Abstract
The dysregulated expression of interleukin 4 (IL-4) can have deleterious effects on the outcome of infectious and allergic diseases. Despite this, the mechanisms by which naive T cells commit to IL-4 expression during differentiation into mature effector cells remain incompletely defined. As compared to cells from most strains of mice, activated CD4+ T cells from BALB mice show a bias towards IL-4 production and T helper 2 commitment in vitro and in vivo. Here, we show that this bias arises not from an increase in the amount of IL-4 produced per cell, but rather from an increase in the proportion of CD4+ T cells that commit to IL-4 expression. This strain-specific difference in commitment was independent of signals mediated via the IL-4 receptor and hence occurred upstream of potential autoregulatory effects of IL-4. Segregation analysis of the phenotype in an experimental backcross cohort implicated a polymorphic locus on chromosome 16. Consistent with a role in differentiation, expression of the phenotype was CD4+ T cell intrinsic and was evident as early as 16 h after the activation of naive T cells. Probabilistic gene activation is proposed as a T cell–intrinsic mechanism capable of modulating the proportion of naive T cells that commit to IL-4 production.
Keywords: interleukin 4, T helper 2, cytokine expression, genetic analysis, BALB/c mice
Control of infectious pathogens requires the maturation of naive CD4+ T cells to effector cells that secrete lymphokines critical to the coordination of the appropriate host immune response. Different pathogens require different responses; intracellular organisms are controlled by CD4+ T cells that secrete inflammatory/activating cytokines such as IFN-γ, designated the Th1 subset, whereas extracellular organisms, such as helminths, are controlled by CD4+ T cells that secrete cytokines such as IL-4 and IL-5, designated the Th2 subset. Left unregulated, such cells can mediate tissue-damaging inflammatory or allergic diseases, respectively. A number of variables influence the type of response which predominates in a given context, including antigen dose and mode of delivery, types of APCs, the nature of stimulatory or costimulatory signals, and the specific cytokines present in the developmental microenvironment (1). Of critical importance in mediating the appearance of Th1 or Th2 cells are the cytokines IL-12 or IL-4, respectively, as demonstrated both in vitro and in vivo using the recombinant cytokines or neutralizing anticytokine antibodies (2–6), and in mice with targeted disruption of these cytokine genes (7, 8) or the transcription factors implicated in their respective signaling pathways (9–13).
During the differentiation of naive, L-selectinhi CD4+ T cells, IL-4 plays a necessary early role in the generation of IL-4–producing effector cells in vitro and in vivo (2, 3, 6). The source of this requisite early IL-4 has been controversial, although increasing evidence in various experimental systems supports a direct contribution from CD4+ T cells themselves (14, 15). Given its critical role in supporting the generation of IL-4–producing effector cells, how expression of this early IL-4 is regulated remains an important question.
Strain-specific differences in the ability to generate cytokine-producing effector CD4+ T cells can be exploited to elucidate developmental control points in the process leading from naive T cells to mature effectors. BALB strain mice have been shown to possess a genetic predisposition towards the development of IL-4–producing effector T cells (4, 5). Comparative studies between BALB and non– IL-4-biased strains of mice have led to the definition of several novel levels of control in the development of mature cytokine-secreting effector cells. Murphy and co-workers have implicated a locus on chromosome 11 in premature loss of functional IL-12 receptor expression as one mechanism underlying the BALB strain bias (16, 17). Since IL-12 can promote the generation of Th1 effector cells and antagonize the Th2-promoting effects of IL-4, the acquired inability to respond to IL-12 favors the generation of IL-4– producing cells. Thus, temporal modulation of sensitivity to cytokine signaling is implicated in effector T cell development. In contrast to strain-specific differences in responsiveness to cytokines, a relatively late event in the differentiative process, we were interested in the early processes that affect how naive T cells initially commit to defined patterns of cytokine expression.
Here, we show that the cellular basis for elevated IL-4 production by BALB strain CD4+ T cells is a novel, genetically regulated developmental process that controls the proportion of naive precursors that commit to IL-4 expression. This process is shown to be influenced, at least in part, by a locus centromeric on chromosome 16. Analysis of reconstituted mixed fetal liver chimeras demonstrated that the process is intrinsic to CD4+ T cells. Thus, T cells maintain a genetically regulated mechanism that affects the probability that naive cells will commit to IL-4 gene expression after their activation. This probability is postulated to be relatively high in BALB-derived cells, resulting in the initial generation of a disproportionately large number of IL-4–expressing cells within populations of activated T cells.
Materials and Methods
Cell Purification.
CD8-depleted spleen cells were prepared by incubation of 2.5–3.0 × 107 spleen cells/ml culture medium (RPMI 1640 with 10% heat-inactivated FCS, 50 μM 2-ME, 2 mM l-glutamine and penicillin/streptomycin) with anti-CD8α mAb (3.155; American Type Culture Collection, Rockville, MD) and a mixture of guinea pig and rabbit complement (Cedarlane Labs. Ltd., Hornby, Ontario, Canada) at 37°C for 45 min. Viable cells were collected by centrifugation over Ficoll. Less than 1% of the remaining T cells were CD8+.
CD4-enriched T cells were prepared by antibody and complement-mediated depletion of CD8+ T cells and MHC class II+ HSA+ B cells, macrophages, and dendritic cells. Briefly, spleen cells in culture medium as described above were incubated with mAbs (all from American Type Culture Collection) against CD8α (3.155), I-Ad,b (BP107.2.2), and heat-stable antigen (J11d) at optimal dilutions as assessed in preliminary experiments, and rabbit and guinea pig complement. After incubation at 37°C for 45 min, viable cells were recovered by centrifugation over Ficoll, washed extensively, and counted before distribution to tissue culture wells. An aliquot of each preparation was analyzed by flow cytometry for the percentage of CD4+ T cells and routinely revealed populations of >85% purity.
Highly purified naive (L-selectinhi) and memory/effector (L-selectinlo) CD4+ T cells were prepared by sterile three-color flow cytometric sorting using a Becton Dickinson Vantage® (Mountain View, CA). CD4-enriched populations as prepared above were incubated at 4.0 × 107 cells/ml in PBS containing 3% FCS at 4°C for 20 min with a cocktail of anti-CD4–PE (YTS191.1; Caltag Laboratories, Inc., South San Francisco, CA), anti-CD8α–FITC (CT-CD8α; Caltag Laboratories, Inc.), anti-HSA–FITC (MI/69; PharMingen, San Diego, CA), and anti– L-selectin–biotin (MEL-14; PharMingen), washed, and then incubated again with streptavidin-APC (Molecular Probes Inc., Eugene, OR). The anti-CD8α antibody used for complement-mediated lysis (3.155) did not block staining by the anti-CD8α immunofluorescent mAb (CT-CD8α) used during sorting. CD4+CD8–L-selectinhi and –L-selectinlo cells were sorted, washed, counted, and resuspended in fresh culture media before distribution to tissue culture wells. Where designated, TCR Vβ8+, Vβ7,+ and Vβ2+ cells were depleted (>99%) using flow cytometry after incubation with the appropriately labeled mAbs (Caltag Laboratories, Inc.). Purity was confirmed by immunofluorescent analysis of sorted cells, and was consistently >98%. Dead cells and debris were electronically excluded using forward- and side-scatter characteristics.
The T cell–depleted APCs were prepared from spleen cell populations by antibody (all from American Type Culture collection) and complement-mediated lysis with anti-Thy 1.2 (J1j.10), anti-CD8α (3.155), and anti-CD4 (RL172). Viable cells were purified over Ficoll and irradiated with 3,000 rad before use. T cell contamination was <1% as assessed by flow cytometric analysis.
T Cell Stimulation with APCs.
