Abstract
In a murine model of respiratory syncytial virus disease, prior sensitization to the attachment glycoprotein (G) leads to pulmonary eosinophilia and enhanced illness. Three different approaches were taken to dissect the region of G responsible for enhanced disease and protection against challenge. First, mutant viruses, containing frameshifts that altered the COOH terminus of the G protein, were used to challenge mice sensitized by scarification with recombinant vaccinia virus (rVV) expressing wild-type G. Second, cDNA expressing these mutated G proteins were expressed by rVV and used to vaccinate mice before challenge with wild-type respiratory syncytial virus (RSV). These studies identified residues 193–205 to be responsible for G-induced weight loss and lung eosinophilia and showed that this region was not was not necessary for induction of protective immunity. Third, mice were sensitized using an rVV that expressed only amino acids 124–203 of the G protein. Upon RSV challenge, mice sensitized with this rVV developed enhanced weight loss and eosinophilia. This is the first time that a region within RSV (amino acids 193–203) has been shown to be responsible for induction of lung eosinophilia and disease enhancement. Moreover, we now show that it is possible to induce protective immunity with an altered G protein without inducing a pathological response.
Keywords: respiratory syncytial virus, G protein, eosinophilia, vaccine, T helper cell
Respiratory syncytial virus (RSV) is the leading cause of bronchiolitis and viral pneumonia among infants and young children (1, 2). Primary infection with RSV usually occurs within the first or second year of life with children between the ages of 6 wk and 9 mo of age at the greatest risk of developing serious illness. RSV bronchiolitis is associated with the development of atopy and asthma in later years (3). Because bronchiolitis is the leading cause of hospital admissions for infants in the western world, development of a safe and effective vaccine is a high priority.
A cautious approach to RSV vaccine development has been taken since the failure of vaccine trials in the 1960's. In these trials children were inoculated intramuscularly with formalin-inactivated RSV. After subsequent natural exposure to RSV, children who had been given the RSV vaccine had greater morbidity and mortality than control vaccinees (4–7). Postmortem examination of these children showed peribronchiolar infiltration and excess eosinophils in the lungs and blood (4, 7).
In a BALB/c mouse model, this type of enhanced pathology can be reproduced by scarifying mice with recombinant vaccinia virus (rVV) expressing only the G protein of RSV followed by intranasal inoculation of infectious RSV (8). The G protein is a highly glycosylated membrane protein thought to be responsible for viral attachment to the host cell. It is produced in both a secreted and membrane-bound form; the function of the secreted form is unknown, but it may function as a decoy for neutralizing antibodies. The COOH terminus of the G protein varies amongst the different strains of RSV, although in human RSV there is a conserved region (amino acids 164–176) that is believed to be the receptor attachment site (9).
In the BALB/c mouse model, vaccination with individual RSV proteins expressed in VVs generates different immunological and pathological responses upon intranasal exposure to RSV (2). Vaccination with rVV expressing the fusion protein (F) of RSV leads to the generation of CTLs and CD4+ cells with a Th1 type of phenotype (i.e., high IFN-γ and no IL-4 or IL-5), whereas rVV expressing the matrix protein (M2) generates a CTL response with little or no T helper response. By contrast, vaccination with rVV expressing the G protein leads to the generation of a Th2 type of response (less IFN-γ and some IL-4 and IL-5) with an eosinophilic influx into the alveolar space (8, 10, 11). This eosinophilic influx and vaccine-enhanced illness resembles the pathology found in the children from the 1960's vaccine trials.
To aid the development of a safe and effective vaccine, this murine model of vaccine-enhanced illness has been used to investigate immunopathological mechanisms involved in RSV illness (2). Since eosinophilia depends on CD4+ T cell recognition of G (12, 13), we mapped G protein regions in BALB/c mice using RSV frameshift mutants and rVV expressing partial or mutant G proteins. These studies identified an 11-amino acid portion of the G protein that is involved in the generation of pulmonary eosinophilia and vaccine-enhanced weight loss, but is not essential for the induction of protective immunity.
Materials and Methods
Viruses.
RSV and rVVs were grown on HEp-2 cells and titered as previously described (8). Frameshift mutants were isolated by selecting escape variants in vitro with anti-G monoclonal antibody (63G) (14). All stocks were free of mycoplasma contamination as determined by DNA hybridization (Genprobe Inc., San Diego, CA).
