Abstract
The β-chemokine RANTES (regulated on activation, normal T cell expressed and secreted) suppresses the infection of susceptible host cells by macrophage tropic strains of HIV-1. This effect is attributed to interactions of this chemokine with a 7-transmembrane domain receptor, CCR5, that is required for virus–cell fusion and entry. Here we identify domains of RANTES that contribute to its biological activities through structure–function studies using a new monoclonal antibody, mAb 4A12, isolated from mice immunized with recombinant human RANTES. This monoclonal antibody (mAb) blocked the antiviral activity of RANTES in infectivity assays with HIV-1Bal, and inhibited the mobilization of intracellular Ca2+ elicited by RANTES, yet recognized this chemokine bound to cell surfaces. Epitope mapping using limited proteolysis, reversed phase high-performance liquid chromatography, and mass spectrometry suggest that residues 55–66 of RANTES, which include the COOH-terminal α-helical region implicated as the glycosaminoglycan (GAG) binding domain, overlap the determinant recognized by mAb 4A12. This is supported by affinity chromatography studies, which showed that RANTES could be eluted specifically by heparin from a mAb 4A12 immunoaffinity matrix. Removal of cell surface GAGs by enzymatic digestion greatly reduced the ability of mAb 4A12 to detect RANTES passively bound on cell surfaces and abrogated the ability of RANTES to elicit an intracellular Ca2+ signal. Taken together, these studies demonstrate that the COOH-terminal α-helical region of RANTES plays a key role in GAG-binding, antiviral activity, and intracellular Ca2+ signaling and support a model in which GAGs play a key role in the biological activities of this chemokine.
Keywords: β-chemokines, human immunodeficiency virus 1, monoclonal antibody, signaling, antiviral effect
The discovery of the antiviral effect of β-chemokines on M-tropic isolates of HIV-1 (1) and the identification of 7-transmembrane domain, G protein–coupled chemokine receptors as entry cofactors for HIV-1 (2–7) set the stage for the development of novel approaches to control HIV-1 infections. Studies using in vitro mutagenesis have identified domains important for coreceptor function (8–25), and revealed that coreceptor activity can be dissociated from receptor activation (8, 22–26), indicating that it might be possible to block virus entry using chemokine analogues without aberrant stimulation of cellular responses.
In addition to the interactions between β-chemokines and the canonical 7-transmembrane domain receptors, two new reports suggest a role for cell surface glycosaminoglycans (GAGs)1 in the ability of RANTES (regulated on activation, normal T cell expressed and secreted) to block infection by HIV-1. In the first of these studies (27), enzymatic removal of cell surface GAGs increased the concentrations of RANTES required to block infection with HIV-1. In the second study (28), β-chemokines complexed with GAGs were found to be secreted by CTL clones and similar complexes formed in vitro with heparan sulfate were more effective than free β-chemokines at blocking the infection of macrophages with M-tropic HIV-1. Our studies, described below, identify a domain of RANTES that contributes to the binding of GAGs, lending further support for a role of GAGs in the biological activities of β-chemokines. We show that a new monoclonal antibody, mAb 4A12, which blocks both the antiviral effect and the mobilization of intracellular Ca2+ elicited by RANTES, recognizes an epitope that overlaps the GAG-binding domain of this β-chemokine. Taken together, our studies strongly support the involvement of cell surface GAGs in both the antiviral effect of RANTES and the ability of RANTES to signal responsive cells.
Materials and Methods
Cell Culture.
PM1 and HUT-78 cells were maintained in culture in RPMI 1640 (GIBCO BRL, Gaithersburg, MD) supplemented with 10% fetal bovine serum (FBS; GIBCO BRL) and 50 μg/ml gentamycin (Sigma Chemical Co., St. Louis, MO), denoted hereafter as complete medium. Human PBMCs were obtained from healthy donors (American Red Cross, Baltimore, MD) by centrifugation on Ficoll-Hypaque density gradients. PHA 5 μg/ml (Sigma Chemical Co.) and 20 U/ml recombinant human IL-2 (Boehringer Mannheim, Indianapolis, IN) were used to activate PBMCs for 3 d in complete medium at 37°C in a humidified incubator at a 7% CO2/air mixture. After activation, the cells were washed and cultured in complete medium containing 20 U/ml IL-2, which was replaced every third day until the cells were used.
Isolation of mAbs.
Murine hybridomas were isolated by fusing the spleens of BALB/cJ mice hyperimmunized with recombinant human RANTES (R&D Systems, Minneapolis, MN) with the P3X63.AG8.653 fusion partner using the standard methods of Lewis and colleagues (29–33). The methods for fusion, screening by ELISA, hybridoma cloning, and hybridoma propagation were as previously described (29–33).
Infectivity Assays.
PM1 cells were infected for 2 h at 37°C with HIV-1BaL at a ratio of 2 × 106 cells to 500 TCID50 in 5 ml culture medium. Cells were then washed to remove virus and placed in tissue culture wells at a density of 2 × 105 cells in 250 μl complete medium. A stock solution of purified mAb 4A12 was diluted in culture medium containing an equal concentration of normal mouse IgG and subsequent serial dilutions were made in this medium to maintain a constant total immunoglobulin concentration of 66 nM. Serial dilutions of mAb 4A12 were preincubated with 10 nM RANTES for 1 h at room temperature and 250 μl of the antibody/chemokine mix then added to the cells to achieve a total culture volume of 500 μl and a final RANTES concentration of 5 nM. The cells were fed 3 d after infection by removing 250 μl of medium and replacing with an equal volume of fresh medium containing the appropriate concentrations of mAb 4A12, RANTES, or the mAb 4A12/RANTES mixture. Levels of infection were determined 7–10 d after infection by measuring HIV-1 P 24 levels using an antigen capture ELISA (DuPont, Inc., Cambridge, MA). All assays were carried out in triplicate.
Flow Cytometry.
PM1 cells were aliquoted at 106 cells/tube using 12 × 75-mm culture tubes. Cells were treated one of three ways: (a) RANTES was added at a concentration of 128 nM; (b) RANTES was preincubated with a fivefold molar excess of mAb 4A12; or (c) RANTES was not included in the cell incubation. In conditions a and c mAb 4A12 was added to the cells. In condition b the mAb 4A12/RANTES complex was added to the cells. Finally, in all three conditions PE-conjugated anti–mouse IgG (anti-IgG-PE; Sigma Chemical Co.) was used to detect the mAb. In all cases cells were incubated on ice for 1 h (45 min for anti-IgG) and kept in PBS supplemented with 2% FBS and 0.1% sodium azide. Cells were washed twice between each incubation. All samples were finally fixed in 1% paraformaldehyde and analyzed on a FACScalibur® flow cytometer (Becton Dickinson, San Jose, CA). To stain for the presence of CCR5, cells were aliquoted as described above. mAb 2D7 (PharMingen, San Diego, CA) was incubated at the concentration suggested by the manufacturer with the cells and binding of this mAb to cells was detected with anti-IgG-PE as above. Similarly, cells were washed twice between incubations, incubated with both antibodies for 45 min each on ice, and kept in PBS supplemented with 2% FBS and 0.1% sodium azide.
Glycanase Treatment of Cells.
PM1 cells or activated PBMCs were incubated with 1 U/ml each of heparinase II, heparinase III, and chondroitinase ABC (Sigma Chemical Co.) for 1 h at 37°C. As a control, untreated PBMCs were simultaneously incubated in culture medium only. After 1 h the cells were washed twice, once in culture medium and once in PBS supplemented with 2% FBS and 0.1% sodium azide. Cells were kept on ice until stained with the indicated antibodies.
Measurement of Intracellular Ca2+ Responses by Flow Cytometry.
PBMCs were isolated from healthy normal donors (described above) and prepared for analysis of Ca2+ signaling as we have described previously (34). RANTES, mAb 4A12, or preincubated mAb 4A12/RANTES (as described above) were used to induce Ca2+ mobilization and analyzed by FACScalibur® (34, 35).
Staphylococcus aureus V8 Protease Digestion of RANTES and Separation of Fragments by Reversed Phase HPLC.
Recombinant human RANTES (carrier free) (R&D Systems) and S. aureus V8 protease (Sigma Chemical Co.) were incubated at 37°C. After 1 h, the digestion mixture was placed on ice and kept until analyzed by HPLC. Undigested as well as digested RANTES were purified by reversed phase HPLC using a C18 column (Waters Instruments, Milford, MA) equilibrated in H2O containing 0.1% TFA. A linear 0–65% gradient of acetonitrile in TFA/H2O was used to elute fragments over a 45 min time course.
Dot Blotting.
Dot blotting was carried out as previously described (33) except that PVDF membranes were used instead of nitrocellulose membranes.
Heparin Elution of RANTES Bound to a mAb 4A12 Immunoaffinity Matrix.
