Abstract
The molecular mechanisms regulating recruitment of intracellular signaling proteins like growth factor receptor–bound protein 2 (Grb2), phospholipase Cγ1, or phosphatidylinositol 3-kinase (PI3-kinase) to the plasma membrane after stimulation of the T cell receptor (TCR)– CD3–ζ complex are not very well understood. We describe here purification, tandem mass spectrometry sequencing, molecular cloning, and biochemical characterization of a novel transmembrane adaptor protein which associates and comodulates with the TCR–CD3–ζ complex in human T lymphocytes and T cell lines. This protein was termed T cell receptor interacting molecule (TRIM). TRIM is a disulfide-linked homodimer which is comprised of a short extracellular domain of 8 amino acids, a 19–amino acid transmembrane region, and a 159–amino acid cytoplasmic tail. In its intracellular domain, TRIM contains several tyrosine-based signaling motifs that could be involved in SH2 domain–mediated protein–protein interactions. Indeed, after T cell activation, TRIM becomes rapidly phosphorylated on tyrosine residues and then associates with the 85-kD regulatory subunit of PI3-kinase via an YxxM motif. Thus, TRIM represents a TCR-associated transmembrane adaptor protein which is likely involved in targeting of intracellular signaling proteins to the plasma membrane after triggering of the TCR.
Keywords: T lymphocytes, T cell receptor, transmembrane adaptor proteins
Acritical question among several unresolved issues regarding the molecular mechanisms leading to T cell activation is how intracellular signaling molecules like growth factor receptor–bound protein 2 (Grb2),1 phospholipase Cγ1, and phosphatidylinositol 3-kinase (PI3-kinase) are recruited to the plasma membrane after antigen recognition by the TCR. A first clue to how the TCR is connected to downstream signaling pathways was provided by the recent cloning of a novel phosphoprotein called linker for activation of T cells (LAT [1]). LAT is a single chain type III transmembrane protein possessing several tyrosine motifs in its intracellular domain that are predicted to mediate associations with src homology 2 (SH2) domains of signaling proteins. Indeed, after T cell activation, LAT associates with a number of intracellular molecules involved in signal transduction, including Grb2, the 85-kD subunit of PI3-kinase, phospholipase Cγ1, SLP76, Sos, and Cbl (1). Thus, tyrosine-phosphorylated LAT likely targets intracellular proteins to the plasma membrane.
Using two-dimensional isoelectric focussing (IEF)/SDS gel electrophoresis, we have previously identified a 29/30-kD disulfide-linked dimer (pp29/30) which is expressed in resting human T lymphocytes, HPB-ALL cells, and NK clones (2). Our data suggested that pp29/30 preferentially associates with the TCR–CD3–ζ complex. This conclusion was drawn from experiments in which coprecipitation of pp29/30 by mAbs directed at signaling receptors expressed on human T lymphocytes (e.g., CD2 and CD4) was strongly impaired when expression of the TCR–CD3–ζ complex was downmodulated by overnight incubation with a CD3-ε mAb.
We describe here the purification, tandem mass spectrometry sequencing, molecular cloning, and biochemical characterization of pp29/30. pp29/30 is a disulfide-linked homodimeric transmembrane molecule with a short extracellular domain of eight amino acids which is expressed exclusively in T lymphocytes and NK cells. In T lymphocytes and T cell lines, pp29/30 coprecipitates and comodulates with the TCR–CD3–ζ complex. Therefore, we termed the protein T cell receptor interacting molecule (TRIM). The 159–amino acid intracellular domain of TRIM contains eight tyrosine residues, three of which are components of peptide sequence motifs that would be predicted to be involved in SH2-mediated interaction with intracellular signaling molecules. After T cell activation, TRIM becomes rapidly tyrosine phosphorylated and is then capable of associating with the 85-kD regulatory subunit (p85) of PI3-kinase via an YxxM motif. Thus, TRIM represents a novel TCR-associated polypeptide likely to be involved in recruitment of intracellular signaling proteins to the plasma membrane after T cell activation.
Materials and Methods
Purification of TRIM.
5 × 109 HPB-ALL cells were washed once in TBS and lysed in 70 ml of 1% Brij58 lysis buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% Brij58 (Pierce Chemical Co., Rockford, IL), 1 mM NaVanadate, 10 mM NaF, 1 mM PMSF, 1 μg/ml aprotinin, and 1 μg/ml leupeptin) at 4°C for 2 h. Postnuclear lysates were consecutively incubated with two aliquots of CNBr-activated Sepharose beads (3 ml packed beads) coupled with the protein A–purified CD3 mAb OKT-3 for 1 h. The immune complexes were subsequently spun down, and the beads from the first and second immunoprecipitations were pooled and washed four times with 10 ml washing buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10 mM NaF, 1 mM PMSF, 0.1% Brij58). An aliquot of 50 μl beads was resuspended in 80 μl kinase buffer (20 mM Tris-HCl, pH 7.5, 10 mM MnCl, 0.1% Brij58) supplemented with 20 μCi [γ-32P]ATP (Amersham Buchler, Braunschweig, Germany), and in vitro phosphorylation was carried out for 20 min at room temperature. The reaction was stopped by the addition of 1 ml stop solution (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 20 mM EDTA, 0.1% Brij58), and the radiolabeled beads were washed six times in this buffer.
Radiolabeled and unlabeled beads were pooled and then incubated for 5 min at room temperature under constant agitation with 5 ml of 100 mM glycine-HCl, pH 2.5 (acidic elution). The solution was neutralized by the addition of 500 μl of 1.5 M Tris-HCl, pH 8.8, the beads were spun down, the supernatant was saved, and the beads were washed once with 4 ml washing buffer. The bead pellet was then resuspended for basic elution in 5 ml of 100 mM triethylamine, pH 11.5, and again incubated for 5 min at room temperature with agitation. This solution was neutralized by the addition of 500 μl of 1 M Tris-HCl, pH 7.5, the beads were spun down, the supernatant was saved, and the beads were washed once with 4 ml washing buffer. Both eluates and both washing supernatants were pooled, and eluted proteins were precipitated overnight at −20°C by addition of 2.5 vol acetone.
After precipitation, proteins were pelleted for 10 min at 10,000 rpm at 4°C. The protein pellet was washed three times with ice-cold methanol and dissolved—aided by sonication—in 100 μl lysis buffer supplemented with 1% Triton X-100. 20 μl of 5× nonreducing sample buffer was added to this solution, which was incubated for 10 min at 37°C, and the insoluble material was removed by centrifugation (5 min, 10,000 rpm). The supernatant was then loaded on the first dimension tube gel of a two-dimensional nonreducing/reducing SDS-PAGE. After electrophoresis, the gel was stained with Coomassie blue solution, destained, and subjected to autoradiography. Protein spots corresponding to pp29/30 were identified, excised from the gels, and further processed as described below.
Peptide Sequencing.
Protein spots corresponding to pp29/30 were excised from four two-dimensional gels stained with Coomassie blue. The spots were in-gel digested with trypsin (3), and the unseparated mixture of recovered tryptic peptides was subjected to nanoelectrospray tandem mass spectrometric sequencing essentially as described (4). Analyses were performed on a triple quadrupole mass spectrometer (API III; PE Sciex Instruments, Ontario, Canada) equipped with the nanoelectrospray ion source developed by our group (5).
cDNA Cloning and Sequencing.
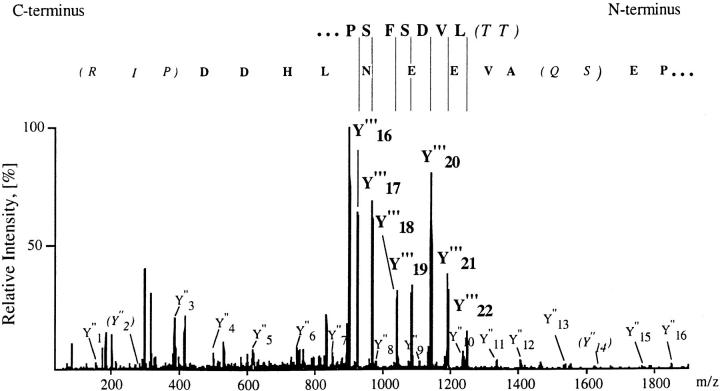
Total RNA was isolated from HPB-ALL cells according to the method of Chomczynski and Sacchi (6). mRNA was enriched using poly(A) quick columns (Stratagene, Heidelberg, Germany) according to the manufacturer's protocol and reverse transcribed with MuLV reverse transcriptase (Gibco, Eggenstein, Germany) and an oligo dT(12–18) primer. This cDNA was used as template for PCR reactions, with fully redundant oligonucleotides deduced from the 25–amino acid pp29/30 peptide [AM(L/I)VDSFSPEASGAVEEN(L/I)HDDthk]. L/I stands for leucine/isoleucine; these isobaric amino acids cannot be distinguished by our method. The underlined amino acids were used for the generation of forward (GTN GAY AGY TTY TCN CCN G) and reverse (YTT RTG NGT RTC RTC RTG) oligonucleotides, respectively. KlenTaq DNA polymerase (Clontech, Palo Alto, CA) was used to amplify the intervening sequence. The samples were amplified on a thermocycler (PTC-200; MJ Research, Watertown, MA) using touchdown programs with annealing temperature ranges of 55–45 or 50–40°C. A fragment of the expected size was excised from a 4% agarose gel, cloned into the pCRII vector (Invitrogen Corp., Carlsbad, CA), and sequenced. The cDNA sequence coding for the internal peptide SQAVEENL was obtained. Comparison of the peptide sequence deduced from the cDNA and the actual mass spectrum of the peptide suggested that two amino acid residues in the internal peptide were likely miscalled because of the very low intensity of the ions of the relevant peptide fragments (see Fig. 1).
Figure 1.
(Top) Nanoelectrospray tandem mass spectrometric sequencing of a 25-residue peptide of pp29/30, showing the tandem mass spectrum acquired from the triply charged ion at m/z = 900.4. Collisional fragmentation of triply charged ions of tryptic peptides typically results in overlapping series of COOH-terminal peptide fragments (Y-ions) bearing two positive charges (Y′′′, bold) or one positive charge (Y′′). The amino acid sequence is deduced from the spectrum by considering the precise mass differences between adjacent peptide fragments in conjunction with the known amino acid molecular weights. The series of doubly charged Y′′′-ions allowed determination of the sequence stretch starting from the NH2 terminus of the peptide (top row). The sequence was then extended to the COOH terminus by following the series of singly charged Y′′-ions (bottom row), which partially overlaps with the series of doubly charged fragments. The interpretation algorithm used here relies heavily on the correct identification of the Y-ions among the other fragment ions in the spectrum. In three regions of the spectrum, the amino acid sequence was called incorrectly, because of the very low intensity of the relevant Y′′-ions (ions Y′′23, Y′′14, and Y′′2). Sequence stretches confirmed after the full-length cloning of the protein are in bold, and the regions where the correct sequence was not obtained are shown in parentheses (the correct sequence determined after the cloning is inside the parentheses in italics). In all cases, the sequence obtained from the cloned protein was consistent with the tandem mass spectrum. Altogether, two contiguous sequence stretches each of nine amino acid residues were accurately called.
