonclassical binding of formylated peptide in crystal structure of the MHC class Ib molecule H2-M3. Cell. 1995;82:655–664. doi: 10.1016/0092-8674(95)90037-3. [DOI] [PubMed] [Google Scholar]
- 34.Vyas JM, Rodgers JR, Rich RR. H-2M3a violates the paradigm for major histocompatibility complex class I peptide binding. J Exp Med. 1995;181:1817–1825. doi: 10.1084/jem.181.5.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith GP, Dabhi VM, Pamer EG, Lindahl KF. Peptide presentation by the MHC class Ib molecule, H2-M3. Int Immunol. 1994;6:1917–1926. doi: 10.1093/intimm/6.12.1917. [DOI] [PubMed] [Google Scholar]
- 36.Gallegos MT, Michan C, Ramos JL. The XylS/AraC family of regulators. Nucleic Acids Res. 1993;21:807–810. doi: 10.1093/nar/21.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramos JL, Rojo F, Zhou L, Timmis KN. A family of positive regulators related to the Pseudomonas putida TOL plasmid XylS and the Escherichia coliAraC activators. Nucleic Acids Res. 1990;18:2149–2152. doi: 10.1093/nar/18.8.2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roche B, Autret S, Levine A, Vannier F, Medina N, Seror SJ. A Bacillus subtilischromosome segment at the 100 degrees to 102 degrees position encoding 11 membrane proteins. Microbiology. 1997;143:3309–3312. doi: 10.1099/00221287-143-10-3309. [DOI] [PubMed] [Google Scholar]
- 39.Yanofsky C. Attenuation in the control of expression of bacterial operons. Nature. 1981;289:751–758. doi: 10.1038/289751a0. [DOI] [PubMed] [Google Scholar]
- 40.Soreq H, Ben-Aziz R, Prody CA, Seidman S, Gnatt A, Neville L, Lieman-Hurwitz J, Lev-Lehman E, Ginzberg D, Lipidot-Lifson Y, et al. Molecular cloning and construction of the coding region for human acetylcholinesterase reveals a G + C-rich attenuating structure. Proc Natl Acad Sci USA. 1990;87:9688–9692. doi: 10.1073/pnas.87.24.9688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kessler M, Ben-Asher E, Resenkov O, Hatini V, Bengal E, Aloni Y. A 21-base pair DNA fragment directs transcription attenuation within the simian virus 40 late leader. J Biol Chem. 1991;266:13019–13027. [PubMed] [Google Scholar]
- 42.Jaeger JA, Turner DH, Zuker M. Predicting optimal and suboptimal secondary structure for RNA. Methods Enzymol. 1990;183:281–306. doi: 10.1016/0076-6879(90)83019-6. [DOI] [PubMed] [Google Scholar]
- 43.Jaeger JA, Turner DH, Zuker M. Improved predictions of secondary structures for RNA. Proc Natl Acad Sci USA. 1989;86:7706–7710. doi: 10.1073/pnas.86.20.7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989;244:48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]
- 45.Pamer EG, Sijts AJAM, Villanueva MS, Busch DH, Sujata V. MHC class I antigen processing of Listeria monocytogenesproteins: implications for dominant and subdominant CTL responses. Immunol Rev. 1997;158:129–136. doi: 10.1111/j.1600-065x.1997.tb00999.x. [DOI] [PubMed] [Google Scholar]
- 46.Marasco WA, Phan SH, Krutzsch H, Showell HJ, Feltner DE, Nairn R, Becker EL, Ward PA. Purification and identification of formyl-methionyl-leucyl-phenylalanine as the major peptide neutrophil chemotactic factor produced by Escherichia coli. . J Biol Chem. 1984;259:5430–5439. [PubMed] [Google Scholar]
- 47.Carp H. Mitochondrial N-formylmethionyl proteins as chemoattractants for neutrophils. J Exp Med. 1982;155:264–275. doi: 10.1084/jem.155.1.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schiffmann E, Corcoran BA, Wahl SM. N-formylmethionyl peptides as chemoattractants for leucocytes. Proc Natl Acad Sci USA. 1975;72:1059–1062. doi: 10.1073/pnas.72.3.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]