Abstract
Genes were isolated using the suppression subtractive hybridization method by stimulation of pro/pre B cells with anti-CD40 and interleukin (IL)-4 to mature Sμ-Sε–switched cells. One of the strongly upregulated genes encodes a novel murine CC chemokine we have named ABCD-1. The ABCD-1 gene has three exons separated by 1.2- and 2.7-kb introns. It gives rise to a 2.2-kb transcript containing an open reading frame of 276 nucleotides. Two polyadenylation sites are used, giving rise to cDNAs with either 1550 or 1850 bp of 3′ untranslated regions. The open reading frame encodes a 24 amino acid–long leader peptide and a 68 amino acid–long mature protein with a predicted molecular mass of 7.8 kD. ABCD-1 mRNA is found in highest quantities in activated splenic B lymphocytes and dendritic cells. Little chemokine mRNA is present in lung, in unstimulated splenic cells, in thymocytes, and in lymph node cells. No ABCD-1 mRNA is detected in bone marrow, liver, kidney, or brain, in peritoneal exudate cells as well as in the majority of all unstimulated B lineage cells tested. It is also undetectable in Concanavalin A–activated/IL-2–restimulated splenic T cells, and in bone marrow–derived IL-2–induced natural killer cells and IL-3–activated macrophages. Recombinant ABCD-1 revealed a concentration-dependent and specific migration of activated splenic T lymphoblasts in chemotaxis assays. FACS® analyses of migrated cells showed no preferential difference in migration of CD4+ versus CD8+ T cell blasts. Murine as well as human T cells responded to ABCD-1. Freshly isolated cells from bone marrow, thymus, spleen, and lymph node, IL-2–activated NK cells, and LPS-stimulated splenic cells, all did not show any chemotactic response. Thus, ABCD-1 is the first chemokine produced in large amounts by activated B cells and acting selectively on activated T lymphocytes. Therefore, ABCD-1 is expected to play an important role in the collaboration of dendritic cells and B lymphocytes with T cells in immune responses.
Keywords: novel chemokine, activated B cells, dendritic cells, chemotaxis, activated T cells
When a T cell–dependent humoral immune response is initiated in a secondary lymphoid organ like the spleen, T and B cells get near to each other in areas which are rich in interdigitating dendritic cells (DC)1 capable of presenting antigen (1–4). While the majority of the clonally expanded B cells initially differentiate into IgM-producing plasma cells, a small number of them migrate into those areas of the spleen in which follicular dendritic cells (FDC) are localized. There they proliferate, hypermutate the variable (V)-region portions of their Ig H- and L-chain genes, and initiate Ig H-chain class switching. The extensive proliferation of B cells and their maturation lead to the formation of germinal centers at the sites of the follicles.
Th cells are essential for the generation of germinal centers, for somatic hypermutation of Ig genes, for Ig H-chain class switching, and for memory B cell formation (2, 5–9). One of the mandatory interactions leading to a germinal center reaction is that of the CD40 ligand on Th cells with CD40 on B cells (10, 11). The stimulatory strength of this interaction can be seen in tissue culture, when either soluble CD40 ligand or CD40-specific mAbs activate B cells to polyclonal proliferation and Ig secretion (for a review, see reference 12). When IL-4 is added, switching to IgG1 and IgE production can be observed (for a review, see reference 13). Such anti-CD40 plus IL-4–induced Sμ-Sε switching can even be observed in vitro in half or more of the differentiated pro B cells from recombination activating gene (RAG)-2−/− mice (14). Since these pro B cells can be expanded to large numbers by growth on stromal cells in the presence of IL-7 (15), they—and their differentiated, Sμ-Sε–switched counterparts—appeared ideal to construct cDNA libraries. The subtraction of genes expressed in the pro B cells from those expressed in the differentiated, Sμ-Sε–switched cells might lead to the isolation of genes involved in germinal center formation, somatic hypermutation, Ig class switching, and memory formation. This has been attempted in this paper.