A forward oligonucleotide (CCA GGC AGT AGA GGA AAA CAT TC) based on the sequenced internal DNA was then used together with a lock-docking reverse oligonucleotide [(T)30VN] in 3′–rapid amplification of cDNA ends (RACE) experiments with cDNA from HPB-ALL poly(A)+ RNA as template and KlenTaq DNA polymerase (Clontech) in a touchdown PCR program with an annealing temperature range of 65–55°C. The resulting DNA fragment was excised from an agarose gel, cloned into the EcoRV site of the BluescriptII vector (Stratagene), and sequenced. It was found to correspond to the 3′ fragment of TRIM, since it contained the nucleotide sequence coding for the second peptide derived by mass spectrometry [(L/ I)FG(L/I)(L/I)R] as well as the sequence for the very COOH-terminal part of the 25–amino acid peptide. The amino acid sequence HDDPIR was deduced from the cDNA and could be verified by a comparison of calculated fragment masses and measured fragment masses in the tandem mass spectrum (see Fig. 1).
The remaining 5′ cDNA was amplified from thymus RACE– ready cDNA (Clontech) with two nested primers deduced from the 3′ sequence (CAC CCT GCT GCC AAG TCC TTC TGC and CAC CAT CCC ATC AAC AAA TGA TCA GG) by 5′-RACE according to the manufacturer's protocols using touch down PCR programs and KlenTaq polymerase (Clontech). The amplified band was purified from an agarose gel and sequenced directly. When the full-length sequence of TRIM was obtained, one peptide that was partially sequenced by tandem mass spectrometry (MQEATPSAQATNETQMCYASLDHSVK) was additionally verified via the peptide sequence tag approach (7). Altogether, 56 amino acids in the deduced sequence of TRIM were verified by tandem mass spectrometry, conclusively confirming the identity of the protein. All sequencing reactions were performed using the dRhodamine dye terminator chemistry (PE Applied Biosystems, Foster City, CA) according to the manufacturer's recommendations and analyzed on an automated sequencer (model 310; PE Applied Biosystems). Computer-aided sequence analysis was performed with the HUSAR software package, which is based on the University of Wisconsin Genetics Computer Group package (8, 9).
Northern Blots.
Human multiple-tissue Northern blots and the human RNA master blot were obtained from Clontech. The membranes were prehybridized and hybridized at 65°C in 50 mM Tris-HCl, pH 7.5, 500 mM NaCl, 1 mM EDTA, 0.1% Na4P2O7, 10% dextran sulfate, 1% casein, 1% SDS, and 250 μg/ml salmon sperm DNA overnight with a TRIM cDNA probe encompassing nucleotides 1–764. Stringent washes were performed twice at 65°C for 20 min in 60 mM Tris-HCl, pH 8.0, 300 mM NaCl, 2 mM EDTA, 1% SDS, and twice at 65°C for 20 min in 20 mM Tris-HCl, pH 8.0, 100 mM NaCl, 0.66 mM EDTA, 0.1% SDS. Blots were then subjected to autoradiography.
cDNA Constructs.
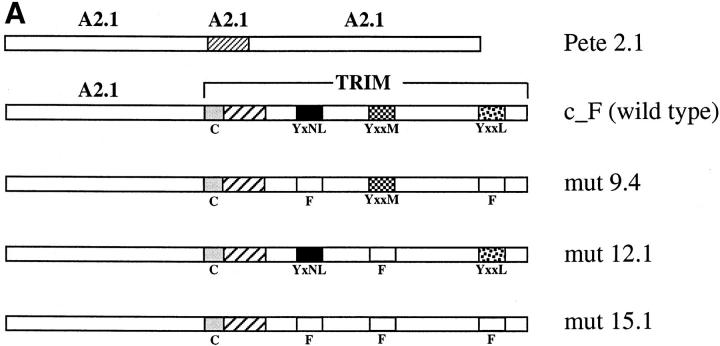
The chimeric HLA-A2.1–TRIM construct (chimera_F) consists of the extracellular domain of the HLA class I allele A2.1 fused to a full-length TRIM molecule (see Fig. 12). It was generated by separately amplifying the extracellular domain of the HLA cDNA with an HLA-A2.1–specific forward primer (ATGGCCGTCATGGCGCCCCG) and a chimeric reverse primer (GGGGCACCCAGAGATTCCTGAGGGCTGGGA-AGACGGCTCCC), and the TRIM fragment with an overlapping chimeric forward primer (GGGAGCCGTCTTCCCAGCCCTCAGGAATCTCTGGGTGCCCC) and a TRIM-specific reverse primer (ATGGTCCAGCTAGTTTATAGG). The amplified fragments were purified from agarose gels and then used together as templates for a chimeric PCR with the HLA-A2.1– specific forward primer and the TRIM–specific reverse primer. The resulting chimeric DNA fragment was gel purified and cloned into the BstXI site of the eukaryotic expression vector pEF-BOS (10). The mutants of chimera_F were produced by site-directed mutagenesis using the Quick Change site-directed mutagenesis kit (Stratagene) according to the manufacturer's protocol. The following mutants were used in this study: c_F,mut9.4 (Y63 to F, Y110 to F), c_F,mut12.1 (Y79 to F), and c_F,mut15.1 (Y63 to F, Y79 to F, Y110 to F). The sequence of all constructs was confirmed by DNA sequencing. Transient and stable transfection of Jurkat cells was performed essentially as described (11).
Figure 12.
Binding of p85 to TRIM requires the YxxM motif. Jurkat T cells were transiently transfected with cDNA constructs encoding chimeric A2.1– TRIM molecules (A). Expression of the chimeras was assessed by indirect immunofluorescence using A2.1 mAb BB7.2. In general, ∼28–34% of the transfected cells expressed the individual constructs (B, left). Equal numbers of alive cells (∼1.5–2 × 106 transfected cells expressing the chimeras) were either left untreated or were stimulated for 2 min with Pervanadate. Subsequently, cells were lysed in NP-40 lysis buffer, precipitated with A2.1 mAb (BB7.2), and subjected to anti–PI3-kinase Western blotting (B, right). Top of lanes, Individual chimeras.
For the transfection experiments in COS cells, we used the following cDNA constructs: wild-type fyn cloned in pSRα expression vector (provided by Dr. A. da Silva, Dana Farber Cancer Institute, Boston, MA) and myc-tagged lck or ZAP70 inserted into pcDNA3 vector (donated by Dr. R. Abraham, The Mayo Clinic, Rochester, MN).
Cells.
Jurkat cells, JRT3-T3.5 cells (a Jurkat variant deficient in expression of the TCR α chain, provided by Dr. A. v. Bonin, Bernard-Nocht Institut für Tropenmedizin, Hamburg, Germany), P116 cells (a Jurkat variant lacking expression of both the ZAP70 and Syk protein tyrosine kinase [PTK], a gift from Dr. R. Abraham [12]), HPB-ALL T cells, and COS cells (provided by Dr. A. da Silva) were maintained in RPMI 1640 (Gibco) supplemented with 10% FCS (Gibco), 1% penicillin-streptomycin (Gibco), and 2% glutamine (Gibco) at 37°C and 100% humidity. The CD8+ cutaneous T cell lymphoma (PBLCPCS) cell line was grown in the same medium with the exception that FCS was replaced by 10% human serum (Sigma, Deisenhofen, Germany) and 40 U/ml of recombinant human IL-2 were added (13). PBLs were prepared by E-rosetting from freshly drawn venous blood of healthy donors as described elsewhere (14). The Jurkat variant Pete 2.1 (stably transfected with a full-length A2.1 allele of the human HLA class I molecule) has been described previously (11).
Transfection.
2.5 × 107 Jurkat cells were transfected as described previously (11). For transfection of COS cells, 2 × 105 exponentially growing cells were incubated overnight in a six-well plate (Nunc, Inc., Roskilde, Denmark). Adherent cells were washed twice with 2 ml of PBS and incubated for 4 h in 1 ml of transfection solution (2 μg/ml DNA, 10 mg/ml DEAE-dextran, 5.6 mg/ml Chloroquine in RPMI 1640 supplemented with 2% FCS). The solution was subsequently removed, and 1 ml of a 10% DMSO (Merck, Darmstadt, Germany) in RPMI was added and allowed to incubate for 2 min. Then cells were washed once with PBS and incubated for 36 h in 2–3 ml of culture medium. Transfected cells were washed in PBS and lysed in 250 μl of lysis buffer.
Abs and Inhibitors.
The following mAbs were used in immunoprecipitation experiments: CD2 (ICRFCD2.1.1A, IgG2a), CD3-ε (OKT-3, IgG2a), CD4 (AICD4.1, IgG1), CD8 (AICD8.1, IgG1), CD45 (AICDCD45.2, IgG1), CD28 (8E3B6, IgG1), HLA class I, allele A2.1 (BB7.2, IgG2a), and antiphosphotyrosine (anti-PTYR) (PY72, IgG1; a gift from Dr. B. Sefton, The Salk Institute, San Diego, CA). Typically, proteins were precipitated from cellular lysates using 25 μl of packed CNBr-Sepharose beads covalently coupled with protein A–purified Abs (6 mg Ab/ml packed beads).
The polyclonal antiserum directed at TRIM was generated by immunizing rabbits with a KLH-coupled synthetic peptide corresponding to the 25–amino acid long peptide (AMLVDSFSPEASGAVEENLHDDTHK) that was determined by nanoelectrospray tandem mass spectrometry sequencing of purified pp29/30. The fact that two 9–amino acid stretches of the 25–amino acid synthetic peptide agreed with the sequence obtained by cDNA sequencing proved to be sufficient for specific recognition of TRIM by the following criteria: (a) ELISA analysis where the antiserum strongly reacted with the wild-type peptide; (b) the specific reactivity of the antiserum with chimeric TRIM molecules (e.g., chimera_F) that were transiently or stably expressed in Jurkat cells; and (c) the specific reactivity of the serum with a wild-type TRIM molecule that was transiently overexpressed in COS cells (see Fig. 10).
Figure 10.
Evidence that TRIM represents a substrate for src-kinases. COS cells were transiently transfected with the cDNA constructs depicted (top). After lysis, 10% of the transfected cells were first analyzed by anti-PYTR Western blotting (A). The membrane was then stripped, and the expression of the individual constructs was assessed using anti-TRIM, anti-lck, anti-fyn, and anti-myc (myc-ZAP70) Abs, respectively (B). The remaining 90% of the lysates were subjected to anti-PTYR immunoprecipitation and analyzed for precipitation of TRIM by anti-TRIM Western blotting (PTyr IP). (C) Wild-type (WT) Jurkat cells or ZAP70/SYK-deficient P116 cells were either left untreated (Med.) or were preincubated for 15 min with the src-kinase inhibitor PP1 (10 μM). Cells were then stimulated for 2 min with anti-TCR mAb C305 in the absence or presence of PP1. Postnuclear lysates were subjected to immunoprecipitation with anti-PTYR mAb PY72, followed by TRIM immunoblotting. As a control, cells were incubated for 2 min with Pervanadate (Perv.).
