Abstract
In mature B lymphocytes, the zinc finger transcription factor early growth response 1 (Egr-1) is one of the many immediate-early genes induced upon B cell antigen receptor engagement. However, its role during earlier stages of lymphopoiesis has remained unclear. By examining bone marrow B cell subsets, we found Egr-1 transcripts in pro/pre-B and immature B lymphocytes, and Egr-1 protein in pro/pre-B–I cells cultivated on stroma cells in the presence of interleukin (IL)-7. In recombinase-activating gene (RAG)-2–deficient mice overexpressing an Egr-1 transgene in the B lymphocyte lineage, pro/pre-B–I cells could differentiate past a developmental block at the B220low BP-1− stage to the stage of B220low BP-1+ pre-B–I cells, but not further to the B220low BP-1+ CD25+ stage of pre-B–II cells. Therefore, during early B lymphopoiesis progression from the B220low BP-1− IL-2R− pro/pre-B–I stage to the B220low BP-1+ IL-2R+ pre-B–II stage seems to occur in at least two distinct steps, and the first step to the stage of B220low BP-1+ pre-B–I cells can be promoted by the overexpression of Egr-1 alone. Wild-type mice expressing an Egr-1 transgene had increased proportions of mature immunoglobulin (Ig)M+ B220high and decreased proportions of immature IgM+ B220low bone marrow B cells. Since transgenic and control precursor B cells show comparable proliferation patterns, overexpression of Egr-1 seems also to promote entry into the mature B cell stage. Analysis of changes in the expression pattern of potential Egr-1 target genes revealed that Egr-1 enhances the expression of the aminopeptidase BP-1/6C3 in pre-B and immature B cells and upregulates expression of the orphan nuclear receptor nur77 in IgM+ B cells.
Keywords: Egr-1, transcription factor, B cell development, BP-1, nur77
Antigen binding to surface (s)Igs in B cells initiates a signal cascade which in the context of secondary signals leads to proliferation and differentiation of mature resting B lymphocytes into plasma or memory cells. Changes in the activity and expression of transcription factors translate activating signals into the modulated expression pattern of downstream genes. One of these transcription factors is called early growth response 1 (Egr-1;1 also known as Krox-24, NGFI-A, Tis-8, zif268, pAT225, or Z-225 [1–3]). Egr-1 is induced very rapidly in many different cell types and tissues, including fibroblasts (1), monocytes (4), lymphocytes (5, 6), kidney (7), neurons (3), and brain (8), in response to a wide range of signals (1–3, 5, 9). In mature B lymphocytes, transient Egr-1 expression is rapidly induced upon stimulation by B cell antigen receptor (BCR) cross-linking (5, 10), whereas signals resulting from Fc receptor cross-linking inhibit induction (11, 12). Thus, the broad spectrum of Egr-1 expression and the diverse modes of Egr-1 induction suggest that Egr-1 functions as a transcriptional regulator that links common biochemical signaling pathways to the rapid modulation of downstream gene expression.
Mature peripheral B lymphocytes originate from bone marrow precursor cells that are ordered according to their phenotype, gene expression, Ig gene rearrangement, and proliferative and developmental potential into the pro-B, pre-B, and immature B lymphocyte subsets (13–17). Transcriptional regulation plays a critical role during B cell development (for a review, see reference 18) as shown by gene targeting of multiple transcription factors. Mutations in these factors that obliterate their activity were shown to arrest B lymphopoiesis at defined stages of maturation (19–29).
Little is known about the expression and function of Egr-1 during early steps of B cell differentiation. Here we report that Egr-1 expression can be detected already in pre-B cells isolated from bone marrow and in fetal liver–derived pre-B cell cultures. These results suggested that Egr-1 might also have a regulatory function in early stages of B lymphopoiesis. However, mice deficient for Egr-1 fail to show defects in lymphocyte or monocyte maturation, most probably because the missing Egr-1 activity is masked by other members of the Egr transcription factor family (30, 31). To bypass the complementing activity of Egr-2, Egr-3, or Egr-4, we studied B lymphocyte differentiation in transgenic mice overexpressing Egr-1 in B cells in normal and recombinase-activating gene (RAG)-2–deficient mice. Since the RAG-2 mutation prevents rearrangement of Ig genes (32), precursor B cells are developmentally arrested in the stage of B220low BP-1− pro/pre-B–I cells (33, 34). Analyzing Egr-1 transgenic RAG-2–deficient mice, we found that pro/pre-B–I cells overcame the RAG-2−/− induced differentiation block at the stage of B220+ BP-1− pro/pre-B–I cells and differentiated into B220+ BP-1+ pre-B–I cells. Comparing B lymphocyte maturation in the bone marrow of normal transgenic and control animals, we found that Egr-1 transgenic mice had increased their fraction of mature cells. Because Egr-1–enhanced progression of developing thymocytes was also found in transgenic mice overexpressing Egr-1 in T cells (35), we propose that Egr-1 activity promotes maturation of B and T lymphocytes.
Materials and Methods
Pre-B Cell Cultures.
Fetal liver cells of day 15–18 embryos were removed and plated onto irradiated ST-2 feeder cells in Iscove's medium containing IL-7 and 10% FCS. Cells were cultured as described previously (36). Cells from transgenic lines were identified by PCR. For further analyses, nonadherent cells were collected and washed twice in ice-cold PBS. Samples from wells containing only ST-2 feeder cells were treated in parallel and served as controls.
Mice.
The detailed description of the generation of Egr-1 transgenic mice using the BALB/c embryonic stem cell line BALB/c-I will be described elsewhere. Egr-1 transgenic mice of the IA7 line were transferred to a special pathogen-free unit by implanting transgenic one-cell embryos into C57BL/6 foster animals kept under specific pathogen–free conditions. The IA7 line was then bred further by mating with wild-type BALB/c mice. RAG-2−/− mice expressing transgenic Egr-1 were obtained by backcrossing female Egr-1 transgenic IA7 mice twice with C57BL/6 RAG-2−/− males. Animals were tested by PCR for the Egr-1 transgene using genomic DNA, 5′-CTTTCGGTTTGGGGCTGGACA-3′ and 5′-CGCTGCTGGTGCTGCTGCTGCTAT-3′, as transgene-specific primer pair. The RAG-2−/− phenotype was verified by FACS® analysis of peripheral blood cells.
RNA Isolation, Northern Blot, and PCR Analysis.
RNA was extracted using the guanidinium isothiocyanate method as described (37). For Northern blotting, 10 μg of total RNA was separated in a 1% agarose gel containing 7% formaldehyde, transferred onto nylon filters, and fixed by UV cross-linking. Filters were prehybridized (50% deionized formamide, 5× SSC, 5× Denhardt's solution, 50 mM NaH2PO4, pH 7, 10 mM Na4P2O7, 0.1% SDS, 0.1 mg/ml denatured salmon sperm DNA) for 2 h at 42°C. For detection of Egr-1–specific transcripts, [α-32P]dATP-labeled probes were prepared from a 1.6-kb EcoRI-HindIII fragment from plasmid 533 (a gift from V. Sukhatme) containing the Egr-1 cDNA by the oligonucleotide priming method (38). The probe was added to the prehybridization and filters were incubated overnight, washed with 0.2× SSC, 0.1% SDS at 42°C, and exposed to X-ray films. Egr-1 expression was analyzed by PCR using cDNA reverse transcribed from total RNA with Superscript II (GIBCO BRL, Eggenstein, Germany) and the Egr-1– specific primers 5′-GCAGATCTCTGACCCGTTCGG-3′ and 5′-CCGAGCGTTTGGCTRGGGATA-3′ as described by T. Miyazaki (35). PCR was performed using Taq polymerase (MBI Fermentas, Inc., Amherst, NY) using 1/25 of the cDNA reaction as template at an annealing temperature of 54°C.
Immunoblot Analysis.