Cells prepared as designated were cultured (using 5 × 106/ml) in triplicate microtiter wells of multiscreen-BV plates (1.2 μm, low protein binding, Durapore membrane; Millipore Corp., Bedford, MA) with anti-TCRβ mAb (H57.597, 12.5 μg/ml), anti-CD28 mAb (37N51.1, 10 μg/ ml), and 50 U/ml recombinant human IL-2. Human CTLA4-Ig (20 μg/ml) was added to block intrinsic CD28/CTLA4 ligands from the APCs. Anti–IL-4 mAb (11B11, 20 μg/ml) was added where indicated to the primary cultures. Where designated, the primary supernatants were collected after 5 d for cytokine analysis. Subsequently, cells were washed extensively in situ using a vacuum manifold that allowed media from each of the 96 wells to be filtered continuously through the microporous bottoms of the wells, leaving the cells intact. These primed cells were then restimulated by the addition of irradiated, T cell–depleted splenocytes (APCs; 5 × 106/well) with fresh anti-TCRβ mAb and IL-2. After 48 h, the supernatants were harvested and analyzed for IL-4 by ELISA. The same results occurred if cells were collected, counted, and redistributed to new wells using the same cell numbers before the secondary stimulation.
T Cell Stimulation without APCs.
Tissue culture wells were precoated with anti-TCR (H57.597, 10 μg/ml) and anti-CD28 (37N51.1, 10 μg/ml) for 3 h at 37°C and then washed extensively with PBS before the addition of highly purified T cells.
Genetic Analysis.
Tail DNA was prepared from a cohort of 63 C57BL/6 × (C57BL/6 × BALB/c) F1 mice using standard methods. A series of DNA marker loci were selected to cover the genome using markers polymorphic between C57BL/6 and BALB/c mice that were evenly spaced over each chromosome as identified using the Whitehead Institute for Biomedical Research/MIT Center for Genome Research (Cambridge, MA) database. Additional markers were used to saturate areas of interest defined by prior studies (16, 18–20). The markers and amplification methods used are available from the authors on request. DNA marker data were assessed in three ways. First, simple t tests contrasting mice with homozygous C57BL/6 markers against heterozygous BALB/c × C57BL/6 markers with respect to IL-4 levels were performed. Second, likelihood-based interval mapping was performed using the Quantitative Trait Locus (QTL)1-Cartographer program. QTL mapping is a technique used to identify the approximate location of genes that influence a trait that exhibits continuous variation (21). It is particularly suited for experimental cross analysis among inbred mouse strains, since strains known to be polymorphic at different loci can be interbred, creating informative recombinations for linkage analysis. Finally, multilocus models were assessed through multiple regression analyses (22, 23). Significance levels for the univariate analysis results were compared according to suggested guidelines (24).
Intracellular Cytokine Determination.
Highly purified CD4 T cells prepared as above from BALB/c, C57BL/6, and (BALB/c × C57BL/6) F1 mice were stimulated in the presence of 25 μg/ml anti–IL-4 receptor mAb M1 (25) for 16 h in wells coated with antibodies to TCRβ (H57.597, 20 μg/ml) and CD28 (37N51.1, 20 μg/ml). 3 h before harvesting, brefeldin A (10 μg/ml; Epicentre Technologies Corp., Madison, WI) was added to promote intracellular accumulation of secreted proteins such as IL-4. Cells were washed twice in PBS/1% FCS and then incubated on ice for 30 min with anti-CD44–CyChrome (IM7; PharMingen). After two washes with PBS, cells were fixed in 4% formaldehyde for 15 min and then washed two more times in PBS/1% FCS. Cells were incubated at room temperature for 10 min in PBS/0.5% saponin and then for 30 min with anti–IL-4–PE (11B11; PharMingen) diluted in PBS/0.5% saponin. Before resuspending in PBS/ 1% FCS for flow cytometric analysis, cells were washed twice in PBS/1% saponin and once in PBS/1% FCS. Controls were stained identically, with the exception of anti–IL-4–PE, which was preincubated for 20 min with recombinant murine IL-4. Analysis was based upon 2 × 105 events live-gated as based on forward- and side-scatter characteristics.
Reverse Transcriptase PCR Analysis.
Highly purified L-selectinhi or L-selectinlo CD4+ T cells from BALB/c or C57BL/6 mice were stimulated by culture in wells precoated with anti-TCRβ mAb (H57.597, 10 μg/ml) and anti-CD28 mAb (37N51.1, 10 μg/ml), in the presence of anti–IL-4R mAb (M1, 25 μg/ml). After 17 h, total RNA was extracted (RNAzol B; Biotecx Laboratories, Houston, TX), reverse transcribed with random hexamers (Clontech Laboratories, Inc., Palo Alto, CA) and Moloney murine leukemia–virus reverse transcriptase (Clontech Laboratories, Inc.), and used to template a competitive PCR assay for the presence of the constitutively expressed gene, hypoxanthine phosphoribosyltransferase (HPRT). Serial twofold cDNA dilutions were performed in the presence of a constant amount of the competitor plasmid, pQRS (26), which contains both HPRT and IL-4 pseudogenes that amplify using the wild-type primers but result in larger, more slowly migrating products after electrophoresis in ethidium bromide–stained 2.5% agarose gels.
Establishment of Hematopoietic Cell Chimeric Mice.
Embryonic day 15 fetuses were recovered from time-mated parental BALB/c and C57BL/6 females. Day 0 was the day of detecting a vaginal plug. Fetal liver cell suspensions from each parental strain were prepared in culture medium, washed, counted, and mixed in a 1:1 ratio. The mixed cells (7 × 106 per mouse) were administered intravenously to reconstitute lethally irradiated (980 rads; BALB/c × C57BL/6) F1 recipient mice. Animals were maintained in a specific pathogen–free facility for 3–6 wk before analysis to allow sufficient time for donor hematopoietic cells to reconstitute the immune system of recipient mice. For analysis, cell suspensions were prepared from the combined spleen and lymph node (cervical, axillary, brachial, inguinal and mesenteric) populations from pools of two to three chimeric mice. In three experiments, reconstitution favored C57BL/6-derived cells (mean percentage C57BL/6-derived CD4+ T cells, 31%; mean percentage BALB/c-derived CD4+ T cells, 8%). CD4+-enriched T cells were prepared as described above and sorted for their expression of H-2Db (mAb KH95; PharMingen) and H-2Dd (mAb 34-2-12; PharMingen) to distinguish C57BL/6-derived, BALB-derived, and F1-derived (host) cells. The purified CD4+ T cell populations (>99%) were activated for 16 h in tissue culture wells precoated with anti-TCR/CD28 mAb and IL-2 in the presence of anti–IL-4R blockade before purification of mRNA and analysis by reverse transcriptase PCR assay for IL-4 expression.
Results
BALB-derived CD4+ T Cells Produce Elevated Levels of IL-4.
CD8-depleted spleen cells that were primed for 5 d with anti-TCR and anti-CD28 mAb in the presence of IL-2, then washed and restimulated under the same conditions for 2 d, produced >40 times more IL-4 when isolated from BALB-, as compared with B10.D2- or C57BL/6-strain mice (Fig. 1). Inclusion of an anti–IL-4 mAb during the priming period abolished the IL-4 recovered during the second stimulation, in agreement with prior studies (2, 3, 6). Similar results were obtained when IL-5 was measured in the secondary reaction, although the increases were more variable (6–40-fold greater amounts from BALB/c T cells in six experiments). NK1.1+ CD4+ T cells were not required for these effects; depletion of Vβ8+, Vβ7+, and Vβ2+ CD4+ T cells, which comprise the majority of these cells (27), did not affect IL-4 production by purified BALB/c or C57BL/6 T cells in this assay (14.4 vs. 0.6 ng/ ml IL-4 in BALB/c or C57BL/6 cultures, respectively). These data indicate that (a) IL-4 was being produced during the priming period and (b) that this IL-4 was required to support the appearance of cells capable of producing IL-4 (and IL-5) upon restimulation. Thus, with this assay, the high IL-4 phenotype of BALB-derived CD4+ T cells could be revealed with nontransgenic T cells from wild-type mice.
Figure 1.