For Western blot analysis, anti-PTYR (4G10), anti-Grb2, anti– mitogen-activated protein (MAP), and anti–PI3-kinase mAbs (all purchased from Upstate Biotechnology Inc., Lake Placid, NY) were used at 1 μg/ml. Anti-p56lck and anti-p59fyn (provided by Dr. A. Veillette, McGill Cancer Center, McGill University, Montreal, Canada) polyclonal Abs were used at 1:10,000 and 1:2,000 vol/vol dilution, respectively. Mouse mAbs directed at ζ (1C10, IgG1) and CD3-ε (SP34, IgG3) were donated by Dr. Marcus Peter, German Cancer Research Center (Heidelberg, Germany), and used at 1:10,000 (ζ) and 1:20,000 vol/vol (CD3-ε) dilutions. Affinity-purified polyclonal anti-TRIM antiserum was prepared as described elsewhere (2) and used at 5–10 μg/ml. For detection of CD2 in cell lysates, protein A–purified CD2 mAb ICRFCD2.5.1B (IgG2a) was used at 10 μg/ml. Secondary horseradish peroxidase– coupled polyclonal antisera were purchased from Dianova GmbH (Hamburg, Germany) and used at a final dilution of 1:20,000 vol/vol in Western blotting experiments. Anti-TCR mAb C305 (IgM; a gift of Dr. A. Weiss, University of California at San Francisco, San Francisco, CA) was used as culture supernatant for the stimulation of Jurkat cells. For inhibition of src-kinase activity, we preincubated cells for 15 min at 37°C with the recently described pyrazolpyrimidine PP1 (reference 15; provided by Dr. S. Ratnofsky, BASF Bioresearch Corp., Worcester, MA) at a final concentration of 10 μM before activation with C305 mAb.
Comodulation Experiment and In Vitro Activation.
To induce modulation of the TCR–CD3 complex, resting T lymphocytes or PBLCPCS cells were cultured for 18 h at 37°C in a 1:2 vol/vol dilution of tissue culture supernatant of anti–CD3-ε mAb (2Ad2A2, IgM; provided by Dr. E. Reinherz, Dana Farber Cancer Institute) at a density of 2 × 106 cells/ml. Expression of cell surface receptors before and after modulation of the TCR–CD3 complex was assessed by indirect immunofluorescence using a standard protocol (14).
In vitro activation of T cells was performed using a mixture of biotinylated anti-CD3 plus anti-CD4 mAbs (10 μg/ml). Cells were incubated for 10 min at 37°C with biotinylated Abs which were then cross-linked for the indicated periods of time by the addition of avidin (final concentration 80 μg/ml; Sigma). For Pervanadate treatment, cells were incubated with 0.1 mM NaVanadate (Merck) plus 1 mM H2O2 (Sigma) for 2 min at room temperature. The cells were washed once in ice-cold TBS, lysed, and then subjected to precipitation experiments (see below).
Precipitation Experiments.
6 × 107 freshly prepared PBLs or 3 × 107 cells of the various T cell lines were washed once in ice-cold TBS and then lysed for 1 h at 4°C in lysis buffer supplemented with 1% of detergent (Brij58, Digitonin, Triton X-100 [Sigma], and NP-40 [Merck]). Postnuclear supernatants were subjected to immunoprecipitation using 25 μl of CNBr-Sepharose beads coupled with protein A–purified mAb as described above. For immunoprecipitation of TRIM, postnuclear lysates were preincubated for 30 min with polyclonal TRIM Ab (1:100 vol/vol), and immune complexes were then collected with protein A–Sepharose (Pharmacia Biotech AB, Uppsala, Sweden). In other experiments, isolated SH2 domains from signaling molecules were expressed as glutathione-S-transferase (GST) fusion protein and used for precipitation experiments as described previously (16, 17). The individual precipitates were washed five times in washing buffer and then either subjected to SDS-PAGE or further processed for two-dimensional gel electrophoresis, in vitro kinase assay, or reprecipitation experiments.
In Vitro Kinase Reaction, Two-dimensional Gel Electrophoresis, Reprecipitation Experiments, and Western Blot Analysis.
In vitro kinase reaction, elution of coprecipitated proteins using 1% Triton X-100 lysis buffer, reprecipitation of in vitro–phosphorylated proteins, two-dimensional gel electrophoresis, and Western blot analysis were performed essentially as described previously (2). For detection of proteins in Western blot experiments, the ECL detection system (Amersham Buchler) was used. When necessary, blots were stripped according to the manufacturer's recommendations and reprobed with additional Abs.
Membrane Preparation.
5 × 107 HPB-ALL cells were washed twice with TBS and incubated for 45 min in 5 ml of hypotonic buffer (10 mM Tris, 10 mM KCl, 1 mM CaCl2, 1 mM MgCl2, 20 μg/ml aprotinin, 20 μg/ml leupeptin, 1 mM PMSF, 1 mM vanadate). Cells were then transferred to a Dounce homogenizer, and membranes were disrupted by applying 30 strokes on ice. The homogenate was spun for 20 min at 15,000 rpm in an Eppendorf microfuge, the supernatant was carefully removed, and the membrane fraction was pelleted by ultracentrifugation (40 min, 33,000 rpm, 4°C) using a swing-out rotor (SW55Ti; Beckman Instruments Inc., Fullerton, CA). The supernatant was removed (cytosolic fraction), and the pellet (membrane fraction) was washed twice with 5 ml of ice-cold TBS and then lysed in 100 μl of lysis buffer supplemented with 1% NP-40.
Confocal Laser Scan Microscopy.
HBP-ALL T cells were cytospun onto 3-aminopropyltriethoxysilane–treated slides (Histo-bond slides; Marienfeld Laboratory Glassware, Bad Mergentheim, Germany) and fixed in cold acetone for 10 min. After drying, the slides were incubated for 1 h with a mixture of affinity-purified anti-TRIM Ab (3 μg/ml) and monoclonal mouse anti–human vimentin (clone VIM 3B4, isotype IgG2a, dilution 1:2; Camon, Wiesbaden, Germany). The Abs were diluted in TBS/0.02% BSA with the addition of 2.5 mg/ml normal human Igs (gamma-venin; Behringwerke AG, Marburg, Germany). TBS/0.02% BSA was used for all washing steps and to dilute the secondary and tertiary reagents. As secondary Abs, a combination of biotinylated goat anti–rabbit IgG (Dianova GmbH; dilution 1:300 vol/vol) and sheep anti–mouse IgG1 (1:50 vol/vol) or IgG2a (1:100 vol/vol) Abs (Serotec Ltd., Kidlington, UK) plus gamma-venin was used, followed by simultaneous incubation with Cy3-conjugated streptavidin (red fluorescence, dilution 1:1,000 vol/vol; Dianova GmbH) and Cy2-conjugated donkey anti–sheep Abs (green fluorescence, dilution 1:100 vol/vol; Dianova GmbH) for 30 min. After final washes, the sections were mounted with histogel mounting medium (Camon). Slides were viewed with a Laserscan microscope (Ernst Leitz GmbH, Wetzlar, Germany) using 570 nm (red emission) and 508 nm (green emission) filters, respectively. Incubations with omission of the primary Abs served as negative controls.
Results
Purification of pp29/30 and Generation of a Polyclonal Antiserum.
To purify pp29/30 (2), CD3-ε immunoprecipitates obtained from Brij58 lysates of HPB-ALL cells were subjected to two-dimensional nonreducing/reducing SDS-PAGE. Off-diagonal protein spots corresponding to pp29/ 30 were excised from the gels and digested with trypsin. The tryptic fragments were subsequently subjected to tandem mass spectrometry sequencing, which revealed that the spots contained several individual proteins. Peptides corresponding to bovine α, β, and κ caseins (likely originating from culture medium) were rapidly identified by searching a comprehensive database with the peptide sequence tags (7). However, searching protein and expressed sequence tag (EST) databases with the sequence tags obtained from the other peptide ions did not identify any known proteins. Therefore, full-length amino acid sequences of these peptides were obtained as described previously (4, 18). Two of the sequenced peptides were encoded by expressed sequence tags found in both public and private databases. Cloning of the corresponding polypeptide resulted in the identification of a novel disulfide-linked dimeric transmembrane adaptor protein (gp30/40) that is exclusively expressed in lymphocytes (Klode, J., A. Marie-Cardine, H. Kirchgessner, E. Bruyns, A. Shevchenko, M. Mann, and B. Schraven, manuscript in preparation). The other two peptides sequenced (see Materials and Methods, and Fig. 7) did not match with gp30/40, indicating that they belonged to an additional protein present in the preparation.
Figure 7.
Amino acid sequence of TRIM. The putative transmembrane region is boxed. The tyrosine-based signaling motifs are depicted underneath the corresponding sequence; the peptide sequences obtained by nanoelectrospray tandem mass spectrometry of purified pp29/30 are double-underlined. Tyrosine residues are underlined. Note that TRIM does not possess a leader sequence, and that a cysteine residue is present in the extracellular domain (C7). The sequence data are available from EMBL/GenBank under accession no. AJ224878.
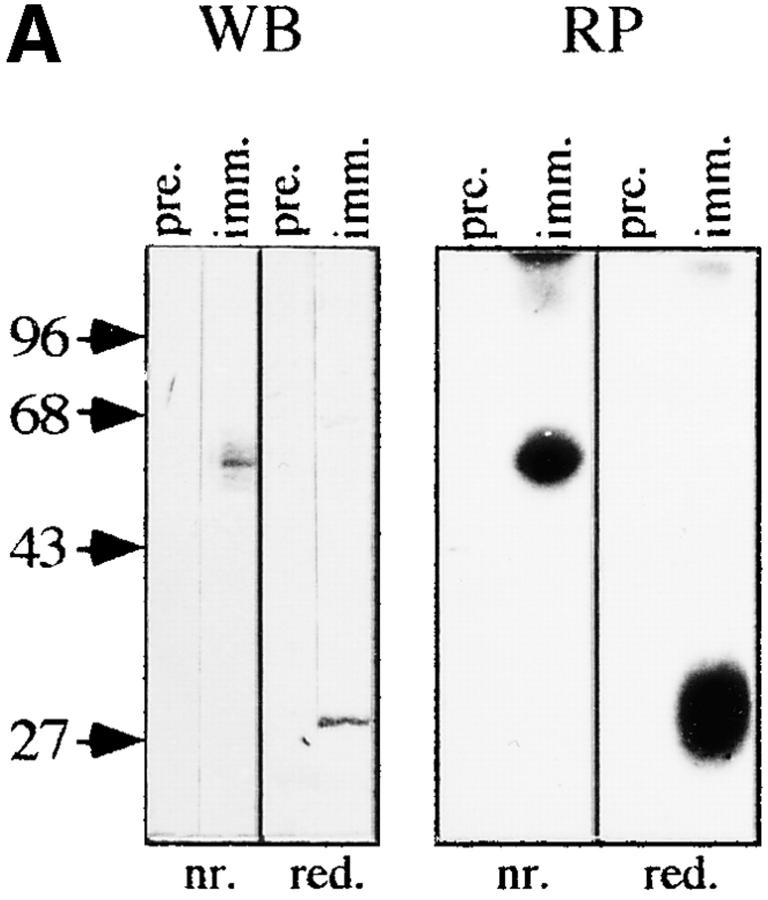
A synthetic peptide corresponding to the 25–amino acid tryptic fragment sequenced by tandem mass spectrometry (Fig. 1) was coupled to KLH and used to prepare a polyclonal rabbit antiserum. Serum obtained from immunized animals but not the preimmune serum reacts in Western blot with a disulfide-linked 60-kD dimer (30 kD reduced) that is expressed in HPB-ALL cells (Fig. 2 A, left). Perhaps more importantly, this antiserum exclusively reprecipitates a disulfide-linked 60-kD dimer from an in vitro–phosphorylated CD3 immunoprecipitate (Fig. 2 A, right) which exactly comigrates with pp29/30 in two-dimensional IEF/SDS gel electrophoresis (Fig. 2 B). Collectively, these data indicate that the antiserum raised against the 25–amino acid peptide detects pp29/30.
