Abstract
Two types of photoreceptors, rods and cones, coexist in the vertebrate retina. An in-depth analysis of the retinal circuitry that transmits rod and cone signals has been hampered by the presence of intimate physical and functional connections between rod and cone pathways. By deleting the cyclic nucleotide-gated channel CNG3 we have generated a mouse lacking any cone-mediated photoresponse. In contrast, the rod pathway is completely intact in CNG3-deficient mice. The functional loss of cone function correlates with a progressive degeneration of cone photoreceptors but not of other retinal cell types. CNG3-deficient mice provide an animal model to dissect unequivocally the contribution of rod and cone pathways for normal retinal function.
Vision of vertebrates is conferred by the concerted action of two phototransduction pathways, the rod and the cone photoreceptor system (1, 2). Rods are responsible for vision at low light intensities, whereas color vision and vision at high light intensities are provided by cones. The neural circuits that transmit information from rods and cones to the ganglion cells are functionally linked to each other in a complex fashion at the level of photoreceptors and bipolar and ganglion cells (3–5). Photoreceptors respond to light by the closure of a cyclic nucleotide-gated (CNG) cation channel (6–8), causing hyperpolarization of the plasma membrane and decrease of the synaptic glutamate release. CNG channels are proposed to have a heterotetrameric structure consisting of α- and β-subunits (6–8). α-Subunits are characterized by their ability to induce the formation of functional channels when expressed alone in heterologous expression systems. In contrast, β-subunits do not give rise to a cGMP-activated current per se, but when coexpressed with α-subunits, they confer channel properties characteristic of native CNG channels such as single-channel flickering and increased sensitivity for cyclic nucleotides and channel blockers. Two types of α-subunits are present in vertebrate retina, CNG1 (9) and CNG3 (10, 11). Protein purification and molecular cloning revealed that CNG1 is expressed in rod outer segment and that it forms a heteromeric channel with the CNG4 β-subunit (9, 12, 13). The CNG3 protein was identified in cone photoreceptors by immunocytochemistry (11). Expression of CNG3 also was detected in sperm, kidney, heart, and brain (6, 8). The low density of cone photoreceptors within the retina of most mammals until now has prevented biochemical purification and, hence, a definite determination of the CNG channel subunits involved in forming the native cone channel. CNG channels also were detected in retinal cells different from photoreceptors such as bipolar cells (14), ganglion cells (15), and Müller glial cells (16). The primary sequences of these channels are not yet known. Similarly, the physiological significance of the CNG channel expression remains to be determined in most of the nonphotoreceptor cells of the retina.
To investigate the specific role of the CNG3 α-subunit within the retinal network in vivo, we have generated CNG3-deficient (CNG3−/−) mice by gene targeting. CNG3−/− mice completely lack cone-mediated light response and suffer from a progressive degeneration of cone photoreceptors. In contrast, rod photoreceptors of CNG3−/− mice are both structurally and functionally intact.
MATERIALS AND METHODS
Gene Targeting.
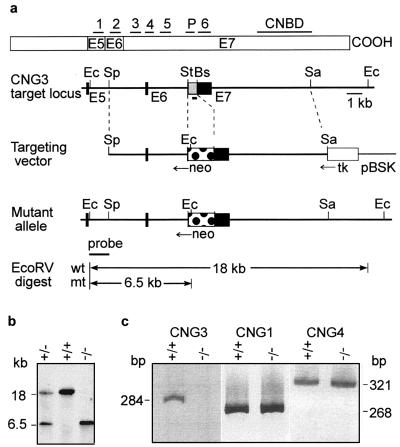
Murine CNG3 DNA was isolated from a genomic P1 library made from 129/Sv mouse tissue (Genome Systems, St. Louis). The exon–intron structure of the gene was resolved partially (Fig. 1a). Three exons were identified corresponding to exons 5–7 of the human CNG3 gene (17). A targeting vector was constructed by cloning a 5.1-kb SphI-StuI and a 7.4-kb BspEI-SacI fragment 5′ and 3′ of the neomycin-resistance expression cassette, respectively. The herpes simplex thymidine kinase cassette was cloned 3′ to the CNG3 sequence. Embryonic stem (ES) cell culture was done as described (18). Briefly, the targeting vector was linearized and electroporated into R1 ES cells. G418/gancyclovir-resistant clones were screened by Southern blot analysis, and ES cells carrying the disrupted allele were microinjected into blastocysts of mouse strain C57BL/6 to produce chimeric mice. Chimeric mice were crossed with strains C57BL/6 (outbred) and 129/Sv (inbred). Heterozygotes then were intercrossed to produce mice homozygous for the CNG3 mutation.