An immunoaffinity matrix was prepared by covalently coupling 1 mg of purified mAb 4A12 IgG2a to activated CH Sepharose 4B (Sigma Chemical Co.) using previously described methods (36). Native RANTES, purified from serum-free culture medium from F3b clone 19 cells (35) and then over a HiTrap heparin affinity FPLC column (Amersham Pharmacia Biotech, Piscataway, NJ) equilibrated in 10 mM Tris HCl, pH 7.6. The bound proteins, including RANTES, eluted with 2 M NaCl in the same buffer. The heparin column eluate was diluted twofold with 10 mM Tris HCl, pH 7.6, and applied over a 1 h period at room temperature to the mAb 4A12 immunoaffinity column equilibrated in the same buffer. The unbound fraction was collected, the matrix was washed with PBS, and heparin sulfate (1 mg/ml) in PBS was run into the column. The column was clamped and allowed to incubate for 30 min at room temperature. The material eluted from the column by the heparin/PBS solution was collected and the column washed again with PBS. The column was subjected to a second round of elution with 0.2 M glycine, pH 2.5, and the eluate fraction collected. The amounts of RANTES in the affinity column load, unbound, and eluate fractions were determined by ELISA (R&D Systems).
Results
mAb 4A12 Blocks the Antiviral Effect of RANTES.
Hybridomas were isolated from BALB/cJ mice hyperimmunized with RANTES as described in Materials and Methods, and three stable clones reactive with RANTES were identified by ELISA. Preliminary studies showed that these mAb are specific for RANTES as judged by the lack of reactivity with macrophage inflammatory protein (MIP)-1α and MIP-1β in ELISA. In additional studies, purified IgG from each of the hybridomas blocked the antiviral effect of RANTES; however, mAb 4A12 was ∼10–20-fold more efficient in this assay than the other two and was thus chosen for further study. A representative experiment showing mAb 4A12 inhibition of antiviral effects is shown in Fig. 1.
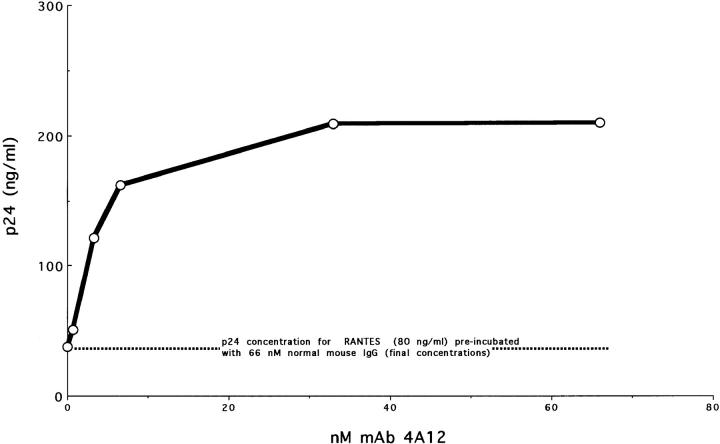
Figure 1.
mAb 4A12 blocks the antiviral effect of RANTES. Purified mAb 4A12 was diluted serially in complete culture medium supplemented with sufficient normal mouse IgG to maintain the total murine Ig concentration at 66 nM. Aliquots of mAb 4A12 were incubated with RANTES at a final concentration of 22 nM for 1 h at ambient temperature. These complexes were added to PM1 cells previously incubated for 2 h at 37°C with 500 TCID50 of HIV-1Bal and were washed before addition of the complexes. The cultures were fed on day 3, and infection was quantified on day 9 by p24 antigen capture ELISA.
In this experiment, incubation of a limiting concentration of RANTES with increasing concentrations of mAb 4A12 resulted in a dose-dependent reversal of the antiviral effect (Fig. 1). No reversal was obtained with normal mouse IgG alone (Fig. 1, 0 nM mAb 4A12). Note that the total IgG concentration remained constant in this assay by dilution of mAb 4A12 into medium containing normal mouse IgG. Similar results were obtained by adding mAb 4A12 and RANTES to PM1 cells infected with HIV-1BaL without preincubation (data not shown).
mAb 4A12 Blocks Intracellular Ca2+ Responses Elicited by RANTES.
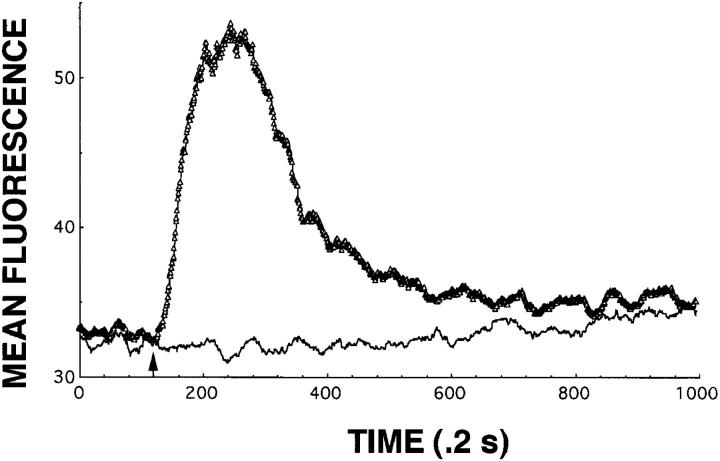
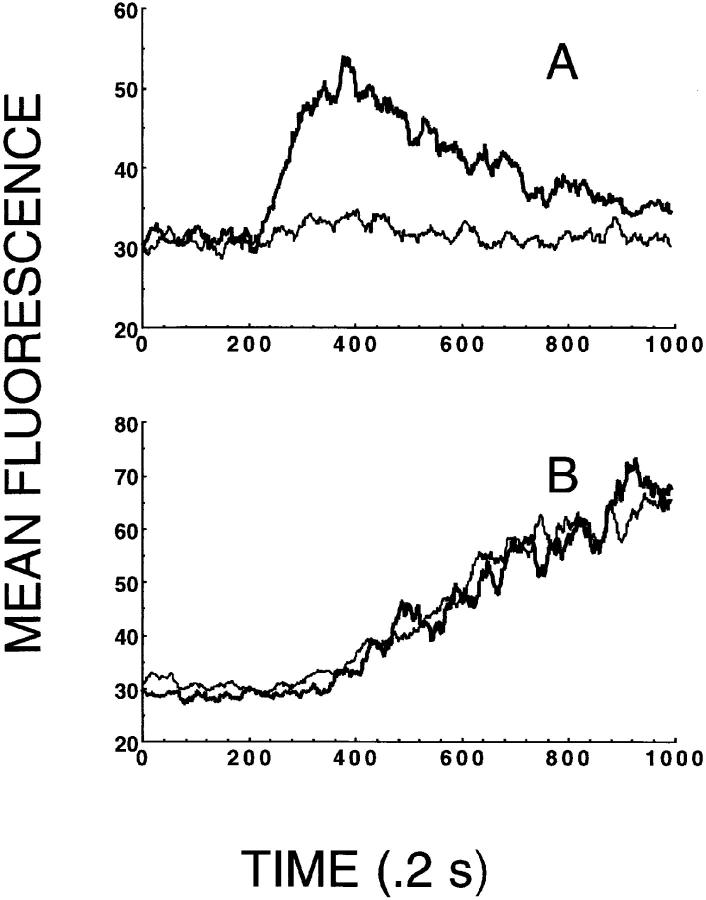
The ability of mAb 4A12 to block intracellular Ca2+ responses elicited by RANTES was evaluated using flow cytometry. These studies used normal human PBMCs activated with PHA plus IL-2 for 3 d followed by expansion with IL-2 alone for 9 d. The activated PBMCs were washed and loaded with the calcium-sensitive indicator dye Fluo-3 and intracellular Ca2+ responses were determined by flow cytometry in response to RANTES as described previously (34, 35). In the experiment shown in Fig. 2, 3 nM RANTES elicited a characteristic intracellular Ca2+ response in the activated PBMCs (triangles). Preincubation of 15 nM mAb 4A12 with 3 nM RANTES abrogated this response (Fig. 2, thin line). In our early experiments, neither preincubation of RANTES with a variety of control IgG2a mAb nor incubation of cells alone with mAb 4A12 yielded unexpected activities, and therefore these controls were excluded from subsequent studies, including the one shown in Fig. 2. Taken together these data show that mAb 4A12 blocks the ability of RANTES to induce intracellular Ca2+ signaling in addition to the antiviral effect of this chemokine.
Figure 2.
mAb 4A12 blocks intracellular Ca2+ responses elicited by RANTES. Normal human PBMCs were activated with PHA plus IL-2 for 3 d followed by expansion with IL-2 alone for 9 d at which time they were used to measure intracellular Ca2+ responses. The cells were washed and loaded with the indicator dye Fluo-3 and responses to RANTES measured as by flow cytometry described in Materials and Methods and references 34 and 35. In this experiment, the cells were stimulated with RANTES at 3 nM or with a mixture of 3 nM RANTES plus 15 nM mAb 4A12. The mixture of RANTES and mAb 4A12 was prepared by incubating these two reagents for 1 h at ambient temperature before use in the assay. RANTES was prepared identically before use in this assay as the positive control. Calcium mobilization is shown as a change in mean fluorescence over time, where 1 time unit is equal to 0.2 s.
mAb 4A12 Recognizes an Epitope Overlapping the GAG Binding Domain of RANTES.