Figure 2.
Characterization of pp29/30. (A) Left, Anti-pp29/30 Western blot analysis (WB) of HPB-ALL lysates (5 × 105/ lane) under nonreducing and reducing conditions. Crude pp29/ 30 antiserum or preimmune serum was used at a 1:500 vol/vol dilution. For detection, we used the ECL system (Amersham Buchler). Right, and B, Reprecipitation experiments (RP). A CD3 (OKT-3) immunoprecipitate was prepared from HPB-ALL cells (107) lysed in Brij58 containing lysis buffer and subjected to in vitro kinase assay. Phosphorylated proteins were subsequently released using Triton X-100 lysis buffer. 10% of the material was directly separated on two-dimensional IEF/SDS-PAGE (B, left). The remaining material was subjected to reprecipitation using preimmune serum or anti-pp29/30 immune serum. The precipitates were run on nonreducing or reducing 12% SDS-PAGE (A, right) or were separated on two-dimensional reducing IEF/SDS-PAGE (B, middle and right). Radiolabeled proteins were visualized by autoradiography.
pp29/30 Coprecipitates and Comodulates with the TCR– CD3–ζ Complex in Human T Cells.
The availability of a polyclonal antiserum directed at pp29/30 allowed us to investigate the molecular relationship between pp29/30 and the TCR–CD3 complex. To this end, HPB-ALL cells were lysed in buffer containing 1% Digitonin. This treatment preserves the integrity of the TCR–CD3–ζ complex while disrupting the weak interactions between the TCR and coreceptor molecules. Lysates were subjected to immunoprecipitation using mAbs directed at different receptor molecules expressed by human T lymphocytes and subsequently analyzed for coprecipitation of pp29/30 by anti-pp29/30 Western blotting. Fig. 3 A demonstrates that under these experimental conditions, pp29/30 exclusively coprecipitates with the CD3-ε molecule but not with the CD2, CD4, CD8, and CD28 coreceptors and also not with the CD45 and HLA class I molecules. Similar results as shown here for HPB-ALL cells were obtained using immunoprecipitates from Digitonin lysates of Jurkat T cells as well as from an IL-2–dependent CD8+ subclone of a recently described cutaneous T cell lymphoma (PBLCPCS T cell line [13]; data not shown). Note that higher molecular weight forms of pp29/30 are detectable in anti-PTYR immunoprecipitates prepared from Pervanadate-treated but not from unstimulated HPB-ALL cells (Fig. 3 A, PTYR lanes). This suggests that pp29/30 represents a potential substrate for PTKs and that its electrophoretic mobility is influenced by its phosphorylation status (see also Fig. 9).
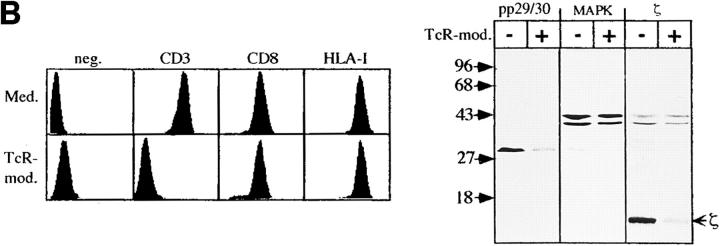
Figure 3.
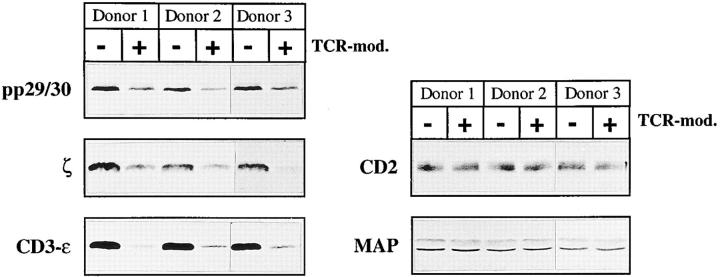
pp29/30 coprecipitates and comodulates with the TCR–CD3–ζ complex. (A) Immunoprecipitates using mAbs directed at the surface molecules depicted (top of lanes) were prepared from 3 × 107 HPB-ALL cells lysed in buffer containing 1% Digitonin. After reducing 14% SDS-PAGE, proteins were blotted onto nitrocellulose, and coprecipitation of pp29/30 was assessed by Western blotting. The first two lanes indicate that pp29/30 is precipitated by an anti-PTYR mAb (PY72) from Pervanadate-treated cells. (B) PBLCPCS cells were incubated overnight with either culture medium or a 1:2 dilution of culture supernatant of CD3-ε mAb 2Ad2A2 (IgM). Expression of CD3-ε (OKT-3), CD8 (AICD8.1), and HLA class I (W6/32) molecules was subsequently determined by indirect immunofluorescence (left). Lysates corresponding to 5 × 105 nonmodulated (Med.) or modulated (TcR-mod.) cells were separated on reducing 14% SDS-PAGE and blotted onto nitrocellulose (right). Western blot analysis was then performed by consecutive incubation of the nitrocellulose membrane with the Abs depicted (top of lanes). MAPK, MAP kinase.
Figure 9.
Tyrosine phosphorylation of TRIM after activation of HPB-ALL cells. (Top) Analysis of TRIM immunoprecipitates prepared from 6 × 107 unstimulated (A and C) or short-term activated (2 min CD3 × CD4 co-cross-linking) HPB-ALL cells (B and D) on two-dimensional IEF/ SDS-PAGE. The immunoprecipitates were first analyzed using anti-PTYR Western blotting (A and B). Membranes were then stripped and incubated with TRIM antiserum (C and D). (Bottom) 3 × 107 HPB-ALL cells were incubated for 10 min at 37°C with a mixture of biotinylated CD3 (OKT-3) and CD4 (AICD4.1) mAbs. Cells were then stimulated for the indicated periods of time by the addition of an appropriate volume of avidin. Lysates corresponding to 5 × 105 cells were analyzed by anti-TRIM Western blotting (right). The remaining lysates were subjected to TRIM immunoprecipitation (IP), resolved on reducing 12% SDS-PAGE, and blotted with anti-PTYR mAb 4G10 (left).
To further substantiate the association between pp29/30 and the TCR–CD3–ζ complex, we analyzed the levels of expression of pp29/30 in untreated PBLCPCS cells and in PBLCPCS cells in which the TCR–CD3–ζ complex had been modulated after overnight incubation with a CD3-ε mAb of the IgM isotype. NP-40 lysates of untreated and TCR-modulated PBLCPCS cells were separated by reducing 14% SDS-PAGE, and were analyzed for expression of pp29/30 using affinity-purified anti-pp29/30 antiserum. Fig. 3 B demonstrates that expression of pp29/30 is strongly impaired in TCR-modulated PBLCPCS compared with untreated cells. Densitometric analysis of the bands corresponding to pp29/30 in untreated versus TCR-modulated cells revealed a 68% reduction of expression in the TCR-modulated state. The extent of pp29/30 loss after modulation of the TCR is similar to that for the ζ chains, which showed a 77% reduction in expression in TCR-modulated cells. In contrast, expression of MAP kinase, CD8, and HLA class I molecules was not altered after modulation of the TCR. A similar downregulation of pp29/30 and ζ as shown for the transformed cell line PBLCPCS was found in TCR-modulated T lymphocytes. In three independent experiments, we observed a 69% average reduction in expression of pp29/30 and ζ after modulation of the TCR–CD3 complex, whereas expression of CD2 and MAP kinase (Fig. 4), as well as of the CD4, CD8, CD45, and HLA class I cell surface molecules, remained unaffected (not shown). In summary, the data shown in Figs. 3 and 4 demonstrate that pp29/30 represents a novel molecule that associates and comodulates with the TCR–CD3–ζ complex in T lymphocytes. Therefore, we termed the protein T cell receptor interacting molecule (TRIM).
Figure 4.
Comodulation of pp29/30 with the TCR–CD3–ζ complex in T lymphocytes. Postnuclear lysates corresponding to 5 × 106 untreated or TCR-modulated (TCR-mod.) T lymphocytes of three individual donors were separated on reducing 14% SDS-PAGE. Western blot analysis was performed by consecutively incubating the nitrocellulose membrane with the indicated Abs.
70% of Cellular TRIM Associate with the TCR–CD3–ζ Complex in PBLCPCS Cells.
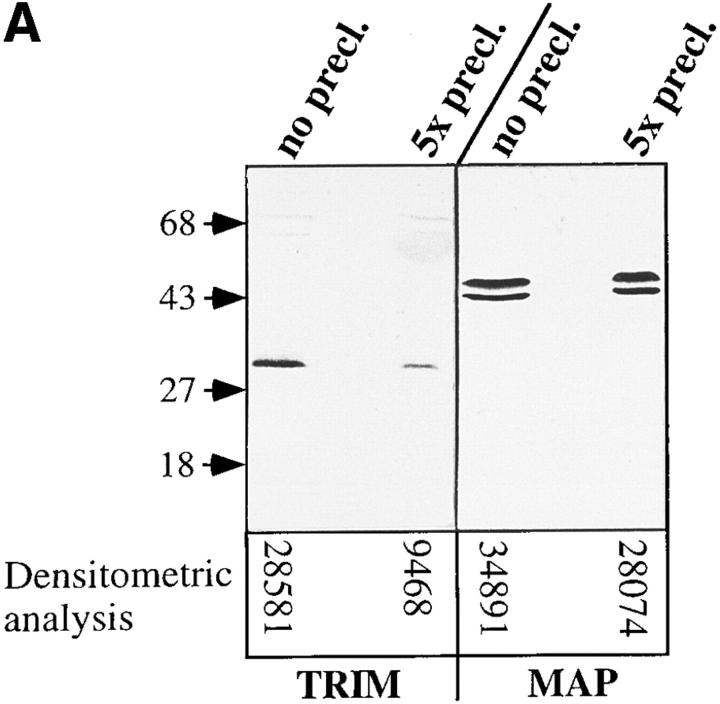
To investigate the stoichiometry of the interaction between TRIM and the TCR– CD3 complex, preclearing experiments were performed. To this end, PBLCPCS cells were lysed in Digitonin and subjected to five consecutive rounds of immunoprecipitation using an Ab directed at CD3-ε. Lysates before and after the fifth preclearing step were separated on reducing 14% SDS-PAGE and analyzed for the presence of TRIM by Western blotting. Densitometric analysis of the TRIM-specific protein bands shown in Fig. 5 A, left, demonstrates that depletion of CD3 molecules results in a 67% reduction of detectable TRIM in cell lysates. In contrast, the amounts of MAP kinase are only slightly reduced after preclearing of CD3-ε. That all CD3-associated TRIM molecules were successfully depleted by this experimental approach was evident from the observation that the CD3 immunoprecipitate corresponding to the third preclearing step was already devoid of any detectable TRIM (and ζ) (Fig. 5 B). Thus, ∼60–70% of all cellular TRIM associate with the TCR– CD3–ζ complex in PBLCPCS cells, in agreement with the comodulation data shown above (Figs. 3 and 4).