Figure 1.
Targeted disruption of the CNG3 gene. (a Upper) Structure of CNG3 protein showing the location of the six transmembrane segments (1–6), the ion-conducting pore (P), and the cyclic nucleotide-binding domain (CNBD). The portion of the protein encoded by exons (E) 5–7 is indicated. (Lower) Restriction maps of the CNG3 target locus, targeting vector, and mutant allele. Exons (E5–E7) and introns are indicated by solid boxes and lines, respectively. The 284-bp fragment amplified by RT-PCR is displayed as a solid box below E7. For gene targeting of the CNG3 locus a portion of E7 (shown in gray) encoding transmembrane segments 3–6 was replaced by the neomycin-resistance cassette (neo). The thymidine kinase (tk) was used for negative selection. Ec, EcoRV; Sp, SphI; St, StuI; Bs, BspEI; Sa, SacI; pBSK, pBluescript SK vector; wt, wild type; mt, mutant. (b) Identification of CNG3+/−, CNG3+/+, and CNG3−/− mice by Southern blot analysis. The wild-type and mutant alleles gave 18- and 6.5-kb fragments, respectively, after EcoRV digestion and hybridization with the external probe. (c) RT-PCR of RNA isolated from retinae of CNG3+/+ and CNG3−/− mice with primers that amplify CNG3, the α-subunit (CNG1), and the β-subunit (CNG4) of rod photoreceptor channel.
Reverse transcription—PCR (RT-PCR) was performed with primer pairs that amplify a 284-bp fragment of CNG3 (5′-TGCACGACTCTCCCGGAAGTACA and 5′-ACCGGATAACCCGAGTCTCCAAG), a 268-bp fragment of CNG1 (5′-CGCGGATCCTATGAGTCGATGCAGCAGAAACT and 5′-GGATGGTACCTCACTGTAGAGAAATTTCCCAGA; ref. 19), and a 321-bp fragment of CNG4 [5′-TCCACTCACTGGGTTTA(T/C)GA(C/T)GG and 5′-GTGCCAGGTGTA(T/C)TC(G/A)TACCA; refs. 13 and 20].
Electroretinography.
Ganzfeld electroretinograms (ERGs) were obtained from anesthetized mice according to described procedures (21). The protocol for the analysis of retinal function resembled an expanded version of the human ERG standard by the International Society for Clinical Electrophysiology of Vision (ISCEV) (22) and included a dark-adapted (scotopic) and a light-adapted (photopic) part. Stimulation and data acquisition were performed with a commercially available ERG setup (Toennies Multiliner Vision; Jaeger/Toennies Höchberg, Germany) featuring a Ganzfeld bowl, a dc amplifier, and a personal computer for stimulus generation and data management. Bandpass filter cutoff frequencies were 1 and 300 Hz for all measurements. The scotopic session started with single flash recordings at light intensities increasing from 10−4 cds/m2 to 10 cds/m2 in logarithmic steps of equal size. Five responses per intensity level were averaged, with an interstimulus interval (ISI) of 5 s (for 0.1, 1, 10, and 100 mcds/m2) or 17 s (for 1, 3, and 10 cds/m2). The value of 3 cds/m2 was included for comparability with the human standard flash (22). In some cases a series of responses to flicker of frequencies ranging from 0.5 to 30 Hz was recorded. After the end of the scotopic session, a homogeneous background illumination of 30 cd/m2 was turned on. After the light adaptation of 10 min, the photopic session began with single flash recordings exactly as above but with a set of higher light intensities (0.1, 0.1, 1, 3, 10, and 25 cds/m2).
