Abstract
Tumor necrosis factor (TNF) and interleukin 1 are known to initiate endothelial vascular cell adhesion molecule (VCAM)-1 transcription primarily by activating nuclear factor (NF)-κB, which translocates to the nucleus. In addition to two NF-κB elements found within the minimal cytokine-inducible VCAM-1 promoter, an interferon-related factor (IRF) element (IRF-1) has been identified close to the transcription initiation site, suggesting that cytokines that induce IRF-1 might affect VCAM-1 expression levels. We therefore investigated the effects of interferons (IFNs), which strongly induce IRF-1, on VCAM-1 transcription and expression. We show that IFN-α and -γ enhance TNF-induced VCAM-1 mRNA transcription and protein expression in human endothelial cells. IFN enhancement of TNF-induced expression is also seen using chloramphenicol acetyl transferase reporter genes linked to the minimal cytokine inducible VCAM-1 promoter. Nuclear IRF-1 is the molecular basis of IFN enhancement, because (a) IFN plus TNF–treated cells displayed increased nuclear IRF-1 levels and increased IRF-1 binding to the VCAM-1 promoter, compared with cells treated with TNF alone; (b) kinetics of nuclear IRF-1 levels correlated with VCAM-1 mRNA levels; (c) transfection with an IRF-1 construct substituted for IFN treatment; and (d) transfection with an expression construct encoding IRF-2, a competitive inhibitor of IRF-1, reduced TNF-induced VCAM-1 expression. Our experiments show that IFN amplifies TNF-induced VCAM-1 expression at the transcriptional level by an IRF-1–dependent pathway.
Keywords: vascular cell adhesion molecule 1, endothelium, interferon, interferon-related factor 1, skin microvasculature
Vascular adhesion molecule (VCAM)-11 (CD106) is a cytokine-inducible molecule expressed on the surface of endothelial cells. Since lymphocyte recruitment largely depends on VCAM-1 expression (1–4), it is of great biological relevance to understand mechanisms regulating the transcription of this particular molecule. So far, TNF (also called TNF-α) and (less potent) IL-1 are known to induce endothelial VCAM-1 expression at the transcriptional level (5). They are thought to drive VCAM-1 gene transcription predominantly by way of nuclear factor (NF)-κB (5–7). NF-κB is translocated from the cytoplasm into the nucleus, where it peaks 30 min after stimulation (8–10). VCAM-1 gene transcription starts as early as 30–60 min after stimulation, but peak levels of VCAM-1 mRNA are not reached before 4 h (11), providing indirect evidence that in addition to NF-κB, another factor(s) may be important in the amplification of VCAM-1 gene transcription in endothelial cells.
Additional evidence for such an “amplification factor” comes from in vivo studies. Endothelial cells of most organs produce high amounts of VCAM-1 in response to TNF or IL-1, but certain vascular beds, such as glomerular or dermal endothelium, respond poorly to these cytokines (12, 13). However, this is not based on a principle inability of glomerular or dermal endothelial cells to produce VCAM-1, because, in the presence of an inflammatory infiltrate, they express high levels of VCAM-1 (13–15), implying that aside from TNF or IL-1, additional factors might be important that drive endothelial VCAM-1 gene expression.
The minimal cytokine-inducible promoter spans the base pairs from −85 to +20 and encloses not only the tandem repeat of the NF-κB site but also a Sp1 and an interferon-related factor (IRF)-1–binding site close to the TATA-Box (11, 16–18). NF-κB is critical for VCAM-1 transcription, since selective overexpression of the NF-κB subunit p65 is sufficient to induce transcription and mutation or deletion of the NF-κB sites abolishes cytokine-inducible gene transcription (7). Sp1 is constitutively bound to its specific binding site on the VCAM-1 promoter, does not interact with NF-κB, and appears not to participate in cytokine-induced VCAM-1 gene transcription (18). IRF-1 is a member of the interferon-related factor family. It is induced after stimulation with TNF, binds to the IRF-1 binding site of the VCAM-1 promoter, and interacts with the p50 subunit of NF-κB (11, 19). However, its role in the transcriptional regulation of the native VCAM-1 gene in endothelial cells is unknown. If IRF-1 is indeed a costimulator of NF-κB–induced VCAM-1 gene transcription in endothelium, it is surprising that IFNs, which strongly induce IRF-1 synthesis, have yet not been identified as costimulators of TNF-induced VCAM-1 gene transcription. In this study, we demonstrate that IFN-α as well as IFN-γ indeed amplify TNF-induced VCAM-1 mRNA transcription and protein expression in human endothelial cells by an IRF-1–dependent pathway.
Materials and Methods
Cells.
Human umbilical vein endothelial cells (HUVEC) were isolated and cultured according to standard procedures as previously described (13). Cells were used at passages 4–6.
Determination of VCAM-1 and Endothelial Leukocyte Adhesion Molecule 1 Surface Expression.
Confluent monolayers of HUVEC were treated according to two different protocols. For the first, HUVEC were treated for the indicated times with TNF (100 U/ml; Genzyme Corp., Cambridge, MA) together with IFN-α (30 ng/ml; Aesca, Vienna, Austria). For the second protocol, HUVEC were treated with IFN-α for indicated time periods, then TNF was added as indicated. In selected experiments, IFN-γ (30 ng/ml; Endogen, Woburn, MA) was used instead of IFN-α. The TNF dosage used has been shown previously to be optimal for the induction of VCAM-1 surface expression (13). The dosage of IFN-α used induced maximal endothelial HLA class I expression and altered cell morphology from a cobblestone to a spindle shape.
Surface expression of VCAM-1 and endothelial leukocyte adhesion molecule (ELAM)-1 was quantified by direct immunofluorescence and flow cytofluorometry using a FACScan® (Becton Dickinson, San Jose, CA) as previously described (13). In brief, single-cell suspensions derived from confluent monolayers were incubated with PE-conjugated mouse anti–human VCAM-1 (1.G11B1), ELAM-1 (1.2B6; both mouse IgG1; Ancell, Läufelingen, Switzerland), or isotype control mAbs (1 μg/ml each). 5,000 cells were counted, and the geometric mean fluorescence levels were determined for each experiment and treatment condition; mean ± SD values from five independent experiments were calculated. Statistics were performed using the Student's t test.