Bone marrow cells from six femurs were isolated and resuspended in FACS buffer (0.1% sodium azide, 3% FCS in PBS). B220-specific biotin-labeled antibody RA3.3A1 (39) was added and incubated for 30 min on ice. Cells were washed, magnetic streptavidin-labeled beads (Dynal, Oslo, Norway) were added, and B cells were isolated. Quality of the sorting process was verified by flow cytometric analysis. The B cells were resuspended in 30 μl lysis buffer (1% NP-40, 150 mM NaCl, 10 mM Tris-HCl, pH 7.0, 0.1 mM PMSF) and incubated on ice for 10 min. Cell debris was removed by centrifugation (10 min, 4°C, 22,000 g), and the extract was separated by SDS-PAGE (8%) and transferred onto nitrocellulose membrane (Hybond C extra; Amersham Pharmacia Biotech, Uppsala, Sweden). Egr-1 was detected using the antiserum C19 (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) at 25 ng/ml followed by peroxidase-conjugated goat anti–rabbit IgG F(ab′)2 (Dianova GmbH, Hamburg, Germany) at 200 ng/ml. Expression of nur77 was analyzed using a mouse IgG anti-nur77 mAb (a gift of B. Osborne, University of Massachusetts, Amherst, MA) followed by peroxidase-conjugated goat anti–mouse IgG (Southern Biotechnology Associates, Inc., Birmingham, AL). IgM was detected by a goat anti–mouse IgM peroxidase-labeled serum (Southern Biotechnology Associates, Inc.). Signals were visualized using an enhanced chemiluminescence (ECL) detection system (Amersham Pharmacia Biotech).
Flow Cytometry.
Flow cytometry was carried out as described previously (40) using the following antibodies: RS3.1-biotin specific for murine IgMa (41), 6C3-biotin for BP-1, 7D4-biotin for IL-2Rα chain, 2B8-biotin for c-kit, S7-biotin for leukosialin, AMS9.1-biotin for IgDa, RA3-6B2–PE for B220, IM7-biotin for Pgp-1, 3E2-PE for intercellular adhesion molecule 1 (ICAM-1) (all from PharMingen Europe, Hamburg, Germany), and biotinylated PB493 (42) to stain immature B lymphocytes. Cells were counterstained using PE- or APC-conjugated streptavidin (PharMingen Europe). Unspecific binding to Fc receptors was blocked by adding unlabeled mouse FcγR-specific mAb 2.4G2. Dead cells were excluded by staining with propidium iodide. Using a FACSCalibur® and CellQuest® software (Becton Dickinson, San Jose, CA), 3–5 × 104 cells were acquired according to their forward/side scatter pattern and analyzed. To analyze Egr-1 expression in bone marrow B cell subsets, 3.4 × 105 pre-B and 7 × 104 immature B cells were isolated from both femurs of a 5-wk-old BALB/c mouse by cell sorting at 4°C according to their IgMa/ PB493 staining pattern using a FACStar® cell sorter and LYSIS II® software (Becton Dickinson).
Bromodeoxyuridine Treatment and Staining.
Bone marrow cells were labeled with bromodeoxyuridine (BrdU; Sigma, Deisenhofen, Germany) starting with a single injection of 1 mg/ml i.p. BrdU and feeding mice continuously with drinking water containing 1 mg/ml BrdU for 48 h as described (43). During the labeling period, the drinking water was protected from light. Simultaneous detection of surface staining and BrdU labeling was done as described (44). After surface marker staining, cells were resuspended in 500 μl 0.15 M NaCl, 1.2 ml ice-cold 95% ethanol was added, and the cells were incubated 30 min on ice. Cells were washed and resuspended in 1 ml fixation buffer (1% paraformaldehyde and 0.01% Tween in PBS). After incubation for 30 min at room temperature, the cells were incubated for 30 min in DNase I solution (50 KU DNase I in 4.2 mM MgCl2/ 0.15 M NaCl, pH 5). Cells were washed, 10 μl anti-BrdU antibody (Becton Dickinson) was added, and the cells were then incubated for 30 min and washed.
Electrophoretic Mobility Shift Assays.
Gel shift was carried out using recombinant Egr-1 as described (45) with double-stranded radiolabeled oligonucleotides from the nur77 and BP-1 promoter regions carrying putative Egr-1 binding sites (bold): 5′-TTCCAAAGTTCCCCCTCAACCCCTC-3′ for BP-1 (position −753 to −729), 5′-GTCAGTGGCGCCCCCGCCCCTCTCCAA-3′ for nur77 (position −66 to −50), and 5′-GGATCCAGCGGGGGCGAGCGGGGGCG-3′ for Egr-1.
Results
Egr-1 Expression in Pre-B and Immature B Cell Precursors.
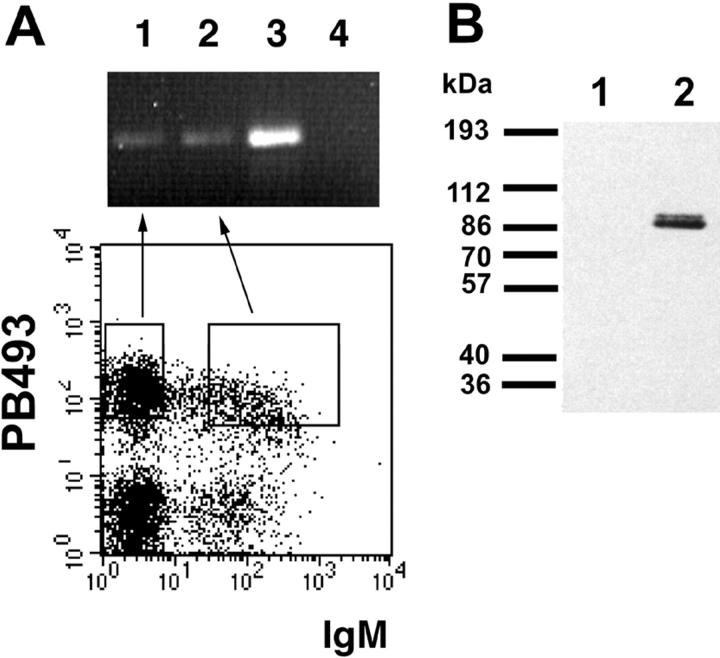
BCR cross-linking has previously been reported to induce Egr-1 expression in mature B cells, but not in immature B lymphocytes or in immature B cell lines (5, 46, 47). We addressed the question of whether unstimulated pre-B and immature B cells express Egr-1 by analyzing sorted B cell subsets from murine bone marrow. Transcription of the Egr-1 gene was found by PCR in both sIgM− (pre-B) and sIgM+ PB493+ immature B cells (Fig. 1 A). Likewise, Egr-1 protein was detected by immunoblotting in sIgM− pre-B cells isolated from fetal liver and expanded in culture on ST-2 stroma cells in the presence of IL-7 (Fig. 1 B). Both results show transcription of the Egr-1 gene and translation of Egr-1 mRNA into detectable amounts of protein as early as the pre-B cell stage before BCR surface expression.
Figure 1.
Expression of Egr-1 during different stages of B cell maturation. (A) Egr-1 RNA expression. Bone marrow cells of a 5-wk-old BALB/c mouse were stained for B220, PB493, and IgMa expression. Pro/pre-B cells (B220low, PB493+, and IgM−) and immature B cells (B220low, PB493+, and IgM+) were sorted and RNA was extracted. Analysis of Egr-1 transcripts by reverse transcription PCR was performed as described by Miyazaki (35). Lane 1 shows expression of Egr-1 in pro/ pre-B cells and lane 2 in immature B cells. Anti-IgM–stimulated splenocytes (lane 3) serve as a positive control. In lane 4, cDNA was omitted from the PCR. (B) Expression of Egr-1 protein. BALB/c fetal liver B cells (day 16) were cultivated in the presence of IL-7 on ST-2 stroma cells. Cellular lysates of 106 cells were examined for Egr-1 expression by immunoblotting using the Egr-1–specific antibody C19 and developed with horseradish peroxidase–coupled goat anti–rabbit IgG. In cultivated pre-B cells, Egr-1 protein expression is easily detected (lane 2). As a negative control an equal amount of ST-2 feeder cells was loaded in lane 1.