Construction of rVVs Expressing G Protein Frameshift Mutants.
pGEM4 derived plasmids encoding the G proteins of frameshift mutants R63/1/2/3 and R63/2/4/8 have been described previously (14). The plasmid encoding the double frameshift G protein mutant R63/2/4/1 was derived from R63/2/4/8; in this case, an extra adenosine was inserted after nucleotide 648 by PCR-based site-directed mutagenesis as described (15). The G protein inserts of the above plasmids were isolated by digestion with HindIII and EcoRI, blunt-ended with Klenow polymerase, and ligated to SmaI linearized pSC11 vector. CV-1 cells were infected with the WR strain of VV and immediately transfected with the pSC11-derived plasmids. Progeny virus was plaque purified three times on HuTK-124B cells in the presence of 15 μg/ml of bromodeoxyuridine to select for thymidine kinase–negative recombinants. The rVVG27 (expressing amino acids 124–203) and rVVTer (empty vector) vaccinia viruses were a gift of Dr. M. Trudel (Centre de Recherche en Virologie, Institut Armand-Frappier, Québec, Canada) and have been described previously (16). rVVG contains the insert for the whole G protein and rVVβgal contains the LacZ gene inserted into the vaccinia vector as a negative control in all experiments (17, 18).
Mouse Infections.
Anesthetized 8–10-wk-old female BALB/c mice (Harlan Olac, Bicester, UK) were scarified with 2 × 106 PFU recombinant vaccinia at the base of the tail. After 2 wk, mice were intranasally inoculated with 1.5 × 106 PFU/mouse of Long and RSV mutants or 3 × 106 PFU/mouse of RSV A2 strain.
Measurement of Eosinophilia in the Bronchoalveolar Lavage.
7 d after RSV infection, bronchoalveolar lavage (BAL) fluid was collected as previously described (19). In brief, individual mice were terminally anesthetized with pentobarbitone and bled via the femoral artery. Lungs were perfused six times with 1 ml of lignocaine in Eagle's media (Sigma Chemical Co., Poole, UK). 200 μl of BAL fluid was cytocentrifuged onto glass slides and stained with Giemsa's reagent for cytological analysis. Eosinophils were counted by flow cytometry as the proportion of the granulocytes compared with total cells and confirmed with microscopic examination of cytospin slides to distinguish between polymorphonuclear cells and eosinophils.
Titration of RSV from Mouse Lungs.
Mice were scarified with recombinant vaccinias 14 d before challenge with whole RSV. 4 d after challenge with RSV, whole lungs were disrupted using glass homogenizers (Jencons, Leighton Buzzard, UK) in 1.3 ml RPMI (Sigma Chemical Co.) supplemented with 2 mM glutamine (GIBCO BRL, Paisley, Scotland), 100 U/ml penicillin, and 100 μg/ml streptomycin on ice. Homogenates were clarified at 10,000 g for 1 min. 50 μl of supernatant and twofold serial dilutions thereof were titrated on HEp-2 monolayers in 96-well plates and plaques were assayed as previously described (8). The theoretical limit of detection for this assay was 5 PFU/lung.
Statistical Analysis.
Kruskal-Wallis tests were used to test for effects between groups and Mann-Whitney U tests were used to perform comparisons between the experimental and control groups. Data analysis was performed using SPSS statistical software.
Results
Mapping of the Eosinophilic Induction Using RSV Mutants.
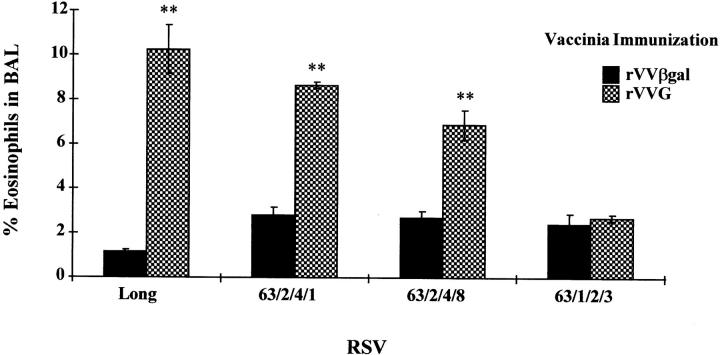
A series of RSV escape mutants were generated using a monoclonal antibody against the G protein (14). These mutants contain frameshift mutations generating G proteins with truncations and/or alterations in the COOH terminus of the protein (Fig. 1). Mice were scarified with rVVG or rVVβgal (control construct) and 2 wk were later intranasally challenged with the parental Long strain or one of the different mutant viruses. 7 d after infection, BAL fluid was collected and the percentage of eosinophils in the BAL was assessed. As in primary infection, mice scarified with rVVβgal and challenged with either Long or mutant viruses showed no eosinophilia. However, mice scarified with rVVG and challenged with whole RSV showed marked pulmonary eosinophilia except for one mutant. Mutant 63/1/2/3 failed to induce eosinophilia. Mice scarified with rVVG and challenged with mutant 63/1/2/3 generated a low level of eosinophilia similar to mice scarified with rVVβgal (P ≥0.86) (Fig. 2). The low level of eosinophilia observed in mice scarified with rVVG after intranasal challenge with 63/1/2/3 was significantly different from mice scarified with rVVG followed by either Long, 63/2/4/1, or 63/2/ 4/8 (P ≤0.01, 0.01, and 0.01, respectively). The differences between mutant 63/1/2/3 and 63/2/4/8 lie between amino acids 193 and 205 (Fig. 1). From these data, the portion of the G protein responsible for eosinophil induction can be localized to this region.