RNA Isolation and Northern Hybridization.
To measure VCAM-1 mRNA levels, HUVEC were preincubated with or without IFN-α for 24 h, followed by TNF stimulation for 1 to 24 h. To measure VCAM-1 mRNA stability, HUVEC were preincubated with or without IFN-α for 24 h, followed by a 4-h TNF pulse. Actinomycin D (2.5 μg/ml; Calbiochem, La Jolla, CA) was then added for 1 to 8 h. After three washes with ice cold PBS, cells were lysed in Hepes buffer A (10 mM, pH 7.8) containing 1.5 mM MgCl, 10 mM KCl, 1 mM dithiothreitol, 0.1% NP-40, and 1 U/ml RNasin (Promega, Madison, WI) for 10 min on ice. Lysates were then centrifuged for 10 min (4°C; 10,000 g), the cytoplasmic supernatants were used for mRNA isolations; and the nuclei were used for preparation of nuclear protein extracts (see below).
Total RNA was isolated by hot phenol extraction as previously described (20). Probes for VCAM-1 (a cocktail of six oligonucleotide antisense probes, R&D Systems, Inc., Minneapolis, MN) and GAPDH (cDNA of 1.1 kb; Clontech, Palo Alto, CA) were labeled with 32P by using the 5′-cDNA labeling kit from Pharmacia Biotech AB (Uppsala, Sweden) according to the manufacturer's instructions. After separation of endothelial RNA on 0.8% agarose gels containing 4 M urea, gels were dried. After prehybridization in ExpressHyb solution (Clontech) for 1 h at 65°C, 32P-labeled VCAM-1 or GAPDH probe (2 ng/ml each) was added and incubated at 65°C for 12 h, followed by washing with 2× SSC (0.3 M NaCl, 30 mM Na3 citrate) first, then in 2× SSC plus 0.1% SDS. The gels were then exposed to Kodak films (Eastman Kodak Co., Rochester, NY) for 1 or 2 d. Northern signals were determined by densitometry (Kodak electrophoresis documentation and analysis system 120) and normalized to their 28S RNA bands.
Western Blots.
Nuclear protein extracts were prepared as previously described (21), normalized according their protein concentrations, and electrophoresed on SDS/8% polyacrylamide gels and blotted onto nitrocellulose in Tris buffer (25 mM) containing 192 mM glycine and 5% methanol at 100 V/15 mA for 1 h. Membranes were incubated with first-step antibodies (rabbit anti–IRF-1, 1:100; rabbit anti–NF-κB/p65, 1:2,000; rabbit anti– Sp-1, 1:500 (all Santa Cruz Biotechnology, Santa Cruz, CA) or control rabbit serum diluted in PBS containing 1% low-fat milk for 2 h at room temperature. After rinsing, blots were incubated with horseradish peroxidase–conjugated goat anti–rabbit secondary antibody (1:100,000; Bio-Rad Laboratories, Hercules, CA), visualized by chemiluminescence (ECL system; Amersham Corp., Buckinghamshire, UK), and recorded on film.
Electrophoretic Mobility Shift Analyses.
Nuclear proteins were extracted as previously described (21). The oligonucleotides used were as follows: VCAM–NF-κB (5′-CTGCCCTGGGTTTCCCCTTGAAGGGATTTCCCTCCGCCTCTGCAACAAGCTCGAGATCCTATG-3′), with the two NF-κB–like sites at positions −77 bp and −63 bp in bold; and VCAM–IRF-1 (5′-AAGCACAGACTTTCTATTTCACTCCGAGATCCTATG-3′), with the IRF-1 site at position −16 bp in bold. The underlined sequence represents an unrelated tail sequence and served as a template for the synthesis of the second strand. Complementary primers were extended with Klenow fragment in the presence of 50 mCi of α-[32P]dCTP (Amersham Corp.), and unlabeled dATP, dGTP, and dTTP, followed by the addition of unlabeled dCTP to ensure completion of the second strand. To generate cold double-stranded probes, unlabeled dCTP instead of labeled dCTP was used. The DNA–protein-binding reaction was performed in a total volume of 20 μl, containing 10 μg of nuclear protein, 50 μg fetal bovine albumin, 105 cpm 32P-labeled probe (∼1 ng), and 2 μg poly[dI:dC] in 4× binding buffer (1× buffer: 12 mM Hepes, pH 7.8; 4 mM Tris; 60 mM KCl; 1 mM EDTA; 12% vol./vol. glycerol; 1 mM dithiothreitol; and 1 mM PMSF). After incubation for 30 min at room temperature, the mixtures were electrophoresed on native 4% polyacrylamide gels for 4 h at 120 V/10–12 mA, dried, and exposed to films. Competitor (in a 100-fold molar excess) or anti–IRF-1 antibody (10 μg), both from Santa Cruz Biotechnology, were added 15 min before the labeled probe.
Transient Transfection with VCAM-1 Promoter CAT Reporter Constructs.
HUVEC at 70–80% confluence in 28 cm2 dishes were transfected with 20 μg of the −85 VCAM-1 CAT promoter construct (16) using standard calcium phosphate precipitation techniques (21). To control for transfection efficiency, 3 μg of a pSV–β-galactosidase control vector (Promega Corp., Madison, WI) was cotransfected in each experiment. 2 d after transfection, cells were stimulated with TNF with or without IFN as indicated. Cells were then harvested by trypsination, pelleted, rinsed, and resuspended in 150 ml of 0.25 M Tris-HCl, pH 8.0. After three cycles of freezing and thawing, and centrifugation at 14,000 g for 10 min, supernatants were collected. β-galactosidase activity was determined by using the Enzyme Assay System from Promega Corp. The amount of cell lysates used for CAT assays were normalized according to their β-galactosidase activity, and variation of β-galactosidase activity within individual experiments was <10%. CAT assays were performed using the CAT Enzyme Assay from Promega Corp. according to the manufacturer's instruction. The reaction was stopped by the extraction of butyrylated [14C]chloramphenicol in 300 μl of mixed xylenes. 200 μl of the resulting xylene phase was mixed with 2 ml scintillation fluid and the activity was measured in a liquid scintillation counter (Wallac, Turku, Finland).