Egr-1 Expression in Transgenic and Normal Mice.
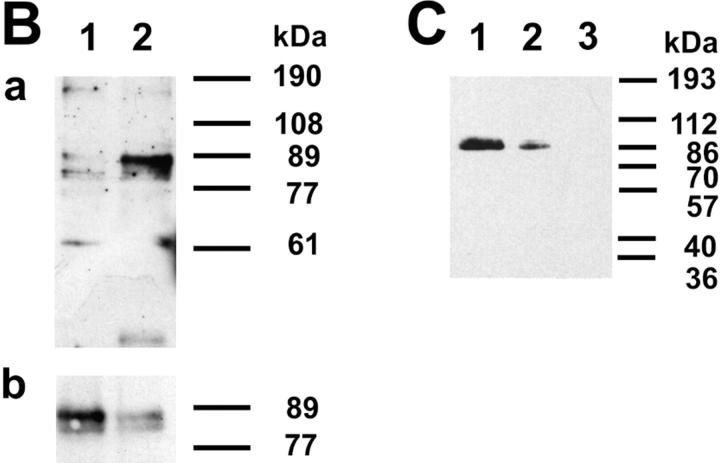
These results suggested that Egr-1 function might also be important during early stages of B lymphopoiesis. To test this hypothesis, we generated transgenic mice expressing Egr-1 specifically in B lymphocytes using an Ig heavy chain promoter/enhancer construct. Four different founder mice showing Egr-1 germline transmission were obtained. By breeding to BALB/c mice, we established the Egr-1 transgenic lines IA7, IB10, IC4, and ID4 (to be published elsewhere). At first, we compared Egr-1 expression between transgenic and BALB/c control mice by Northern and immunoblotting (Fig. 2). Spleen cells of the line IA7 expressed 10-fold more Egr-1 mRNA than the control littermates, whereas the other lines showed Egr-1 expression levels of about two- to threefold above unstimulated spleen cells (Fig. 2 A). Since transgenic IC4 mice expressed only low levels of Egr-1 they were abandoned. Carrying out most of the experiments with mice from lines IA7, IB10, and ID4, we found only small variations between these transgenic lines. Testing Egr-1 protein expression, we found high levels in purified B220+ bone marrow B cells as well as in cultivated pre-B cells isolated from fetal liver (Fig. 2, B and C, respectively).
Figure 2.
Expression of transgenic Egr-1. (A) Comparison of Egr-1 mRNA levels in BALB/c and Egr-1 transgenic spleen cells. RNA was extracted from splenocytes, and Northern blot analysis was performed using a probe specific for endogenous and recombinant Egr-1 mRNA. Because the endogenous and transgenic Egr-1 mRNA species migrate with different electrophoretic mobilities, they are easily identified on Northern blots (data not shown). To standardize for the amounts of mRNA, filters were rehybridized with a GAPDH-specific probe. The relative intensity of the Egr-1 expression in BALB/c and in four transgenic lines IA7, IB10, IC4, and ID4 was determined using a PhosphorImager (Molecular Dynamics, Sunnyvale, CA). (B) Expression of Egr-1 protein in B cells of the bone marrow. B220+ B cells were purified from the bone marrow of three BALB/c and three ID4 transgenic mice using streptavidin-loaded magnetic beads and the B220-specific, biotinylated antibody RA3.3A1. Whole cell lysates with equal amounts of protein were analyzed for Egr-1 expression by immunoblotting using the Egr-1–specific antibody C-19 (a). Lane 1 shows Egr-1 expression in BALB/c bone marrow cells, lane 2 in ID4 mice. In parallel, samples were stained with a horseradish peroxidase–conjugated goat anti–mouse IgM antibody to standardize for the different amounts of protein loaded per lane (b). (C) Increased expression of Egr-1 protein in transgenic pre-B cells. Pre-B cell cultures and immunoblots were performed as described for Fig. 1 B. Transgenic pre-B cells (IA7, lane 1) express higher levels of Egr-1 than an equal amount of wild-type BALB/c pre-B cells (lane 2). Egr-1 protein is undetectable in whole cell lysates of corresponding numbers of ST-2 feeder cells (lane 3).
These results show that transgenic Egr-1 is expressed during similar stages of B cell maturation, but at far higher levels than endogenous Egr-1.
Egr-1 Expression Promotes At Least Two Different Stages of B Cell Development.
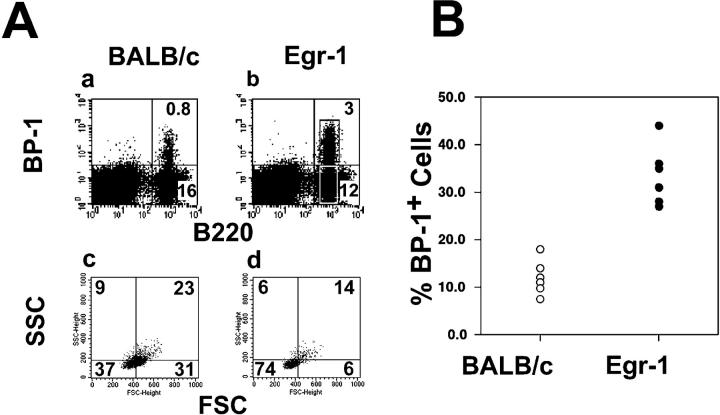
To examine whether enhanced Egr-1 expression has an effect on early stages of B cell development, we backcrossed the IA7 transgenic mice to a RAG-2–deficient background. The RAG-2 mutation prevents rearrangement of the Ig genes (32) and therefore blocks B cell maturation at the pro/pre-B–I cell stage (16, 34). These cells carry the surface markers c-kit and CD43; <5– 15% express BP-1 and <1% express the IL-2Rα chain (data not shown). Phenotypically, these pro/pre-B–I cells correspond to fraction B as classified by Hardy et al. (13). FACS® analysis of control and Egr-1 transgenic RAG-2– deficient mice revealed an unchanged expression pattern for c-kit and CD43, but a three- to fourfold increase in the fraction of BP-1+ cells, compared with control littermates (Fig. 3). Phenotypically, these BP-1+ c-kit+ CD43+ pre-B lymphocytes are defined as fraction C cells (13), and progression into this stage normally requires RAG-2 expression and Ig heavy chain gene rearrangement (16, 34), suggesting that Egr-1 might support the maturation of fraction B pre-B cells even in the absence of RAG-2 activity. The reduced cell size of transgenic BP-1+ B220+ lymphocytes as reflected by changes in forward/side scatter (Fig. 3 A, c and d) also supports the Egr-1–induced progression of B cell development. Since we did not detect cell surface markers characteristic for pre-B–II/fraction D cells, i.e., IL-2Rα and high levels of heat-stable antigen, the expression of the Egr-1 transgene seems to promote the transition of pre-B–I cells from fraction B to fraction C but not further.
Figure 3.
Egr-1 induces development of BP-1+ B220low pre-B cells. Bone marrow cells of IA7 transgenic mice with a RAG-2 background and control littermates were stained with BP-1– and B220-specific antibodies and analyzed by flow cytometry. Dead cells were excluded by gating for propidium iodide–negative cells, and 5 × 104 cells were acquired according to their forward/side scatter (FSC, SSC) profile. As shown for one individual example in A, IA7 mice (b) have about three times more BP-1+ cells than control littermates (a). Changes in the forward/side scatter pattern document that most of the B220+ BP-1+ cells (74% small cells, d) are smaller than the majority of BP-1− cells (37% small and 54% large cells, c). B compiles the BP-1 staining pattern for six RAG-2–deficient IA7 mice and six control littermates.