IL-4 production by CD4-enriched spleen cell populations. Spleen cells from BALB/c (H-2d), BALB.B (H-2b), BALB.K (H-2k), and B10.D2 (H-2d) mice were depleted of CD8+ T cells and were activated with anti-TCRβ and anti-CD28 mAbs in the presence of IL-2, and in the absence (left) or presence (right) of anti–IL-4 mAb. After 5 d, cells were restimulated with fresh APCs, anti-TCRβ mAb, and IL-2. After 48 h, the supernatants were harvested and analyzed for IL-4 by ELISA. Results depict means ± SEM of triplicate determinations. C57BL/6 cells gave results comparable to B10.D2 cells.
The recovery of higher amounts of IL-4 after restimulation could reflect augmented transduction of signals through the IL-4R in BALB-derived T cells, since IL-4 might enhance its own production via Stat6-mediated transcriptional activation (28). To assess this possibility, naive, L-selectinhi CD4+ T cells were purified from BALB/c and C57BL/6 mice in which the IL-4 gene was inactivated (29). Cells were primed in the presence of titrating amounts of exogenous IL-4, and, after restimulation, assayed for IL-5 production as an independent means of assessing IL-4 responsiveness (1). Across the range of IL-4 doses tested, IL-4–deficient BALB/c cells made slightly less, and certainly no more, IL-5 than did cells from IL-4– deficient C57BL/6 mice (Fig. 2 A). These data suggest that differences in IL-4R signaling efficiency were not sufficient to explain the augmented levels of IL-4 (and IL-5) produced by restimulated BALB/c CD4+ T cells.
Figure 2.
Defining the high IL-4 phenotype in BALB/c CD4+ T cells. (A) Highly purified L-selectinhi CD4+ T cells from 10th-backcross BALB/c or C57BL/6 IL-4–deficient mice were incubated with anti-TCRβ mAb, anti-CD28 mAb, IL-2, CTLA4-Ig, and BALB/c APCs. Murine recombinant IL-4 was added at the designated concentrations. After 5 d, cells were washed extensively, counted, and recultured in the presence of fresh anti-TCRβ mAb, IL-2, and APCs. After 2 d, the supernatants were collected and analyzed for IL-5 by ELISA. Results depict the mean ± SEM of triplicate determinations and are representative of three comparable experiments. (B) Highly purified L-selectinhi CD4+ T cells from BALB/c or B10.D2 mice were stimulated with anti-TCRβ mAb, anti-CD28 mAb, IL-2, CTLA4-Ig, and BALB/c APCs, but in the absence or presence of anti–IL-4R mAb as designated. After 5 d, primary supernatants were collected, and the cells were washed, counted, and recultured with anti-TCRβ mAb, IL-2, and APCs. After 2 d, the secondary supernatants were collected. Both primary and secondary supernatants were analyzed for IL-4 by ELISA. Results depict means ± SEM of triplicate determinations and are representative of three comparable experiments.
Independent of positive feedback through IL-4R signaling, TCR/CD28-mediated activation might directly stimulate augmented IL-4 production from naive BALB/c CD4+ T cells. To address this possibility, naive, L-selectinhi CD4+ T cells were primed with anti-TCR/CD28 in the presence of blocking anti–IL-4R mAb. The efficacy of the IL-4R blockade was demonstrated by the dose-dependent abrogation of IL-4 production upon restimulation, which was complete at 25 μg/ml mAb. In a reciprocal fashion, IL-4R blockade now allowed the detection of IL-4 that accumulated during priming, presumably due to the inhibition of receptor-mediated uptake of secreted IL-4. More importantly, compared with B10.D2 or C57BL/6, BALB/c-derived CD4+ T cells produced substantially more IL-4 (Fig. 2 B and data not shown). Thus, differential IL-4 production that occurred in direct response to activation during priming and that was independent of signals mediated by the IL-4R correlated with the ability of BALB/c, but not C57BL/6 or B10.D2, CD4+ T cells to differentiate into high IL-4 producers upon secondary stimulation. This was further supported by an analysis of IL-4 transcripts present in these cells as early as 16 h after stimulation. Even in the presence of saturating amounts of anti–IL-4R mAb, highly purified naive (L-selectinhi) BALB/c CD4+ T cells contained 8–16-fold more IL-4 mRNA as compared to C57BL/6 cells (Bix, M., and R.M. Locksley, unpublished observations; data not shown, and see below). Since these cells were activated by cross-linking antibodies in the absence of APCs or exogenous cytokines, the high IL-4 production phenotype was intrinsic to L-selectinhi CD4– enriched T cells.
IL-4 Production Is Distinct from Loss of IL-12 Responsiveness.
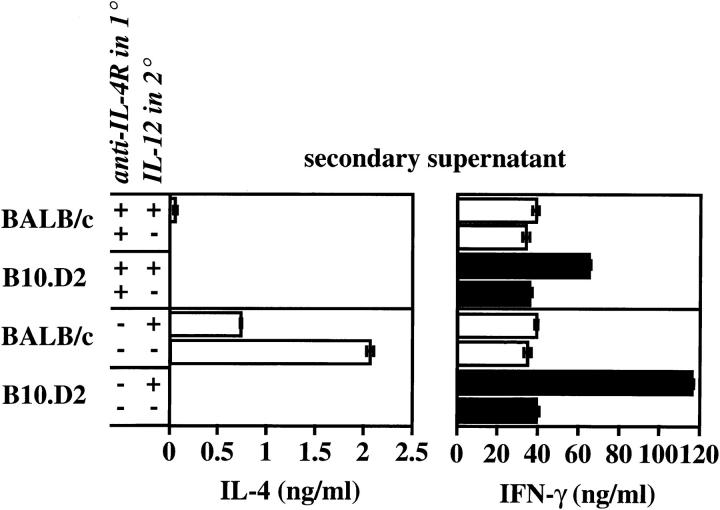
Prior studies of the Th subset bias of BALB/c T cells suggested that premature loss of IL-12 responsiveness was responsible for the relatively low amounts of IFN-γ recovered from these cells after restimulation in vitro (17). To investigate the potential relatedness of this phenomenon to the high IL-4 phenotype, highly purified naive, L-selectinhi CD4+ T cells from BALB/c and B10.D2 mice were primed with anti-TCR/CD28 mAb in the presence or absence of IL-4 receptor blockade (25 μg/ml anti–IL-4R mAb). After restimulation in the presence of IL-12, the relative capacity of the cells to respond with increased production of IFN-γ was then assessed. Additional cultures were primed twice sequentially to create more completely differentiated Th subset populations (30). In both singly and doubly primed cultures, responsiveness to IL-12 was impaired among BALB/c cells, even in the presence of IL-4R blockade (Fig. 3 and data not shown). Similarly, the IL-12 responsiveness of B10.D2 cells remained intact, even in the presence of IL-4R blockade. These data indicate that the response or nonresponse to IL-12 was expressed independently of IL-4 produced during priming, suggesting that the high IL-4 production phenotype represented a distinct aspect of Th subset development.
Figure 3.
Loss of IL-12 responsiveness segregates independently from IL-4 production. Highly purified L-selectinhi CD4+ T cells from BALB/c and B10.D2 mice at 106 cells/ml were stimulated with anti-TCRβ and anti-CD28 mAbs and IL-2 in the presence of irradiated BALB/c APC and human CTLA4-Ig (20 μg/ml) to block intrinsic CD28/CTLA4 ligands, and in the presence or absence of 25 μg/ml anti–IL-4R mAb as designated. After 5 d, cells were washed extensively and recultured for 2 d at 106 cells/ml with fresh APCs, anti-TCRβ mAb, and IL-2, in the presence or absence of recombinant IL-12 (10 ng/ml). After 2 d, the supernatants were collected and analyzed for IL-4 and IFN-γ using ELISA. Results depict the means ± SEM of triplicate determinations and are representative of two comparable experiments.