Transient Transfection with IRF-1 and -2 Plasmids.
IRF-1, IRF-2, plasmids or the appropriate parent control vector have been described previously (22). It has been demonstrated previously that proteins derived from these mouse IRF genes transactivate human and monkey genes (22–24). Transfection was performed by standard calcium phosphate precipitation techniques as described elsewhere (21). 72 h later, cells were treated with TNF as indicated and harvested by trypsination, and ELAM-1 and VCAM-1 surface expressions were determined by FACS® as described above. All plasmids were tested for LPS contamination. To control for transfection efficiency, cells were transfected with green fluorescent protein (pEGFP; Clontech) and green fluorescent cells were determined by FACS® 2–4 d later. After excitation with 488 nm, ∼10% at day 2, ∼20% at day 3, and up to 25% of total cells at day 4 emitted green fluorescence. The mean fluorescence of positive cells was ∼10–100-fold above the negative control.
Skin Organ Culture and Immunohistochemistry.
Normal human skin was obtained from discarded human tissue according to a protocol approved by the University of Vienna Ethics Committee (Vienna, Austria). Organ cultures were performed as previously described (13). In brief, human skin was placed into RPMI 1640 culture medium (GIBCO BRL, Gaithersburg, MD) supplemented with 10% (vol/vol) FCS (GIBCO BRL). Where indicated, the medium was supplemented with recombinant TNF (1,000 U/ml) with or without IFN-α or IFN-γ (100 ng/ml each). After incubation in humidified 5% CO2/95% air at 37°C for 24 h, explants were harvested, snap frozen, and cryosectioned. Immunohistochemistry was performed by using mAb H18/7 (mouse IgG1 anti–ELAM-1, 1 μg/ml), mAb E1/6 (mouse IgG1 anti–VCAM-1, 1 μg/ml; both from Becton Dickinson), or mouse IgG1 isotype control as first-step reagents as previously described (13). Within the papillary dermis, numbers of ELAM-1– or VCAM-1–positive vessels were counted in two different sections in each specimen by viewing a total of six randomly selected high power fields (×40 objective) per specimen and treatment condition by two observers blinded to the treatment conditions. Values are given as mean ± SD.
Results and Discussion
Interferon Enhances TNF-induced VCAM-1 Protein Surface Expression.
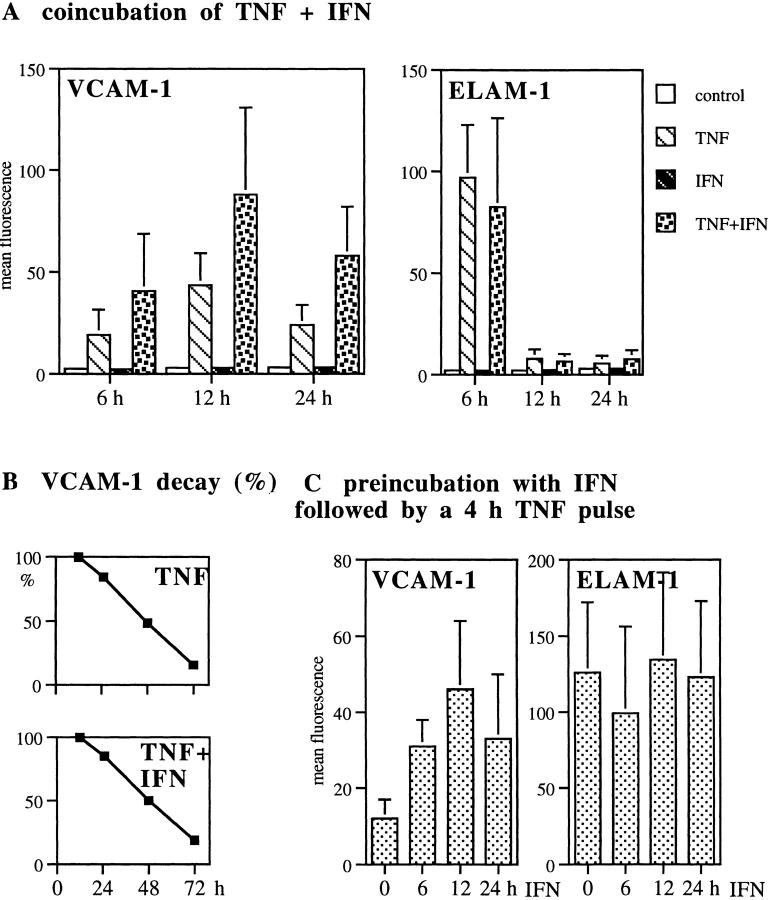
VCAM-1 expression levels on HUVEC stimulated with IFN plus TNF significantly exceeded those induced by TNF alone at each time point (P <0.05; Fig. 1 A). VCAM-1 expression peaked 12–13 h after TNF treatment in both treatment protocols (Fig. 1 A). The decay of VCAM-1 expression was followed for up to 72 h. In both treatment protocols, VCAM-1 expression continuously decreased, with parallel curve slopes (data not shown). When the decay was expressed as a percentage of their respective 12-h peak levels, the decay curves overlapped (Fig. 1 B). VCAM-1 expression was absent in controls as well as in IFN-treated cells at all time points (Fig. 1 A).
Figure 1.
IFN enhances TNF-induced VCAM-1 protein surface expression. VCAM-1 and ELAM-1 surface expression levels were determined by FACS® as described above. (A) HUVEC were treated with TNF or with IFN-α plus TNF for the indicated times. Mean ± SD values of five independent experiments are given. (B) VCAM-1 surface protein decay. 12-h peak levels were considered 100% and VCAM-1 expression at later times were calculated as the percentage of their respective peak levels. Results reflect one out of three representative experiments. (C) HUVEC were preincubated with IFN-α followed by TNF pulse for 4 h. The difference between VCAM-1 geometric mean fluorescence levels in TNF versus TNF plus IFN-α groups was significant at each time point in A and B; P <0.05.