Figure 1.
The primary structure of the G protein from RSV (Long strain) and the mutants used in this study (14). The stippled region indicates the transmembrane domain and hashed boxes denote amino acids changed due to frameshift mutations. The location of the intracellular (IC), transmembrane (TM), mucinoid I, mucinoid II, and conserved domains are indicated.
Figure 2.
Eosinophilic influx in the BAL fluid following scarification with rVVβgal (filled bars) or rVVG (checked bars) after intranasal inoculation with mutant RSV. Data shown is representative of four experiments and each bar represents the mean percentage of individual eosinophil counts in each group ± SEM (n = 4). Significant differences (P ≤0.05) between rVVG and rVVβgal using Mann-Whitney paired comparisons test are indicated by **. The significance levels between rVVG and rVVβgal vaccinated mice are as follows: Long, P ≤0.03; 63/2/4/1, P ≤0.03; 63/ 2/4/8, P ≤0.03; 63/1/2/3, P ≤0.86.
A possible explanation for this finding is that the large deletion in 63/1/2/3 altered viral infectivity and subsequent eosinophilia generation. Viral lung titers on days 4 and 7 after infection showed no differences between the parental Long strain virus and viral mutants (data not shown). Despite the large alteration of the COOH terminus of the G protein, all mutant viruses infected the lungs efficiently and replicated as well as Long strain virus and all were cleared by day 7.
Mapping of the Eosinophilic Antigen Using rVVs.
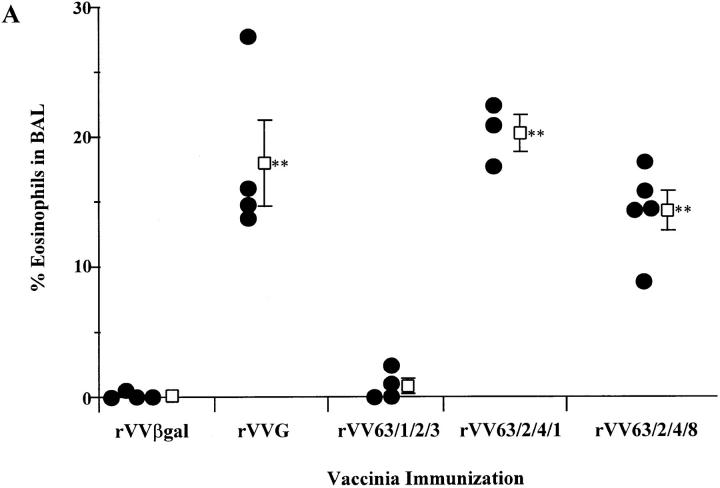
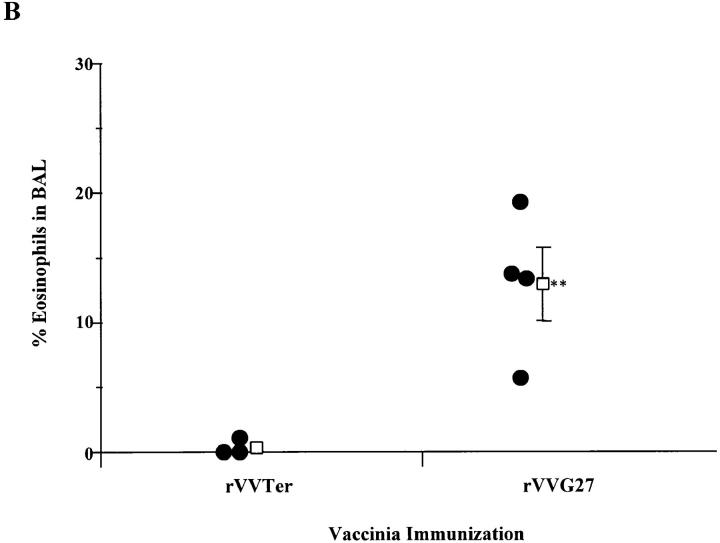
To complement the studies described above and ensure that the eosinophilia was due only to sensitization to the attachment protein G, rVVs expressing mutant G proteins were used to sensitize mice. Mice were scarified with recombinant vaccinia and challenged intranasally with wild-type RSV (A2 strain) after 14 d. The G protein of the A2 strain is 95% homologous to Long strain and is identical within region 193– 205. Mirroring the first set of studies, no eosinophilia was observed in the BAL of mice scarified with rVV63/1/2/3 (Fig. 3 A). The level of eosinophilia in rVV63/1/2/3-vaccinated mice was not significantly different from rVVβgal-primed mice (P ≥0.556) and was significantly reduced compared with that seen in mice primed with rVVG, rVV63/ 2/4/1, or rVV63/2/4/8 (P ≤0.02, 0.02, and 0.05, respectively). To confirm that the lack of eosinophilia observed in mutant rVV63/1/2/3 mice was not due to a loss of stability or secondary structures, mice were scarified with recombinant vaccinia (rVVG27) expressing 80 amino acids of the G protein (124–203) (Fig. 1). After intranasal RSV challenge, marked eosinophilia was observed, similar to that of mice primed with rVVG (P ≥0.29) and significantly greater than in control mice primed with rVVTer (the control construct) (P ≤0.03) (Fig. 3 B). This confirms that the eosinophilic region lies within this 80-amino acid portion of the G protein. Together, these data using mutant viruses and rVV with inserted mutant G proteins or portions of the G protein, suggest that amino acids 193–203 contain the critical region responsible for lung eosinophilia.