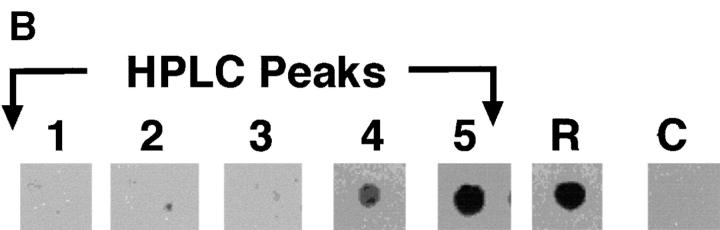
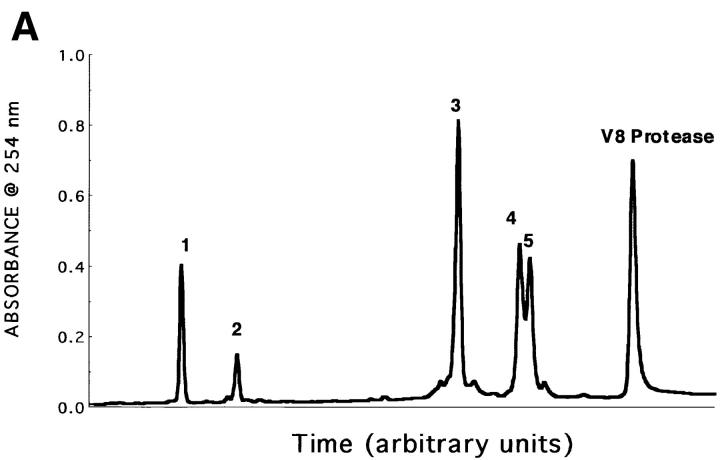
Because mAb 4A12 blocks both the antiviral effect and intracellular Ca2+ signaling elicited by RANTES, it is likely that the epitope recognized by this mAb overlaps a structure that plays a key role in these biological responses. Accordingly, epitope mapping was undertaken to identify this structure. Early mapping studies carried out using synthetic overlapping peptides failed to identify a reactive peptide, suggesting the epitope recognized by mAb 4A12 is discontinuous. Therefore, a different strategy was taken in which RANTES was digested with S. aureus V8 protease that cleaves selectively at glutamic and aspartic acid residues. After digestion, five distinct polypeptide peaks were obtained by reversed phase HPLC (Fig. 3 A). Dot blot analyses revealed that the peaks 1, 2, and 3 were not reactive with mAb 4A12, whereas peaks 4 and 5 were (Fig. 3 B).
Figure 3.
HPLC profile and mAb 4A12 reactivity of V8 protease digests of RANTES. (A) Reversed phase HPLC profile of RANTES digest products. RANTES was digested with 10 U S. aureus V8 protease as described in Materials and Methods. The digestion mixture was purified by reversed phase HPLC using a C18 column (Waters Instruments) equilibrated in H2O containing 0.1% TFA. A linear gradient of TFA/H2O and acetonitrile/H2O was used to elute fragments over 45 min. (B) Dot blot. Fractions from reversed-phase HPLC were pulled onto a PVDF membrane using a vacuum manifold and washed three times with Tris-buffered saline. The blot was then blocked with BLOTTO (33) and incubated with 5 μg/ml mAb 4A12. Binding was detected using anti–mouse IgG alkaline phosphatase and visualized by chemiluminescence as described previously (33). R, RANTES (positive control); C, irrelevant peptide (negative control).
Sequence analyses by either mass spectrometry or NH2-terminal Edman degradation were used to identify the peptides in each peak as listed in Table 1. Peak 1 mapped to residues 55–60 in the COOH terminus of RANTES. The observed molecular mass was 846, which corresponds well to the predicted mass of 845. Peak 2 also mapped to the COOH terminus of RANTES, in this case to residues 61– 66. The observed mass of 738.8 was in agreement with the predicted mass of 737.9 for this peptide. NH2-terminal sequencing of peak 3 revealed two peptides; one peptide sequence corresponded to the first nine residues in the NH2 terminus of RANTES, whereas the second peptide sequence corresponded to residues 27–35. Assignment of the penultimate residue in the latter peptide was ambiguous and most likely corresponds to the cysteine at position 34 in RANTES. The observed mass for the NH2-terminal peptide was 2,922, which is consistent with the predicted mass of 2,924.8 for a fragment that includes residues 1–26 of RANTES. As expected, this fragment terminates with a glutamic acid, which is the recognition site of V8 protease. The observed mass for the second peptide was 6,101, which is consistent with the predicted mass of 6,104.7 for a fragment corresponding to residues 27–54 of RANTES. The 1–26 and 27–54 peptides include cysteines at positions 10 and 34 that are disulfide bonded in native RANTES. Thus, peak 3 contains a polypeptide that is comprised of two disulfide-bonded peptides corresponding to residues 1–54 of RANTES with a single cleavage at the glutamic acid located at position 26.
Table 1.
RANTES Cleavage Products after Digestion with S. aureus V8 Protease
| Fraction number | Corresponding residues | MS/MS | NH2-terminal sequence | Molecular masses by mass spectrometry | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Theoretical | Observed | |||||||||
| dalton | ||||||||||
| 1 | 55–60 | KKWVRE | NT | 845 | 846 | |||||
| 2 | 61–66 | YINSLE | NT | 737.9 | 738.8 | |||||
| 3 | 1–54 | NT | SPYSSDTTP/YFYTSGK*S | 2924.8/6104.7 | 2922/6101 | |||||
| 4§ | 1–66 | ND | ND | ND | ||||||
| 5 | 1–68 | NT | SPYSS | 7851.9 | 7846 | |||||
NT, not tested; ND, not detected.
Fraction 4 was identified by amino acid analysis.
Peak 4, as determined by amino acid analysis, produced results consistent with a fragment of RANTES corresponding to residues 1–66. Such a product is predicted to occur after V8 digestion of RANTES due to a single cleavage at glutamic acid residue 66 that removes the last two residues (methionine and serine) in the COOH terminus of the molecule. Peak 5 contained undigested RANTES as shown by NH2-terminal sequencing and mass spectrometry. The NH2-terminal sequence was SPYSS, which corresponds to the first five residues of RANTES, and the observed mass was 7,846, which corresponds to the predicted mass of 7,851.9 for uncleaved RANTES.
Because mAb 4A12 reacted with the fragment corresponding to residues 1–66 but not with that corresponding to residues 1–54, it is probable that epitope recognized by this antibody overlaps residues 55–66 of RANTES although the epitope must involve additional residues as the peptides corresponding to this region were not reactive with mAb 4A12 (our unpublished data). Residues 55–66 include the α-helical region of RANTES (Fig. 4) and the corresponding region has been implicated as the GAG-binding domain for several (37–39) but not all (40) chemokines. This raised the possibility that mAb 4A12 might recognize an epitope associated with the GAG-binding domain of RANTES.
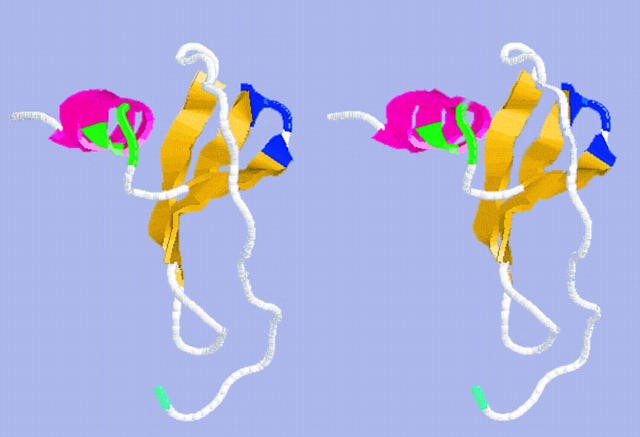
Figure 4.
Stereo drawing of RANTES depicting α-helical region and cationic sites. This structure was generated using the averaged nuclear magnetic resonance coordinates for the file 1RTO.PDB obtained from the Protein Data Bank at Brookhaven National Laboratories (Brookhaven, NY). These coordinates were described originally in reference 48. The drawing is rendered in “cross-eyed” stereo and uses the following color scheme: yellow, β-sheets; pink, α-helix; blue, cationic site 1 (Arg 44, Lys 45, Arg 47); green, cationic site 2 (Lys 55, Lys 56, Arg 59); cyan, NH2 terminus of RANTES (Ser 1); white, all other residues of RANTES.