We next tested whether preincubation of HUVEC with IFN would enhance the ascent in VCAM-1 expression in response to a 4-h TNF pulse. TNF alone for 4 h induced minimal VCAM-1 expression, not significantly above the level of the negative control. In contrast, IFN preincubation for 6–24 h followed by a TNF pulse for 4 h produced significant VCAM-1 expression levels, which equaled VCAM-1 peak levels obtained by a 12-h TNF pulse (Fig. 1 C).
Thus, the combination of IFN-α plus TNF had two effects: first, coincubation experiments indicate that IFN acts in synergy with TNF to ‘superinduce' VCAM-1 expression without altering VCAM-1 decay; and second, preincubation with IFN appears to prime cells to respond more rapidly and more efficiently to TNF exposure. Dose titration revealed that IFN effects are dose dependent (data not shown). IFN-γ (30 ng/ml) plus TNF was as effective as IFN-α plus TNF in increasing VCAM-1 surface expression (data not shown). Most importantly, ELAM-1 expression, which served as a negative control for IFN effects in each experiment, was readily induced by TNF but remained unaffected by IFN co- or preincubation (Fig. 1, A and C).
Interferon Enhances TNF-induced VCAM-1 mRNA.
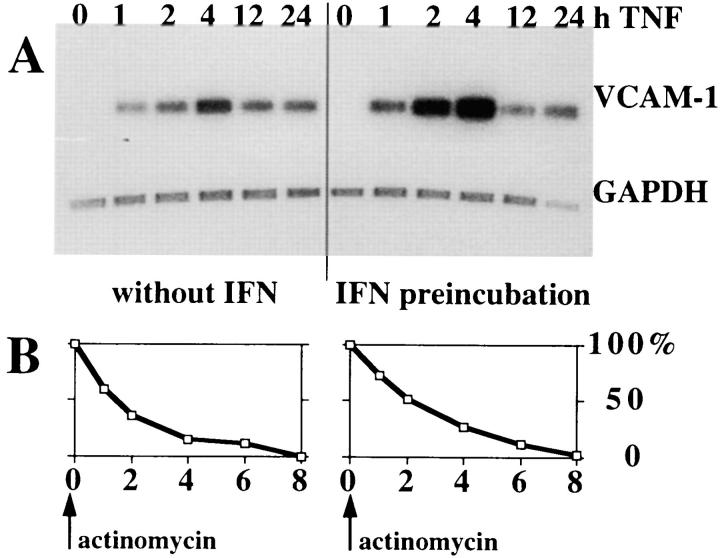
By Northern hybridization, IFN pretreatment before TNF pulse resulted in a significant increase in VCAM-1 mRNA levels as compared with TNF alone. Strong induction was already seen between 1 and 2 h after stimulation (up to four times above the level induced with TNF alone), but VCAM-1 mRNA expression peaked 4 h after stimulation in both treatment protocols. VCAM-1 mRNA was undetectable in unstimulated HUVEC as well as in cells treated with IFN alone (Fig. 2 A) To determine the effects of IFN on VCAM-1 mRNA stability, IFN-pretreated or untreated HUVEC were pulsed with TNF for 4 h. After transcription was stopped by actinomycin D, VCAM-1 mRNA gradually decreased and was virtually absent 8 h after actinomycin treatment in both TNF- and IFN plus TNF– treated cells. Decay rates were equal in both treatment protocols (Fig. 2 B), with VCAM-1 mRNA half-life time being 90 and 120 min, respectively (difference not significant). Therefore, we conclude that IFN enhances TNF- induced VCAM-1 expression at the transcriptional level.
Figure 2.
IFN enhances TNF-induced VCAM-1 mRNA expression without effecting the mRNA stability. (A) HUVEC were preincubated with or without IFN-α followed by TNF stimulation. Total RNA was isolated at the indicated time points. VCAM-1 mRNA expression was determined by Northern hybridization (one out of two independent experiments). (B) VCAM-1 mRNA decay; mRNA production was stopped after a 4-h TNF pulse by the addition of actinomycin D and mRNA was isolated at indicated times thereafter. 4-h peak mRNA levels were considered 100%, and VCAM-1 mRNA at later time points were calculated as the percentage of their respective 4-h peak levels; mean of three experiments shown.
Interferon Enhances TNF-induced VCAM-1 Reporter Gene Expression.
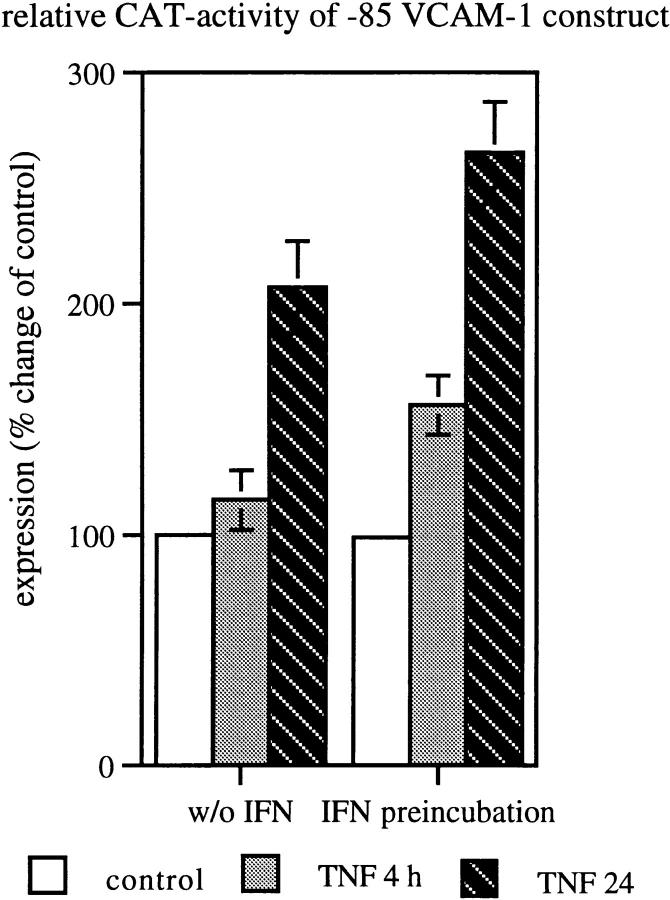
Since previous experiments using sequential deletion of the VCAM-1 promoter identified the region spanning the bps −85 to +20 of the 5′ flanking region as the minimal cytokine inducible promoter element that confers TNF response, we investigated whether this short element is sufficient to mediate the synergistic effect of TNF and IFN. We transiently transfected HUVEC with the −85 VCAM-CAT construct encompassing a tandem repeat of the NF-κB binding site, an Sp1 site, and an IRF-1 site. In contrast to TNF alone, IFN preincubation followed by a 4- or 24-h TNF pulse induced a significant increase of CAT activity as early as 4 h after stimulation (P <0.05). Comparable to previous results (11, 17), TNF alone induced only minimal CAT activity at this time point. Moreover, peak CAT activity seen at 24 h after stimulation was significantly higher in IFN plus TNF as compared with TNF alone (P <0.05; Fig. 3). These results demonstrate that the cooperative effects of IFN on TNF-induced VCAM-1 gene expression is mediated by transcription factors binding to this particular region.