Figure 3.
Eosinophilia in the BAL of mice primed with rVVs expressing mutant G proteins (A) or portions of the G protein (B) followed by intranasal inoculation with A2 RSV. 2 wk after scarification with rVVβgal, rVVG, rVV63/1/2/3, rVV63/2/ 4/1, or rVV63/2/4/8 (A), or rVVTer or rVVG27 (B), mice were challenged intranasally with wild-type RSV (strain A2). Representative data of 3 experiments are shown where circles represent single mice and open squares represent the mean percentage of eosinophils in the BAL fluid from each group ± SEM. Significant differences between the mean eosinophilia-generated rVVβgal are indicated by **.
Deletion of the Portion of G Protein Responsible for G-enhanced Weight Loss.
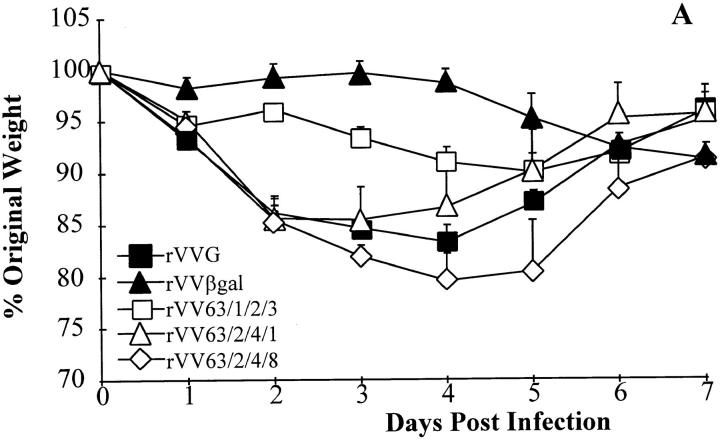
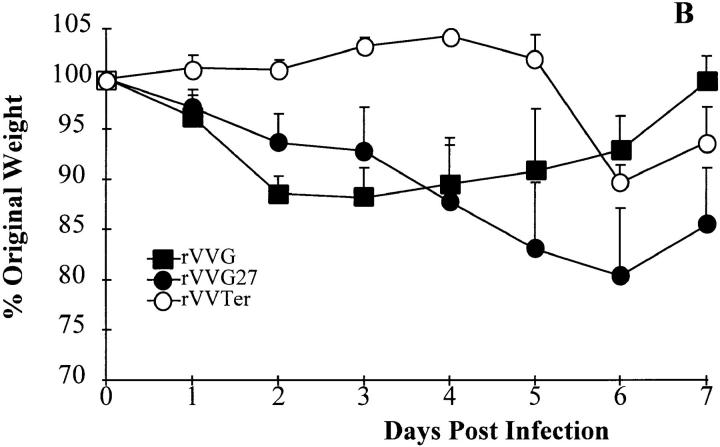
The level of eosinophilia in the lungs is only one measure of vaccine-enhanced illness; another is weight loss. Each mouse was weighed daily after intranasal challenge with RSV. Sensitization with the G protein from mutant virus 63/1/2/3 did not significantly enhance weight loss during RSV challenge compared with control vaccination with rVVβgal, whereas rVVG or any of the other mutants did enhance weight loss (Fig. 4 A). To confirm that this region contains an epitope that is responsible for vaccine-enhanced weight loss, recombinant vaccinia expressing amino acids 124–203 of the G protein (rVVG27) was used to prime mice followed by intranasal challenge with live RSV (Fig. 4 B). Mice sensitized with this portion of the G protein suffered marked weight loss equal to that caused by wild-type G. These data illustrate that the portion of the G protein responsible for pulmonary eosinophilia is also responsible for vaccine-enhanced weight loss.
Figure 4.
Weight loss after vaccination with rVVG mutants. Weights were taken daily after intranasal challenge with wild-type A2. Data are expressed as a mean percentage (3–4 per group) of the original weight (day = 0) + SEM. Representative experiment from three experiments.