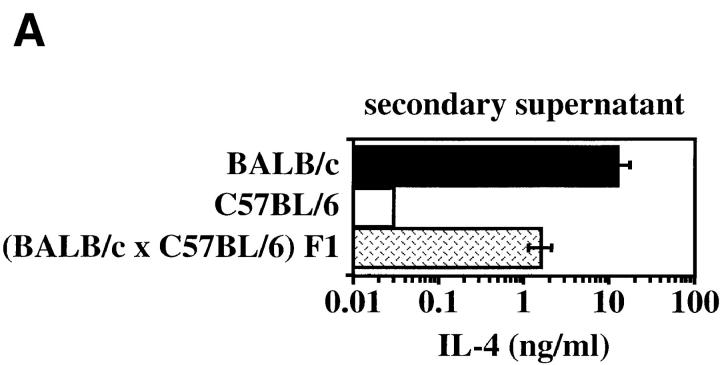
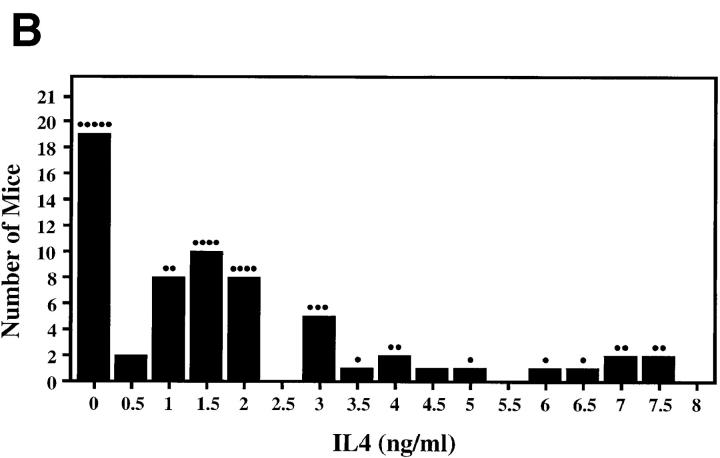
Previous genetic analysis mapped the IL-12 unresponsiveness phenotype to chromosome 11 near the IL-4 gene (16). To determine whether the high IL-4 phenotype revealed in our studies would be amenable to similar genetic analysis, CD4+ T cells from (BALB/c × C57BL/6) F1 and parental mice were compared. The restimulated F1 cells produced levels of IL-4 that were intermediate to those from the parental animals (Fig. 4 A). (BALB/c × C57BL/6) F1 mice were then backcrossed to the low IL-4–producing C57BL/6 parent strain, and CD4+ T cells from the 63 progeny were scored for levels of IL-4 production (Fig. 4 B). The IL-4 distribution did not conform to that predicted for a simple monogenic trait (50% representation by each parental phenotype). Despite this, the bimodal distribution of IL-4 levels about the C57BL/6 (IL-4 levels <0.5) and (BALB/c × C57BL/6) F1 (IL-4 levels ∼1) parental levels suggested that a small number of genes might be implicated, and a genome-wide approach was undertaken using QTL analysis.
Figure 4.
Genotypic analysis of the high IL-4 phenotype. (A) CD4-enriched spleen cells from BALB/c, C57BL/6, and (BALB/c × C57BL/6) F1 mice were cultured with anti-TCR and anti-CD28 mAbs with IL-2. The supernatants were harvested 48 h after the second stimulation and were analyzed for IL-4 by ELISA. Results depict means ± SEM of triplicate determinations and are representative of five experiments. (B) Distribution of IL-4 production during secondary stimulation by CD4+ T cells from 63 progeny of C57BL/6 × (BALB/c × C57BL/6) F1 mice. Individual mice displaying similar IL-4 levels are grouped together in bins represented as vertical columns in increasing increments of 0.5 ng/ml IL-4 (e.g., mice in the column above 1 had IL-4 levels >0.5 and ≤1.0). Individual mice that inherited the BALB/c D16Mit81 allele are indicated as discrete dots (•) above each vertical column. (C) Interval mapping with markers along the centromeric region of chromosome 16. A cohort of 63 C57BL/6 × (BALB/c × C57BL/6) F1 progeny was typed for its IL-4 phenotype and for inheritance of parental polymorphic simple sequence length markers that spanned the mouse genome. The likelihood of inheriting a BALB/c allele at D16Mit81 correlated with the high IL-4 phenotype, with a peak lod score of 3.8 (P < 0.004). Locations of polymorphic markers are shown at the bottom, with black horizontal bar depicting 95% confidence interval.
QTL mapping relies on the cosegregation of polymorphic alleles at known locations throughout the genome with hypothetical alleles at neighboring loci that influence the trait of interest. If evidence for such cosegregation can be found, then one can infer the existence of a trait-influencing locus near the implicated marker loci (31). QTL mapping strategies make no assumptions about the ultimate number or location of loci influencing the trait, although the size of the effect of each locus on the trait of interest, or “percentage of variation explained,” will dictate the number of individuals and the number of markers required to map the trait with some power of confidence (32). Thus, QTL mapping designs can accommodate phenotypes of some complexity, making it an ideal approach for many biologic phenotypes.
DNA was prepared from each of the 63 backcrossed mice and used to screen for the inheritance of a representative set of simple sequence length DNA markers selected to span the mouse genome. Markers were selected based on polymorphisms between the BALB/c and C57BL/6 alleles, permitting the rapid identification of mice that had inherited the BALB/c allele at a given locus. By simple univariate t tests, only three markers showed evidence for significant or suggestive linkage. Two adjacent markers on chromosome 16 (D16Mit81 and D16Mit73) and one on chromosome 7 (D7Mit103) showed strong genotypic associations with IL-4 levels. Interval mapping with 10 markers spanning chromosome 16 resulted in a peak lod score of 3.8 (95% support interval = 2.5 centimorgans) at the D16Mit81 locus (Fig. 4 C). This locus alone could account for 27% of the variation in IL-4 levels among the backcross progeny. Multiple regression analysis involving D16Mit81 and D7Mit73 suggested that the putative D7Mit73-linked locus had a much smaller effect on IL-4 levels than the chromosome 16 locus (r 2 for D16Mit81 locus = 26.75; r 2 for D7Mit73 = 5.76; Table 1). Taken together, these two loci could account for one-third of the variance in IL-4 levels produced by CD4+ T cells in vitro. A model positing interaction between the two loci produced nonsignificant results for the interaction term. No linkage to chromosome 11 (or any other chromosome) was apparent, confirming the independence of this phenotype from the previously described IL-12 unresponsiveness trait. Thus, a novel genetic locus that contributes to Th subset bias has been identified on the centromeric portion of chromosome 16. A second locus on chromosome 7 was suggested, albeit with less statistical confidence.
Table 1.
Multiple Regression Analysis of Linked Markers with IL-4 as the Dependent Variable
| Model | Term | Coeff. | S.E. | F-stat. | P value | r 2 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Noninteracting | Y-intercept | 3.516 | 0.364 | 93.18 | <0.001 | — | ||||||
| D16Mit81 | 1.646 | 0.544 | 9.15 | 0.004 | 26.75 | |||||||
| D7Mit103 | 1.075 | 0.568 | 3.58 | 0.065 | 5.76 | |||||||
| Interacting | Y-intercept | 3.726 | 0.378 | 97.36 | <0.001 | — | ||||||
| D16Mit81 | 2.253 | 0.643 | 12.28 | 0.001 | 27.32 | |||||||
| D7Mit103 | 2.279 | 0.905 | 6.33 | 0.016 | 6.78 | |||||||
| Interaction | −1.932 | 1.147 | 2.84 | 0.100 | 2.77 |
Two models were fit to the data. First, one in which no interaction was assumed between the chromosome 16 and chromosome 7 loci (noninteracting) and second, one in which an interaction was assumed (interacting). Coeff., regression coefficient; S.E., standard error of the regression coefficient; F-stat., F-statistic measuring the contribution of the term to the IL-4 variance explained; P value, probability level associated with the F-statistic; r 2, percentage of the IL-4 variance explained.
BALB/c-specific High IL-4 Production Is Due to an Increase in the Proportion of CD4+ T Cells That Commit to IL-4 Expression.