These results indicated that Egr-1 expression might also change maturation of later stage bone marrow B cell subsets. According to the expression pattern of B220 and IgM, three main subsets corresponding to three consecutive stages of maturation can be distinguished: IgM− B220low pre-B cells, B220low IgM+ IgD− immature B cells, and finally the B220high IgM+ IgD+ mature B cells (16, 48). The proportion of mature B cells in the bone marrow increases with the age of the animal. Therefore, mice at 4 wk of age have fewer mature B cells than older animals. Comparing 23 control littermates with 23 IA7 transgenic mice between 4 and 40 wk of age, we found in general fewer immature and more mature B cells in the IA7 bone marrow than in age-matched controls (Fig. 4). These results suggest that Egr-1 also promotes the differentiation of immature B cells into mature B cells.
Figure 4.
Higher frequency of mature and lower frequency of immature bone marrow B cells in Egr-1 transgenic mice. (A) Compares the differences between the B220low IgM+ (immature) and B220high IgM+ (mature) B cell subsets of control littermates and transgenic mice. Bone marrow cells of 4–40-wk-old BALB/c littermates (n = 23) and of IA7 transgenic mice (n = 23) were stained for IgM (RS3.1) and B220 (RA3-6B2) and analyzed by flow cytometry after acquisition of 3 × 104 cells gated according to their forward/side scatter profile. The first diagram (a) compares the changes in the percentage of immature IgM+ B220low cells between age-matched BALB/c littermates (open circles) and IA7 mice (filled circles) over a period of 36 wk. P values <0.05 corresponding to the individual time points indicate statistically significant differences between transgenic and control mice. The second diagram (b) shows the increase in the percentage of mature IgM+ B220high cells in older mice. Although transgenic IA7 mice tend to have more mature bone marrow B cells, they do not show the statistically significant differences seen for the immature B cells. (B) Two individual examples of a BALB/c littermate (a) and an IA7 transgenic mouse (b) at 18 wk. The numbers indicate the percentage of B220+ cells in each subset. Similar analyses of bone marrow B cells from the IB10 and ID4 transgenic mice showed almost identical results.
Normal Proliferation of B Cells in Egr-1 Transgenic Mice.
The enlarged population of BP-1+ pre-B cells in transgenic RAG-2–deficient mice and the higher percentage of mature cells in transgenic RAG-2+ animals could result from an increased proliferative potential of B cell precursors. Because dividing B lymphocytes can be traced by incorporation of the nucleotide analogue BrdU (49), we compared the proportion of BrdU+ cells in the pre-B, immature B, and mature B cell subsets between control littermates and transgenic mice after 2 d of in vivo labeling. The similar percentages of BrdU+ cells in all B cell subsets of both groups of mice suggest that the expression of transgenic Egr-1 does not change the proliferative potential of B cells (Fig. 5).
Figure 5.
Similar proliferation of IA7 and BALB/c bone marrow B cell subsets. To determine the fraction of cycling bone marrow B cells, nine BALB/c and eight IA7 littermates were fed with BrdU for 2 d. Bone marrow cells were analyzed flow cytometrically by acquiring 3 × 104 cells stained for B220 and IgM as described in the legend to Fig. 2, and after permeabilization with an FITC-labeled anti-BrdU antibody. In both groups of mice pre-B (IgM− B220low), immature (IgM+ B220low) BALB/c, and mature (IgM+/low B220high) bone marrow B cells incorporated similar amounts of BrdU, indicating comparable proliferation rates. Similar results were obtained when we used ID4 instead of IA7 transgenic mice.
Regulation of Downstream Genes.
The increased maturation of transgenic B cells would be expected to correlate with changes in the expression of genes controlled directly or indirectly by Egr-1. Using a panel of antibodies, we screened transgenic bone marrow B cells for alterations in the expression pattern of B cell–specific surface markers by flow cytometry. Changes were not observed except for a slight increase in the percentage of BP-1+ cells and in BP-1 expression levels in all transgenic lines (Fig. 6 A). As discussed above, the proportion of BP-1+ cells might relate to the Egr-1–enhanced transition of fraction B pre-B cells into the fraction C stage, but it would not explain higher BP-1 surface levels. Therefore, we speculated that Egr-1 might regulate the transcription of the BP-1 gene and screened the BP-1 promoter for potential Egr-1 binding sites. Finding a (5′-GAGGGGGAA) sequence ∼1.6 kb upstream of the mRNA start (50) resembling an Egr-1 binding site (5′-GCGGGGGCG), we analyzed by an electrophoretic mobility shift assay (EMSA) if recombinant Egr-1 binds to an oligonucleotide containing the putative Egr-1 recognition site from the BP-1 promoter. As shown in Fig. 6 C, labeled oligonucleotides containing a cognate Egr-1 binding site (lane 1) or the binding site from the BP-1 promoter (lane 15) produced a shifted DNA–protein complex with identical electrophoretic mobility. Their intensities were reduced only by adding an excess of unlabeled oligonucleotides with an Egr-1 binding site but not by competing with an Sp-1 binding site (lanes 2–5 and 16–19, respectively). Likewise, only the addition of Egr-1– but not of Sp-1–specific antibodies retarded the migration of the complex (lanes 6 and 7, 20 and 21). Therefore, the forced expression of Egr-1 in transgenic B cells may not only help pre-B cells to proceed from fraction B into fraction C but may also enhance the expression of BP-1, which is normally upregulated during this transition.
Figure 6.
Expression of downstream genes depending on Egr-1 activity. (A) Upregulated BP-1 expression in transgenic mice. Bone marrow cells from a BALB/c control littermate and an IA7 mouse were analyzed flow cytometrically by acquiring 3 × 104 live cells stained for B220, IgM, and BP-1. The dot plots are gated B220+ cells and illustrate increased levels and a higher percentage of BP-1 staining in both IgM− and IgM+ subsets. IB10 and ID4 mice showed very similar changes in BP-1 expression. (B) Induction of nur77 in transgenic bone marrow B cells. Using ID4 and BALB/c mice with comparable percentages of mature bone marrow B cells, B220+ cells were purified from the bone marrow as described in the legend to Fig. 2. In the BALB/c sample, protein extracts from 3.3 × 106 B220low and 1.3 × 105 B220high cells (as determined by FACS® analysis) were loaded; for the ID4 sample, the respective amounts were 3 × 106 B220low and 2 × 105 B220high cells. Nur77 expression was detected by Western blotting using a nur77-specific antibody. The amount of protein loaded per lane was controlled by reprobing the blots with a goat anti–mouse IgM antibody. The staining shows elevated nur77 expression in ID4. Size markers (in kD) as indicated were run in parallel to the samples. (C) Binding of recombinant Egr-1 to sequences present in the BP-1 and nur77 promoters. Recombinant Egr-1 was incubated with radioactively labeled oligonucleotides carrying a cognate Egr-1 binding site (lanes 1–7, Egr-1), with an oligonucleotide from the nur77 promoter (lanes 8–14, nur77), or with an oligonucleotide from the BP-1 promoter (lanes 15–21, BP-1) and analyzed by EMSA as described previously (reference 45). The sequences of the respective Egr-1 binding sites are shown (top). Specific binding was proven first by competing either with an excess of an unlabeled oligonucleotide carrying a cognate Egr-1 binding site (lanes 4, 5; 11, 12; 18, 19) or by using an oligonucleotide with an Sp-1 site (lanes 2, 3; 9, 10; 16, 17), and second by inducing a “supershift” by adding the Egr-1–specific antibody C19 to the binding reaction (lanes 6, 13, and 20). Replacing the Egr-1–specific antibody with an Sp-1–specific antibody had no effect on the migration of the DNA–Egr-1 complex (lanes 7, 14, and 21).