Figure 5.
70% of pp29/30 associates with the TCR–CD3–ζ complex in PBLCPCS cells. Digitonin lysates corresponding to 3 × 107 PBLCPCS cells were consecutively incubated five times with 50 μl of packed CNBr-Sepharose beads coupled with purified OKT-3. (A) Lysates corresponding to 5 × 105 cells before the first (no precl.) and after the fifth (5x precl.) preclearing step were separated by reducing 14% SDS-PAGE and analyzed by anti-pp29/30 and anti–MAP kinase Western blotting. The densitometric analysis of the individual lanes is indicated. (B) Immunoprecipitates corresponding to the first (no precl.), the third (3x precl.), and the fifth (5x precl.) preclearing were blotted onto nitrocellulose and analyzed for coprecipitation of TRIM and ζ chains by Western blotting. The protein bands migrating at 25 and 50 kD (right) represent the heavy and light chains of the precipitating mouse mAb.
The Association between TRIM and Components of the TCR– CD3–ζ Complex Occurs at the Cell Membrane.
To obtain information regarding the subcellular localization of TRIM, membrane and cytosolic fractions of HPB-ALL cells were analyzed for the presence of TRIM and, as a control, MAP kinase by Western blotting. As shown in Fig. 6 A, TRIM is exclusively detectable in the membrane fraction, whereas MAP kinase is only found in the cytosol. Thus, TRIM clearly represents a polypeptide associated with cellular membranes. To address whether TRIM associates with the TCR at the plasma membrane or exclusively intracellularly (as described for the TCR–CD3-associated CD3-ω chain [19–22]), a reprecipitation experiment was performed. To this end, wild-type Jurkat cells and HPB-ALL cells were incubated on ice for 30 min with a CD3-ε mAb of the IgG2a isotype. Cells were then washed with ice-cold buffer (to remove all unbound CD3 mAbs) and lysed in Digitonin. After depletion of the nuclei, the preformed immune complexes were collected using protein A–Sepharose and subjected to in vitro kinase assay. The in vitro–labeled proteins were subsequently released from the primary immunoprecipitates using SDS-containing buffer and subjected to a second round of immunoprecipitation using CD3-ε mAb, ζ mAb, and TRIM antiserum, respectively. These secondary immunoprecipitates were separated on reducing 14% SDS-PAGE and analyzed by autoradiography. As a positive control, we used HPB-ALL cells that were first lysed in Digitonin-containing buffer and then subjected to CD3 immunoprecipitation, whereas the Jurkat variant JRT3-T3.1, which is deficient in expression of the TCR α chain and therefore does not express a CD3 complex on the cell surface (but expresses identical amounts of CD3-ε as wild-type Jurkat cells intracellularly; data not shown), was used as a negative control.
Figure 6.


TRIM associates with the TCR–CD3–ζ complex at the cell membrane. (A) Membrane (lanes 1 and 3) and cytosolic (lanes 2 and 4) fractions were prepared from HPB-ALL cells, separated on reducing 14% SDS-PAGE, and blotted with anti-TRIM polyclonal Ab (lanes 1 and 2). The blot was then stripped and reincubated with anti– MAP kinase Ab (lanes 3 and 4). (B) 3 × 107 intact Jurkat cells (top left), TCR α chain–deficient JRT3-T3.5 cells (top right), or HPB-ALL cells (bottom left) were incubated for 30 min on ice with protein A–purified CD3 mAb (OKT-3). Cells were washed three times with ice-cold TBS to remove unbound Abs and then lysed in 1% Digitonin. Preformed immunocomplexes were collected with protein A–Sepharose and subjected to in vitro kinase assay. In vitro–phosphorylated proteins were released from the primary immunoprecipitates and subjected to reprecipitation using protein A–Sepharose alone (lanes 1 and 5), TRIM antiserum (lanes 2 and 6), anti-ζ mAb (lanes 3 and 7), or CD3-ε mAb (lanes 4 and 8). Reprecipitated proteins were visualized by autoradiography. Bottom right, The same experiment with the exception that cells (HPB-ALL) were first lysed in Digitonin and then subjected to CD3-ε immunoprecipitation. (C) Confocal laser scan microscopy demonstrating plasma membrane localization of TRIM in HPB-ALL cells. The subcellular localization of TRIM was assessed using affinity-purified polyclonal anti-TRIM Ab (Cy3, red). The green fluorescence (Cy2) depicts the localization of vimentin. Original magnifications ×600.
Fig. 6 B, top left and bottom left, clearly demonstrates that TRIM coprecipitates with cell surface–expressed CD3-ε in wild-type Jurkat cells and HPB-ALL cells. This association seems to be identical to the association detectable in HPB-ALL cells that are first lysed and then subjected to CD3 immunoprecipitation (bottom right). No in vitro–labeled components of the TCR–CD3–ζ complex were reprecipitated from JRT3-T3.1 cells (top right) under identical experimental conditions, ruling out the possibility that the externally applied CD3-ε mAb passed the cell membrane. These data indicate that TRIM associates with the TCR–CD3–ζ complex at the plasma membrane.
The subcellular localization of TRIM was further assessed by immunofluorescence analysis (Fig. 6 C) of HPB-ALL cells. To this end, acetone-permeabilized cells were incubated with TRIM antiserum (red) or an Ab directed at the cytoskeleton protein vimentin (green). Confocal laser scan microscopy confirmed the reprecipitation data shown in Fig. 6 B, namely that a fraction of TRIM localizes at the plasma membrane. In addition, the molecule seems to accumulate in a perinuclear region within the cell. Interestingly, a similar subcellular distribution has previously been described for the transmembrane adaptor molecule LAT (1).
Molecular Cloning of TRIM cDNA.
Further analysis of TRIM required molecular cloning of its corresponding cDNA. To this end, cDNA prepared from poly(A)+ RNA of HPB-ALL cells was used as template for touchdown PCR reactions, with fully redundant oligonucleotides deduced from the sequence of the 25–amino acid peptide obtained by tandem mass spectrometry to amplify an internal fragment of TRIM (coding for SQAVEENI). This sequence information was used to clone the rest of the gene by 5′ and 3′ RACE. The amino acid sequence of TRIM is depicted in Fig. 7.
An open reading frame of 558 bp (not shown) codes for a leaderless transmembrane protein with a calculated molecular mass of 21 kD that possesses a short 8–amino acid extracellular domain without potential glycosylation sites, a 19–amino acid transmembrane region lacking charged residues that presumably serves as signal anchor sequence for transmembrane transport (23), and a 159–amino acid cytoplasmic tail. Transient expression of the TRIM cDNA in COS cells resulted in the generation of a protein which exactly comigrates with endogenous TRIM derived from HPB-ALL cells (data not shown, and see Fig. 10). Thus, the 9-kD difference between the calculated molecular mass of TRIM and its apparent molecular mass on SDS-PAGE likely results from posttranslational modifications or insufficient binding of SDS.
The extracellular domain of TRIM contains one cysteine residue (C7) which is most likely responsible for formation of the interchain disulfide bond. In the intracellular portion, there are two potential phosphorylation sites for protein kinase C (S116 and S132) and two potential phosphorylation sites for casein kinase 2 (T50 and T151). Note that S116 is followed by a short stretch of amino acids enriched in positively charged residues (K118–K127).
Importantly, each chain of TRIM possesses eight tyrosine residues in its intracellular portion, at least three of which could be involved in SH2-mediated interactions with other signaling proteins. Thus, Y63 and Y110 are part of YxxL sequences that could bind to the SH2 domains of src-PTKs, whereas Y79 belongs to a YxxM motif which has the potential to bind to the SH2 domain of the 85-kD regulatory subunit of PI3-kinase (24). In summary, the structural analysis suggests that TRIM could be involved in recruitment of SH2 domain–containing signaling proteins to the plasma membrane after TCR-mediated activation of T lymphocytes. Computer-assisted analysis of the TRIM cDNA and amino acid sequence in all available databases revealed no obvious homology to any known gene.
TRIM Is Preferentially Expressed in T Lymphocytes and NK Cells.
Expression of the TRIM gene was determined by Northern blot analysis using a TRIM cDNA probe. The multiple-tissue Northern blots shown in Fig. 8 demonstrate that the TRIM transcript is strongly expressed in thymus and to a lesser extent in spleen, LNs, and PBLs. Note that TRIM mRNA is already strongly expressed in fetal thymus at weeks 17–24 of gestation (not shown). In contrast, it is undetectable in bone marrow and fetal liver, strongly suggesting that expression of TRIM is restricted to the T cell compartment. This assumption was confirmed by anti-TRIM Western blot analysis of cellular lysates prepared from different hematopoietic cell lines. There, TRIM protein was only detectable in T cell lines but not in B cell lines or in the erythroleukemic cell line K562 or the monocytic cell line U937 (not shown). In agreement with our previous data (2), low amounts of TRIM are detectable in NK cells (not shown). Lack of expression of TRIM in B lymphocytes was also confirmed by double staining of histological sections prepared from human tonsils using TRIM antiserum and a mAb directed at the CD20 glycoprotein (not shown). In summary, expression of TRIM is restricted to T and NK cells.
Figure 8.

Northern blot analysis demonstrating expression of TRIM in the hematopoietic system. Northern blot analysis was performed on blots containing poly(A)+ RNA from the indicated tissues. The blots were hybridized with a radiolabeled TRIM cDNA probe encompassing nucleotides 1–764.
Tyrosine Phosphorylation of TRIM in Response to T Cell Activation.
As reported above (Fig. 7), the intracellular portion of TRIM possesses several tyrosine-based amino acid motifs which could recruit SH2 domain–containing proteins to the plasma membrane after T cell activation. One requirement for such a model is that TRIM represents an in vivo substrate for PTKs, becoming phosphorylated on tyrosine residues after triggering of the TCR. To assess this possibility, HPB-ALL cells were either left unstimulated or were activated for increasing periods of time using a combination of biotinylated CD3 and CD4 mAbs which were cross-linked on the cell surface with avidin. A representative anti-PTYR Western blot of anti-TRIM immunoprecipitates prepared from NP-40 lysates of unstimulated or CD3 × CD4–activated HPB-ALL cells is shown in Fig. 9, bottom left. It demonstrates that tyrosine phosphorylation of TRIM is already detectable after 30 s of T cell activation, reaches a maximum after 1 min of stimulation, and then slowly declines within the next 40 min. At all time points, comparable amounts of the TRIM protein are expressed in the cells (bottom right). This rules out the possibility that the differences between resting and activated HPB-ALL cells are due to altered levels of protein expression rather than elevated levels of tyrosine phosphorylation. Note that tyrosine phosphorylation of TRIM after T cell activation was also observed in freshly prepared human T lymphocytes as well as in the Jurkat T cell line stimulated with biotinylated OKT-3 or anti-TCR mAb C305 alone (not shown, and see Fig. 10).