Histology.
The polyclonal antibody (AbmCG3) directed against the COOH-terminal 20-mer peptide of murine CNG3, RRCCGFSPDRENSEDASKTD, was generated in rabbit by standard techniques (23). AbmCG3 specifically detected a protein of the predicted size (80 kDa) in membrane extracts of HEK293 cells transfected with an expression vector carrying the CNG3 cDNA (not shown). Immunohistochemistry was carried out with frozen sections of paraformaldehyde-fixed retinae as described (24). For light-microscopic immunohistochemistry, 12-μm sections were incubated with the antibody AbmCG3 diluted 1:200–500. Cones were identified by biotin-conjugated peanut agglutinin (lectin PNA) (25). The immunoreaction was visualized by using the ABC method and a nickel-enhanced diaminobenzidine (DAB-Ni) reaction. For electron microscopic immunohistochemistry, retinae were cut into 100-μm vertical sections. After cryoprotection, the specimens were freeze-thawed several times to enhance antibody penetration. They then were blocked in 20% normal goat serum and incubated with AbmCG3 (1:50 dilution). The immunoreaction was visualized by using the ABC method and DAB-Ni. Subsequently, the specimens were postfixed in 2.5% glutaraldehyde and the DAB reaction was silver-intensified. After 1-h OsO4 fixation, the tissue was embedded in Epon and ultrathin sections were cut and examined in the electron microscope. Conventional electron microscopy was carried out on retinae fixed in 1% paraformaldehyde/2.5% glutaraldehyde according to standard protocols.
RESULTS
CNG3−/− mice were produced by targeted deletion of exon 7 of the CNG3 gene (Fig. 1a). The deletion of CNG3 was confirmed by Southern blot analysis (Fig. 1b) and RT-PCR by using CNG3-specific primers (Fig. 1c). In contrast to CNG3−/− mice, the CNG3 transcript was detected by RT-PCR in mice that were heterozygous for the CNG3 deletion (CNG3+/− mice) (not shown). The amount of mRNA for rod CNG channel α- (CNG1) and β-subunits (CNG4) was comparable in CNG3−/− and wild-type (CNG3+/+) mice (Fig. 1c), indicating that the deletion of CNG3 did not affect the expression level of the rod channel. Heterozygous matings of outbred and inbred mice produced viable pups with the expected Mendelian frequency (148:273:137). CNG3−/− mice were normal in their appearance and body weight. At present, the oldest CNG3−/− mice have reached an age of 12 months. Up to this age there was no significant difference in mortality between wild-type and knockout mice. Knockout mice of both sexes were fertile and produced offspring of the same litter size (6.7 ± 1.9 pups per litter, n = 12, strain 129/Sv) as CNG3+/+ mice (6.1 ± 2.1 pups per litter, n = 15, strain 129/Sv).