Protection against RSV Challenge.
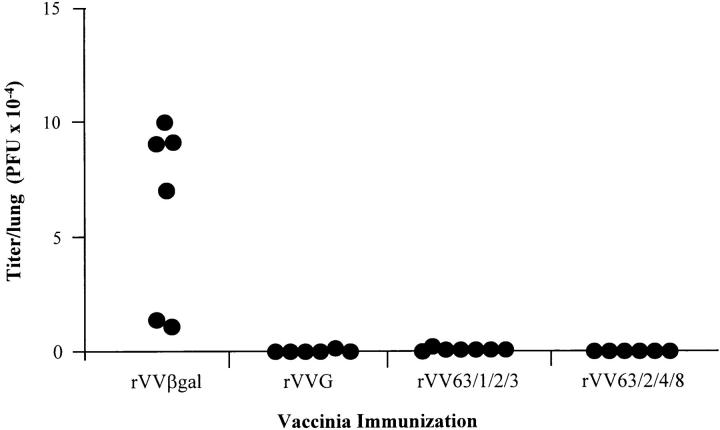
To examine whether rVV63/1/2/3 could still provide protection even though it does not induce eosinophilia, mice were scarified with recombinant vaccinias and challenged intranasally with wild-type RSV. All mice vaccinated with either of the frameshift mutants (rVV63/2/4/8 or rVV63/1/2/3) or with wild-type G (rVVG) were protected from RSV infection at day 4 (Fig. 5) and no virus was recovered from the lungs of any mice at day 7 (data not shown). These results confirm that it is possible to protect mice from subsequent RSV infection without inducing lung eosinophilia.
Figure 5.
RSV titers from the lungs of mice vaccinated with rVVs. Mice were scarified with rVVs expressing either βgal, wild-type G, or mutated G proteins (rVV63/2/4/8 or rVV63/1/2/3) and challenged intranasally with whole RSV. 4 d after challenge, lungs were homogenized and assayed for RSV. Titers from individual mice are shown from one of two separate experiments which gave similar results.
Discussion
This is the first study to show that a single region within the G protein is responsible for enhanced illness and lung eosinophilia and that in its absence it is possible to induce protective immunity without pathology. The critical importance of this region for lung eosinophilia was evident both at the priming (rVV63/1/2/3) and challenge (63/1/ 2/3) phases of the immune response. Mapping eosinophilia induction with the mutant G proteins and rVVG27 locates the critical region to be amino acids 193–203. There are two possible explanations for this finding. First, the mutation in 63/1/2/3 could have eliminated all CD4+ T cell recognition of the G protein. Since G-induced RSV disease is mainly a T cell–mediated, immunopathological process, a reduction in the CD4+ T cell response would lead to less cellular infiltration into the lungs, less eosinophilia, no RSV-specific antibody response, and less weight loss. A second possibility is that the mutation introduced into 63/ 1/2/3 does not eliminate the entire CD4+ response but rather a vigorous, eosinophil-inducing component thereof. We believe this to be the case since priming with rVV63/ 1/2/3 generates an appreciable antibody response and a pulmonary lymphocytic infiltrate similar to that seen in wild-type G-sensitized mice, but with a significant decrease in the ratio of CD4+:CD8+ cells (data not shown). This suggests that a strong CD4+ T cell response is abolished by the alteration of residues 193–203 and that this response is essential for induction of lung eosinophilia. Recently, using overlapping peptides, a T cell epitope that induces a Th2 response has been mapped to amino acids 184–198 (19a). This peptide includes six residues from the critical region mapped in our study (KPGKKT). Using a predictive algorithm for class II MHC binding peptides (strong and weak anchor positions: 1, 4, 9: I, K or R, K, respectively) (20), we have identified two possible I-Ed epitopes, both of which would be disrupted in the 63/1/2/3 mutant. These potential epitopes are located at amino acids 185-193 (ICKRIPNKK) and 189–197 (IPNKKPGKK) (bold type indicates anchor and subanchor residues).
In other viral systems, single epitopes have been shown to be responsible for Th2-like CD4+ responses. Proteins from mouse hepatitis virus (JHM; reference 21), hepatitis B virus (22), and retroviruses (23) have been shown to generate Th2 responses and have been mapped using peptides and T cell clones and lines. Unlike our study, in none of these systems has it been possible to examine the effects of altering these epitopes upon the host immune response and induction of pathology. Interestingly, the critical epitope in JHM virus is also I-Ed–restricted (ILNKPRQKR) and contains at least four identical or similar residues to the proposed critical motifs of RSV G. It is possible that these homologous epitopes have similar binding affinities for MHC class II molecules, thus skewing towards a Th2-type response (24).