Interestingly, the anti-TRIM Ab precipitates two tyrosine-phosphorylated 29–30-kD bands from activated HPB-ALL cells which migrate with slightly different molecular weights on one-dimensional SDS-PAGE (Fig. 9, bottom left). This could indicate that several of the eight individual tyrosine residues in the intracellular portion of TRIM represent targets for TCR-activated PTKs. That both bands likely correspond to differentially phosphorylated forms of TRIM was concluded from the two-dimensional anti-PTYR Western blot analysis of anti-TRIM immunoprecipitates prepared from short-term (2 min) activated HPB-ALL cells shown in Fig. 9, top, B. There, at least three distinct tyrosine-phosphorylated spots are detected by anti-PTYR mAb. Moreover, reprobing the TRIM immunoprecipitates with anti-TRIM antiserum revealed the appearance of at least two slightly more acidic TRIM spots in the activated state (Fig. 9 D, top) compared with the resting state (C, top). Finally, when anti-PTYR immunoprecipitates prepared from Pervanadate-treated HPB-ALL cells or from CD3 × CD4–activated human T lymphocytes were analyzed by anti-TRIM Western blotting, several differentially tyrosine-phosphorylated forms of TRIM were identified (Fig. 4, and data not shown).
Evidence that TRIM Becomes Phosphorylated by Src-kinases.
The rapid phosphorylation of TRIM on tyrosine residues suggested that a membrane-proximal tyrosine kinase is responsible for its phosphorylation after TCR stimulation. To analyze which tyrosine kinase can phosphorylate TRIM in vivo, TRIM was transiently expressed in COS cells together with vectors coding for fyn, lck, or ZAP70 or with a combination of lck and ZAP70. Lysates of the transfected cells were analyzed by anti-PTYR Western blotting. In parallel, anti-PTYR immunoprecipitates were prepared from the transfected cells and investigated for the presence of tyrosine-phosphorylated TRIM by anti-TRIM Western blot. Fig. 10, A and B, demonstrates that under these experimental conditions, TRIM becomes tyrosine phosphorylated by lck and fyn but not by ZAP70. In addition, concomitant expression of lck and ZAP70 does not induce higher levels of TRIM tyrosine phosphorylation than expression of lck alone. Also, coexpression of TRIM and Syk in COS cells did not result in tyrosine phosphorylation of TRIM (not shown). These data indicate that TRIM could represent a protein that is preferentially phosphorylated by tyrosine kinases of the src family. To further substantiate this assumption, TCR-mediated tyrosine phosphorylation of TRIM was analyzed in wild-type Jurkat cells and in a Jurkat variant lacking expression of both ZAP70 and Syk tyrosine kinases (P116 cells [12]). As shown in Fig. 10 C, TRIM becomes strongly tyrosine phosphorylated in P116 cells after engagement of the TCR or upon Pervanadate stimulation. In addition, preincubation of P116 cells with the recently described inhibitor for src-kinases, PP1 (15), completely abrogates TCR-mediated tyrosine phosphorylation of TRIM. Thus, tyrosine phosphorylation of TRIM can occur independently of expression of members of the Syk family PTKs and can be prevented by an inhibitor with a proposed specificity for src-kinases.
Association of Tyrosine-phosphorylated TRIM with Signaling Molecules.
To assess whether tyrosine-phosphorylated TRIM could interact with other signaling components of T cells, the isolated SH2 domains derived from the src-kinases lyn, fyn, src, and lck, the p85 regulatory subunit of PI3-kinase (p85), ZAP70, Shc, SLP76, Grb2, SHP1, and SHP2 were expressed as GST fusion proteins, coupled to glutathione-Sepharose beads, and incubated with an NP-40 lysate of short-term (2 min CD3 × CD4)-treated HPB-ALL cells. Bound proteins were resolved by SDS-PAGE, transferred onto nitrocellulose, and probed with anti-TRIM antiserum. Fig. 11 A demonstrates that tyrosine-phosphorylated TRIM strongly binds to the isolated SH2 domains of the src-PTKs lyn, fyn, src, and lck. In addition, TRIM binds to the SH2 domains of the p85 regulatory subunit of PI3-kinase. A hardly detectable signal is obtained when Grb2 is used for precipitation, whereas no signal above background levels is detectable with the SH2 domains of ZAP70, Syk, Shc, SLP76, SHP1, and SHP2. In addition, no binding of TRIM to any of the above SH2 domains was observed in nonstimulated cells (not shown). Thus, as predicted from the amino acid sequence analysis, TRIM in its tyrosine-phosphorylated state has the capacity to interact with SH2 domains of particular signaling molecules, namely those of src-PTKs, the p85 regulatory subunit of PI3-kinase, and potentially Grb2.
Figure 11.
Interaction of TRIM with intracellular signaling molecules. (A) 107 HPB-ALL cells per lane were activated for 2 min by cross-linking their CD3 and CD4 molecules using biotinylated Abs. Postnuclear lysates of the stimulated cells were incubated with the recombinant GST–SH2 domain fusion proteins depicted (top of lanes). Precipitates were run on reducing 14% SDS-PAGE and subjected to TRIM Western blotting. PI3K, PI3-kinase. N + C, NH- and COOH-terminal SH2 domains of the corresponding fraction. (B) HPB-ALL cells were either left unstimulated or were stimulated for 2 min by CD3 × CD4 cross-linking as described above. Lysates corresponding to 5 × 105 cells were analyzed by anti-PTYR Western blotting (left), and the remaining material was subjected to TRIM immunoprecipitation (IP). Precipitates were run on reducing 12% SDS-PAGE and subjected to Western blotting using anti-Grb2 or anti-p85 mAbs (right). Note that the Grb2 blot had to be overexposed (five times longer compared with the p85 blot) to visualize coprecipitation of Grb2.
To analyze whether tyrosine-phosphorylated TRIM associates with one of the above identified signaling molecules after T cell activation in vivo, anti-TRIM immunoprecipitates were obtained from resting or short-term activated HPB-ALL cells and investigated further by Western blotting using monoclonal or polyclonal Abs directed at p56lck, p59fyn, p85, and Grb2. As shown in Fig. 11 B, TRIM Ab coprecipitates the 85-kD subunit of PI3-kinase after T cell activation. Moreover, low amounts of Grb2 were observed in TRIM immunoprecipitates prepared from either nonstimulated or TCR-activated HPB-ALL cells, whereas we were unable to detect lck or fyn in TRIM immunoprecipitates obtained from either resting or activated lymphocytes (not shown). Collectively, these data demonstrate that TRIM possesses the capacity to recruit at least the regulatory p85 subunit of PI3-kinase to the cell surface after triggering of the TCR.
The Interaction of TRIM with p85 Is Mediated via the YxxM Motif of TRIM.
To investigate whether the interaction between TRIM and p85 involves one of the three tyrosine-based signaling motifs of TRIM, we transiently transfected Jurkat cells with chimeric TRIM molecules that were either composed of the complete wild-type TRIM molecule linked to the extracellular domain of the A2.1 allele of the human HLA class I molecule (chimera_F), or a mutant thereof in which the tyrosine residues of the Y63xxL, the Y79xxM, and the Y110xxL motifs of TRIM were mutated to phenylalanine (chimera_F,mut.15). After overnight incubation, the transfection efficiency was first assessed by indirect immunofluorescence (Fig. 12 B, left). Then one half of the transfected cells was left unstimulated, and the other half was incubated for 2 min with Pervanadate. Subsequently, anti-A2.1 immunoprecipitates were prepared from NP-40 lysates of the transfectants and subjected to Western blot analysis using PI3-kinase Abs. As a positive control, we used a Jurkat variant that was stably transfected with chimera_F, whereas a Jurkat variant that stably expresses a wild-type A2.1 molecule (Pete 2.1 cell line [11]) served as negative control. Fig. 12 B, top right, demonstrates that p85 coprecipitates with the wild-type chimera but not with the TRIM chimera in which tyrosines 63, 79, and 110 were mutated to phenylalanine.
To further identify the site within TRIM that is responsible for its binding to p85, A2.1/TRIM chimeras were transiently expressed in Jurkat cells in which either both Y63 and Y110 (c_F,mut9.4) or Y79 alone (c_F,mut12.1) were mutated to phenylalanine (Fig. 12 A). After Pervanadate treatment of the transfectants, coprecipitation of PI3-kinase with the chimeric molecules was assessed. Fig. 12 B, bottom right, depicts that p85 binds to c_F,mut9.4 but not to c_F,mut12.1. This indicates that the interaction between TRIM and p85 depends on the presence of the YxxM motif within the intracytoplasmic tail of TRIM.
Discussion
Signals initiated by binding of MHC-associated peptides to the TCR are transduced to the intracellular environment via the TCR-associated CD3 molecules and the ζ chains (25–27). Here we describe the molecular cloning and biochemical characterization of a novel TCR-associated protein, termed TRIM, which likely participates in TCR-mediated signal transduction.
The molecular cloning of TRIM was unusually difficult for a number of reasons. First, the amounts of pp29/30 protein that could be purified from HPB-ALL cells were rather low compared with other proteins we have purified recently (e.g., lymphocyte phosphatase–associated protein [LPAP], src kinase–associated protein of 55 kD [SKAP55], and SKAP55 homologue [references 17 and 28, and Marie-Cardine, A., A.M. Verhagen, C. Eckerskorn, and B. Schraven, manuscript submitted for publication]). Second, only separation of the CD3 immunoprecipitates on nonreducing/reducing SDS-PAGE allowed the identification of TRIM as a Coomassie blue–stained protein; this was not possible when the samples were analyzed on two-dimensional IEF/ SDS-PAGE (not shown). This is most likely due to the fact that tyrosine-phosphorylated TRIM migrates at a different position in IEF than nonphosphorylated TRIM (see, for example, Fig. 9).
The lower resolution of two-dimensional nonreducing/ reducing SDS-PAGE compared with IEF/SDS-PAGE could be responsible for the fact that the preparation of pp29/30 protein turned out to be composed of a mixture of at least five polypeptides (three caseins, gp30/40, and TRIM). Third, of the two tandem mass spectrometry– derived peptides finally found to belong to TRIM (Figs. 1 and 7), only the 25–amino acid tryptic peptide was suitable for designing oligonucleotides that could be used in reverse transcription PCR reactions. Peptides of this size lead to complicated mass spectra and ambiguities in sequence interpretation (Fig. 1). In the case of TRIM, even though this was the only suitable peptide in a background of at least four other proteins, successful cloning was still possible. Three peptides covering altogether 56 amino acids were sequenced by tandem mass spectrometry and were retrospectively matched to the novel protein. Thus, undoubtedly, the TRIM molecule described in this paper is identical to the previously identified pp29/30 polypeptide that associates with the TCR (2, 29). As we have demonstrated recently, “de novo” sequencing of large tryptic peptides will now be much facilitated by a novel hybrid quadrupole-time-of-flight tandem mass spectrometer that was not available to us for this investigation (30).