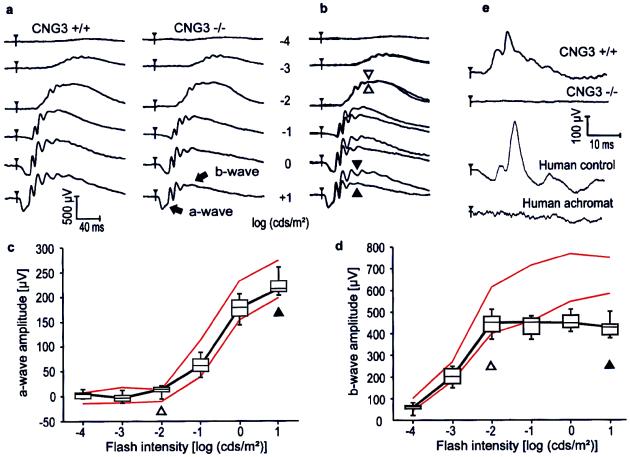
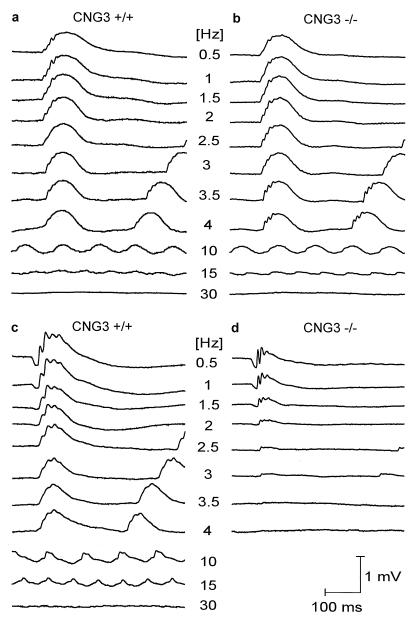
To investigate the effect of the CNG3 deletion on retinal function, Ganzfeld ERGs were recorded for 2-month-old CNG3+/+, CNG3+/−, and CNG3−/− mice at increasing stimulus intensities under dark-adapted (Fig. 2a–d) and light-adapted (Fig. 2e) conditions. The results of the heterozygotes are not shown because they were not distinguishable from those of the CNG3+/+ mice. Although appearing similar in shape, the ERGs of wild-type and knockout mice recorded under dark-adapted (scotopic) conditions revealed characteristic differences (Fig. 2 a and b). The b-wave, the large, positive deflection reflecting bipolar cell activity (26), had similar thresholds in both knockouts and controls, and no differences were observed up to approximately 0.01 cds/m2 intensity (open arrowheads in Fig. 2 b and d). However, unlike in controls, b-wave amplitudes of CNG3−/− mice did not increase further with flash intensity (solid arrowheads in Fig. 2 b and d). In contrast to the b-wave, the scotopic a-wave, which reflects predominantly the photoreceptor response (27), did not show any difference between knockout and control animals over the entire intensity range (Fig. 2 b and c). This result strongly suggests that photoreceptors expressing CNG3 (cones) do not contribute substantially to the scotopic a-wave under the given conditions. The normal amplitude of the a-wave at all light intensities together with the specific lack of amplitude increase of the b-wave at high light intensities in CNG3−/− mice suggested that the deletion of CNG3 specifically disrupted cone function whereas the rod pathway was intact. To confirm that CNG3−/− mice have a specific defect in the cone pathway, ERGs were recorded from light-adapted mice by using a background of 30 cd/m2 (Fig. 2e, top traces). Under bright-light adaptation the rod pathway is suppressed and only cells in the cone pathway contribute to the ERG (28, 29). Whereas the CNG3+/+ mice showed a normal response, there was no perceivable cone response in the knockouts. Recently, multiple mutations were found in the CNG3 gene of patients suffering from total colorblindness (achromatopsia) (30). Fig. 2e Lower shows ERGs measured under light-adapted conditions from a human control and a human achromat. The human ERGs are strikingly similar to the respective ERGs of wild-type and knockout mice. Another typical finding in human achromats is a reduced flicker fusion frequency to stimuli that induce a mixed rod–cone response, whereas the flicker fusion frequency to stimuli that are too dim to induce a substantial cone contribution is normal (31). Fig. 3 shows scotopic ERGs induced by trains of flashes (flicker). At low stimulus intensity (0.01 cds/m2) causing a pure rod response, no difference was found between the control (Fig. 3a) and the CNG3−/− mouse (Fig. 3b). At high stimulus intensity (3 cds/m2), the ERG response of the CNG3−/− mouse (Fig. 3d) was less than that of the control (Fig. 3c), even at the lowest frequency. Under these conditions the flicker fusion frequency was only about 3–3.5 Hz, which is approximately 10% of the control value.
Figure 2.