In this and in previous studies, induction of eosinophilia is indicative of a Th2-type response (10, 11, 25). The recruitment of eosinophils into the alveolar space could be due to IL-5 production (26) or the induction of chemokines such as eotaxin (27) or RANTES (regulated on activation, T cell expressed and secreted; reference 28). In our study, this same region is also responsible for the increased weight loss that is observed upon challenge. Weight loss, however, is not a specific indicator of Th2-type, eosinophilic pathology since mice lose weight late in a primary infection and may show disease enhancement due to sensitization to other RSV proteins (10). Hussell et al. have shown that IL-12 treatment of mice primed with rVVG induces a Th1-type response and diminishes eosinophilia but weight loss is unaffected or in some cases enhanced (29). Our data indicate that region 193–203 of the G protein not only induces eosinophilia (probably via the Th2 pathway), but also induces an immune response (such as TNF-α; reference 30) that is responsible for increased weight loss.
These findings provide a framework which may allow the development of an RSV vaccine without vaccine-enhanced illness. In the mouse model of RSV, vaccine-enhanced illness presents two possible pathways for eosinophil induction: first, a CD4+ response to eosinophilic epitopes within the primary structure of the G protein such as described here, or second, the elimination of the CD8+ response that controls the Th2 response (31). We have recently shown that it is possible to generate eosinophilia to the F protein of RSV when CD8+ cells are deleted. Normally, priming with the F protein of RSV generates a Th1-type response and virus-specific CTLs that are an abundant source of IFN-γ. When CD8+ T cells are deleted, the response is switched to a Th2-type response that generates lung eosinophilia. It appears that in the F response, CD8+ cells are responsible for controlling the Th2-type response, most likely through the production of IFN-γ (13). Similarly, the failure of the formalin-inactivated RSV vaccine trials could have been due to the lack of CD8+ T cell induction since soluble antigen is processed and presented via the MHC class II pathway. Both aspects of the immune response need to be considered for the development of future RSV vaccines.
Our results suggest that it is possible to induce a protective immune response with the G protein without inducing a detrimental eosinophilic response. Since the G protein does not induce a CTL response in BALB/c mice (32), eosinophil induction could in part be due to the lack of a CD8+ response. In spite of this, we have shown that it is possible to induce protective immunity with the G protein if the eosinophilogenic portion has been altered. Vaccines need to induce a CD4+ response to promote B cell production of protective antibody, but IFN-γ production by CD8+ cells may also be essential to protect against augmentation of illness. However, induction of a strong and unopposed CD4+ response can also be detrimental. Sensitization with the M2 protein alone leads to enhanced illness, and transfer of isolated CD4+ T cells causes augmented disease as well (10, 33). An ideal vaccine should strike a balance between inducing CD4+ and CD8+ T cell responses. Vaccination that stimulates either a CD4+ or CD8+ T cell response alone might lead to pathological consequences (34). Whether these tenets for vaccine development will hold true in an outbred human population remains to be tested.
Acknowledgments
We would like to thank Dr. M. Trudel for providing vaccinia recombinants rVVG27 and rVVTer. We are also grateful to Dr. B.A. Askonas for critical review of this manuscript, Dr. E.T. Bergman for statistical analysis of this data, and Teresa Delgado for technical assistance with the generation of vaccinia recombinants.
This work was supported by the Wellcome Trust and J.A. Melero was supported by grant PM96-0025 from Dirección General de Enseñanza Superior, MEC.
Footnotes
T.E. Sparer and S. Matthews contributed equally to this work.
References
- 1.Anderson LJ, Heilman CA. Protective and disease-enhancing immune responses to respiratory syncytial virus. J Infect Dis. 1995;171:1–7. doi: 10.1093/infdis/171.1.1. [DOI] [PubMed] [Google Scholar]
- 2.Openshaw PJM. Immunopathological mechanisms in respiratory syncytial virus disease. Semin Immunopathol. 1995;17:187–201. doi: 10.1007/BF00196165. [DOI] [PubMed] [Google Scholar]
- 3.Sigurs N, Bjarnason R, Sigurbergsson F, Kjellman B, Bjorksten B. Asthma and immunoglobulin E antibodies after respiratory syncytial virus bronchiolitis: a prospective cohort study with matched controls. Pediatrics. 1995;95:500–505. [PubMed] [Google Scholar]
- 4.Kim HW, Canchola JG, Brandt CD, Pyles G, Chanock RM, Jensen K, Parrott RH. Respiratory syncytial virus disease in infants despite prior administration of antigenic inactivated vaccine. Am J Epidemiol. 1969;89:422–434. doi: 10.1093/oxfordjournals.aje.a120955. [DOI] [PubMed] [Google Scholar]