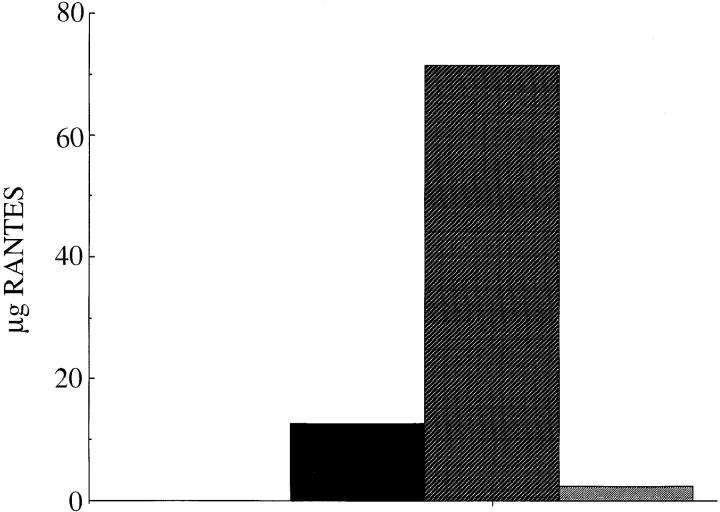
This possibility was tested by affinity chromatography in which heparin was used to elute RANTES from the mAb 4A12 immunoaffinity matrix. A preparation of native RANTES (∼100 μg), purified from the culture supernatant of a transformed human T cell (F3b clone 19; reference 35) was captured by the mAb 4A12 affinity column. The unbound fraction was then collected, and the column was washed (flow-through fraction; Fig. 5, black bar) and then treated for 30 min at ambient temperature with 1 mg/ ml heparin in PBS (Fig. 5, hatched bar). To ensure that heparin removed all of the bound RANTES, the column was then washed and again treated with 0.2 M glycine, pH 2.5 (Fig. 5, stippled bar), which we routinely use to elute RANTES from mAb 4A12 immunoaffinity columns. As shown in Fig. 5, most of the RANTES captured by the mAb 4A12 affinity column was eluted with heparin. In this experiment, 17.5 μg RANTES was found in the flow-through fraction and 71.2 μg RANTES was eluted with heparin. By contrast, only 2.3 μg RANTES was eluted using the pH 2.5 buffer. These results demonstrate competitive binding for RANTES between heparin and mAb 4A12, which is consistent with the epitope mapping data suggesting that this mAb recognizes an epitope in the GAG-binding domain of RANTES.
Figure 5.
Elution of RANTES by heparin from mAb 4A12 column. Native RANTES purified from the supernatant of the HTLV-1 transformed cell line, F3b (35), was adsorbed onto an affinity matrix of mAb 4A12. The column was washed and sequentially eluted with heparin (1 mg/ml in PBS) and 0.2 M glycine, pH 2.5. RANTES was quantified by capture ELISA as described in Materials and Methods. Black bar, flow through; hatched bar, heparin eluate; stippled bar, pH 2.5 eluate.
mAb 4A12 Displays Two Patterns of Reactivity with RANTES and the Binding of this Chemokine to Cell Surface GAGs.
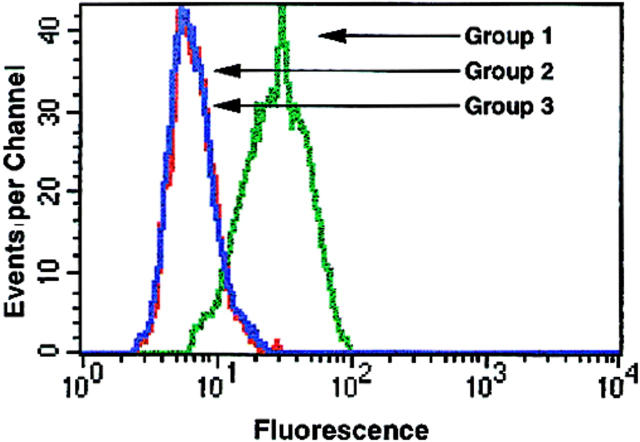
Because the above studies suggest that mAb 4A12 recognizes an epitope overlapping the GAG-binding domain of RANTES, we used flow cytometry to investigate the ability of this mAb to detect RANTES bound to GAGs on the cell surface. In one experimental condition, (Fig. 6, Group 1) PM1 cells were incubated with RANTES (128 nM) followed by exposure to mAb 4A12. Finally, mAb 4A12 binding was detected by flow cytometry using anti-IgG-PE. In a second experimental condition (Fig. 6, Group 2) PM1 cells were treated identically except that RANTES and a fivefold molar excess of mAb 4A12 were mixed for 1 h before incubation with the cells. Comparison of the flow histograms for groups 1 and 2 in Fig. 6 shows that mAb 4A12 detects RANTES prebound to PM1 cells. No signal was observed when mAb 4A12 was excluded from the assay (data not shown). Thus, once RANTES is bound to the cell surface, it can be detected by mAb 4A12. However, if RANTES was admixed with mAb 4A12 before exposure to PM1 cells, no signal was obtained (Fig. 6; compare flow histograms for groups 2 and 3). Similar results have been obtained using 125I-RANTES (our unpublished results). Thus, preincubation with mAb 4A12 blocks the binding of RANTES to cell surfaces. This paradoxical result is explained most readily by the recent observation that binding to cell surface GAGs induces oligomerization of RANTES (reference 41 and see Discussion). Therefore, mAb 4A12 is recognizing the GAGs binding domain of a molecule that is not directly binding to GAGs but instead to other RANTES molecules. Two additional series of experiments were carried out to examine this possibility further.
Figure 6.
Detection of RANTES bound to cell surfaces by flow cytometry. PM1 cells in Group 1 (green line) were treated with 128 nM RANTES followed by a 45-min incubation with a fivefold molar excess of mAb 4A12 as described in Materials and Methods. Cells in Group 3 (red line) were not incubated with RANTES, but treated with 4A12 alone. Cells in Group 2 (blue line) were treated with RANTES and 4A12 that had been previously incubated together. In all conditions 4A12 binding was visualized with anti-IgG-PE and the fluorescence intensity was determined by flow cytometry.
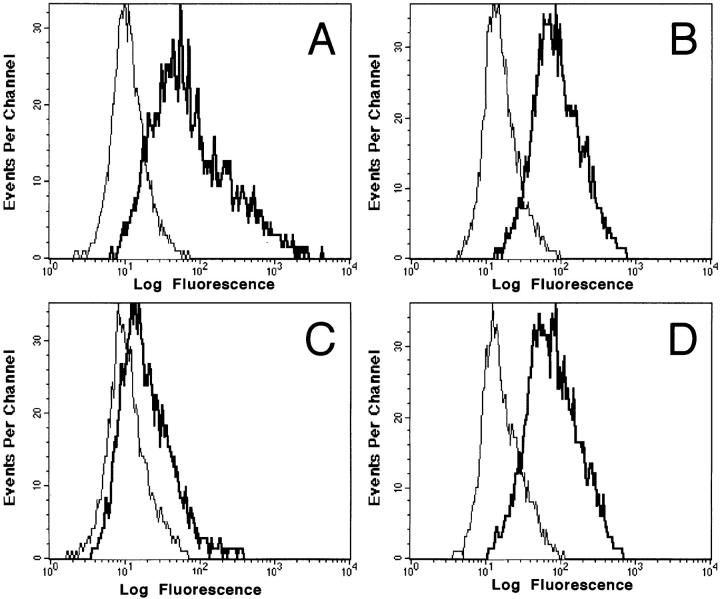
First, cell surface GAGs were removed enzymatically from cells to establish that the binding of RANTES detected by mAb 4A12 is dependent on GAGs. In these studies, we used activated PBMCs that were treated either with medium alone or with a mixture of glycanases including heparinase II and III and chondroitinase ABC as described in Materials and Methods. The treated cells were incubated with RANTES and cell surface binding was detected with mAb 4A12 followed by anti-IgG-PE. As shown in Fig. 7 a, incubation of activated PBMCs with medium alone before exposure to RANTES produced the expected signal with mAb 4A12 (thick line), whereas no signal was observed in the absence of RANTES (thin line). By contrast, the signal was reduced markedly when activated PBMCs were incubated with the glycanase mixture before exposure to RANTES followed by detection with mAb 4A12 and anti-IgG-PE (Fig. 7 c). Background signals were not changed by removal of cell surface GAGs (compare thin lines in a and c). Thus, the removal of cell surface GAGs from the activated PBMCs dramatically reduced the binding of RANTES detected by mAb 4A12. The removal of GAGs had no effect on CCR5 expression (Fig. 7, b and d) as detected by anti-CCR5 mAb 2D7. This result shows that the effect of glycanase treatment is specific for RANTES and not due to a nonspecific removal of other cell surface markers. Furthermore, glycanase treatment had no effect on other cell surface markers including CD3, CD8, CXCR4, and CD4 (data not shown). Thus, most of the RANTES binding detected by mAb 4A12 and flow cytometry is dependent on cell surface GAGs. We have obtained identical results for biotinylated MIP-1α and MIP-1β when PM1 cells are glycanase treated and binding is detected by flow cytometry using FITC-streptavidin as well as with the binding of 125I-RANTES to glycanase-treated PBMCs (our unpublished results).