Only limited information exists concerning the molecular mechanisms that regulate the selective migration of T and B cells into the areas of a follicle, and their residence at the sites where interactions between T and B cells can take place which generate a germinal center. It is evident that chemokines and their receptors play an important role in the collaboration of T and B cells and in the formation of a germinal center (16).
Chemokines comprise a group of secreted proteins whose best described biological function is to selectively control the migration of certain leukocyte populations to localized sites of inflammatory or immune reactions (16– 19). Chemokines have been classified in four families based on the relative position of their first two of four critical and conserved cysteine residues. In the CXC or α family, the two NH2-terminal cysteine residues are separated by one amino acid; in the CC or β family, they are adjacent; in the C or γ family, only one of the two cysteine residues is conserved (20); and finally, in the CX3C family, the cysteine residues are separated by three amino acids (21). Chemokines act on all different types of cellular targets; however, the CC chemokines tend to act usually on monocytes, T lymphocytes, and in some cases eosinophils, basophils, and mast cells. The repertoire of CC chemokines has expanded rapidly to include macrophage-inflammatory protein (MIP)-1α, -1β, -3α, also reported as exodus or liver and activation– regulated chemokine (LARC), and -3β or EBV-induced molecule 1–ligand chemokine (ELC), RANTES (regulated upon activation, normal T cell expressed and secreted), I-309, monocyte chemotactic protein (MCP)-1, -2, -3, and -4, eotaxin, thymus and activation–regulated chemokine (TARC), B lymphocyte chemoattractant (BLC), also reported as B cell– attracting chemokine (BCA)-1 in humans, and DC-CK-1. These molecules are usually 70–100 amino acids long and have 25–70% homology to each other. They act through G protein–coupled seven transmembrane domain receptors (22).
FDC are suspected to be the source of BLC/BCA-1, a chemokine recognized on B cells by the Burkitt's lymphoma receptor (BLR)-1/CXCR5 chemokine receptor (23, 24). BLC/BCA-1 attracts B cells into the follicles, a process that becomes defective in BLR-1−/− mice (16). Antigen-pulsed DC produce DC-CK-1, which attracts naive T cells to be activated near T cell–rich zones of the spleen (25). This, in turn, sets the stage for activated T cells to be attracted into the follicles for collaboration with B cells.
One of the highly expressed genes identified in the subtraction library encodes a novel CC chemokine. This chemokine, named ABCD-1 by us, is expressed by activated B cells and DC, and attracts activated T cells. This study describes the isolation and structural characterization of the gene, its expression pattern, and the chemoattractive targets of its protein. It discusses the possible function of this novel chemokine within a T cell–dependent humoral response of B cells.
Materials and Methods
Mice and Cells.
C57Bl/6 mice or RAG-2–deficient mice, originally obtained from F. Alt (Children's Hospital, Boston, MA), were maintained at the animal facilities of the Basel Institute for Immunology and Umeå University. RAG-2–deficient, bcl-2 transgenic pro B cells (R2BFL) and the normal, bcl-2 transgenic fetal liver pre B cell line (bcl-2-5.8) were established, maintained in culture, and induced to differentiate by anti-CD40 mAb plus IL-4 as described previously (14, 15, 26).
The B lineage cell lines 63-12 (27), 38B9 (28, 29), NFS-5 (30), 70Z/3 (31), A20 (32), WEHI-231 (33), J558 (34), X63 (35), and Sp2/0 (36), the T lineage cell lines BW5147 (37, 38) and EL-4 (38), ST-2 stromal cells (39), and ex vivo cells were cultured in IMDM (GIBCO BRL, Gaithersburg, MD) containing 5 × 10−5 M 2-ME, 1× nonessential amino acids (GIBCO BRL), 0.03% primatone (Quest International, Naarden, The Netherlands), 100 μg/ml kanamycin (GIBCO BRL) (complete medium), 2% FCS (BioConcept, Allschwil, Switzerland). The cytotoxic T cell line CTLL (40) was grown in complete medium, 2% FCS, and 103 U/ml recombinant (r)IL-2. All cytokines used in this study were obtained from an X63 cell line transfected with the appropriate cytokine cDNA (41).