The nur77 gene (also called NGFI-B [3] or N10 [51]) encoding an orphan nuclear receptor represents one of the transcription factors found to be induced by Egr-1 (52). Analyzing nur77 expression in purified B220+ bone marrow cells of transgenic (ID4) and control littermates by Western blotting, we found nur77 to be expressed only by transgenic but not by control B cells (Fig. 6 B). Since upregulated nur77 expression was not found in cultivated transgenic sIg− pre-B cells (data not shown), the induction of the nur77 gene seems to be confined to pre-B–II or sIgM+ bone marrow cells. To analyze if Egr-1 could directly induce nur77 transcription, we tested by EMSA the binding of recombinant Egr-1 to an oligonucleotide of the nur77 promoter (position −74 to −50) containing a cognate Egr-1 binding site. As shown in Fig. 6 C, binding of recombinant Egr-1 produced a DNA–protein complex (lane 8). The specificity of Egr-1 binding was demonstrated by a competition assay using an excess of an oligonucleotide carrying an Egr-1 binding site (lanes 11 and 12) and by the decreased mobility of the complex upon the addition of an Egr-1–specific antibody (lane 13).
Since it was shown recently that nur77 activity is involved in the induction of apoptosis during negative selection of thymocytes (53–56) and in the upregulation of CD95L expression (57), we looked for enhanced CD95L expression in Egr-1 transgenic mice compared with control littermates. In contrast to the results reported for nur77– expressing T cells, we did not find increased CD95L expression in Egr-1 transgenic B cells (data not shown).
In response to BCR-derived signals, Egr-1 is thought to modulate the expression pattern of downstream genes that promote further activation and differentiation of B lymphocytes. Using variants of the B cell line WEHI-231, it was shown that Egr-1 induces the expression of CD44 and of intracellular adhesion molecule 1 (ICAM-1 [58, 59]). Therefore, we tested the expression pattern of both surface markers in our transgenic mice, but did not detect differences when compared with BALB/c controls (data not shown).
Discussion
Egr-1 Accelerates B Cell Maturation.
Mature B cells respond to signals resulting from antigen receptor engagement by immediately inducing Egr-1 transcription (5), but the role of Egr-1 in earlier stages of B cell development has not been defined. The different stages and the order of B cell development are well characterized, allowing the precise typing of bone marrow B cells according to the expression of characteristic cell surface markers, the rearrangement of Ig genes, and the proliferative and differentiation potential of B cell precursors (13, 14, 16, 60, 61). By analyzing Egr-1 expression in bone marrow–derived B lymphocyte subsets and by testing Egr-1 expression in cultivated, fetal liver–derived pre-B cells, we have shown that Egr-1 is also expressed in pre-B cells lacking sIgM as well as in immature sIgM+ B cells in the absence of sIgM-induced signals. These observations suggest that Egr-1 might also have a regulatory function in pre-B cell development. By studying transgenic mice overexpressing Egr-1 from the pre-B stage on, we have found higher proportions of mature B cells and fewer immature B cells in transgenic animals than in control littermates. To identify if early stages of B lymphopoiesis are sensitive to Egr-1 activity, we arrested B cell development at the stage of pro/pre-B–I cells by backcrossing the Egr-1 transgenic line IA7 to mice deficient in RAG-2. Since the null mutation in the RAG-2 gene prevents rearrangement of Ig genes (32), B cell precursors do not receive stimulating signals required for developmental progression beyond the stage of B220low CD43+ BP-1− pro/pre-B cells (16, 62), also defined as fraction B (13). Comparing the phenotype of bone marrow pro/pre-B cells from transgenic and control mice, we found a three- to fourfold increased population of BP-1+ pre-B cells in Egr-1 transgenic mice. Since the transcription activation function of Egr-1 seems to enhance BP-1 expression in more mature B cell subsets, the increase in BP-1+ pre-B cells could also reflect the induction of BP-1 expression only and not Egr-1–induced differentiation. However, this seems to be less likely because transgenic BP-1+ cells were found to be smaller than BP-1− cells, consistent with further maturation. Therefore, these results suggest that forced expression of Egr-1 in BP-1− pro/pre-B cells induces progression into the stage of BP-1+ pre-B cells (fraction C). Since these cells failed to upregulate the IL-2Rα chain and heat-stable antigen, two markers characteristic for pre-B–II cells (fraction C′ and D [13, 16]), overexpression of Egr-1 in pro/pre-B–I cells seems to be sufficient to induce differentiation to fraction C, but not to more mature stages of B lymphopoiesis.
Progression of pro/pre-B cells developmentally arrested by a mutation in the RAG-2 gene into more mature pre-B cell stages is also induced by in vivo cross-linking of the Ig-α/Ig-β heterodimer using Ig-β–specific mAbs (63). Under those conditions, anti–Ig-β–treated pro/pre-B–I cells become smaller in size and acquire IL-2Rα expression in addition to BP-1. Since they also downregulate c-kit (CD117) and CD43, they are considered as small pre-B–II cells. In the same report, it was shown that Ig-β cross-linking stimulates tyrosine phosphorylation of several substrate proteins, including Ig-α, Syk, and Vav, and the activation of mitogen-activated protein kinase extracellular signal– regulated kinase (ERK)1. Based on these results, Nagata et al. (63) proposed that the signal cascade initiated by Ig-β activation evokes differentiation signals similar to those delivered by the pre-BCR in normal B cell development. For mature B cells it is known that BCR engagement upregulates Egr-1 transcription through a signal cascade including p21/ras and mitogen-activated protein kinase (ERK [10, 64]), and for other cell types it has been shown that ERK activation induces Egr-1 transcription (65). Since RAG-2– deficient pro/pre-B–I cells overexpressing Egr-1 do not reach the same developmental stage as anti–Ig-β–stimulated cells, it seems likely that Egr-1 activity substitutes only part of the differentiation signal originating from the pre-BCR.
Analyzing later stages of B cell development in RAG-2+/+ Egr-1 transgenic mice, we observed lower proportions of immature and increased proportions of mature bone marrow B cells compared with their wild-type littermates, whereas there was no increased proliferation of transgenic pre-B or immature B cells detectable. These findings are consistent with the current model of the development from immature to mature B cells (66, 67). Immature B cells leave the bone marrow and enter the spleen where about half of them reach the mature stage (42). Mature bone marrow B cells are thought to be part of the recirculating pool. This would suggest that Egr-1 influences this migration at one or several steps.
Egr-1 expression was also found in CD4−CD8− double negative thymocytes by Miyazaki (35). Overexpression of transgenic Egr-1 in a RAG-2–deficient background allowed thymocytes to bypass the RAG-2–dependent block at the IL-2R+ Pgp-1− double negative stage and develop into immature CD8 single-positive cells, but not further to the CD4+CD8+ double-positive cell stage. In cortical CD4+CD8+ thymocytes, Egr-1 expression was reported by Shao et al. (68) to be dependent on TCR engagement, suggesting that high level expression of Egr-1 in the thymus might be a consequence of thymocyte selection. The high coincidence of Egr-1 expression in analogous B and T cell precursor subsets and the increased differentiation of pro/pre-B–I cells and thymocytes in Egr-1 transgenic mice suggest that Egr-1 activity regulates similar functions in both types of lymphocytes.
Downstream Target Genes.
Searching for potential downstream target genes responding to Egr-1, we found increased expression of the nuclear orphan receptor nur77 in bone marrow B cells from transgenic mice but not in cultivated transgenic pro/pre-B–I cells. Since we also could demonstrate binding of recombinant Egr-1 to a cognate Egr-1 binding site present in the nur77 promoter, it seems likely that Egr-1 directly induces nur77 expression in B lymphocytes before the mature B cell stage. It was reported that nur77 activity is involved in the regulation of thymocyte apoptosis (53, 54) by inducing CD95L expression (57). However, in the Egr-1 transgenic mice, we could detect neither upregulation of CD95L expression by bone marrow B cells nor an increased frequency of apoptotic cells (Warnatz, K., unpublished results). On the other hand, cellular responses other than apoptosis may be linked to nur77 function, since it is also induced upon antigen receptor ligation in B and T cells during proliferative responses (52, 69). Besides nur77 promoter, we also found enhanced expression of BP-1 in pre-B–II and in IgM+ bone marrow cells. Similar to the nur77, we could also demonstrate binding of recombinant Egr-1 to a sequence from the BP-1 promoter resembling an Egr-1 binding site. Therefore, it seems that Egr-1 activity promotes not only development to the stage of BP-1+ cells (fraction C), but also an increased surface expression of BP-1 on BP-1+ B cells. Although it is known that BP-1 acts as an aminopeptidase catalyzing the hydrolysis of acidic amino acid residues from the NH2 termini of proteins (70), its role in B lymphopoiesis remains to be clarified (71, 72).