Figure 3.
IFN enhances TNF-induced VCAM-1 reporter gene expression. 72 h after transfection, HUVEC were treated with TNF for 4 or 24 h or preincubated with IFN-α before TNF stimulation. Mean ± SD values of three independent experiments are given, P <0.05 between TNF and TNF plus IFN groups.
The Amount of Nuclear IRF-1 Correlates with VCAM-1 Expression Levels.
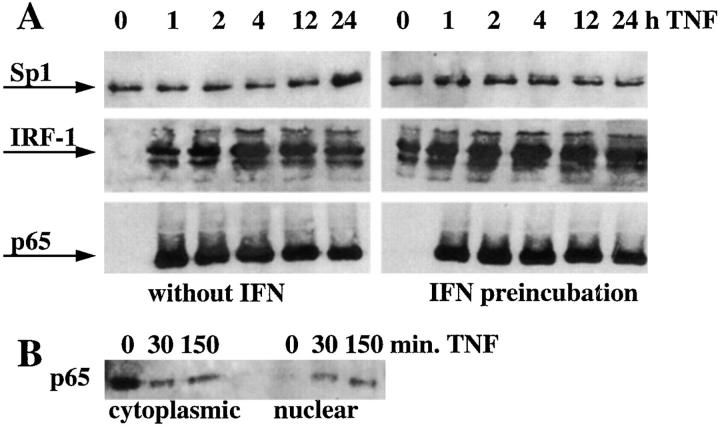
The minimal cytokine inducible promoter element of VCAM-1 contains identified binding sites for three transcription factors, NF-κB, Sp1, and IRF-1. Assuming that the synergistic effect of IFN plus TNF on VCAM-1 gene transcription depends on one of these factors, we measured nuclear protein levels of p65, Sp1, and IRF-1. Nuclear p65 is virtually undetectable in unstimulated cells but peaked as early as 30 min after TNF stimulation (Fig. 4 B), and is continuously detectable within the nucleus at later time points in TNF as well as after IFN plus TNF–treated cells in comparable amounts (Fig. 4 A). Nuclear Sp1 is continuously detectable within the nucleus in untreated as well as treated cells in equal amounts (Fig. 4 A), which is consistent with the fact that Sp1 is a constitutive transcription factor (25). Nuclear IRF-1 was undetectable in unstimulated cells and peaked 4 h after TNF stimulation. Most importantly, in HUVEC preincubated with IFN, nuclear IRF-1 was induced even before the addition of TNF. A subsequent TNF pulse caused a steep and strong rise in nuclear IRF-1 levels, three to four times above the amount of that seen in cells treated with TNF alone (Fig. 4 A).
Figure 4.
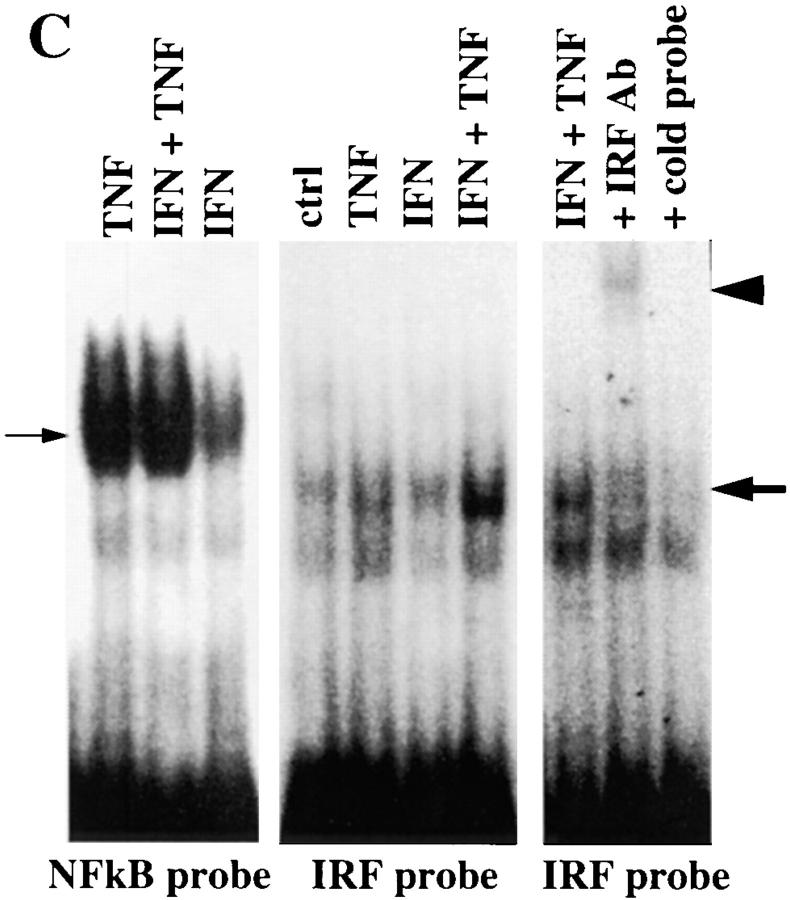
IFN enhances nuclear IRF-1 but does not affect nuclear NF-κB or Sp-1 in TNF-treated cells. (A) Nuclear transcription factors were examined by Western blotting at baseline or after stimulation with TNF for 1, 2, 4, 12, and 24 h or preincubated with IFN-α before TNF stimulation (representative result out of three independent experiments). (B) p65 levels within the cytoplasm continuously decrease and nuclear p65 proportionally increase in response to TNF, confirming efficient separation of cytoplasmic from nuclear proteins. (C) Electromobility shift assay using oligonucleotide probes for the IRF-1– or the NF-κB–binding sites of the VCAM-1 gene. Nuclear extracts were isolated from HUVEC, preincubated with or without IFN-α for 24 h, and pulsed with TNF for 1 h. Small arrow, NF-κB complex; large arrow, IRF-1 complex; arrowhead, supershifted IRF-1 complex; one out of three experiments shown.