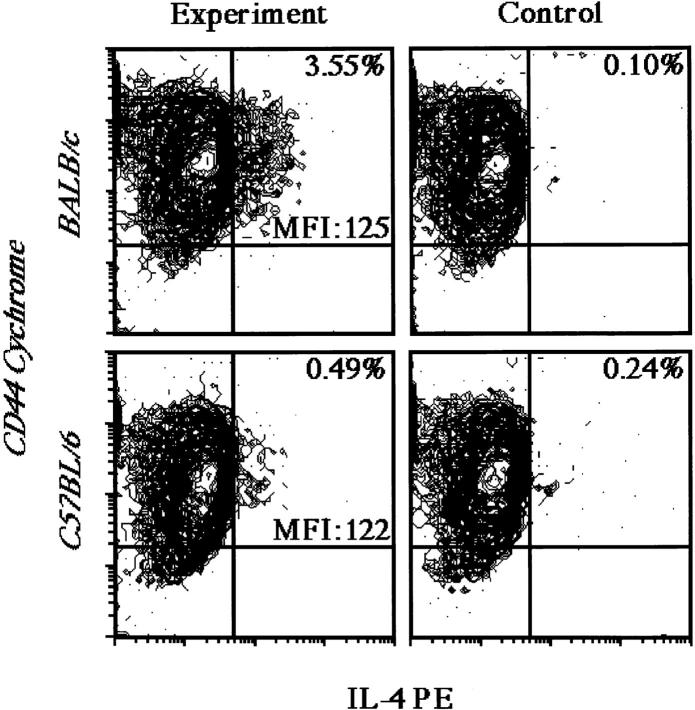
Prior studies have indicated that <5% of CD4+ T cells begin to express IL-4 after primary activation (33). Therefore, the increased production of IL-4 by BALB/c T cells could reflect an elevation in the proportion of cells induced to express IL-4 and/or increased production from the cells that commit to IL-4 expression. To address these possibilities, CD4-enriched T cells were activated for 16 h in the presence of anti–IL-4R mAb and examined for intracellular IL-4 content as described previously (34). Activated, CD44+, BALB/c, and C57BL/6 CD4+ T cells expressed comparable levels of IL-4, as judged by the mean fluorescence intensity of the positive cells. However, the frequency of IL-4–containing cells was up to 10-fold higher (range: 3–10-fold; n = 8; P < 0.05) for BALB/c as compared with C57BL/6 cells (Fig. 5). Furthermore, analysis of activated CD4+ T cells from (BALB/c × C57BL/6) F1 mice revealed an intermediate frequency of IL-4–producing cells (Bix, M., unpublished observations; data not shown), concordant with the earlier ELISA measurements of IL-4 production after restimulation. These data suggest that the high IL-4 phenotype of BALB/c mice stems not from the elevated production of IL-4 per cell, but rather from an increase in the proportion of naive T cells that commit to IL-4 expression after TCR activation.
Figure 5.
Intracellular IL-4 expression in stimulated CD4+ T cells. CD4-enriched T cells from BALB/c and C57BL/6 mice were stimulated in the presence of anti–IL-4R mAb (M1, 25 μg/ml) and IL-2 in wells precoated with anti-TCRβ mAb and anti-CD28 mAb. After 13 h, 10 μg/ml brefeldin A was added to promote the accumulation of secreted proteins. After an additional 3 h of culture, the cells were stained for the activation marker CD44 and, after permeabilization, for intracellular IL-4 content. Controls were handled identically, except that anti–IL-4–PE mAb was incubated with recombinant murine IL-4 before use in the cell-staining assays. Viable CD44+ T cells were gated using forward and side-scatter characteristics for their intensity of IL-4 staining (left) as compared to staining in the presence of control reagents (right). Percentages reflect IL-4 positive cells. MFI, mean fluorescence intensity of the IL-4 gate. Results from one of four comparable experiments are shown.
BALB/c CD4+ T Cells in Both the Naive, L-selectinhi and Memory, L-selectinlo Populations Exhibit the High IL-4 Phenotype.
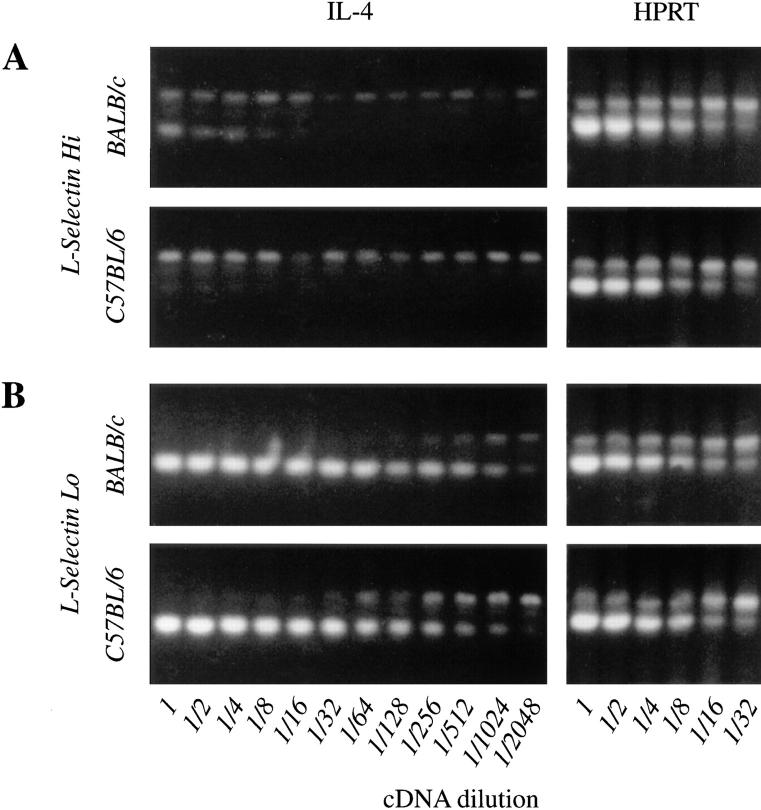
The majority of helper cells that produce IL-4 express an L-selectinlo phenotype, consistent with their existence as memory or effector populations. The IL-4 produced by these cells is thought to promote the generation of IL-4– producing cells from naive, L-selectinhi CD4+ T cells (35). To determine whether the BALB strain-specific high IL-4 phenotype was indeed confined to the L-selectinlo pool, we used a sensitive semiquantitative PCR assay to analyze the IL-4 mRNA recovered 16 h after activation of highly purified L-selectinlo and L-selectinhi CD4+ T cells (>98% purity) from BALB/c and B10.D2 mice. Consistent with prior reports, L-selectinlo cells expressed 2,000-fold more IL-4 transcripts than L-selectinhi cells. Despite the relative paucity of IL-4 transcripts among the L-selectinhi cells, the BALB strain-specific high IL-4 phenotype was evident nonetheless in both populations as defined by their L-selectin phenotypes (Fig. 6). Two observations suggested that the detection of IL-4 transcripts in activated L-selectinhi cells was not simply due to contamination by L-selectinlo cells. First, even an unlikely 2% contamination would predict a maximal 50-fold, rather than a 2,000-fold, difference in the levels of IL-4 transcripts recovered between the two cell populations. Second, the relative difference in IL-4 transcript levels between BALB/c and B10.D2 cells was consistently higher among cells that were L-selectinhi (8– 16-fold) as opposed to L-selectinlo (4–8-fold). Together, these results suggest that naive, L-selectinhi CD4+ T cells can be activated by TCR/CD28 cross-linking to express the IL-4 gene and, moreover, that the BALB strain-specific high IL-4 phenotype is manifest within the population of cells thought to be enriched for naive T cells.
Figure 6.
Expression of the high IL-4 phenotype in highly purified L-selectinhi and L-selectinlo CD4+ T cells populations. Highly purified L-selectinhi (A) and L-selectinlo (B) CD4+ T cells were incubated in wells precoated with anti-TCRβ and anti-CD28 mAbs, IL-2, and anti–IL-4R mAb. After 16 h, mRNA was purified, reverse transcribed, and analyzed for the presence of IL-4 transcripts in relation to the expression of constitutively expressed HPRT transcripts using competitive PCR. Upper bands in each gel denote the amplified competitor product, lower bands denote the authentic transcript prepared from the cells indicated.
The High IL-4 Phenotype Develops as a T Cell–autonomous Trait.