Conclusions.
Here we provide evidence that Egr-1 supports at least two distinct steps of B cell maturation, the progression into the pre-B and into the mature B cell stage. Since Egr-1 activity is also sufficient to promote the development of double negative thymocytes into immature single-positive CD8low cells (35), as well as macrophage in vitro differentiation (73, 74), this transcription factor seems to play an important role in the differentiation of three major hematopoietic cell types.
Acknowledgments
We are grateful to Petra Fiedler for excellent technical assistance. We thank Barbara Osborne (University of Massachusetts, Amherst, MA) for generously providing nur77-specific antibodies and probes, Toru Miyazaki (Basel Institute for Immunology) for helpful discussions and for communicating unpublished results, and Harald Illges (University of Konstanz, Germany) and Mary O'Riordan (University of California, San Francisco, CA) for critically reading the manuscript.
Abbreviations used in this paper
- BCR
B cell antigen receptor
- BrdU
bromodeoxyuridine
- Egr-1
early growth response 1
- EMSA
electrophoretic mobility shift assay
- ERK
extracellular signal–regulated kinase
- RAG
recombinase-activating gene
Footnotes
Part of this work was supported through Deutsche Forschungsgemeinschaft grants Ei235/3-1 and Ei235/4-1 (to H. Eibel).
References
- 1.Sukhatme VP, Kartha S, Toback FG, Taub R, Hoover RG, Tsai-Morris CH. A novel early growth response gene rapidly induced by fibroblast, epithelial cell and lymphocyte mitogens. Oncogene Res. 1987;1:343–355. [PubMed] [Google Scholar]
- 2.Lemaire P, Revelant O, Bravo R, Charnay P. Two mouse genes encoding potential transcription factors with identical DNA-binding domains are activated by growth factors in cultured cells. Proc Natl Acad Sci USA. 1988;85:4691–4695. doi: 10.1073/pnas.85.13.4691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Milbrandt J. A nerve growth factor-induced gene encodes a possible transcriptional regulatory factor. Science. 1987;238:797–799. doi: 10.1126/science.3672127. [DOI] [PubMed] [Google Scholar]
- 4.Kharbanda S, Nakamura T, Stone R, Hass R, Bernstein S, Datta R, Sukhatme VP, Kufe D. Expression of the early growth response 1 and 2 zinc finger genes during induction of monocytic differentiation. J Clin Invest. 1991;88:571–577. doi: 10.1172/JCI115341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seyfert VL, Sukhatme VP, Monroe JG. Differential expression of a zinc finger-encoding gene in response to positive versus negative signaling through receptor immunoglobulin in murine B lymphocytes. Mol Cell Biol. 1989;9:2083–2088. doi: 10.1128/mcb.9.5.2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zipfel PF, Irving SG, Kelly K, Siebenlist U. Complexity of the primary genetic response to mitogenic activation of human T cells. Mol Cell Biol. 1989;9:1041–1048. doi: 10.1128/mcb.9.3.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ouellette AJ, Malt RA, Sukhatme VP, Bonventre JV. Expression of two “immediate early” genes, Egr-1 and c-fos, in response to renal ischemia and during compensatory renal hypertrophy in mice. J Clin Invest. 1990;85:766–771. doi: 10.1172/JCI114502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Watson MA, Milbrandt J. Expression of the nerve growth factor-regulated NGFI-A and NGFI-B genes in the developing rat. Development (Camb) 1990;110:173–183. doi: 10.1242/dev.110.1.173. [DOI] [PubMed] [Google Scholar]
- 9.Sukhatme VP, Cao XM, Chang LC, Tsai-Morris CH, Stamenkovich D, Ferreira PC, Cohen DR, Edwards SA, Shows TB, Curran T, et al. A zinc finger-encoding gene coregulated with c-fos during growth and differentiation, and after cellular depolarization. Cell. 1988;53:37–43. doi: 10.1016/0092-8674(88)90485-0. [DOI] [PubMed] [Google Scholar]
- 10.McMahon SB, Monroe JG. Activation of the p21ras pathway couples antigen receptor stimulation to induction of the primary response gene egr-1 in B lymphocytes. J Exp Med. 1995;181:417–422. doi: 10.1084/jem.181.1.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klaus SJ, Phillips NE, Parker DC. Effects of IL-4 and Fc gamma receptor II engagement on Egr-1 expression during stimulation of B lymphocytes by membrane immunoglobulin crosslinking. Mol Immunol. 1993;30:1553–1558. doi: 10.1016/0161-5890(93)90463-l. [DOI] [PubMed] [Google Scholar]
- 12.Gottschalk AR, Joseph LJ, Quintans J. Fc gamma RII cross-linking inhibits anti-Ig-induced Egr-1 and Egr-2 expression in B cells. J Immunol. 1994;152:2115–2122. [PubMed] [Google Scholar]
- 13.Hardy RR, Carmack CE, Shinton SA, Kemp JD, Hayakawa K. Resolution and characterization of pro-B and pre–pro-B cell stages in normal mouse bone marrow. J Exp Med. 1991;173:1213–1225. doi: 10.1084/jem.173.5.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li YS, Hayakawa K, Hardy RR. The regulated expression of B lineage associated genes during B cell differentiation in bone marrow and fetal liver. J Exp Med. 1993;178:951–960. doi: 10.1084/jem.178.3.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rolink A, Melchers F. B lymphopoiesis in the mouse. Adv Immunol. 1993;53:123–156. doi: 10.1016/s0065-2776(08)60499-x. [DOI] [PubMed] [Google Scholar]
- 16.Rolink A, Grawunder U, Winkler TH, Karasuyama H, Melchers F. IL-2 receptor alpha chain (CD25, TAC) expression defines a crucial stage in pre-B cell development. Int Immunol. 1994;6:1257–1264. doi: 10.1093/intimm/6.8.1257. [DOI] [PubMed] [Google Scholar]
- 17.Osmond DG, Rolink A, Melchers F. Murine B lymphopoiesis: towards a unified model. Immunol Today. 1998;19:65–68. doi: 10.1016/s0167-5699(97)01203-6. [DOI] [PubMed] [Google Scholar]
- 18.Henderson A, Calame K. Transcriptional regulation during B cell development. Annu Rev Immunol. 1998;16:163–200. doi: 10.1146/annurev.immunol.16.1.163. [DOI] [PubMed] [Google Scholar]
- 19.Bain G, Maandag EC, Izon DJ, Amsen D, Kruisbeek AM, Weintraub BC, Krop I, Schlissel MS, Feeney AJ, van Roon M, et al. E2A proteins are required for proper B cell development and initiation of immunoglobulin gene rearrangements. Cell. 1994;79:885–892. doi: 10.1016/0092-8674(94)90077-9. [DOI] [PubMed] [Google Scholar]