To demonstrate that nuclear IRF-1 is indeed able to interact with the IRF-1–binding site of the VCAM-1 promoter, electromobility shift assays were performed (Fig. 4 C). Nuclear extracts isolated from HUVEC pretreated with IFN and pulsed with TNF for 1 h showed two to four times enhanced DNA–protein complex formation as compared with cells treated with TNF or IFN alone (Fig. 4 C). This complex was supershifted by the addition of anti– IRF-1 Ab. Specificity of the DNA–protein interaction was confirmed by competition with unlabeled VCAM-1 IRF probe in 100 M excess (Fig. 4 C), whereas competition with a nonspecific cold oligonucleotide had no effect (data not shown). Only traces of DNA–protein complexes were seen when nuclear extracts of untreated HUVEC were assayed.
In contrast to the situation seen with IRF-1, nuclear extracts from TNF- and from IFN plus TNF–treated cells interacted equally with the NF-κB–binding site of the VCAM-1 promoter (Fig. 4 C). From these results, we conclude that the synergistic effect of IFN plus TNF on VCAM-1 gene expression is independent from nuclear NF-κB levels, but correlates with nuclear IRF-1 levels. If true, overexpression of IRF-1 should have the same effect on TNF-induced VCAM-1 expression as does cytokine treatment with IFN. On the other hand, IRF-2, a competitive inhibitor of IRF-1, should reduce TNF-induced VCAM-1 expression.
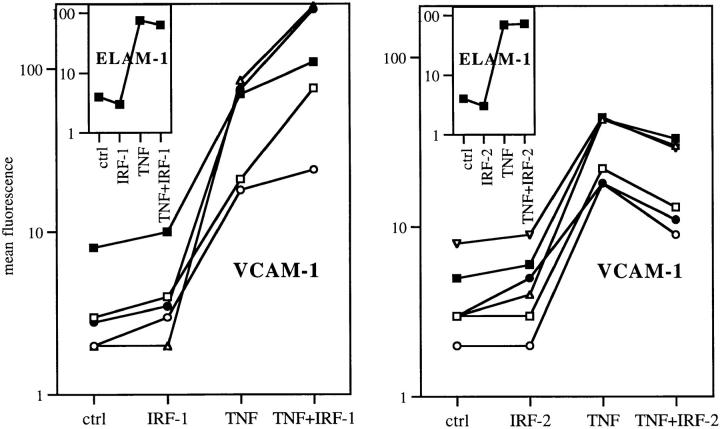
IRF-1 Transfection Enhances and IRF-2 Diminishes TNF-induced VCAM-1 Transcription.
72 h after transfection with IRF-1, IRF-2, or control vectors, HUVEC were pulsed with TNF for 4 h and VCAM-1 or ELAM-1 surface expression was measured by FACS®. Indeed, TNF produced significantly elevated amounts of VCAM-1 in IRF-1 transfectants and significantly reduced amounts of VCAM-1 in IRF-2 transfectants (P <0.05; Fig. 5). It should be noted that the synergistic effect of IRF-1 overexpression on TNF-induced VCAM-1 expression was less pronounced than that observed by treatment of cells with IFN plus TNF. However, keeping in mind that the transfection efficiency ranged from 10 to 20%, the effects of IRF-1 overexpression on TNF-induced VCAM-1 expression was well within the range of that seen in IFN-treated cells. Overexpression of IRF-1 per se did not affect VCAM-1 or ELAM-1 protein expression (Fig. 5), just as treatment with IFN did not induce VCAM-1 (Fig. 1). Moreover, neither transfection with IRF-1 nor with IRF-2 had any influence on TNF-induced ELAM-1 expression (Fig. 5).
Figure 5.
IRF-1 transfection enhances and IRF-2 diminishes TNF-induced VCAM-1 expression. 72 h after transfection, cells were pulsed with TNF for 4 h and protein surface expression was quantified by FACS®. The difference in VCAM-1 expression after TNF treatment was P <0.05 between controls and IRF-1, as well as between controls and IRF-2–transfected cells. Each line represents one independent experiment. Insets show ELAM-1 expression (mean of five experiments).
An alternative experimental approach to test the role of IRF-1 in VCAM-1 transcription is to use VCAM-1 reporter constructs bearing mutated IRF-1–binding motifs. However, such constructs are not reactive to any stimulus when placed into endothelium (11). It is not clear whether this is due to a technical problem or to an endothelial-specific phenomenon. Since mechanisms of VCAM-1 gene transcription in endothelial cells are different from, for example, smooth muscle cells or P19 carcinoma cells (26), results obtained with such nonendothelial cells cannot be easily used to predict mechanisms of the transcriptional regulation of the native VCAM-1 gene in endothelial cells. Our approach using IRF-1 or IRF-2 overexpression in endothelial cells clearly demonstrates that these two factors modulate TNF-induced transcription of the native VCAM-1 gene, even without using mutated constructs.
IFN Enhances TNF-induced VCAM-1 Expression In Vivo.