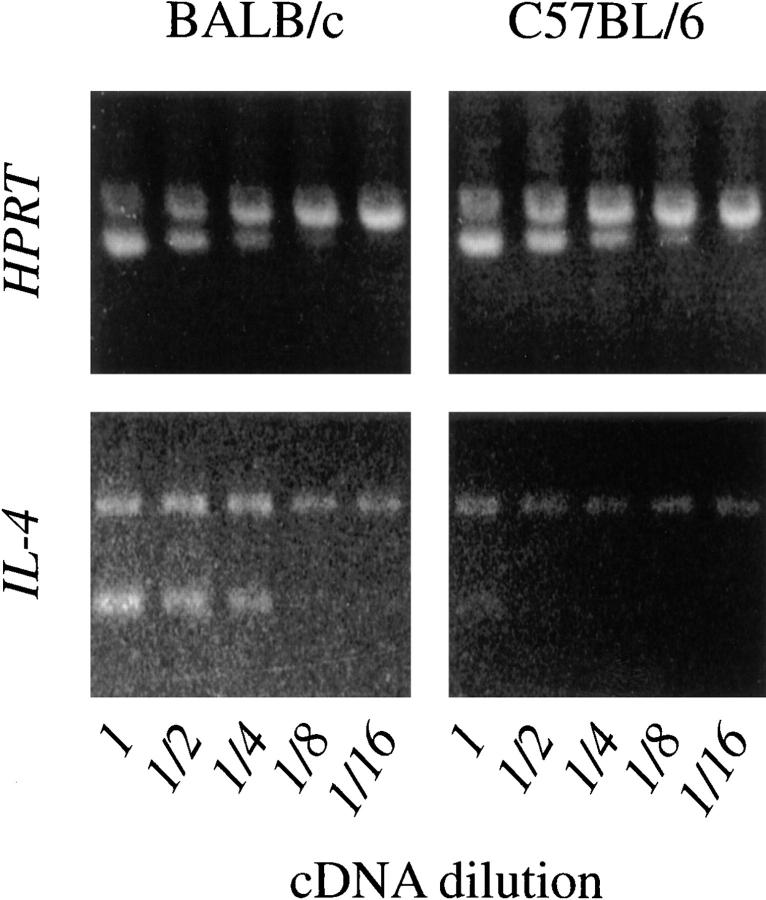
The BALB strain-specific high IL-4 phenotype could develop in a cell-autonomous fashion or arise from transinteractions, which impart the phenotype to BALB cells. Such interactions might include exposure to BALB-specific self-antigens or APC-mediated signals (36). To distinguish between these possibilities, mixed hematopoietic cell chimeras were constructed and analyzed. Such chimeras allowed donor-derived hematopoietic precursor cells from two genetically distinct parents to develop in the same animal with equivalent exposure to transinteractions. Therefore, any strain-specific properties that persist in the donor-derived cells developing in the chimeric animals must reflect cell-autonomous expression of their distinct genetic origins. BALB/c and C57BL/6 embryonic day 15 fetal liver cells were used as a rich source of hematopoietic precursors devoid of mature T cells. The liver cells were mixed and transferred to lethally irradiated (BALB/c × C57BL/6) F1 mice. Animals were analyzed between 3 and 6 wk after reconstitution. Donor-derived H-2Db+d− C57BL/6 and H-2Db−d+ BALB/c CD4+ T cells were sorted from host-derived H-2Db+d+ cells using flow cytometry, and the purified cells were activated with anti-TCR/CD28 mAb for 16 h in the presence of anti–IL-4R mAb. Relative IL-4 mRNA content of the activated cells was then compared using the semiquantitative PCR assay. The chimera-derived CD4+ T cells of BALB/c origin produced 8–16-fold more IL-4 mRNA after activation than did chimera-derived CD4+ T cells of C56BL/6 origin, a difference that was similar to that seen using cells derived from unmanipulated mice from the two parental backgrounds (Fig. 7). These data suggest that the BALB strain-specific high IL-4 phenotype was acquired in a CD4+ T cell–autonomous fashion, consistent with a process involved with a T cell intrinsic program of differentiation.
Figure 7.
Expression of the high IL-4 phenotype in BALB/c-origin CD4+ T cells from F1 chimeric mice. T cells from irradiated, reconstituted, (BALB/c × C57BL/6) F1 chimeric mice that contained a mixture of parentally derived lymphoid cells were sorted for expression of CD4+ and either H-2Db (C57BL/6 origin; right) or H-2Dd (BALB/c origin; left), but not both (F1 origin). Purified cells were incubated in wells precoated with anti-TCRβ and anti-CD28 mAbs, IL-2, and anti–IL-4R mAb. After 16 h, the RNA was purified and analyzed for IL-4 expression using the competitive PCR assay.
Discussion
We have identified a novel, genetically regulated developmental process that controls the proportion of naive CD4+ T cells that commit to IL-4 expression. This process is influenced, at least in part, by a locus centromeric on chromosome 16 which we have designated dice (determinant of IL-4 commitment). Evidence for additional genetic complexity to this phenotype was suggested by a lesser association with a second locus on chromosome 7 (dice-2). Together, these two genetic regions could account for 33% of the increased IL-4 phenotype of BALB/c CD4+ T cells. dice's influence on IL-4 levels was independent of signals mediated via the IL-4 receptor, and hence upstream of potential autoregulatory effects of IL-4. Consistent with a putative role in effector cell differentiation, dice acted in a CD4+ T cell–intrinsic manner that was evident as early as 16 h after activation.
As shown here and elsewhere (Fig. 1) (2–6), the IL-4 produced during priming was required for the subsequent appearance of IL-4–producing cells, and thus for Th2 lineage commitment. Since dice influences the number of cells that become IL-4 producers, and hence the total amount of IL-4 produced by a population of CD4+ T cells, dice influences the balance among Th subsets. We propose the existence of a mechanism, controlled in part by dice, that regulates the probability that activated naive, L-selectinhi CD4+ T cells will commit to IL-4 gene expression. This probability is postulated to be relatively high in BALB-derived cells, resulting in the generation of a disproportionately large number of IL-4–expressing cells within activated T cell populations. Although this mechanism remains speculative, these data provide evidence that the amounts of IL-4 produced by naive T cells can be genetically regulated by controlling the proportion of cells that commit to its expression. By influencing the balance of Th subsets that might potentially be generated, such a process could modulate host susceptibility to infectious and/or autoimmune diseases.
Several transcription factors have been implicated in Th subset commitment. Both GATA-3 and c-maf transcripts were present in Th2, but not Th1 clones and cells, and were shown to activate expression of the IL-4 promoter when overexpressed in non-T cell lines, or, in the case of GATA-3, in Th1 cells (37, 38). These factors were identified by subtractive methods comparing cells enriched for Th1 or Th2 commitment. Since the IL-4 commitment phenotype (as mediated by dice, dice-2, and potentially other genes not revealed in this analysis) was expressed independently of input from either the IL-4R or IL-12R, we conclude that it operates upstream of events mediated by these cytokine-regulated pathways.
Recent studies of the acquisition of cytokine profiles during the activation of naive helper T cells have revealed multiple patterns of cytokine gene expression that were distinct from the canonical Th1 and Th2 subsets (33, 39– 41). When cloned under nonselective conditions (e.g., in the absence of exogenous IL-4 or IL-12), CD4+ T cells could be identified that expressed IL-3, IL-4, IFN-γ, and GM-CSF in a variety of combinatorially assorted patterns (31). Similar conclusions were made in an analysis of IL-2, IL-4, IL-5, IL-10, and IFN-γ (32). Studies using TCR transgenic CD4+ T cells activated in vitro (33, 42) or in vivo (43) showed that only a fraction of activated cells expressed any given cytokine, and that again substantial heterogeneity occurred in the patterns of cytokines expressed by individual cells. With repeated activation, the overall diversity of cytokine profiles diminished as the proportion of cells expressing similar patterns increased, and these patterns became increasingly refractory to alterations induced by exogenous cytokines (32). These and other studies suggest that polarization of responses into effector Th1 and Th2 phenotypes may reflect the selective expansion of cells that express cytokine patterns probabilistically acquired during early activation–induced differentiation. According to such a scheme, the cytokine expression profile of mature populations may be constrained by the initial probabilities of expression for each particular cytokine. In accordance with such a model, conditions that favor the outgrowth of new populations of Th effector cells with their own distinct cytokine profiles continue to be discovered (44, 45).