Figure 7.
Glycanase treatment blocks RANTES binding. PBMCs were treated a mixture of glycanases consisting of 1 U/ml each heparinase II, heparinase III, and chondroitinase ABC or medium alone. Cells were stained for RANTES binding as detected with 4A12 or CCR5 expression as detected with 2D7. Fluorescence intensity was analyzed by flow cytometry. A and C represent RANTES binding (thick line) as compared with background (thin line) in untreated and glycanase treated cells, respectively. B and D represent 2D7 anti-CCR5 (thick line) staining as compared with isotype control (thin line) in untreated and glycanase-treated cells, respectively.
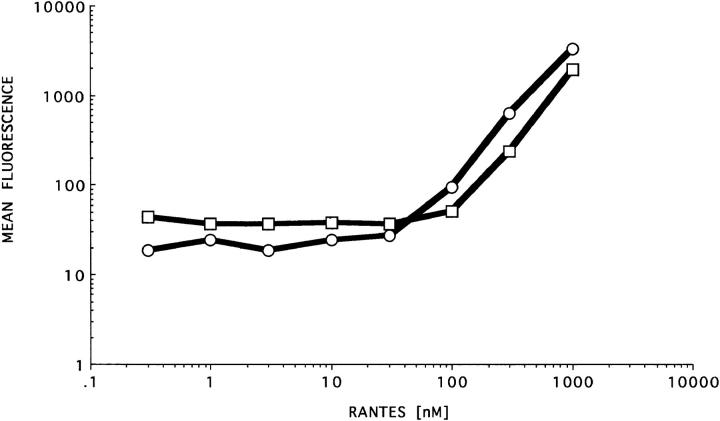
Second, a titration curve was constructed to determine whether the observed binding is saturable (Fig. 8). In these experiments, both PM1 cells and their parental cell line, HUT-78, were used. PM1 cells express the 7-transmembrane domain receptors that bind RANTES, whereas the HUT-78 cells do not (42). The cells were incubated with RANTES at concentrations ranging from 0.1–1,000 nM and binding was detected as described in Fig. 5 using mAb 4A12. A plot of the mean fluorescence intensity determined for each histogram versus RANTES concentration is shown in Fig. 8. Binding was detected only at RANTES concentrations >30 nM and increased exponentially above this concentration without an obvious plateau even at 1,000 nM, which was the highest concentration tested. This pattern of binding is consistent with the selective detection of “low affinity” interactions between RANTES and GAGs the cell surface. This binding is at least 100-fold higher than the 0.3 nM binding constant described for binding of 125I-RANTES to activated lymphocytes (reference 43 and our manuscript in preparation). The exponential binding observed by flow cytometry at concentrations of RANTES >30 nM and the lack of saturation is consistent with the “recruitment” of RANTES monomers onto GAG-bound RANTES to form oligomers as reported (41).
Figure 8.
Titration curve of RANTES on PM1 and HUT-78 cells. Hut-78 cells (□) and their derivative cell line PM1 (○) were incubated with serial dilutions of RANTES ranging from 0.3 nM to 1 μM. After incubation, mAb 4A12 (33 μM) was added to all conditions and binding was detected with anti-IgG-PE. Fluorescence intensity was determined by flow cytometry as described in Materials and Methods.
Intracellular Signaling Induced by RANTES Is Blocked by Removal of Cell Surface GAGs.
The studies described above suggest that mAb 4A12 blocks the antiviral effect of RANTES by inhibiting the interaction between this chemokine and cell surface GAGs. Unfortunately, this hypothesis is difficult to test directly, as removal of cell surface GAGs reduces infection by HIV-1 (27). Thus, we sought additional relationships between cell surface GAGs and the biological activity of RANTES. The ability of RANTES to elicit intracellular Ca2+ signals is well known (44) and we used this response to determine whether the removal of cell surface GAGs affects the biological activity of RANTES. We were not able to elicit Ca2+ signals in PM1 cells using any of a variety of agonists (including RANTES and anti-CD3), and therefore the experiment was carried out using primary PHA-activated lymphocytes grown for 9 d on IL-2 and treated with the glycanase mixture as described for Fig. 7. Control populations of the same cells were sham-treated with medium alone. Both sham- and glycanase-treated cells were loaded with the Ca2+ indicator Fluo-3, and Ca2+ mobilization was measured in response to RANTES as described previously (34, 35). As a specificity control for the effect of glycanase treatment on the response elicited by RANTES, both sham- and glycanase-treated cells were stimulated with anti-CD3.
As shown in Fig. 9 a, sham-treated cells responded with a rise in intracellular Ca2+ when stimulated with 3 nM RANTES. By contrast, no increase in intracellular Ca2+ was observed in glycanase-treated cells in response to 3 nM RANTES. This effect was not due to a generalized decrease in cellular response in the glycanase-treated cells because essentially identical Ca2+ signals were observed when both sham- and glycanase-treated cells were stimulated with an anti-CD3 mAb (clone UCHT-1, 66 nM; Fig. 9 b). Thus, cell surface GAGs are required for Ca2+ signaling induced by RANTES in primary peripheral blood T cells. We have made similar observations for a wide array of both CC and CXC chemokines in PBMCs (Burns, J., R.C. Gallo, G.K. Lewis, and A.L. DeVico, manuscript in preparation), suggesting that cell surface GAGs play a key role the Ca2+ responses elicited by these agonists as well.
Figure 9.
Cell surface GAGs are required for RANTES to elicit intracellular Ca2+ signals. Primary human lymphocytes activated with PHA for 3 d followed by expansion in IL-2 for 9 d were either treated with a mixture of glycanases consisting of 1 U/ml each of heparinase II, heparinase III, and chondroitinase ABC (thin lines) or medium alone (thick lines), as described in Materials and Methods. The cells were loaded with the Ca2+ indicator dye Fluo-3, and intracellular Ca2+ responses were elicited by either 3 nM RANTES (A) or 66 nM anti-CD3 (clone UCHT-1; B) and recorded by flow cytometry as previously described (34, 35).
Discussion
In this report, we have shown that mAb 4A12 blocks two of the biological responses elicited by RANTES, the antiviral effect against HIV-1 and intracellular Ca2+ signaling, implying that the epitope recognized by this mAb overlaps a structure that plays a role in these responses. Epitope mapping studies suggest the GAG-binding domain of RANTES as this structure. This suggestion was confirmed by studies in which heparin was used to elute RANTES from mAb 4A12 bound to a solid surface and by the ability of mAb 4A12 to block the binding of RANTES to cell surface GAGs. Taken together, our studies lend further support to the emerging importance of GAG-binding by RANTES in its biological activities (27, 28).
The first indication of a role for GAG-binding in the biological activity of RANTES came from studies in which removal of cell surface proteoglycans from PM1 cells reversed the antiviral effect of RANTES (27). Those studies also suggested that the relative resistance of macrophages to the antiviral activity of β-chemokines is due to the absence of an appropriate cell surface proteoglycan. Most recently, it was shown that the β-chemokines, RANTES, MIP-1α, and MIP-1β, are secreted as proteoglycan complexes from activated CD8+ CTLs (28). Those studies also showed that the ability of RANTES to inhibit HIV-1 infection of primary monocytes is dependent on interactions with GAGs. These studies suggest a model for the biological activity of RANTES in which cell surface proteoglycans play a key role. Therefore, it is surprising that this domain has not been mapped for RANTES. Our epitope mapping studies with mAb 4A12 add new information in this regard by implicating residues 55–66 in the α-helical region of RANTES as a component of the GAG-binding domain of RANTES.
The first identification of GAG-binding domains of chemokines came from studies of the α-chemokines PF4 (37) and IL-8 (38). It is known that the interaction between proteins and GAGs is primarily electrostatic in nature (for review see reference 45), and so cationic patches on the surfaces of chemokines are likely regions for GAG-binding. Alignment of α-chemokine sequences reveals a conserved cluster of basic amino acids in the COOH-terminal α-helices of these molecules (for an example see reference 39). Mutagenesis of the four lysines at positions 61, 62, 65, and 66 in PF4 showed that positive charges in this region are critical for heparin binding (37). Similarly, successive deletion of the COOH-terminal α-helical residues of IL-8 abrogated its ability to bind heparin (38). There is evidence that positively charged residues interspersed among the basic residues of the α-helix may affect the fine specificity of these chemokines for GAGs (39). Although these studies strongly suggest that the conserved COOH-terminal α-helix of α-chemokines comprises the GAG-binding domain, the picture is more complex for β-chemokines.