- 5.Fulginiti VA, Eller JJ, Sieber OF, Joyner JW, Minamitani M, Meiklejohn G. Respiratory virus immunization. I. A field trial of two inactivated respiratory virus vaccines: an aqueous trivalent parainfluenza virus vaccine and an alum-precipitated respiratory syncytial virus vaccine. Am J Epidemiol. 1969;89:436–448. doi: 10.1093/oxfordjournals.aje.a120956. [DOI] [PubMed] [Google Scholar]
- 6.Kapikian AZ, Mitchell RH, Chanock RM, Shvedoff RA, Stewart CE. An epidemiologic study of altered clinical reactivity to respiratory syncytial (RS) virus infection in children previously vaccinated with an inactivated RS virus vaccine. Am J Epidemiol. 1969;89:405–421. doi: 10.1093/oxfordjournals.aje.a120954. [DOI] [PubMed] [Google Scholar]
- 7.Chin J, Magoffin RL, Shearer LA, Schieble JH, Lennette EH. Field evaluation of a respiratory syncytial virus vaccine and a trivalent parainfluenza virus vaccine in a pediatric population. Am J Epidemiol. 1969;89:449–463. doi: 10.1093/oxfordjournals.aje.a120957. [DOI] [PubMed] [Google Scholar]
- 8.Openshaw PJM, Clarke SL, Record FM. Pulmonary eosinophilic response to respiratory syncytial virus infection in mice sensitized to the major surface glycoprotein G. Int Immunol. 1992;4:493–500. doi: 10.1093/intimm/4.4.493. [DOI] [PubMed] [Google Scholar]
- 9.Levine S, Klaiber R, Franco, Paradiso PR. Demonstration that glycoprotein G is the attachment protein of respiratory syncytial virus. J Gen Virol. 1987;68:2521–2524. doi: 10.1099/0022-1317-68-9-2521. [DOI] [PubMed] [Google Scholar]
- 10.Alwan WH, Kozlowska WJ, Openshaw PJM. Distinct types of lung disease caused by functional subsets of antiviral T cells. J Exp Med. 1994;179:81–89. doi: 10.1084/jem.179.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Srikiatkhachorn A, Braciale TJ. Virus-specific memory and effector T lymphocytes exhibit different cytokine responses to antigens during experimental murine respiratory syncytial virus infection. J Virol. 1997;71:678–685. doi: 10.1128/jvi.71.1.678-685.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graham BS, Bunton LA, Wright PF, Karzon DT. Role of T lymphocyte subsets in the pathogenesis of primary infection and rechallenge with respiratory syncytial virus in mice. J Clin Invest. 1991;88:1026–1033. doi: 10.1172/JCI115362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hussell T, Baldwin CJ, O'Garra A, Openshaw PJM. CD4+T-cells control Th2-driven pathology during pulmonary respiratory syncytial virus infection. Eur J Immunol. 1997;27:3341–3349. doi: 10.1002/eji.1830271233. [DOI] [PubMed] [Google Scholar]
- 14.Garcia Barreno, B., A. Portela, T. Delgado, J.A. Lopez, and J.A. Melero. Frame shift mutations as a novel mechanism for the generation of neutralization resistant mutants of human respiratory syncytial virus. EMBO (Eur Mol Biol Organ) J. 1990;9:4181–4187. doi: 10.1002/j.1460-2075.1990.tb07642.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arbiza J, Taylor G, Lopez JA, Furze J, Wyld S, Whyte P, Stott J, Wertz G, Sullender W, Trudel M, Melero JA. Characterization of two antigenic sites recognized by neutralizing monoclonal antibodies directed against the F glycoprotein of human respiratory syncytial virus. J Gen Virol. 1992;73:2225–2234. doi: 10.1099/0022-1317-73-9-2225. [DOI] [PubMed] [Google Scholar]
- 16.Simard C, Nadon F, Sèguin C, Trudel M. Evidence that the amino acid region 124–203 of glycoprotein G from the respiratory syncytial virus (RSV) constitutes a major part of the polypeptide domain that is involved in the protection against RSV infection. Antiviral Res. 1995;28:303–315. doi: 10.1016/0166-3542(95)00053-4. [DOI] [PubMed] [Google Scholar]
- 17.Stott EJ, Ball LA, Young KK, Furze J, Wertz GW. Human respiratory syncytial virus glycoprotein G expressed from recombinant vaccinia virus vector protects mice against live virus challenge. J Virol. 1986;60:607–613. doi: 10.1128/jvi.60.2.607-613.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wertz GW, Stott EJ, Young KKJ, Anderson K, Ball LA. Expression of the fusion protein of human respiratory syncytial virus from recombinant vaccinia virus vectors and protection of vaccinated mice. J Virol. 1987;61:293–301. doi: 10.1128/jvi.61.2.293-301.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hussell T, Spender LC, Georgiou A, O'Garra A, Openshaw PJM. Th1 and Th2 cytokine induction in pulmonary T-cells during infection with respiratory syncytial virus. J Gen Virol. 1996;77:2447–2455. doi: 10.1099/0022-1317-77-10-2447. [DOI] [PubMed] [Google Scholar]
- 19a.Tebbey, P.W., and G.E. Hancock. 1997. Characterization of immune responses to the attachment (G) protein of respiratory syncytial virus in inbred mice. 10th International Conference on Negative Strand Viruses. A152 (Abstr.).