Increasingly, probabilistic gene expression has been implicated in mediating alternative pathways of cellular growth and differentiation (46). Single-cell analysis of IL-2 expression revealed a bimodal all-or-none pattern of transcription initiation, consistent with threshold effects that must be overcome and that are achieved only by subgroups within the population (47). Recent reports document the apparent independent regulation of the IL-4 alleles within CD4+ T cells, consistent with probabilistic activation of this locus (48, 49). The identification of genetic loci that affect expression of this fundamental switch point in CD4+ T cell biology may lead to a systematic dissection of the mechanisms regulating cytokine gene expression in developing T cells, and provide cellular targets that might affect this expression at early periods before effector functions become invariant.
Acknowledgments
The authors thank Robert Coffman, DNAX Research Institute, for mAbs and the IL-4 −/− mice; James Allison, University of California, Berkeley, for mAbs to CD28; Peter Linsley, Bristol-Myers Squibb, for CTLA4-Ig; Eric Weider, Gladstone Institute of Virology, University of California, San Francisco, for assistance with flow cytometry; Fred Finkelman, University of Cincinnati, for mAb M1 to the IL-4 receptor; Anne O'Garra, DNAX Research Institute, for help with the intracellular cytokine method; and Nigel Killen and Art Weiss, University of California, San Francisco, for critical reading of the manuscript.
This work was supported by National Institutes of Health grants AI26918, HL56385, HL94-011, and RR03655. M. Bix was supported by a grant from the Irvington Institute and a Burroughs Wellcome Biomedical Science Career Award.
Footnotes
Abbreviations used in this paper: dice, determinant of IL-4 commitment; HPRT, hypoxanthine phosphoribosyltransferase; QTL, quantitative trait locus.
References
- 1.Abbas AK, Murphy KM, Sher A. Functional diversity of helper T lymphocytes. Nature. 1996;383:787–793. doi: 10.1038/383787a0. [DOI] [PubMed] [Google Scholar]
- 2.Seder RA, Paul WE, Davis MM, Fazekas de St B, Groth The presence of IL-4 during in vivo priming determines the lymphokine-producing potential of CD4+T cells from T cell receptor transgenic mice. J Exp Med. 1992;176:1091–1098. doi: 10.1084/jem.176.4.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hsieh C-S, Heimberger AB, Gold JS, O'Garra A, Murphy KM. Differential regulation of T helper phenotype development by interleukins 4 and 10 in an αβ-T cell receptor transgenic system. Proc Natl Acad Sci USA. 1992;89:6065–6069. doi: 10.1073/pnas.89.13.6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hsieh C-S, Macatonia SE, O'Garra A, Murphy KM. T cell genetic background determines default T helper phenotype development in vitro. J Exp Med. 1995;181:713–721. doi: 10.1084/jem.181.2.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guery J-C, Galbiati F, Smiroldo S, Adorini L. Selective development of T helper (Th) 2 cells induced by continuous administration of low dose soluble proteins to normal and β2-microglobulin–deficient BALB/c mice. J Exp Med. 1996;183:485–496. doi: 10.1084/jem.183.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hsieh C-S, Macatonia SE, Tripp CS, Wolf SF, O'Garra A, Murphy KM. Development of Th1 CD4+ T cells through IL-12 produced by Listeria-induced macrophages. Science. 1993;260:547–549. doi: 10.1126/science.8097338. [DOI] [PubMed] [Google Scholar]
- 7.Magram J, Connaughton SE, Warrier RR, Carvajal DM, Wu C-Y, Ferrante J, Stewart C, Sarmiento U, Faherty DA, Gately MK. IL-12–deficient mice are defective in IFN-γ production. Immunity. 1996;4:471–481. doi: 10.1016/s1074-7613(00)80413-6. [DOI] [PubMed] [Google Scholar]
- 8.Kopf M, Le Gros G, Bachmann M, Lamers MC, Bluethmann H, Kohler G. Disruption of the murine IL-4 gene blocks Th2 cytokine responses. Nature. 1993;362:245–248. doi: 10.1038/362245a0. [DOI] [PubMed] [Google Scholar]
- 9.Takeda K, Tanaka T, Shi W, Matsumoto M, Minami M, Kashiwamura S-I, Nakanishi K, Yoshida N, Kishimoto T, Akira S. Essential role for Stat6 in IL-4 signalling. Nature. 1996;380:628–630. doi: 10.1038/380627a0. [DOI] [PubMed] [Google Scholar]
- 10.Shimoda K, van Deursen J, Sangster MY, Sarawar SR, Carson RT, Tripp RA, Chu C, Quelle FW, Nosaka T, Vignali DAA, et al. Lack of IL-4–induced Th2 response and IgE class switching in mice with disrupted Stat6 gene. Nature. 1996;380:630–633. doi: 10.1038/380630a0. [DOI] [PubMed] [Google Scholar]
- 11.Kaplan MH, Schindler U, Smiley ST, Grusby MJ. Stat6 is required for mediating responses to IL-4 and for the development of Th2 cells. Immunity. 1996;4:313–319. doi: 10.1016/s1074-7613(00)80439-2. [DOI] [PubMed] [Google Scholar]
- 12.Kaplan MH, Sun Y-L, Hoey T, Grusby MJ. Impaired IL-12 responses and enhanced development of Th2 cells in Stat4-deficient mice. Nature. 1996;382:174–177. doi: 10.1038/382174a0. [DOI] [PubMed] [Google Scholar]
- 13.Thierfelder WE, van Deursen JM, Yamamoto K, Tripp RA, Sarawar SR, Carson RT, Sangster MY, Vignali DAA, Doherty PC, Grosveld GC, Ihle JN. Requirement for Stat4 in IL-12–mediated responses of natural killer and T cells. Nature. 1996;382:171–174. doi: 10.1038/382171a0. [DOI] [PubMed] [Google Scholar]
- 14.Schmitz J, Thiel A, Kuhn R, Rajewsky K, Muller W, Assenmacher M, Radbruch A. Induction of interleukin-4 (IL-4) expression in T helper (Th) cells is not dependent on IL-4 from non-Th cells. J Exp Med. 1994;179:1349–1353. doi: 10.1084/jem.179.4.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Launois P, Maillard I, Pingel S, Swihart KG, Xenarios I, Acha-Orbea H, Diggelmann H, Locksley RM, MacDonald HR, Louis JA. IL-4 rapidly produced by Vβ4Vα8 CD4+ T cells instructs Th2 development and susceptibility to Leishmania majorin BALB/c mice. Immunity. 1997;6:541–549. doi: 10.1016/s1074-7613(00)80342-8. [DOI] [PubMed] [Google Scholar]
- 16.Gorham JD, Guler ML, Steen RG, Mackey AJ, Daly MJ, Frederick K, Dietrich WF, Murphy KM. Genetic mapping of a murine locus controlling development of T helper 1/T helper 2 type responses. Proc Natl Acad Sci USA. 1996;93:12467–12472. doi: 10.1073/pnas.93.22.12467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guler ML, Jacobson NG, Gubler U, Murphy KM. T cell genetic background determines maintenance of IL-12 signaling. Effects on BALB/c and B10.D2 T helper cell type 1 phenotype development. J Immunol. 1997;159:1767–1774. [PubMed] [Google Scholar]
- 18.Roberts M, Mock BA, Blackwell JM. Mapping of genes controlling Leishmania majorinfection in CXS recombinant inbred mice. Eur J Immunogenet. 1993;20:349–362. doi: 10.1111/j.1744-313x.1993.tb00154.x. [DOI] [PubMed] [Google Scholar]
- 19.Roberts LJ, Baldwin TM, Curtis JM, Handman E, Foote SJ. Resistance to Leishmania majoris linked to the H2 region on chromosome 17 and to chromosome 9. J Exp Med. 1997;185:1705–1710. doi: 10.1084/jem.185.9.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beebe AM, Mauze S, Schork NJ, Coffman RL. Serial backcrossing mapping of multiple loci associated with resistance to Leishmania majorin mice. Immunity. 1997;6:551–557. doi: 10.1016/s1074-7613(00)80343-x. [DOI] [PubMed] [Google Scholar]
- 21.Liu, B.H. 1988. Statistical Genomics: Linkage Mapping and QTL Analysis. CRC Press, Inc., New York. 645 pp.