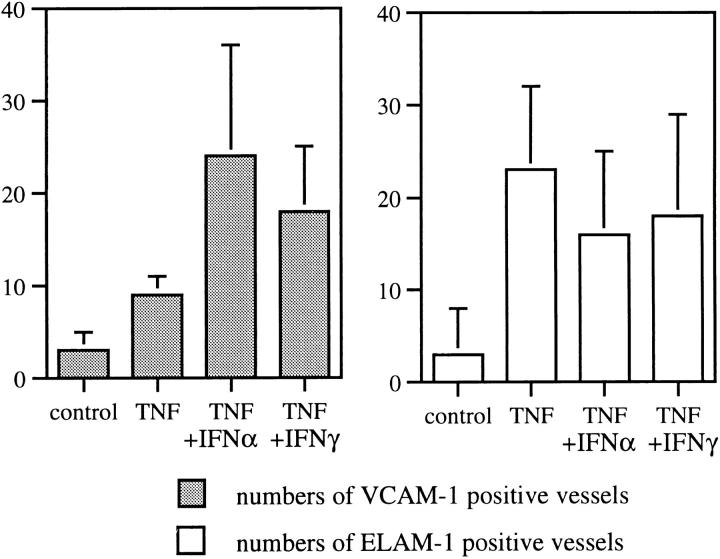
Under normal conditions, TNF induces only minimal amounts of VCAM-1 on endothelial cells of certain vascular beds, such as skin, glomeruli, or large arteries (12, 13, 15, 27). However, under conditions of inflammation, TNF strongly induces VCAM-1 expression, e.g., on skin endothelial cells (13, 15), indicating the existence of an additional factor(s) that amplifies sensitivity to TNF. By using skin organ culture, we show that in contrast to TNF alone, the combination of IFN plus TNF effectively induced VCAM-1 expression of skin endothelial cells even in noninflamed skin (Fig. 6), suggesting that IFN provides the missing costimulation.
Figure 6.
IFN increases TNF-induced VCAM-1 expression on superficial vascular endothelium of human skin. After 24 h of organ culture, human skin was subjected to immunohistochemistry, and ELAM-1– and VCAM-1–positive vessels were quantified as described in Materials and Methods. Mean ± SD values of six independent experiments are given. The difference between numbers of VCAM-1–positive vessels in TNF versus IFN-α or -γ plus TNF was significant, P <0.05.
Conclusion.
We thus conclude that IFN-α and IFN-γ are able to (a) prime endothelial cells to respond more rapidly to a subsequent TNF exposure and (b) amplify TNF-driven VCAM-1 expression in endothelial cells. This costimulation is regulated at the transcriptional level and depends on the nuclear transcription factor IRF-1.
Since lymphocytes are the major source of such IFNs, this IRF-1–dependent costimulatory pathway may become operative when lymphocytes respond to a local stimulus such as antigen recognition and release IFNs. Moreover, this pathway appears to be of particular importance for the regulation of VCAM-1 expression by skin endothelial cells. The skin is a site naturally exposed to high amounts of toxins or antigens. The need for IFN in addition to TNF for efficient VCAM-1 expression in this particular vascular bed may represent a protective mechanism to raise the threshold for chronic lymphocytic infiltration.
Acknowledgments
The authors wish to thank Dr. D.C. Dean for providing the VCAM-1 CAT plasmids, and Markus Milacek and Ingrid Hammer for their excellent technical assistance.
This work was supported by grants P10317MED and P12240-MED from the Austrian Science Foundation, Vienna, Austria (to P. Petzelbauer).
Abbreviations used in this paper
- ELAM
endothelial leukocyte adhesion molecule
- HUVEC
human umbilical endothelial vein endothelial cells
- IRF
interferon-related factor
- NF
nuclear factor
- VCAM
vascular cell adhesion molecule
References
- 1.Pelletier RP, Ohye RG, Vanbuskirk A, Sedmak DD, Kincade P, Ferguson RM, Orosz CG. Importance of endothelial VCAM-1 for inflammatory leukocytic infiltration in vivo. J Immunol. 1992;149:2473–2481. [PubMed] [Google Scholar]
- 2.Orosz CG, Ohye RG, Pelletier RP, Van BA, Huang E, Morgan C, Kincade PW, Ferguson RM. Treatment with anti–vascular cell adhesion molecule 1 monoclonal antibody induces long-term murine cardiac allograft acceptance. Transplantation (Baltimore) 1993;56:453–460. doi: 10.1097/00007890-199308000-00039. [DOI] [PubMed] [Google Scholar]
- 3.Isobe M, Suzuki J, Yagita H, Okumura K, Yamazaki S, Nagai R, Yazaki Y, Sekiguchi M. Immunosuppression to cardiac allografts and soluble antigens by anti–vascular cellular adhesion molecule-1 and anti–very late antigen-4 monoclonal antibodies. J Immunol. 1994;153:5810–5818. [PubMed] [Google Scholar]
- 4.Nakajima H, Sano H, Nishimura T, Yoshida S, Iwamoto I. Role of vascular cell adhesion molecule 1/very late activation antigen 4 and intercellular adhesion molecule 1/lymphocyte function–associated antigen 1 interactions in antigen-induced eosinophil and T cell recruitment into the tissue. J Exp Med. 1994;179:1145–1154. doi: 10.1084/jem.179.4.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Osborn L, Hession C, Tizard R, Vassallo C, Luhowskyj S, Chi RG, Lobb R. Direct expression cloning of vascular cell adhesion molecule 1, a cytokine-induced endothelial protein that binds to lymphocytes. Cell. 1989;59:1203–1211. doi: 10.1016/0092-8674(89)90775-7. [DOI] [PubMed] [Google Scholar]
- 6.Ahmad M, Marui N, Alexander RW, Medford RM. Cell type–specific transactivation of the VCAM-1 promoter through an NF-kappa B enhancer motif. J Biol Chem. 1995;270:8976–8983. doi: 10.1074/jbc.270.15.8976. [DOI] [PubMed] [Google Scholar]
- 7.Shu HB, Agranoff AB, Nabel EG, Leung K, Duckett CS, Neish AS, Collins T, Nabel GJ. Differential regulation of vascular cell adhesion molecule 1 gene expression by specific NF-kappa B subunits in endothelial and epithelial cells. Mol Cell Biol. 1993;13:6283–6289. doi: 10.1128/mcb.13.10.6283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Read MA, Neish AS, Luscinskas FW, Palombella VJ, Maniatis T, Collins T. The proteasome pathway is required for cytokine-induced endothelial-leukocyte adhesion molecule expression. Immunity. 1995;2:493–506. doi: 10.1016/1074-7613(95)90030-6. [DOI] [PubMed] [Google Scholar]