- 20.Rammensee HG, Friede T, Stevanoviic S. MHC ligands and peptide motifs: first listing. Immunogenetics. 1995;41:178–228. doi: 10.1007/BF00172063. [DOI] [PubMed] [Google Scholar]
- 21.Van der Veen RC. Immunogenicity of JHM virus proteins: characterization of a CD4+T cell epitope on nucleocapsid protein which induces different T-helper cell subsets. Virology. 1996;225:339–346. doi: 10.1006/viro.1996.0608. [DOI] [PubMed] [Google Scholar]
- 22.Milich DR, Peterson DL, Schodel F, Jones JE, Hughes JL. Preferential recognition of hepatitis B nucleocapsid antigens by Th1 or Th2 cells is epitope and major histocompatibility complex dependent. J Virol. 1995;69:2776–2785. doi: 10.1128/jvi.69.5.2776-2785.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haraguchi, S., R.A. Good, M.J. Yarish, G.J. Cianciolo, and N.K. Day. 1995. Differential modulation of Th1- and Th2-related cytokine mRNA expression by a synthetic peptide homologous to a conserved domain within retroviral envelope protein. Proc. Natl. Acad. Sci. USA. 92:3611–3615. (See published erratum 92:9009.) [DOI] [PMC free article] [PubMed]
- 24.Pfeiffer C, Stein J, Southwood S, Ketelaar H, Sette A, Bottomly K. Altered peptide ligands can control CD4 T lymphocyte differentiation in vivo. J Exp Med. 1995;181:1569–1574. doi: 10.1084/jem.181.4.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alwan WH, Record FM, Openshaw PJM. Phenotypic and functional characterization of T cell lines specific for individual respiratory syncytial virus proteins. J Immunol. 1993;150:5211–5218. [PubMed] [Google Scholar]
- 26.Graham BS, Henderson GS, Tang YW, Lu X, Neuzil KM, Colley DG. Priming immunization determines T helper cytokine mRNA expression patterns in lungs of mice challenged with respiratory syncytial virus. J Immunol. 1993;151:2032–2040. [PubMed] [Google Scholar]
- 27.Collins PD, Marleau S, Griffiths DA, Johnson, Jose PJ, Williams TJ. Cooperation between interleukin-5 and the chemokine eotaxin to induce eosinophil accumulation in vivo. J Exp Med. 1995;182:1169–1174. doi: 10.1084/jem.182.4.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saito T, Deskin RW, Casola A, Haeberle H, Olszewska B, Ernst PB, Alam R, Ogra PL, Garofalo R. Respiratory syncytial virus induces selective production of the chemokine RANTES by upper airway epithelial cells. J Infect Dis. 1997;175:497–504. doi: 10.1093/infdis/175.3.497. [DOI] [PubMed] [Google Scholar]
- 29.Hussell T, Khan U, Openshaw PJM. IL-12 treatment attenuates Th2 and B cell responses but does not improve vaccine-enhanced lung illness. J Immunol. 1997;159:328–334. [PubMed] [Google Scholar]
- 30.Orange JS, Salazar TP, Mather, Opal SM, Spencer RL, Miller AH, McEwen BS, Biron CA. Mechanism of interleukin 12-mediated toxicities during experimental viral infections: role of tumor necrosis factor and glucocorticoids. J Exp Med. 1995;181:901–914. doi: 10.1084/jem.181.3.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Srikiatkhachorn A, Braciale TJ. Virus specific CD4+T lymphocytes downregulate T helper cell type 2 cytokine secretion and pulmonary eosinophilia during experimental murine respiratory syncytial virus infection. J Exp Med. 1997;186:421–432. doi: 10.1084/jem.186.3.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pemberton RM, Cannon MJ, Openshaw PJM, Ball LA, Wertz GA, Askonas BA. Cytotoxic T-cell specificity for respiratory syncytial virus proteins: fusion protein is an important target antigen. J Gen Virol. 1987;68:2177–2182. doi: 10.1099/0022-1317-68-8-2177. [DOI] [PubMed] [Google Scholar]
- 33.Alwan WH, Openshaw PJM. Distinct patterns of T and B cell immunity to respiratory syncytial virus induced by individual proteins. Vaccine. 1993;11:431–437. doi: 10.1016/0264-410x(93)90284-5. [DOI] [PubMed] [Google Scholar]
- 34.Alwan WH, Record FM, Openshaw PJM. CD4+ T cells clear virus but augment disease in mice infected with respiratory syncytial virus: comparison with the effects of CD4+cells. Clin Exp Immunol. 1992;88:527–536. doi: 10.1111/j.1365-2249.1992.tb06482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]