- 22.Zeng ZG. Precision mapping of quantitative trait loci. Genetics. 1994;136:1457–1468. doi: 10.1093/genetics/136.4.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schork NJ, Kreiger JE, Trolliet MR, Franchini KG, Koike G, Krieger EM, Lander ES, Dzau VJ, Jacob HJ. A biometrical genome search in rats reveals the multigenic basis of blood pressure variation. Genome Res. 1995;5:164–172. doi: 10.1101/gr.5.2.164. [DOI] [PubMed] [Google Scholar]
- 24.Lander ES, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- 25.Beckmann MP, Schooley KA, Gallis B, Vanden T, Bos, Friend D, Alpert R, Raunio R, Prickett KS, Baker PE, Park LS. Monoclonal antibodies block murine IL-4 receptor function. J Immunol. 1990;144:4212–4219. [PubMed] [Google Scholar]
- 26.Reiner SL, Zheng S, Corry DB, Locksley RM. Constructing polycompetitor cDNAs for quantitative PCR. J Immunol Methods. 1993;165:37–46. doi: 10.1016/0022-1759(93)90104-f. [DOI] [PubMed] [Google Scholar]
- 27.Bendelac A, Rivera MN, Park S-H, Roark JH. Mouse CD1-specific NK1 T cells: development, specificity and function. Annu Rev Immunol. 1997;15:535–562. doi: 10.1146/annurev.immunol.15.1.535. [DOI] [PubMed] [Google Scholar]
- 28.Lederer JA, Perez VL, DesRoches L, Kim SM, Abbas AK, Lichtman AH. Cytokine transcriptional events during helper T cell subset differentiation. J Exp Med. 1996;184:397–406. doi: 10.1084/jem.184.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuhn R, Rajewsky K, Muller W. Generation and analysis of interleukin 4–deficient mice. Science. 1991;254:707–710. doi: 10.1126/science.1948049. [DOI] [PubMed] [Google Scholar]
- 30.Murphy E, Shibuya K, Hosken N, Openshaw P, Maino V, Davis K, Murphy K, O'Garra A. Reversibility of T helper 1 and 2 populations is lost after long-term stimulation. J Exp Med. 1996;183:901–913. doi: 10.1084/jem.183.3.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schork, N., and Z. Chakravarti. 1996. A nonmathematical overview of modern gene mapping techniques applied to human diseases. In Molecular Genetics and Gene Therapy of Cardiovascular Disease. S. Mockrin, editor. Marcel Dekker, Inc., New York. 79–109.
- 32.Lander ES, Botstein D. Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics. 1989;121:185–199. doi: 10.1093/genetics/121.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bucy RP, Karr L, Huang G-Q, Li J, Carter D, Honjo K, Lemons JA, Murphy KM, Weaver CT. Single cell analysis of cytokine gene coexpression during CD4+ T-cell phenotype development. Proc Natl Acad Sci USA. 1995;92:7565–7569. doi: 10.1073/pnas.92.16.7565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Openshaw P, Murphy EE, Hosken NA, Maino V, Davis K, Murphy K, O'Garra A. Heterogeneity of intracellular cytokine synthesis at the single-cell level in polarized T helper 1 and T helper 2 populations. J Exp Med. 1995;182:1357–1367. doi: 10.1084/jem.182.5.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gollob KJ, Coffman RL. A minority subpopulation of CD4+ T cells directs the development of naive CD4+ T cells into IL-4-secreting cells. J Immunol. 1994;152:5180–5188. [PubMed] [Google Scholar]
- 36.Gieni RS, Fang Y, Trinchieri G, Umetsu DT, DeKruyff RH. Differential production of IL-12 in BALB/c and DBA/2 mice controls IL-4 versus IFN-gamma synthesis in primed CD4 lymphocytes. Int Immunol. 1996;8:1511–1520. doi: 10.1093/intimm/8.10.1511. [DOI] [PubMed] [Google Scholar]
- 37.Zheng W, Flavell RA. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell. 1997;89:587–596. doi: 10.1016/s0092-8674(00)80240-8. [DOI] [PubMed] [Google Scholar]
- 38.Ho I-C, Hodge MR, Rooney JW, Glimcher LH. The proto-oncogene c-maf is responsible for tissue-specific expression of interleukin-4. Cell. 1996;85:973–983. doi: 10.1016/s0092-8674(00)81299-4. [DOI] [PubMed] [Google Scholar]
- 39.Kelso A, Gough NM. Coexpression of granulocyte-macrophage colony-stimulating factor, γ interferon, and interleukins 3 and 4 is random in murine alloreactive T-lymphocyte clones. Proc Natl Acad Sci USA. 1988;85:9189–9193. doi: 10.1073/pnas.85.23.9189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Assenmacher M, Schmitz J, Radbruch A. Flow cytometric determination of cytokines in activated murine T helper lymphocytes: expression of interleukin-10 and interferon-γ in interleukin-4–expressing cells. Eur J Immunol. 1994;24:1097–1101. doi: 10.1002/eji.1830240513. [DOI] [PubMed] [Google Scholar]
- 41.Kelso A, Groves P, Troutt AB, Francis K. Evidence for the stochastic acquisition of cytokine profile by CD4+ T cells activated in a T helper type 2–like response in vivo. Eur J Immunol. 1995;25:1168–1175. doi: 10.1002/eji.1830250506. [DOI] [PubMed] [Google Scholar]
- 42.Bucy RP, Panoskaltsis-Mortari A, Huang G, Li J, Karr L, Russell JH, Murphy KM, Weaver CT. Heterogeneity of single cell cytokine gene expression in clonal T cell populations. J Exp Med. 1994;180:1251–1262. doi: 10.1084/jem.180.4.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rogers WO, Weaver CT, Kraus LA, Li J, Li L, Bucy RP. Visualization of antigen-specific T cell activation and cytokine expression in vivo. J Immunol. 1997;158:649–657. [PubMed] [Google Scholar]
- 44.Chen Y, Kuchroo VK, Inobe J-I, Hafler DA, Weiner HL. Regulatory T cell clones induced by oral tolerance: suppression of autoimmune encephalomyelitis. Science. 1994;265:1237–1240. doi: 10.1126/science.7520605. [DOI] [PubMed] [Google Scholar]
- 45.Groux H, O'Garra A, Bigler M, Rouleau M, Antonenko S, de Vries JE, Roncarolo MB. A CD4+ T cell subset inhibits antigen-specific T cell responses and prevents colitis. Nature. 1997;389:737–742. doi: 10.1038/39614. [DOI] [PubMed] [Google Scholar]
- 46.McAdams HH, Arkin A. Stochastic mechanisms in gene expression. Proc Natl Acad Sci USA. 1997;94:814–819. doi: 10.1073/pnas.94.3.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fiering S, Northrop JP, Nolan GP, Mattila PS, Crabtree GR, Herzenberg LA. Single cell assay of a transcription factor reveals a threshold in transcription activated by signals emanating from the T cell antigen receptor. Genes Dev. 1990;4:1823–1834. doi: 10.1101/gad.4.10.1823. [DOI] [PubMed] [Google Scholar]
- 48.Bix M, Locksley RM. Independent and epigenetic regulation of the interleukin-4 alleles in CD4+ T cells. Science. 1998;281:1352–1354. doi: 10.1126/science.281.5381.1352. [DOI] [PubMed] [Google Scholar]
- 49.Riviere I, Sunshine MJ, Littman DR. Regulation of IL-4 expression by activation of individual alleles. Immunity. 1998;9:217–228. doi: 10.1016/s1074-7613(00)80604-4. [DOI] [PubMed] [Google Scholar]