- 20.Zhuang Y, Soriano P, Weintraub H. The helix-loop-helix gene E2A is required for B cell formation. Cell. 1994;79:875–884. doi: 10.1016/0092-8674(94)90076-0. [DOI] [PubMed] [Google Scholar]
- 21.Urbanek P, Wang ZQ, Fetka I, Wagner EF, Busslinger M. Complete block of early B cell differentiation and altered patterning of the posterior midbrain in mice lacking Pax5/BSAP. Cell. 1994;79:901–912. doi: 10.1016/0092-8674(94)90079-5. [DOI] [PubMed] [Google Scholar]
- 22.Lin H, Grosschedl R. Failure of B-cell differentiation in mice lacking the transcription factor EBF. Nature. 1995;376:263–267. doi: 10.1038/376263a0. [DOI] [PubMed] [Google Scholar]
- 23.Kaneko H, Ariyasu T, Inoue R, Fukao T, Kasahara K, Teramoto T, Matsui E, Hayakawa S, Kondo N. Expression of Pax5 gene in human haematopoietic cells and tissues: comparison with immunodeficient donors. Clin Exp Immunol. 1998;111:339–344. doi: 10.1046/j.1365-2249.1998.00509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bain G, Robanus EC, Maandag, te Riele HP, Feeney AJ, Sheehy A, Schlissel M, Shinton SA, Hardy RR, Murre C. Both E12 and E47 allow commitment to the B cell lineage. Immunity. 1997;6:145–154. doi: 10.1016/s1074-7613(00)80421-5. [DOI] [PubMed] [Google Scholar]
- 25.Nutt SL, Urbanek P, Rolink A, Busslinger M. Essential functions of Pax5 (BSAP) in pro-B cell development: difference between fetal and adult B lymphopoiesis and reduced V-to-DJ recombination at the IgH locus. Genes Dev. 1997;11:476–491. doi: 10.1101/gad.11.4.476. [DOI] [PubMed] [Google Scholar]
- 26.Schilham MW, Oosterwegel MA, Moerer P, Ya J, de Boer PA, van de Wetering M, Verbeek S, Lamers WH, Kruisbeek AM, Cumano A, Clevers H. Defects in cardiac outflow tract formation and pro-B-lymphocyte expansion in mice lacking Sox-4. Nature. 1996;380:711–714. doi: 10.1038/380711a0. [DOI] [PubMed] [Google Scholar]
- 27.Kim U, Qin XF, Gong S, Stevens S, Luo Y, Nussenzweig M, Roeder RG. The B-cell-specific transcription coactivator OCA-B/OBF-1/Bob-1 is essential for normal production of immunoglobulin isotypes. Nature. 1996;383:542–547. doi: 10.1038/383542a0. [DOI] [PubMed] [Google Scholar]
- 28.Nielsen PJ, Georgiev O, Lorenz B, Schaffner W. B lymphocytes are impaired in mice lacking the transcriptional co-activator Bob1/OCA-B/OBF1. Eur J Immunol. 1996;26:3214–3218. doi: 10.1002/eji.1830261255. [DOI] [PubMed] [Google Scholar]
- 29.Schubart DB, Rolink A, Kosco-Vilbois MH, Botteri F, Matthias P. B-cell-specific coactivator OBF-1/ OCA-B/Bob1 required for immune response and germinal centre formation. Nature. 1996;383:538–542. doi: 10.1038/383538a0. [DOI] [PubMed] [Google Scholar]
- 30.Lee SL, Tourtellotte LC, Wesselschmidt RL, Milbrandt J. Growth and differentiation proceeds normally in cells deficient in the immediate early gene NGFI-A. J Biol Chem. 1995;270:9971–9977. doi: 10.1074/jbc.270.17.9971. [DOI] [PubMed] [Google Scholar]
- 31.Lee SL, Wang Y, Milbrandt J. Unimpaired macrophage differentiation and activation in mice lacking the zinc finger transcription factor NGFI-A (EGR1) Mol Cell Biol. 1996;16:4566–4572. doi: 10.1128/mcb.16.8.4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shinkai Y, Rathbun G, Lam KP, Oltz EM, Stewart V, Mendelsohn M, Charron J, Datta M, Young F, Stall AM, et al. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell. 1992;68:855–867. doi: 10.1016/0092-8674(92)90029-c. [DOI] [PubMed] [Google Scholar]
- 33.Rolink A, Karasuyama H, Haasner D, Grawunder U, Martensson IL, Kudo A, Melchers F. Two pathways of B-lymphocyte development in mouse bone marrow and the roles of surrogate L chain in this development. Immunol Rev. 1994;137:185–201. doi: 10.1111/j.1600-065x.1994.tb00665.x. [DOI] [PubMed] [Google Scholar]
- 34.Chen J, Ma A, Young F, Alt FW. IL-2 receptor alpha chain expression during early B lymphocyte differentiation. Int Immunol. 1994;6:1265–1268. doi: 10.1093/intimm/6.8.1265. [DOI] [PubMed] [Google Scholar]
- 35.Miyazaki T. Two distinct steps during thymocyte maturation from CD4−CD8− to CD4+CD8+distinguished in the early growth response (Egr)-1 transgenic mice with a recombinase-activating gene–deficient background. J Exp Med. 1997;186:877–885. doi: 10.1084/jem.186.6.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rolink A, Kudo A, Karasuyama H, Kikuchi Y, Melchers F. Long-term proliferating early pre B cell lines and clones with the potential to develop to surface Ig-positive, mitogen reactive B cells in vitro and in vivo. EMBO (Eur Mol Biol Organ) J. 1991;10:327–336. doi: 10.1002/j.1460-2075.1991.tb07953.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook, J., E.J. Fritsch, and T. Maniatis. 1989. Molecular Cloning: A Laboratory Manual. 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 39.Coffman RL, Weissman IL. B220, a B cell specific member of the T200 glycoprotein family. Nature. 1981;289:681–683. doi: 10.1038/289681a0. [DOI] [PubMed] [Google Scholar]
- 40.Brombacher F, Köhler G, Eibel H. B cell tolerance in mice transgenic for anti-CD8 immunoglobulin μ chain. J Exp Med. 1991;174:1335–1346. doi: 10.1084/jem.174.6.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schüppel R, Wilke E, Weiler E. Monoclonal anti-allotype antibody towards BALB/c IgM. Analysis of specificity and site specific crossover in recombinant strain BALB-IghVa/Igh-Cb . Eur J Immunol. 1987;17:553–561. doi: 10.1002/eji.1830170527. [DOI] [PubMed] [Google Scholar]
- 42.Rolink A, Andersson J, Melchers F. Characterization of immature B cells by a novel monoclonal antibody, by turnover and by mitogen reactivity. Eur J Immunol. 1998;28:3738–3748. doi: 10.1002/(SICI)1521-4141(199811)28:11<3738::AID-IMMU3738>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 43.Schittek B, Rajewsky K, Forster I. Dividing cells in bone marrow and spleen incorporate bromodeoxyuridine with high efficiency. Eur J Immunol. 1991;21:235–238. doi: 10.1002/eji.1830210136. [DOI] [PubMed] [Google Scholar]
- 44.Coligan, J.E., A.M. Kruisbeek, D.H. Margulies, E.M. Shevach, and W. Strober. 1992. Current Protocols in Immunology. Vols. 1–3. John Wiley & Sons, Inc., New York.