ERG responses from 2-month-old CNG3+/+ and CNG3−/− mice. (a) Dark-adapted, single-flash ERG series in a CNG3+/+ and in a CNG3−/− mouse. Note the similar appearance except for smaller amplitudes at higher flash intensities in the CNG3−/− mouse. (b) Superposition of the responses from a. Up to 0.01 cds/m2, the responses do not differ in b-wave amplitude (open arrowheads). With a further increase in stimulus luminance, the difference in b-wave but not a-wave amplitude becomes obvious (solid arrowheads). (c) a-Wave amplitude from four CNG3−/− and four CNG3+/+ littermates as a function of the logarithm of the flash intensity. Boxes indicate the 25–75% quantile range, the bars indicate the 5 and 95% quantiles, and the central bar indicates the median of the CNG3−/− data. The red lines delimit the normal range given by the 5 and 95% quantile of the CNG3+/+ littermates. No differences between control and knockout mice were detectable, pointing to a pure rod origin of the a-wave at the tested intensity range. (d) b-Wave amplitude from four CNG3−/− and four CNG3+/+ littermates as a function of the logarithm of the flash intensity. At intensities higher than 0.01 cds/m2, the b-wave of the CNG3−/− mice does not increase further in amplitude, presumably because of the lack of cone contribution. (e) Light-adapted single-flash ERG in a CNG3+/+ and a CNG3−/− mouse, a human control, and an achromat by using flashes of 3-cds/m2 intensity. No stimulus-related activity can be recorded in the CNG3−/− mouse as in the human achromat.
Figure 3.
ERG responses from 2-month-old dark-adapted CNG3+/+ and CNG3−/− mice to trains of flashes (flicker) of a fixed intensity, but varying frequency. (a and b) Scotopic flicker ERG series in a CNG3+/+ (a) and in a CNG3−/− (b) mouse obtained with a flash intensity of 0.01 cds/m2. Note the similar appearance and the normal flicker fusion frequency in the CNG3−/− mouse. (c and d) Scotopic flicker ERG series in a CNG3+/+ (c) and in a CNG3−/− (d) mouse obtained with a flash intensity of 3 cds/m2. Because of the high flash intensity, the ERG response of the CNG3−/− mouse is smaller even at the lowest frequency, and the flicker fusion frequency is only about 10% of normal.
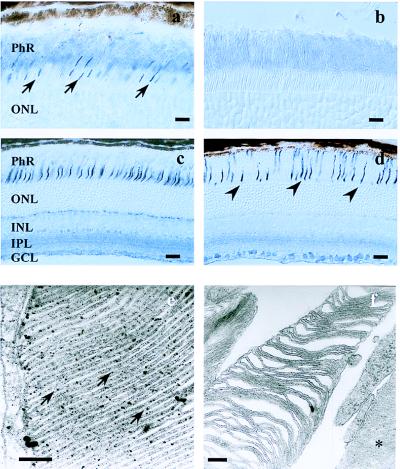
In humans, loss of cone or rod photoreceptor function has been shown to be linked to the development of retinal degenerations (32, 33). Therefore, we asked whether or not the deletion of CNG3 also was associated with the induction of degenerative processes in the murine retina. An antibody directed against CNG3 specifically labeled the outer segments of the cone photoreceptors in CNG3+/+ mice (Fig. 4a). No cells other than cone photoreceptors were marked by the antibody in the retina. The CNG3 channel was localized ultrastructurally on the membranous disks of cones (Fig. 4e). In contrast, no CNG3 protein was detectable in the retina of CNG3−/− mice (Fig. 4b). The general structure of CNG3−/− retinae was not affected significantly, as compared with wild-type retinae (Fig. 4 a–d). In particular, the photoreceptor layer of CNG3−/− mice was developed normally, and neither its thickness nor the number of rod photoreceptors was reduced. The expression of rhodopsin in rod outer segments also was not disturbed (not shown). The structural integrity of the major retinal cell layers including the rod photoreceptors is in agreement with the ERG data showing that rod-mediated light transduction is functionally normal in CNG3−/− mice. Staining of the cone photoreceptors with peanut agglutinin, a lectin that selectively binds the extracellular matrix surrounding the cones, revealed that the number of cones was reduced in CNG3−/− mice as compared with their wild-type littermates (Fig. 4 c and d). In 2-month-old CNG3−/− mice the number of cones was reduced to approximately 10–20% of the control value. No cone photoreceptors were detected in mice older than 8 months. Transmission electron microscopy revealed that the disks of cone outer segments of CNG3−/− mice were partially disorganized (Fig. 4f), whereas the disks of rod outer segments were no different from the wild-type mice.