- 9.Lederer JA, Liou JS, Kim S, Rice N, Lichtman AH. Regulation of NF-kappa B activation in T helper 1 and T helper 2 cells. J Immunol. 1996;156:56–63. [PubMed] [Google Scholar]
- 10.Read MA, Whitley MZ, Williams AJ, Collins T. NF-κB and IκBα: an inducible regulatory system in endothelial activation. J Exp Med. 1994;179:503–512. doi: 10.1084/jem.179.2.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neish AS, Read MA, Thanos D, Pine R, Maniatis T, Collins T. Endothelial interferon regulatory factor 1 cooperates with NF-kappa B as a transcriptional activator of vascular cell adhesion molecule 1. Mol Cell Biol. 1995;15:2558–2569. doi: 10.1128/mcb.15.5.2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Savage CO, Brooks CJ, Adu D, Richards G, Howie AJ. Cell adhesion molecule expression within human glomerular and kidney organ culture. J Pathol. 1997;181:111–115. doi: 10.1002/(SICI)1096-9896(199701)181:1<111::AID-PATH698>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 13.Petzelbauer P, Bender JR, Wilson J, Pober JS. Heterogeneity of dermal microvascular endothelial cell antigen expression and cytokine responsiveness in situ and in cell culture. J Immunol. 1993;151:5062–5072. [PubMed] [Google Scholar]
- 14.Pall AA, Howie AJ, Adu D, Richards GM, Inward CD, Milford DV, Richards NT, Michael J, Taylor CM. Glomerular vascular cell adhesion molecule-1 expression in renal vasculitis. J Clin Pathol. 1996;49:238–242. doi: 10.1136/jcp.49.3.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petzelbauer P, Pober JS, Keh A, Braverman IM. Inducibility and expression of microvascular endothelial adhesion molecules in lesional, perilesional, and uninvolved skin of psoriatic patients. J Invest Dermatol. 1994;103:300–305. doi: 10.1111/1523-1747.ep12394720. [DOI] [PubMed] [Google Scholar]
- 16.Iademarco MF, McQuillan JJ, Rosen GD, Dean DC. Characterization of the promoter for vascular cell adhesion molecule-1 (VCAM-1) J Biol Chem. 1992;267:16323–16329. [PubMed] [Google Scholar]
- 17.Neish AS, Williams AJ, Palmer HJ, Whitley MZ, Collins T. Functional analysis of the human vascular cell adhesion molecule 1 promoter. J Exp Med. 1992;176:1583–1593. doi: 10.1084/jem.176.6.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neish AS, Khachigian LM, Park A, Baichwal VR, Collins T. Sp1 is a component of the cytokine-inducible enhancer in the promoter of vascular cell adhesion molecule-1. J Biol Chem. 1995;270:28903–28909. doi: 10.1074/jbc.270.48.28903. [DOI] [PubMed] [Google Scholar]
- 19.Ten RM, Blank V, Le BO, Kourilsky P, Israel A. Two factors, IRF1 and KBF1/NF-kappa B, cooperate during induction of MHC class I gene expression by interferon alpha beta or Newcastle disease virus. C R Acad Sci III. 1993;316:496–501. [PubMed] [Google Scholar]
- 20.Kunstfeld R, Lechleitner S, Gröger M, Wolff K, Petzelbauer P. HECA-452+T cells migrate through superficial vascular plexus but not through deep vascular plexus endothelium. J Invest Dermatol. 1997;108:343–348. doi: 10.1111/1523-1747.ep12286483. [DOI] [PubMed] [Google Scholar]
- 21.Gille J, Swerlick RA, Lawley TJ, Caughman SW. Differential regulation of vascular cell adhesion molecule-1 gene transcription by tumor necrosis factor alpha and interleukin-1 alpha in dermal microvascular endothelial cells. Blood. 1996;87:211–217. [PubMed] [Google Scholar]
- 22.Harada H, Willison K, Sakakibara J, Miyamoto M, Fujita T, Taniguchi T. Absence of the type I IFN system in EC cells: transcriptional activator (IRF-1) and repressor (IRF-2) genes are developmentally regulated. Cell. 1990;63:303–312. doi: 10.1016/0092-8674(90)90163-9. [DOI] [PubMed] [Google Scholar]
- 23.Fujita T, Kimura Y, Miyamoto M, Barsoumian EL, Taniguchi T. Induction of endogenous IFN-alpha and IFN-beta genes by a regulatory transcription factor, IRF-1. Nature. 1989;337:270–272. doi: 10.1038/337270a0. [DOI] [PubMed] [Google Scholar]
- 24.Harada H, Fujita T, Miyamoto M, Kimura Y, Maruyama M, Furia A, Miyata T, Taniguchi T. Structurally similar but functionally distinct factors, IRF-1 and IRF-2, bind to the same regulatory elements of IFN and IFN-inducible genes. Cell. 1989;58:729–739. doi: 10.1016/0092-8674(89)90107-4. [DOI] [PubMed] [Google Scholar]
- 25.Saffer JD, Jackson SP, Annarella MB. Developmental expression of Sp1 in the mouse. Mol Cell Biol. 1991;11:2189–2199. doi: 10.1128/mcb.11.4.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iademarco MF, McQuillan JJ, Dean DC. Vascular cell adhesion molecule 1: contrasting transcriptional control mechanisms in muscle and endothelium. Proc Natl Acad Sci USA. 1993;90:3943–3947. doi: 10.1073/pnas.90.9.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hauser IA, Johnson DR, Madri JA. Differential induction of VCAM-1 on human iliac venous and arterial endothelial cells and its role in adhesion. J Immunol. 1993;151:5172–5185. [PubMed] [Google Scholar]