- 45.Dinkel A, Aicher WK, Haas C, Zipfel PF, Peter HH, Eibel H. Transcription factor Egr-1 activity down-regulates Fas and CD23 expression in B cells. J Immunol. 1997;159:2678–2684. [PubMed] [Google Scholar]
- 46.Seyfert VL, McMahon SB, Glenn WD, Yellen AJ, Sukhatme VP, Cao XM, Monroe JG. Methylation of an immediate-early inducible gene as a mechanism for B cell tolerance induction. Science. 1990;250:797–800. doi: 10.1126/science.2237429. [DOI] [PubMed] [Google Scholar]
- 47.Yellen AJ, Glenn W, Sukhatme VP, Cao XM, Monroe JG. Signaling through surface IgM in tolerance-susceptible immature murine B lymphocytes. Developmentally regulated differences in transmembrane signaling in splenic B cells from adult and neonatal mice. J Immunol. 1991;146:1446–1454. [PubMed] [Google Scholar]
- 48.Rolink A, Melchers F. Generation and regeneration of cells of the B-lymphocyte lineage. Curr Opin Immunol. 1993;5:207–217. doi: 10.1016/0952-7915(93)90006-e. [DOI] [PubMed] [Google Scholar]
- 49.Förster I, Rajewsky K. The bulk of the peripheral B-cell pool in mice is stable and not rapidly renewed from the bone marrow. Proc Natl Acad Sci USA. 1990;87:4781–4784. doi: 10.1073/pnas.87.12.4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang J, Walker H, Lin Q, Jenkins N, Copeland NG, Watanabe T, Burrows PD, Cooper MD. The mouse BP-1 gene: structure, chromosomal localization, and regulation of expression by type I interferons and interleukin-7. Genomics. 1996;33:167–176. doi: 10.1006/geno.1996.0180. [DOI] [PubMed] [Google Scholar]
- 51.Ryseck RP, Macdonald-Bravo H, Mattei MG, Ruppert S, Bravo R. Structure, mapping and expression of a growth factor inducible gene encoding a putative nuclear hormonal binding receptor. EMBO (Eur Mol Biol Organ) J. 1989;8:3327–3335. doi: 10.1002/j.1460-2075.1989.tb08494.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mittelstadt PR, DeFranco AL. Induction of early response genes by cross-linking membrane Ig on B lymphocytes. J Immunol. 1993;150:4822–4832. [PubMed] [Google Scholar]
- 53.Liu ZG, Smith SW, McLaughlin KA, Schwartz LM, Osborne BA. Apoptotic signals delivered through the T-cell receptor of a T-cell hybrid require the immediate-early gene nur77. Nature. 1994;367:281–284. doi: 10.1038/367281a0. [DOI] [PubMed] [Google Scholar]
- 54.Woronicz JD, Calnan B, Ngo V, Winoto A. Requirement for the orphan steroid receptor Nur77 in apoptosis of T-cell hybridomas. Nature. 1994;367:277–281. doi: 10.1038/367277a0. [DOI] [PubMed] [Google Scholar]
- 55.Zhou T, Cheng J, Yang P, Wang Z, Liu C, Su X, Bluethmann H, Mountz JD. Inhibition of Nur77/ Nurr1 leads to inefficient clonal deletion of self-reactive T cells. J Exp Med. 1996;183:1879–1892. doi: 10.1084/jem.183.4.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Calnan BJ, Szychowski S, Chan FK, Cado D, Winoto A. A role for the orphan steroid receptor Nur77 in apoptosis accompanying antigen-induced negative selection. Immunity. 1995;3:273–282. doi: 10.1016/1074-7613(95)90113-2. [DOI] [PubMed] [Google Scholar]
- 57.Weih F, Ryseck RP, Chen L, Bravo R. Apoptosis of nur77/N10-transgenic thymocytes involves the Fas/ Fas ligand pathway. Proc Natl Acad Sci USA. 1996;93:5533–5538. doi: 10.1073/pnas.93.11.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Maltzman JS, Carman JA, Monroe JG. Role of EGR1 in regulation of stimulus-dependent CD44 transcription in B lymphocytes. Mol Cell Biol. 1996;16:2283–2294. doi: 10.1128/mcb.16.5.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maltzman JS, Carmen JA, Monroe JG. Transcriptional regulation of the ICAM-1 gene in antigen receptor– and phorbol ester–stimulated B lymphocytes: role for transcription factor EGR1. J Exp Med. 1996;183:1747–1759. doi: 10.1084/jem.183.4.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Osmond DG. B cell development in the bone marrow. Semin Immunol. 1990;3:173–180. [PubMed] [Google Scholar]
- 61.Lu L, Smithson G, Kincade PW, Osmond DG. Two models of murine B lymphopoiesis: a correlation. Eur J Immunol. 1998;28:1755–1761. doi: 10.1002/(SICI)1521-4141(199806)28:06<1755::AID-IMMU1755>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 62.Young F, Ardman B, Shinkai Y, Lansford R, Blackwell TK, Mendelsohn M, Rolink A, Melchers F, Alt FW. Influence of immunoglobulin heavy- and light-chain expression on B-cell differentiation. Genes Dev. 1994;8:1043–1057. doi: 10.1101/gad.8.9.1043. [DOI] [PubMed] [Google Scholar]
- 63.Nagata K, Nakamura T, Kitamura F, Kuramochi S, Taki S, Campbell KS, Karasuyama H. The Igα/ Igβ heterodimer on μ-negative proB cells is competent for transducing signals to induce early B cell differentiation. Immunity. 1997;7:559–570. doi: 10.1016/s1074-7613(00)80377-5. [DOI] [PubMed] [Google Scholar]
- 64.McMahon SB, Monroe JG. The role of early growth response gene 1 (egr-1) in regulation of the immune response. J Leukocyte Biol. 1996;60:159–166. doi: 10.1002/jlb.60.2.159. [DOI] [PubMed] [Google Scholar]
- 65.Cohen DM. Urea-inducible Egr-1 transcription in renal inner medullary collecting duct (mIMCD3) cells is mediated by extracellular signal-regulated kinase activation. Proc Natl Acad Sci USA. 1996;93:11242–11247. doi: 10.1073/pnas.93.20.11242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lortan JE, Roobottom CA, Oldfield S, MacLennan IC. Newly produced virgin B cells migrate to secondary lymphoid organs but their capacity to enter follicles is restricted. Eur J Immunol. 1987;17:1311–1316. doi: 10.1002/eji.1830170914. [DOI] [PubMed] [Google Scholar]
- 67.Allman DM, Ferguson SE, Lentz VM, Cancro MP. Peripheral B cell maturation. II. Heat-stable antigenhisplenic B cells are an immature developmental intermediate in the production of long lived marrow-derived B cells. J Immunol. 1993;151:4431–4444. [PubMed] [Google Scholar]
- 68.Shao H, Kono DH, Chen LY, Rubin EM, Kaye J. Induction of the early growth response (Egr) family of transcription factors during thymic selection. J Exp Med. 1997;185:731–744. doi: 10.1084/jem.185.4.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Garcia I, Pipaon C, Alemany S, Perez-Castillo A. Induction of NGFI-B gene expression during T cell activation. Role of protein phosphatases. J Immunol. 1994;153:3417–3425. [PubMed] [Google Scholar]
- 70.Wu Q, Lahti J, Air G, Burrows P, Cooper M. Molecular cloning of the murine BP-1/6C3 antigen: a member of the zinc-dependent metallopeptidase family. Proc Natl Acad Sci USA. 1990;87:993–997. doi: 10.1073/pnas.87.3.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lin Q, Taniuchi I, Kitamura D, Wang J, Kearney JF, Watanabe T, Cooper MD. T and B cell development in BP-1/6C3/aminopeptidase A-deficient mice. J Immunol. 1998;160:4681–4687. [PubMed] [Google Scholar]
- 72.Wang J, Lin Q, Wu Q, Cooper MD. The enigmatic role of glutamyl aminopeptidase (BP-1/6C3 antigen) in immune system development. Immunol Rev. 1998;161:71–77. doi: 10.1111/j.1600-065x.1998.tb01572.x. [DOI] [PubMed] [Google Scholar]
- 73.Nguyen HQ, Hoffman-Liebermann B, Liebermann DA. The zinc finger transcription factor Egr-1 is essential for and restricts differentiation along the macrophage lineage. Cell. 1993;72:197–209. doi: 10.1016/0092-8674(93)90660-i. [DOI] [PubMed] [Google Scholar]
- 74.Krishnaraju K, Nguyen HQ, Liebermann DA, Hoffman B. The zinc finger transcription factor Egr-1 potentiates macrophage differentiation of hematopoietic cells. Mol Cell Biol. 1995;15:5499–5507. doi: 10.1128/mcb.15.10.5499. [DOI] [PMC free article] [PubMed] [Google Scholar]