Figure 4.
Retinal morphology of 2-month-old CNG3−/− and CNG3+/+ mice. (a) Cryosection of the retina of a CNG3+/+ mouse incubated with the CNG3 antibody. The antibody specifically labels the outer segments of cone photoreceptors (arrows). (b) A section of a CNG3−/− retina incubated with the CNG3 antibody reveals no labeling. (c and d) Detection of cone photoreceptors with the lectin peanut agglutinin in CNG3+/+ (c) and CNG3−/− (d) mice. Note the reduced number of cones in CNG3−/− mice (arrows). [Bar = 10 μm (a and b) and 20 μm (c and d).] The sections (a–d) were taken from the central region of the inferior-nasal retina. PhR, inner and outer segments of the photoreceptors; ONL, outer nuclear layer; INL, inner nuclear layer; IPL, inner plexiform layer; GCL, ganglion cell layer. (e and f) Electron microscopy photomicrographs. (e) Ultrastructural localization of CNG3 on the membranous disks of a cone outer segment of a CNG3+/+ mouse (arrows). (f) Cone outer segment of a CNG3−/− mouse revealing partial degeneration of disk structure. A rod outer segment next to the cone outer segment is marked by an asterisk. [Bar = 0.2 μm (e and f).]
DISCUSSION
In this study we report the genetic deletion of the CNG channel CNG3 in mice. CNG3-deficient mice show no gross abnormalities, indicating that CNG3 is not necessary for normal mouse development. Surprisingly, CNG3-deficient mice of both sexes are fertile and reproduce at a normal rate. This finding argues against the notion that CNG3 plays an important role in the control of sperm motility and, therefore, fertility (11, 34).
The major effect of the CNG3-deletion is in retinal function. CNG3-deficient mice completely lack cone photoreceptor-mediated vision. CNG3 thus is indispensable for cone phototransduction and cannot be substituted by other CNG channels such as the rod photoreceptor or olfactory CNG channel. There is sufficient evidence that rod and cone pathways are anatomically and functionally linked in the vertebrate retina (3–5). The finding that the rod-mediated vision is unaffected in CNG3-deficient mice indicates that there is a distinct rod pathway in the retina that is completely independent of the presence of functional cones. This result is supported by recent data obtained from mice that contained a strongly diminished number of cones because of the cone-specific expression of diphtheria toxin (35). The retinal phenotype of CNG3-deficient mice is reminiscent of human total colorblindness (achromatopsia). Both human achromats and CNG3−/− mice specifically lack functional cones whereas the rod pathway is intact (31). Missense mutations were identified recently in the CNG3 gene of achromats (30). Our results clearly confirm the idea that a nonfunctional CNG3 channel is the primary cause of achromatopsia in these patients.
The functional ablation of cone function in CNG3−/− mice concurs with a progressive degeneration of the cone photoreceptors. The molecular events leading to the degeneration of cones in CNG3−/− mice are not known at present. However, the finding that the retinae of young CNG3−/− mice still contain a substantial number of cones indicates that the degeneration may not immediately follow from the absence of the CNG3 channel as such but could be due to a secondary process that is induced by the absence of the CNG3 channel. Because CNG3 is a major entry pathway for calcium in cone photoreceptors (6, 8), the complete lack of this channel would result in a decrease of cellular calcium concentration. At low calcium concentration the cone membrane guanylate cyclase may be stimulated permanently (36), leading to elevated levels of cGMP, which may trigger the degeneration. Such a mechanism is supported by the finding that several types of rod or cone degenerations have been linked to mutations in those photoreceptor proteins that regulate either synthesis or hydrolysis of cGMP (33, 37).
In summary, CNG3−/− mice provide a valuable model to functionally dissect rod from cone photoreceptor-mediated signaling pathways in vivo. Finally, CNG3−/− mice will allow studies into the pathogenesis and therapy of human achromatopsia in an animal model.