On SDS-PAGE, TRIM migrates with an apparent molecular mass of 29–30 kD (Fig. 2), which differs from the calculated mass of 21 kD. This could be the result of posttranslational modifications of TRIM, or might reflect insufficient binding of SDS, which has been reported for some proteins with acidic isoelectric points. The possibility that the amino acid sequence reported here is not complete can be excluded, since transfection of a TRIM cDNA into COS cells resulted in the expression of a protein that migrates at the same position as endogenous TRIM on SDS-PAGE (not shown, and Fig. 10).
TRIM is a disulfide-linked dimeric transmembrane protein which is exclusively expressed in T lymphocytes and NK cells. It possesses a short extracellular domain of 8 amino acids, a 19–amino acid transmembrane region, and a 159–amino acid intracytoplasmic tail (Fig. 7). With regard to its expression and its overall structure, TRIM shares some characteristics with the ζ chains. Thus, both proteins are disulfide-linked dimers with very short extracellular domains. Moreover, both molecules are exclusively expressed in T lymphocytes and NK cells. The most significant structural difference between TRIM and ζ is that TRIM, even though it contains several consensus tyrosine-phosphorylation motifs, does not possess immunoreceptor tyrosine–based activation motifs (Fig. 7).
TRIM also shares structural homology with the recently described LAT protein (1). Both molecules are type III transmembrane molecules that have only a very short extracellular domain and possess a number of potential tyrosine-phosphorylation sites in their intracytoplasmic tails that could mediate binding to SH2 domains of intracellular signaling molecules. Thus, LAT and TRIM (as well as gp30/40; Klode, J., A. Marie-Cardine, H. Kirchgessner, E. Bruyns, A. Shevchenko, M. Mann, and B. Schraven, manuscript in preparation) define a novel family of transmembrane molecules that are likely involved in recruitment of intracellular signaling molecules to the plasma membrane after TCR stimulation. We suggest terming this novel family of signaling proteins transmembrane adaptor proteins (TRAPs).
Coprecipitation experiments performed under conditions that preserve the integrity of the TCR–CD3–ζ complex (Fig. 3 A) confirmed our previous assumption that TRIM physically associates with the TCR. The interaction between TRIM and the TCR–CD3–ζ complex was further substantiated by our observation that 60–70% of the TRIM molecules comodulate with the TCR–CD3 complex in T cell lines and PBLs (Fig. 3 B, and Fig. 4). The loss of detectable TRIM in TCR-modulated cells is quantitatively similar to that of ζ. Whether TRIM becomes internalized and degraded in TCR-modulated T cells or whether it is shed from the cell surface in TCR-modulated cells is not yet clear.
The specificity of TRIM comodulation with the TCR was further proven by comparative analysis of the expression levels of CD2, CD8, HLA class I molecules, and MAP kinase, which clearly did not change upon TCR modulation (Fig. 3 B, and Fig. 4). That the majority of TRIM indeed is included in a complex with TCR–CD3–ζ was shown in a preclearing experiment where depletion of CD3-ε resulted in an ∼70% reduction of detectable TRIM in lysates of PBLCPCS cells (Fig. 5). Thus, TRIM represents a transmembrane adaptor molecule that associates with the TCR. The association between TRIM and the TCR occurs at the level of the plasma membrane, as judged from laser scan microscopy and from coprecipitation experiments in which TRIM was found to associate with cell surface–expressed CD3-ε (Fig. 6, B and C). However, these experiments do not exclude the possibility that TRIM already associates with components of the TCR–CD3–ζ complex in the cytoplasm.
The molecular mechanism underlying the interaction between TRIM and the TCR complex has not yet been clarified. The CD3 molecules associate with the TCR via salt bridges formed between oppositely charged amino acids in the transmembrane regions of the individual proteins (31). Since TRIM does not possess charged residues in its transmembrane domain, it has to interact with the TCR– CD3–ζ complex via a different molecular mechanism.
It is necessary to mention that despite the fact that TRIM comodulates with the TCR complex in PBLs, so far we have not been able to demonstrate coprecipitation of TRIM with CD3-ε in Digitonin lysates of such cells. Whether this reflects differences in the architecture of the TCR–CD3–ζ–TRIM complexes in resting versus proliferating cells resulting in different susceptibilities toward detergents has to be investigated in further studies. However, given the high reproducibility of TRIM comodulation with the TCR in T lymphocytes, the lack of TRIM coprecipitation with CD3 does not question its association with the TCR. In this regard, it has recently been demonstrated that the ζ chains associate more loosely with the TCR in thymocytes than in mature T lymphocytes (32, 33). Moreover, in contrast to T cell hybridomas and T cell lines, the turnover of ζ seems to occur independently of the expression of the other components of the TCR in PBLs (34). It is possible that a similar situation holds true for the association between TRIM and the TCR in various types of cells.
Potential structural differences in the architecture of the TCR–CD3–ζ–TRIM complex could also be important in light of the fact that TRIM is expressed at all stages of T cell development (ranging from fetal thymocytes of week 14–17 of embryonic development, to adult thymocytes and mature T cells). If the association between TRIM and the TCR becomes altered during T cell development and/or T cell activation, this could result in qualitatively or quantitatively different T cell responses after engagement of the TCR. Additional studies are required to investigate these possibilities.
TRIM becomes tyrosine phosphorylated within seconds after TCR-mediated T cell activation (Fig. 9). Coexpression studies performed in COS cells revealed that members of the src-kinases such as lck and fyn are capable of phosphorylating TRIM, whereas ZAP70 and Syk had no influence on its tyrosine-phosphorylation status under the same experimental conditions (Fig. 10). Together with the findings that almost identical levels of tyrosine phosphorylation of TRIM as found in wild-type Jurkat cells are inducible in a Jurkat variant lacking expression of both Syk and ZAP70 (P116 cells; Fig. 10), and that the recently described src-kinase inhibitor PP1 (15) completely inhibited TCR-mediated tyrosine phosphorylation of TRIM, our data strongly suggest that TRIM could represent a specific substrate for src-kinases. However, we certainly cannot exclude the possibility that particular tyrosine residues of TRIM become phosphorylated by Syk and/or ZAP70 after T cell activation.
Three out of the eight potential tyrosine residues in the intracytoplasmic tail of TRIM are part of sequence motifs (two YxxL motifs and one YxxM motif [Fig. 7]) which could interact in the tyrosine-phosphorylated state with the SH2 domains of intracellular signaling molecules such as src-kinases and the 85-kD regulatory subunit of PI3-kinase. Indeed, in short-term activated HPB-ALL cells, tyrosine-phosphorylated TRIM is coprecipitated by the isolated SH2 domains of src-kinases and by the SH2 domains of p85 (Fig. 11 A).
Western blot analysis of TRIM immunoprecipitates prepared from TCR-stimulated HPB-ALL cells revealed an association between TRIM and p85 that depends on the activation status of the cells (Fig. 11 B). These data suggest that TRIM is capable of recruiting p85 to the plasma membrane after TCR-mediated activation of T lymphocytes.
Using chimeric constructs composed of TRIM and the extracellular domain of the HLA class I molecule A2.1 (chimera_F), we confirmed that the YxxM motif of TRIM is responsible for its interaction with p85 (Fig. 12). Thus, a wild-type A2.1–TRIM chimera associated with p85 upon Pervanadate treatment of Jurkat cells, whereas both a chimera carrying point mutations in all three tyrosine motifs (chimera_F,mut15) and a chimera in which tyrosine 79 was mutated to phenylalanine (chimera_F,mut12.1) failed to coprecipitate p85 (Fig. 12 B). Our results collectively suggest that the association between TRIM and p85 is direct and involves the tyrosine-phosphorylated YxxM motif of TRIM and the SH2 domains of p85. However, formal proof for this assumption requires coexpression studies in heterologous systems or the use of the yeast two-hybrid system.
In contrast to p85, we could not detect either lck or fyn in TRIM immunoprecipitates obtained from resting or activated T cells, although the isolated SH2 domains of the kinases bound tyrosine-phosphorylated TRIM in vitro. The latter result is not entirely unexpected, since we have recently observed that tyrosine-phosphorylated adaptor proteins like SKAP55 bind to the isolated SH2 domain of lck, whereas they do not associate with the kinase in intact cells (17, 29, 35).
It is noteworthy to mention that Y63 is not only a component of a signaling motif that potentially binds to SH2 domains of src-kinases; it could also mediate binding to the SH2 domain of Grb2 (because of the presence of an asparagine in the +2 position). However, although low amounts of Grb2 consistently coprecipitated with TRIM (see Fig. 11 B, and data not shown), this interaction seems to be of low stoichiometry and is apparently independent of the activation status of the cells. In addition, anti-Grb2 Western blot analysis of A2.1 immunoprecipitates prepared from Jurkat cells transiently transfected with the above A2.1–TRIM chimeras indicated that Grb2 also binds to the triple tyrosine mutant (not shown). This suggests that the Y63xNL motif does not mediate binding between TRIM and Grb2. However, we cannot exclude the possibility that the SH2 domain of Grb2 binds to one of the other tyrosine residues expressed in the intracytoplasmic tail of TRIM, or that Grb2 binds to TRIM via a sequence motif that does not involve its SH2 domain. Further studies are required to assess whether Grb2 represents a second intracellular signaling molecule that binds to TRIM.
The function of TRIM has not yet been defined. Transient overexpression of chimera_F or its mutated versions in Jurkat cells did not influence TCR-mediated induction of the nuclear factor of activated T cells (NFAT) and/or the transcriptional activity of the IL-2 promoter (our unpublished observations). These preliminary data could indicate that TRIM is involved in TCR-mediated signal transduction pathways that do not converge at the level of the IL-2 gene. Nevertheless, our data suggest that TRIM represents a novel TCR-associated molecule capable of connecting the TCR to the intracellular signaling machinery. Identification of the signaling pathways in which TRIM participates will provide novel insights into the membrane proximal signaling processes involved in T cell growth and differentiation.
Acknowledgments
We would like to thank Drs. R. Abraham, A. v. Bonin, S. Courtneidge, S. Fischer, G. Koretzky, U. Moebius, B. Neel, M. Peter, S. Ratnofsky, E. Reinherz, A. da Silva, B. Sefton, E. Vivier, A. Veillette, A. Weiss, and J. Wienands for kindly providing our laboratory with reagents. We also thank Drs. B. Schaefer and J. Braunstein for their help with laser scan microscopy.