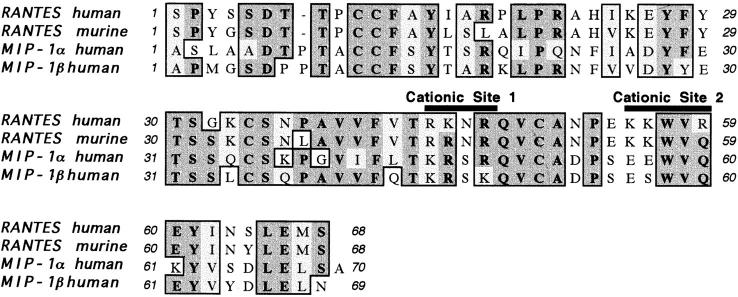
Alignment of the α-helical region of RANTES, MIP-1α, and MIP-1β shows that only RANTES has a patch of basic residues at positions 55, 58, and 60 in its COOH terminus (Fig. 10). MIP-1α has a single lysine at position 61 and MIP-1β lacks any basic residues in this region altogether. Thus, other residues must contribute to GAG-binding in these molecules. Recent mutagenesis of MIP-1α has shown that changing the lysine at position 61 to an alanine has no apparent effect on heparin binding (40). By contrast, changing the arginines to alanines at positions 18, 46, and 48, one at a time, abrogated heparin binding while changing the lysine at position 46 to an alanine altered the affinity of MIP-1α for heparin (42). This region is denoted as cationic site 1 in Fig. 10 as the cationic character of this site is highly conserved among RANTES, MIP-1α, and MIP-1β. The conservation of this site among these chemokines contrasts sharply with the cationic site 2 shown in Fig. 10 that exists only in RANTES in this comparison. Cationic site 2 is located in the α-helical region of RANTES and this site is located at ∼45° to the highly conserved cationic site 1 in the structure shown in Fig. 4. Our studies are most consistent with mAb 4A12 recognizing an epitope that overlaps cationic site 2, implicating this site in the recognition of GAGs by RANTES. Three observations support this conclusion.
Figure 10.
Multiple alignment of β-chemokine sequences. Multiple alignment of RANTES, MIP-1α, and MIP-1β was carried out using the default settings for the ClustalW subroutine of MacVector (version 6.01; Oxford Molecular, Inc., Oxford, England, UK). The following sequences were obtained from the specified databases via NCBI Entrez and used for the alignments: Swiss-Prot accession number P13501, human RANTES; Swiss-Prot accession number P30882, murine RANTES; Genbank accession number L10918, human MIP-1α; and Swiss-Prot accession number P13236, human MIP-1β.
First, removal of residues 55–66 abrogates reactivity with mAb 4A12. Analysis of Peak 4 in Fig. 3 showed that the epitope recognized by mAb 4A12 is independent of residues 67 and 68. Furthermore, the molecule found in Peak 3 (Fig. 3) was unreactive with mAb 4A12 and this was comprised of disulfide bonded fragments of RANTES corresponding to residues 1–54 with a cleavage at the glutamic acid located at position 26. Thus, it is possible that removal of residues 55–68, cleavage at glutamic acid 26, or both, abrogates the binding of mAb 4A12. Our data are most consistent with the involvement of residues 55–66 in the epitope recognized by mAb 4A12. Glutamic acid 26 is conserved between human and murine RANTES and studies have shown that mAb 4A12 does not recognize the latter (data not shown). Therefore, it is unlikely that the epitope recognized by mAb 4A12 includes this residue. Furthermore, glutamic acid 26 is located on the back surface of RANTES in Fig. 4 and is away from the α-helix or cationic site 1, making it unlikely that cleavage of this residue would contribute significantly to the loss of reactivity for this epitope. By contrast, residues 55–66 overlap cationic site 2 and harbor amino acids that are not conserved between human and murine RANTES or among human RANTES and MIP-1α or MIP-1β.
Second, as mentioned above, studies have shown that mAb 4A12 does not recognize murine RANTES. Except for a conserved lysine to arginine substitution at position 45, cationic site 1 is completely conserved between these two species of RANTES. By contrast, there is an arginine to glutamine change at position 59 in cationic site 2 and a nonconservative serine to tyrosine change at position 64 that is located on the same face of the α-helix as cationic site 2 when comparing human and murine RANTES. Thus, our data are most consistent with mAb 4A12 recognizing a discontinuous epitope that overlaps the α-helix of RANTES, which harbors cationic site 2. This epitope would be critically dependent on the residue at position 64.
Third, preliminary studies showed that mAb 4A12 does not recognize either MIP-1α or MIP-1β. These β-chemokines also have nonconservative amino acid changes at position 64. Both MIP-1α and MIP-1β have an aspartic acid at position 64 compared with the serine at this position in RANTES, consistent with the loss of reactivity of mAb 4A12 with murine RANTES that has a tyrosine at this position. Taken together, our data strongly suggests that mAb 4A12 recognizes an epitope that overlaps the α-helix of RANTES. Furthermore, these data suggest that this region contributes to the GAG-binding domain of this chemokine.
In the discussion above, it should be noted that the residues implicated in the binding of mAb 4A12 and GAGs do not overlap those reported previously to affect the binding of RANTES to CCR1, CCR3, and CCR5 or to affect the ability of RANTES to signal via these receptors (46). In those studies, residues 1–4, 6, 7, 12, 14, 15, and 17 contributed to the interaction between RANTES and CCR1, CCR3, or CCR5. The precise roles of these residues varied with the assay (binding or signaling) and with the receptor (CCR1, CCR3, or CCR5), but the results clearly implicate the first 17 amino acids in the interaction of RANTES with its canonical 7-transmembrane domain G protein–coupled receptors. Although it can not completely be ruled out that the epitope recognized by mAb 4A12 could overlap these residues instead of residues 55–66, we consider this unlikely for the following reasons. Our data shows that mAb 4A12 recognizes an epitope overlapping the GAG-binding domain. This is shown by the ability of mAb 4A12 to block the binding of RANTES to cell surface GAGs and by the ability of heparin to elute RANTES from immobilized mAb 4A12. The data reported in reference 48 are not consistent with first 17 residues of RANTES affecting GAG-binding. Some of the mutations described in reference 48 differentially affected the binding of RANTES to 3T3 cells transfected with either CCR3 or CCR5. Since 3T3 cells were used as the parent in both transfections, the observed differences are due to the 7-transmembrane receptor and not to GAG-binding. Thus, the residues of RANTES involved in receptor binding versus GAG-binding must lie in different regions of the molecule.
Our data and the reports cited above strongly suggest that the GAG-binding domain and the 7-transmembrane binding domain of RANTES are not congruent; however, as pointed out above, they could overlap with one another and with the epitope recognized by mAb 4A12. In this regard, it should be noted that a mutant MIP-1α that lacks the ability to bind heparin also failed to bind to CCR1 (47). Although our results clearly show that the ability to bind GAGs is necessary for the antiviral effect against HIV-1 and the ability of RANTES to elicit intracellular Ca2+ signaling, they do not speak directly to the potential overlap between the functional domains of RANTES. This possibility must be kept in mind in any analysis of the mechanisms by which β-chemokines bind to cell surfaces to elicit specific biological responses. At the minimum, our data argue strongly that 7-transmembrane receptors alone are not sufficient for the two responses evaluated in this report, the antiviral effect and Ca2+ signaling.
Although our data show that the epitope recognized by mAb 4A12 overlaps the GAG-binding domain of RANTES, the paradoxical pattern of binding determined by the order of reagent addition requires additional comment. This binding pattern is most consistent with the ability of cell surface GAGs to induce the oligomerization of RANTES (41). In our studies, preincubation of RANTES with mAb 4A12 blocked the binding of RANTES to cell surfaces as judged by flow cytometry. It is likely that preincubation of the reagents works this way because mAb 4A12 blocks the initial “seeding” of the cell surface GAGs with RANTES, thereby preventing oligomerization. On the other hand, preincubation of the cells with RANTES followed by detection with mAb 4A12 allows the concentration-dependent oligomerization of RANTES by cell surface GAGs. This would allow some GAG-binding sites on the ends of the oligomers to remain free and detectable by mAb 4A12. Consistent with this observation, the epitope recognized by mAb 4A12 is exposed on the free ends of oligomeric RANTES as judged by molecular modeling. Our studies showing that removal of GAGs by glycanase treatment reduces the signal in assays where RANTES is preincubated with cells before detection by mAb 4A12 are consistent with this interpretation. This conclusion is also supported by our flow cytometry studies in which we were unable to obtain saturable binding by RANTES detected by mAb 4A12. Thus, most of the binding seen by flow cytometry when cells are preincubated with RANTES is dependent upon GAG binding and not binding to a 7-transmembrane receptor. In our hands, it requires >30 nM RANTES to begin to detect a signal by flow cytometry, which is 30– 100-fold above the dissociation constant reported for the binding of RANTES to the 7-transmembrane receptors CCR1, CCR3, and CCR5 (46). Subsequent studies in our group using 125I-RANTES allows detection of binding in the range of 0.1–1.5 nM, but even at these concentrations >50% of the binding is sensitive to removal of GAGs (our manuscript in preparation). Thus, any binding experiment done using RANTES must take into account the interaction between the chemokine and GAGs in addition to the binding due to the interaction of this chemokine with the 7-transmembrane receptors.