Acknowledgments
We thank P. Mayr, S. Erhard, G. Härer, and K. Mai for technical support. Research was supported by grants from the Deutsche Forschungsgemeinschaft (SFB 391 and 430), Fond der Chemischen Industrie, Bundesministerium für Bildung und Forschung (Fö. 01KS9602), Interdisziplinäres Klinisches Forschungszentrum (IKFZ) Tübingen, and Fortüne Grant 517 der Universität Tübingen.
ABBREVIATIONS
- CNG
cyclic nucleotide gated
- ERG
electroretinogram
- RT-PCR
reverse transcription–PCR
Footnotes
References
- 1.Sterling P. In: The Synaptic Organization of the Brain. Sepherd G M, editor. New York: Oxford Univ. Press; 1990. pp. 170–213. [Google Scholar]
- 2.Baylor D. Proc Natl Acad Sci USA. 1996;93:560–565. doi: 10.1073/pnas.93.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mills S L, Massey S C. Nature (London) 1995;377:734–737. doi: 10.1038/377734a0. [DOI] [PubMed] [Google Scholar]
- 4.Schneeweis D M, Schnapf J L. Science. 1995;268:1053–1056. doi: 10.1126/science.7754386. [DOI] [PubMed] [Google Scholar]
- 5.Smith R G, Freed M A, Sterling P. J Neurosci. 1986;6:3505–3517. doi: 10.1523/JNEUROSCI.06-12-03505.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Finn J T, Grunwald M E, Yau K W. Annu Rev Physiol. 1996;58:395–426. doi: 10.1146/annurev.ph.58.030196.002143. [DOI] [PubMed] [Google Scholar]
- 7.Zagotta W N, Siegelbaum S A. Annu Rev Neurosci. 1996;19:235–263. doi: 10.1146/annurev.ne.19.030196.001315. [DOI] [PubMed] [Google Scholar]
- 8.Biel M, Zong X, Ludwig A, Sautter A, Hofmann F. Rev Physiol Biochem Pharmacol. 1999;135:151–171. doi: 10.1007/BFb0033672. [DOI] [PubMed] [Google Scholar]
- 9.Kaupp U B, Niidome T, Tanabe T, Terada S, Bönigk W, Stühmer W, Cook N J, Kangawa K, Matsuo H, Hirose T, et al. Nature (London) 1989;342:762–766. doi: 10.1038/342762a0. [DOI] [PubMed] [Google Scholar]
- 10.Biel M, Zong X, Distler M, Bosse E, Klugbauer N, Murakami M, Flockerzi V, Hofmann F. Proc Natl Acad Sci USA. 1994;91:3505–3509. doi: 10.1073/pnas.91.9.3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weyand I, Godde M, Frings S, Weiner J, Müller F, Altenhofen W, Hatt H, Kaupp U B. Nature (London) 1994;368:859–863. doi: 10.1038/368859a0. [DOI] [PubMed] [Google Scholar]
- 12.Cook N J, Hanke W, Kaupp U B. Proc Natl Acad Sci USA. 1987;84:585–589. doi: 10.1073/pnas.84.2.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Körschen H G, Illing M, Seifert R, Sesti F, Williams A, Gotzes S, Colville C, Müller F, Dose A, Godde M, et al. Neuron. 1995;15:627–636. doi: 10.1016/0896-6273(95)90151-5. [DOI] [PubMed] [Google Scholar]
- 14.Nawy S, Jahr C E. Nature (London) 1990;346:269–271. doi: 10.1038/346269a0. [DOI] [PubMed] [Google Scholar]
- 15.Ahmad I, Leinders-Zufall T, Kocsis J D, Shepherd G M, Zufall F, Barnstable C J. Neuron. 1994;12:155–165. doi: 10.1016/0896-6273(94)90160-0. [DOI] [PubMed] [Google Scholar]