Abbreviations used in this paper
- Grb2
growth factor receptor–bound protein 2
- GST
glutathione-S-transferase
- IEF
isoelectric focussing
- LAT
linker for activation of T cells
- MAP
mitogen-activated protein
- PI3-kinase
phosphatidylinositol 3-kinase
- PTK
protein tyrosine kinase
- PTYR
phosphotyrosine
- RACE
rapid amplification of cDNA ends
- SH2
src homology 2
- SKAP55
src kinase–associated phosphoprotein of 55 kD
- TRIM
T cell receptor interacting molecule(s)
Footnotes
A. Marie-Cardine is a recipient of a fellowship from the Training and Mobility of Researchers program of the European Community (ERBFMBICT950472). This work was supported in part by the Deutsche Forschungs Gemeinschaft grants SCHR/533/1-1 and SFB 405/A5 (B. Schraven) and B9 (F. Autschbach). The work in M. Mann's laboratory was supported in part by a generous grant from the German Technology Ministry (BMBF).
M. Mann's present address is Center of Experimental Bioinformatics, Odense University, Staermosegaadsvej 16, DK-5230 Odense M, Denmark.
E. Bruyns, A. Marie-Cardine, and H. Kirchgessner contributed equally to this work.
References
- 1.Zhang W, Sloan-Lancaster J, Kitchen J, Trible RP, Samelson LE. LAT: the ZAP-70 tyrosine kinase substrate that links T cell receptor to cellular activation. Cell. 1998;92:83–92. doi: 10.1016/s0092-8674(00)80901-0. [DOI] [PubMed] [Google Scholar]
- 2.Schraven B, Ratnofsky S, Gaumont Y, Lindegger H, Kirchgessner H, Bruyns E, Moebius U, Meuer SC. Identification of a novel dimeric phosphoprotein (pp29/30) associated with signaling receptors in human T lymphocytes and natural killer cells. J Exp Med. 1994;180:897–906. doi: 10.1084/jem.180.3.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins from silver stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 4.Wilm M, Shevchenko A, Houthaeve T, Breit S, Schweigerer L, Fotsis T, Mann M. Femtomole sequencing of proteins form polyacrylamide gels by nano electrospray mass spectrometry. Nature. 1996;379:466–469. doi: 10.1038/379466a0. [DOI] [PubMed] [Google Scholar]
- 5.Wilm M, Mann M. Analytical properties of the nano electrospray ion source. Anal Chem. 1996;68:1–8. doi: 10.1021/ac9509519. [DOI] [PubMed] [Google Scholar]
- 6.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 7.Mann M, Wilm MS. Error-tolerant identification of peptides in sequence databases by peptide sequence tags. Anal Chem. 1994;66:4390–4399. doi: 10.1021/ac00096a002. [DOI] [PubMed] [Google Scholar]
- 8.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Senger M, Glatting KH, Ritter O, Suhai S. X-HUSAR, an X-based graphical interface for the analysis of genomic sequences. Comput Methods Programs Biomed. 1995;46:131–141. doi: 10.1016/0169-2607(94)01610-r. [DOI] [PubMed] [Google Scholar]
- 10.Mizushima S, Nagata S. pEF-BOS, a powerful mammalian expression vector. Nucleic Acids Res. 1990;18:5322–5325. doi: 10.1093/nar/18.17.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bruyns E, Kirchgessner H, Meuer S, Schraven B. Biochemical analysis of the CD45-p56lckcomplex in Jurkat T cells lacking expression of lymphocyte phosphatase-associated phosphoprotein. Int Immunol. 1998;10:185–194. doi: 10.1093/intimm/10.2.185. [DOI] [PubMed] [Google Scholar]
- 12.Williams BL, Schreiber KL, Zhang W, Wange RL, Samelson LE, Leibson PJ, Abraham RT. Genetic evidence for differential coupling of Syk family kinases to the T-cell receptor: reconstitution studies in a ZAP-70-deficient Jurkat T-cell line. Mol Cell Biol. 1998;18:1388–1399. doi: 10.1128/mcb.18.3.1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bagot M, Echchakir H, Mami-Chouaib F, Charue D, Bernheim A, Chouaib S, Boumsell L, Bensussan A. Isolation of tumor-specific cytotoxic CD4+ and CD4+ CD8dim+ T-cell clones infiltrating a cutaneous T cell lymphoma. Blood. 1998;91:4331–4341. [PubMed] [Google Scholar]
- 14.Schraven B, Roux M, Hutmacher B, Meuer SC. Triggering of the alternative pathway of human T cell activation involves members of the T200 family of glycoproteins. Eur J Immunol. 1989;19:397–403. doi: 10.1002/eji.1830190226. [DOI] [PubMed] [Google Scholar]
- 15.Hanke JH, Gardner JP, Dow RL, Changelian PS, Brisette WH, Weringer EJ, Pollok BA, Connelly PA. Discovery of a novel, potent, and src-family-selective tyrosine kinase inhibitor. J Biol Chem. 1996;271:695–701. doi: 10.1074/jbc.271.2.695. [DOI] [PubMed] [Google Scholar]
- 16.Marie-Cardine A, Kirchgessner H, Eckerskorn C, Meuer SC, Schraven B. Human T lymphocyte activation induces tyrosine phosphorylation of α-tubulin and its association with the SH2 domain of the p59fyn protein tyrosine kinase. Eur J Immunol. 1995;25:3290–3297. doi: 10.1002/eji.1830251214. [DOI] [PubMed] [Google Scholar]
- 17.Marie-Cardine A, Bruyns E, Verhagen AM, Eckerskorn C, Kirchgessner H, Meuer SC, Schraven B. Molecular cloning of SKAP55, a novel protein that associates with the protein tyrosine kinase p59fyn in human T-lymphocytes. J Biol Chem. 1997;272:16077–16080. doi: 10.1074/jbc.272.26.16077. [DOI] [PubMed] [Google Scholar]
- 18.Shevchenko A, Wilm M, Mann M. Peptide sequencing by mass spectrometry for homology searches and cloning of genes. J Protein Chem. 1997;16:481–490. doi: 10.1023/a:1026361427575. [DOI] [PubMed] [Google Scholar]
- 19.Pettey CL, Alarcon B, Malin R, Weinberg K, Terhorst C. T3-p28 is a protein associated with the delta and epsilon chains of the T cell receptor-T3 antigen complex during biosynthesis. J Biol Chem. 1987;262:4854–4859. [PubMed] [Google Scholar]
- 20.Alarcon B, Berkhout B, Breitmeyer J, Terhorst C. Assembly of the human T cell receptor-CD3 complex takes place in the endoplasmic reticulum and involves intermediary complexes between the CD3-γ.δ.ε core and single T cell receptor α or β chains. J Biol Chem. 1988;263:2953–2961. [PubMed] [Google Scholar]
- 21.Bonifacino JS, Lippincott-Schwartz J, Chen C, Antusch D, Samelson LE, Klausner RD. Association and dissociation of the murine T cell receptor associated protein (TRAP). Early events in the biosynthesis of a multisubunit receptor. J Biol Chem. 1988;263:8965–8971. [PubMed] [Google Scholar]
- 22.Neisig A, Vangsted A, Zeuthen J, Geisler C. Assembly of the T-cell antigen receptor. Participation of the CD3 ω chain. J Immunol. 1993;151:870–879. [PubMed] [Google Scholar]
- 23.Andrews DW, Johnson AE. The translocon: more than a hole in the ER membrane. TIBS (Trends Biochem Sci) 1996;21:365–369. [PubMed] [Google Scholar]
- 24.Songyang Z, Shoelson SE, Chaudhuri M, Gish G, Pawson T, Haser WG, King F, Roberts T, Ratnofsky S, Lechleider RJ, et al. SH2 domains recognize specific phosphopeptide sequences. Cell. 1993;72:767–778. doi: 10.1016/0092-8674(93)90404-e. [DOI] [PubMed] [Google Scholar]
- 25.Irving BA, Weiss A. The cytoplasmic domain of the T cell receptor zeta chain is sufficient to couple to receptor-associated signal transduction pathways. Cell. 1991;64:891–901. doi: 10.1016/0092-8674(91)90314-o. [DOI] [PubMed] [Google Scholar]
- 26.Wegener A-MK, Letourneur F, Hoeveler A, Brocker T, Luton F, Malissen B. The T cell receptor/CD3 complex is composed of at least two autonomous transduction modules. Cell. 1992;68:83–95. doi: 10.1016/0092-8674(92)90208-t. [DOI] [PubMed] [Google Scholar]
- 27.Letourneur F, Klausner RD. Activation of T cells by a tyrosine kinase activation domain in the cytoplasmic tail of CD3 ε. Science. 1992;255:79–82. doi: 10.1126/science.1532456. [DOI] [PubMed] [Google Scholar]
- 28.Schraven B, Schoenhaut D, Bruyns E, Koretzky G, Eckerskorn C, Wallich R, Kirchgessner H, Sakorafas P, Labkovsky B, Ratnofsky S, Meuer S. LPAP, a novel 32-kDa phosphoprotein that interacts with CD45 in human lymphocytes. J Biol Chem. 1994;269:29102–29111. [PubMed] [Google Scholar]
- 29.Schraven, B., E. Bruyns, H. Kirchgessner, S. Meuer, and A. Marie-Cardine. 1997. Identification and molecular characterisation of novel phosphoproteins associated with signaling receptor complexes in human T-lymphocytes. In Kinases and Phosphatases in Lymphocyte and Neuronal Signaling. H. Yakura, editor. Springer-Verlag, Berlin. 100–111.
- 30.Shevchenko A, Chernushevich I, Standing KG, Thompson B, Wilm M, Mann M. Rapid “de novo” peptide sequencing by a combination of nanoelectrospray, isotopic labeling and a quadrupole/time-of-flight mass spectrometer. Rapid Commun Mass Spectrom. 1997;11:1015–1024. doi: 10.1002/(SICI)1097-0231(19970615)11:9<1015::AID-RCM958>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 31.Klausner RD, Lippincott-Schwartz J, Bonifacino JS. The T cell antigen receptor: insights into organelle biology. Annu Rev Cell Biol. 1990;6:403–431. doi: 10.1146/annurev.cb.06.110190.002155. [DOI] [PubMed] [Google Scholar]
- 32.Kuwabara I, Ohno H, Punt JA, Hashimoto Y, Saito T. Transition from TCR-beta dimer to TCR-alpha beta-expressing cells by introduction of an alpha-chain in an immature thymocyte cell line. J Immunol. 1994;152:2148–2156. [PubMed] [Google Scholar]
- 33.Takase K, Wakizaka K, von Boehmer H, Wada I, Moriya S, Saito T. A new 12 kilodalton dimer associated with pre-TCR complex and clonotype-independent CD3 complex on immature thymocytes. J Immunol. 1997;159:741–747. [PubMed] [Google Scholar]
- 34.Ono S, Ohno H, Saito T. Rapid turnover of the CD3 zeta chain independent of the TCR-CD3 complex in normal T cells. Immunity. 1995;2:639–644. doi: 10.1016/1074-7613(95)90008-x. [DOI] [PubMed] [Google Scholar]
- 35.Schraven B, Marie-Cardine A, Koretzky G. Molecular analysis of the fyn complex: cloning of SKAP55 and SLAP-130, two novel adaptor proteins which associate with fyn and may participate in the regulation of T cell receptor-mediated signaling. Immunol Lett. 1997;57:165–169. doi: 10.1016/s0165-2478(97)00053-9. [DOI] [PubMed] [Google Scholar]