The recognition of the GAG-binding domain by mAb 4A12 raises the question of how this mAb blocks the antiviral effect of RANTES. The best model based on current information would be that only GAG–RANTES complexes interact with the 7-transmembrane receptor in a way that prevents the ability of HIV-1 to use it as a coreceptor. In this case, mAb 4A12 would block the antiviral effect by simply blocking the formation of these complexes. This model is consistent with the studies showing that cell surface GAGs enhance the anti-viral effect (27) of RANTES in lymphoid cells and that soluble GAG-RANTES complexes are strongly anti-viral for peripheral blood macrophages (28). Although this model is currently under evaluation in our group, the data presented above show that mAb 4A12 blocks the intracellular Ca2+ response elicited by RANTES in addition to blocking the antiviral effect. This suggests that GAGs play a role in the ability of RANTES to signal cells as well. This has been confirmed by experiments in which removal of cell surface GAGs abrogates the intracellular Ca2+ response elicited by RANTES, arguing that GAGs play a much broader role than previously suspected in the biological responses to this chemokine. In summary, the data presented above strongly support a role for GAGs in the biological activities of RANTES and suggest that GAGs must be considered along with the canonical 7-transmembrane domain receptors in studies attempting to understand the biology of this chemokine.
Acknowledgments
This work was supported by National Institutes of Health (NIH) grants to A.L. DeVico and G.K. Lewis, as well as by the Institute of Human Virology. Jennifer Burns was supported by NIH predoctoral training grant T32 AI07540.
Abbreviations used in this paper
- FBS
fetal bovine serum
- GAG
glycosaminoglycan
- MIP
macrophage inflammatory protein
- RANTES
regulated on activation, normal T cell expressed and secreted
- TCID50
tissue culture dose that infects 50% of cultures
Footnotes
The skillful technical assistance of Mr. Robert Tuskan and Ms. Beverly Lamb is genuinely appreciated. We thank Dr. Roberta Kamin-Lewis for critical comments. We thank Dr. K. Swiderek, Beckman Research Institute of the City of Hope, City of Hope, CA for sequence analysis.
A.L. DeVico and G.K. Lewis contributed equally to this work.
This work was submitted by J.M. Burns in partial fulfillment of the requirements of a Ph.D. thesis in the Department of Microbiology and Immunology, University of Maryland School of Medicine, Baltimore, MD 21201.
References
- 1.Cocchi F, DeVico AL, Garzino-Demo A, Arya SK, Gallo RC, Lusso P. Identification of RANTES, MIP-1 alpha, and MIP-1 beta as the major HIV-suppressive factors produced by CD8+ T cells. Science. 1995;270:1811–1815. doi: 10.1126/science.270.5243.1811. [DOI] [PubMed] [Google Scholar]
- 2.Feng Y, Broder CC, Kennedy PE, Berger EA. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 3.Dragic T, Litwin V, Allaway GP, Martin SR, Huang YX, Nagashima KA, Cayanan C, Maddon PJ, Koup RA, Moore JP, Paxton WA. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 4.He J, Chen Y, Farzan M, Choe H, Ohagen A, Gartner S, Busciglio J, Yang X, Hofmann W, Newman W, et al. CCR3 and CCR5 are co-receptors for HIV-1 infection of microglia. Nature. 1997;385:645–649. doi: 10.1038/385645a0. [DOI] [PubMed] [Google Scholar]
- 5.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath PD, Wu L, Mackay CR, LaRosa G, Newman W, et al. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 6.Alkhatib G, Combadiere C, Broder CC, Feng Y, Kennedy PE, Murphy PM, Berger EA. CC CKR5: a RANTES, MIP-1α, MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 7.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton RE, Hill CM, et al. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 8.Farzan M, Choe H, Martin KA, Sun Y, Sidelko M, Mackay CR, Gerard NP, Sodroski J, Gerard C. HIV-1 entry and macrophage inflammatory protein-1β-mediated signaling are independent functions of the chemokine receptor CCR5. J Biol Chem. 1997;272:6854–6857. doi: 10.1074/jbc.272.11.6854. [DOI] [PubMed] [Google Scholar]
- 9.Alkhatib G, Ahuja SS, Light D, Mummidi S, Berger EA, Ahuja SK. CC chemokine receptor 5-mediated signaling and HIV-1 co-receptor activity share common structural determinants. Critical residues in the third extracellular loop support HIV-1 fusion. J Biol Chem. 1997;272:19771–19776. doi: 10.1074/jbc.272.32.19771. [DOI] [PubMed] [Google Scholar]
- 10.Alkhatib G, Berger EA, Murphy PM, Pease JE. Determinants of HIV-1 coreceptor function on CC chemokine receptor 3. Importance of both extracellular and transmembrane/cytoplasmic regions. J Biol Chem. 1997;272:20420–20426. doi: 10.1074/jbc.272.33.20420. [DOI] [PubMed] [Google Scholar]
- 11.Frade JMR, Llorente M, Mellado M, Alcami J, Gutierrez-Ramos JC, Zaballos A, Real G, Martinez AC. The amino-terminal domain of the CCR2 chemokine receptor acts as coreceptor for HIV-1 infection. J Clin Invest. 1997;100:497–502. doi: 10.1172/JCI119558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farzan M, Choe H, Vaca L, Martin K, Sun Y, Desjardins E, Ruffing N, Wu L, Wyatt R, Gerard N, et al. A tyrosine-rich region in the N terminus of CCR5 is important for human immunodeficiency virus type 1 entry and mediates an association between gp120 and CCR5. J Virol. 1998;72:1160–1164. doi: 10.1128/jvi.72.2.1160-1164.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dragic T, Trkola A, Lin SW, Nagashima KA, Kajumo F, Zhao L, Olson WC, Wu L, Mackay CR, Allaway GP, et al. Amino-terminal substitutions in the CCR5 coreceptor impair gp120 binding and human immunodeficiency virus type 1 entry. J Virol. 1998;72:279–285. doi: 10.1128/jvi.72.1.279-285.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Picard L, Simmons G, Power CA, Meyer A, Weiss RA, Clapham PR. Multiple extracellular domains of CCR-5 contribute to human immunodeficiency virus type 1 entry and fusion. J Virol. 1997;71:5003–5011. doi: 10.1128/jvi.71.7.5003-5011.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuhmann SE, Platt EJ, Kozak SL, Kabat D. Polymorphisms in the CCR5 genes of African green monkeys and mice implicate specific amino acids in infections by simian and human immunodeficiency viruses. J Virol. 1997;71:8642–8656. doi: 10.1128/jvi.71.11.8642-8656.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benkirane M, Jin DY, Chun RF, Koup RA, Jeang KT. Mechanism of transdominant inhibition of CCR5-mediated HIV-1 infection by ccr5Δ32. J Biol Chem. 1997;272:30603–30606. doi: 10.1074/jbc.272.49.30603. [DOI] [PubMed] [Google Scholar]
- 17.Samson M, LaRosa G, Libert F, Paindavoine P, Detheux M, Vassart G, Parmentier M. The second extracellular loop of CCR5 is the major determinant of ligand specificity. J Biol Chem. 1997;272:24934–24941. doi: 10.1074/jbc.272.40.24934. [DOI] [PubMed] [Google Scholar]
- 18.Wu L, LaRosa G, Kassam N, Gordon CJ, Heath H, Ruffing N, Chen H, Humblias J, Samson M, Parmentier M, et al. Interaction of chemokine receptor CCR5 with its ligands: multiple domains for HIV-1 gp120 binding and a single domain for chemokine binding. J Exp Med. 1997;186:1373–1381. doi: 10.1084/jem.186.8.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ross TM, Bieniasz PD, Cullen BR. Multiple residues contribute to the inability of murine CCR-5 to function as a coreceptor for macrophage-tropic human immunodeficiency virus type 1 isolates. J Virol. 1998;72:1918–1924. doi: 10.1128/jvi.72.3.1918-1924.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brelot A, Heveker N, Pleskoff O, Sol N, Alizon M. Role of the first and third extracellular domains of CXCR-4 in human immunodeficiency virus coreceptor activity. J Virol. 1997;71:4744–4751. doi: 10.1128/jvi.71.6.4744-4751.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Picard L, Wilkinson DA, McKnight A, Gray PW, Hoxie JA, Clapham PR, Weiss RA. Role of the amino-terminal extracellular domain of CXCR-4 in human immunodeficiency virus type 1 entry. Virology. 1997;231:105–111. doi: 10.1006/viro.1997.8506. [DOI] [PubMed] [Google Scholar]