- 16.Kusaka S, Dabin I, Barnstable C J, Puro D G. J Physiol. 1996;497:813–824. doi: 10.1113/jphysiol.1996.sp021812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wissinger B, Müller F, Weyand I, Schuffenhauer S, Thanos S, Kaupp U B, Zrenner E. Eur J Neurosci. 1997;9:2512–2521. doi: 10.1111/j.1460-9568.1997.tb01680.x. [DOI] [PubMed] [Google Scholar]
- 18.Pfeifer A, Aszódi A, Seidler U, Ruth P, Hofmann F, Fässler R. Science. 1996;275:2082–2086. doi: 10.1126/science.274.5295.2082. [DOI] [PubMed] [Google Scholar]
- 19.Pittler S J, Lee A K, Altherr M R, Howard T A, Seldin M F, Hurwitz R L, Wasmuth J J, Baehr W. J Biol Chem. 1992;267:6257–6262. [PubMed] [Google Scholar]
- 20.Biel M, Zong X, Ludwig A, Sautter A, Hofmann F. J Biol Chem. 1996;271:6349–6355. doi: 10.1074/jbc.271.11.6349. [DOI] [PubMed] [Google Scholar]
- 21.Ruether K, van de Pol D, Jaissle G, Berger W, Tornow R-P, Zrenner E. Invest Ophthalmol Visual Sci. 1997;38:710–718. [PubMed] [Google Scholar]
- 22.Marmor M, Zrenner E. Doc Ophthalmol. 1995;89:199–210. doi: 10.1007/BF01203373. [DOI] [PubMed] [Google Scholar]
- 23.Ludwig A, Flockerzi A, Hofmann F. J Neurosci. 1997;17:1339–1349. doi: 10.1523/JNEUROSCI.17-04-01339.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cellerino A, Kohler K. J Comp Neurol. 1995;386:149–160. [PubMed] [Google Scholar]
- 25.Blanks J C, Johnson L V. J Comp Neurol. 1983;221:31–41. doi: 10.1002/cne.902210103. [DOI] [PubMed] [Google Scholar]
- 26.Stockton R A, Slaughter M M. J Gen Physiol. 1989;93:101–122. doi: 10.1085/jgp.93.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Penn R D, Hagins W A. Nature (London) 1969;223:201–205. doi: 10.1038/223201a0. [DOI] [PubMed] [Google Scholar]
- 28.Aguilar M, Stiles W S. Opt Acta. 1954;1:59–63. [Google Scholar]
- 29.Peachey N S, Goto Y, al-Ubaidi M R, Naash M I. Neurosci Lett. 1993;162:9–11. doi: 10.1016/0304-3940(93)90547-x. [DOI] [PubMed] [Google Scholar]
- 30.Kohl S, Marx T, Giddings I, Jägle H, Jacobson S G, Apfelstedt-Sylla E, Zrenner E, Sharpe L T, Wissinger B. Nat Genet. 1998;19:257–259. doi: 10.1038/935. [DOI] [PubMed] [Google Scholar]
- 31.Sharpe L T, Nordby K. In: Night Vision: Basic, Clinical and Applied Aspects. Hess R F, Sharpe L T, Nordby K, editors. Cambridge, U.K.: Cambridge Univ. Press; 1990. pp. 335–389. [Google Scholar]
- 32.Dryja T P, Finn J T, Peng Y-W, McGee T L, Berson E L. Proc Natl Acad Sci USA. 1995;92:10177–10181. doi: 10.1073/pnas.92.22.10177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shastry B S. Cell Mol Life Sci. 1997;53:419–429. doi: 10.1007/s000180050050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wiesner B, Weiner J, Middendorff R, Hagen V, Kaupp U B, Weyand I. J Cell Biol. 1998;142:473–484. doi: 10.1083/jcb.142.2.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soucy E, Wang Y, Nirenberg S, Nathans J, Meister M. Neuron. 1998;21:481–493. doi: 10.1016/s0896-6273(00)80560-7. [DOI] [PubMed] [Google Scholar]
- 36.Koch K-W. Rev Physiol Biochem Pharmacol. 1994;125:150–192. doi: 10.1007/BFb0030910. [DOI] [PubMed] [Google Scholar]
- 37.Dizhoor A M, Boikov S G, Olshevskaya E V. J Biol Chem. 1998;273:17311–17314. doi: 10.1074/jbc.273.28.17311. [DOI] [PubMed] [Google Scholar]