- 22.Doranz BJ, Lu ZH, Rucker J, Zhang TY, Sharron M, Cen YH, Wang ZX, Guo HH, Du JG, Accavitti MA, et al. Two distinct CCR5 domains can mediate coreceptor usage by human immunodeficiency virus type 1. J Virol. 1997;71:6305–6314. doi: 10.1128/jvi.71.9.6305-6314.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gosling J, Monteclaro FS, Atchison RE, Arai H, Tsou CL, Goldsmith MA, Charo IF. Molecular uncoupling of C-C chemokine receptor 5-induced chemotaxis and signal transduction from HIV-1 coreceptor activity. Proc Natl Acad Sci USA. 1997;94:5061–5066. doi: 10.1073/pnas.94.10.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Atchison RE, Gosling J, Monteclaro FS, Franci C, Digilio L, Charo IF, Goldsmith MA. Multiple extracellular elements of CCR5 and HIV-1 entry: dissociation from response to chemokines. Science. 1996;274:1924–1926. doi: 10.1126/science.274.5294.1924. [DOI] [PubMed] [Google Scholar]
- 25.Cocchi F, DeVico AL, Garzino-Demo A, Cara A, Gallo RC, Lusso P. The V3 domain of the HIV-1 gp120 envelope glycoprotein is critical for chemokine-mediated blockade of infection. Nat Med. 1996;2:1244–1247. doi: 10.1038/nm1196-1244. [DOI] [PubMed] [Google Scholar]
- 26.Alkhatib G, Locati M, Kennedy PE, Murphy PM, Berger EA. HIV-1 coreceptor activity of CCR5 and its inhibition by chemokines: independence from G protein signaling and importance of coreceptor downmodulation. Virology. 1997;234:340–348. doi: 10.1006/viro.1997.8673. [DOI] [PubMed] [Google Scholar]
- 27.Oravecz T, Pall M, Wang J, Roderiquez G, Ditto M, Norcross MA. Regulation of anti-HIV-1 activity of RANTES by heparan sulfate proteoglycans. J Immunol. 1997;159:4587–4592. [PubMed] [Google Scholar]
- 28.Wagner L, Yang OO, Garcia-Zepeda EA, Ge Y, Kalams SA, Walker BD, Pasternack MS, Luster AD. Beta-chemokines are released from HIV-1-specific cytolytic T-cell granules complexed to proteoglycans. Nature. 1998;391:908–911. doi: 10.1038/36129. [DOI] [PubMed] [Google Scholar]
- 29.Hornbeck PV, Lewis GK. Idiotype connectance in the immune system. I. Expression of a cross-reactive idiotype on induced anti-p-azophenylarsonate antibodies and on endogenous antibodies not specific for arsonate. J Exp Med. 1983;157:1116–1136. doi: 10.1084/jem.157.4.1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Evan GI, Lewis GK, Ramsay G, Bishop JM. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hornbeck PV, Lewis GK. Idiotype connectance in the immune system. II. A heavy chain variable region idiotope that dominates the antibody response to the p-azobenzenearsonate group is a minor idiotope in the response to trinitrophenyl group. J Exp Med. 1985;161:53–71. doi: 10.1084/jem.161.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morita CT, Godfrey WL, Goodman JW, Lewis GK. Arsonate-specific murine T cell clones. III. Correlation between clonotype expression and fine specificity for analogs of L-tyrosine-p-azobenzenearsonate. J Immunol. 1986;137:2139–2144. [PubMed] [Google Scholar]
- 33.Abacioglu YH, Fouts TR, Laman JD, Claassen E, Pincus SH, Moore JP, Roby CA, Kamin-Lewis R, Lewis GK. Epitope mapping and topology of baculovirus-expressed HIV-1 gp160 determined with a panel of murine monoclonal antibodies. AIDS Res Hum Retroviruses. 1994;10:371–381. doi: 10.1089/aid.1994.10.371. [DOI] [PubMed] [Google Scholar]
- 34.Burns, J.M., and G.K. Lewis. 1997. Improved measurement of calcium mobilization by flow cytometry. Biotechniques. 23: 1022–1024, 1026. [DOI] [PubMed]
- 35.Pal R, Garzino-Demo A, Markham PD, Burns J, Brown M, Gallo RC, DeVico AL. Inhibition of HIV-1 infection by the beta-chemokine MDC. Science. 1997;278:695–698. doi: 10.1126/science.278.5338.695. [DOI] [PubMed] [Google Scholar]
- 36.Devico AL, Rahman R, Sarngadharan MG, Veronese FD. Mechanism of enzyme inhibition mediated by anti-reverse transcriptase antibodies from HIV type 1-infected individuals. AIDS Res Hum Retroviruses. 1994;10:953–960. doi: 10.1089/aid.1994.10.953. [DOI] [PubMed] [Google Scholar]
- 37.Maione TE, Gray GS, Hunt AJ, Sharpe RJ. Inhibition of tumor growth in mice by an analogue of platelet factor 4 that lacks affinity for heparin and retains potent angiostatic activity. Cancer Res. 1991;51:2077–2083. [PubMed] [Google Scholar]
- 38.Webb LM, Ehrengruber MU, Clark-Lewis I, Baggiolini M, Rot A. Binding to heparan sulfate or heparin enhances neutrophil responses to interleukin 8. Proc Natl Acad Sci USA. 1993;90:7158–7162. doi: 10.1073/pnas.90.15.7158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Witt DP, Lander AD. Differential binding of chemokines to glycosaminoglycan subpopulations. Curr Biol. 1994;4:394–400. doi: 10.1016/s0960-9822(00)00088-9. [DOI] [PubMed] [Google Scholar]
- 40.Koopmann W, Krangel MS. Identification of a glycosaminoglycan-binding site in chemokine macrophage inflammatory protein-1alpha. J Biol Chem. 1997;272:10103–10109. doi: 10.1074/jbc.272.15.10103. [DOI] [PubMed] [Google Scholar]
- 41.Hoogewerf AJ, Kuschert GS, Proudfoot AE, Borlat F, Clark-Lewis I, Power CA, Wells TN. Glycosaminoglycans mediate cell surface oligomerization of chemokines. Biochemistry. 1997;36:13570–13578. doi: 10.1021/bi971125s. [DOI] [PubMed] [Google Scholar]
- 42.Lusso P, Cocchi F, Balotta C, Markham PD, Louie A, Farci P, Pal R, Gallo RC, Reitz MS., Jr Growth of macrophage-tropic and primary human immunodeficiency virus type 1 (HIV-1) isolates in a unique CD4+ T-cell clone (PM1): failure to downregulate CD4 and to interfere with cell-line-tropic HIV-1. J Virol. 1995;69:3712–3720. doi: 10.1128/jvi.69.6.3712-3720.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Utsunomiya I, Tani K, Gong W, Oppenheim JJ, Wang JM. Differential expression of binding sites for chemokine RANTES on human T lymphocytes. Eur J Immunol. 1997;27:1406–1412. doi: 10.1002/eji.1830270617. [DOI] [PubMed] [Google Scholar]
- 44.Jackson RL, Busch SJ, Cardin AD. Glycosaminoglycans: molecular properties, protein interactions, and role in physiological processes. Physiol Rev. 1991;71:481–539. doi: 10.1152/physrev.1991.71.2.481. [DOI] [PubMed] [Google Scholar]
- 45.Bacon KB, Premack BA, Gardner P, Schall TJ. Activation of dual T cell signaling pathways by the chemokine RANTES. Science. 1995;269:1727–1730. doi: 10.1126/science.7569902. [DOI] [PubMed] [Google Scholar]
- 46.Pakianathan DR, Kuta EG, Artis DR, Skelton NJ, Hebert CA. Distinct but overlapping epitopes for the interaction of a CC-chemokine with CCR1, CCR3 and CCR5. Biochemistry. 1997;36:9642–9648. doi: 10.1021/bi970593z. [DOI] [PubMed] [Google Scholar]
- 47.Graham GJ, Wilkinson PC, Nibbs RJ, Lowe S, Kolset SO, Parker A, Freshney MG, Tsang ML, Pragnell IB. Uncoupling of stem cell inhibition from monocyte chemoattraction in MIP-1α by mutagenesis of the proteoglycan binding site. EMBO (Eur Mol Biol Organ) J. 1996;15:6506–6515. [PMC free article] [PubMed] [Google Scholar]
- 48.Skelton NJ, Aspiras F, Ogez J, Schall TJ. Proton NMR assignments and solution conformation of RANTES, a chemokine of the C-C type. Biochemistry. 1995;34:5329–5342. doi: 10.1021/bi00016a004. [DOI] [PubMed] [Google Scholar]