For stimulation, cells were washed three times and cultured in complete medium, 2% FCS, and different stimuli. Anti-CD40 mAb (14) was used at 10 μg/ml; rIL-4 at 100 U/ml; Escherichia coli LPS (a gift from Drs. C. Galanos and O. Luderitz, Max-Planck-Institut, Freiburg, Germany) at 10 μg/ml; and anti-μ (clone M41 [42]) coupled to Sepharose beads at 200 μg/ml.
T lymphoblasts were obtained by culturing splenic cells in complete medium, 10% FCS in the presence of 2.5 μg/ml Con A (Sigma Chemical Co., St. Louis, MO) for 2 d. To remove Con A, cells were incubated on ice for 30 min with 50 mM methyl-α-d-mannopyranoside (Sigma Chemical Co.) and washed once with complete medium, 2% FCS. Cells were restimulated with 103 U/ml rIL-2 for 1 or 2 d. The population obtained after 2-d culture in the presence of rIL-2 was 43% CD4+ single positive, 50% CD8+ single positive, and 7% CD4−CD8− double negative (data not shown).
NK cells were obtained by growing RAG-2–deficient bone marrow cells in complete medium, 2% FCS, and 103 U/ml rIL-2 as described previously (43). After 6 d of culture, proliferating cells were granulous and had the phenotype of NK1.1+, IL-2Rα−, IL-2Rβ+ NK cells (reference 43, and data not shown). At this time point, the cells were harvested and used for RNA isolation or for chemotaxis assays.
Macrophages were obtained by culture of RAG-2–deficient bone marrow cells in complete medium, 2% FCS in the presence of 103 U/ml rIL-3. At the end of the 6-d culture period, the cells that adhered to the plastic culture dish and showed morphology of macrophages were harvested, RNA was extracted, and cDNA was prepared.
Mesenteric LN CD11c+ DC were purified as described elsewhere (44). Maturation was induced in vitro by culture of the cells for 24 h in the presence of 10 μg/ml anti-CD40 mAb or 20 ng/ml GM-CSF and 50 ng/ml TNF-α. Cells were then harvested, total RNA was isolated, and cDNA was prepared.
Human neonatal leukocytes were isolated from cord blood by the standard Ficoll-Paque method. CD4-enriched T cells were obtained by negative sorting with anti-CD8 mAb (clone OKT8), and stimulated with plate-bound anti-CD3 mAb (clone TR66, 5 μg/ml [45]) and anti-CD28 mAb (clone CD28.6, 2 μg/ml; provided by D. Olive, INSERM, Marseille, France) and either with human rIL-12 (2 ng/ml; Hoffmann-La Roche, Nutley, NJ) plus neutralizing mAb to IL-4 (200 ng/ml; PharMingen, San Diego, CA) or with human rIL-4 (200 U/ml; PharMingen) plus neutralizing mAb to human IL-12 (2 μg/ml; R&D Systems, Inc., Minneapolis, MN) in RPMI 1640 medium supplemented with 2 mM l-glutamine, 1% nonessential amino acids, 1% pyruvate, 5 × 10−5 M 2-ME, 50 μg/ml kanamycin, and 5% human serum (Swiss Red Cross, Bern, Switzerland). Human rIL-2 (500 U/ml) was added at day 3. After 1 wk, the cultures were restimulated in the same polarizing conditions and analyzed after 10 d of culture.
Chemotaxis Assay.
Cell migration was evaluated using 5-μm pore filters (Transwell, 24-well cell cluster; Costar Corp., Cambridge, MA). Cells were washed once, adjusted to 3 × 106 cells/ml, and incubated in complete medium, 2% FCS for 10–15 min at 37°C in 10% CO2. 5 × 105 cells were placed onto the Transwell filter which had been preincubated with complete medium, for 10 min. Filters were then transferred to another well containing 500 μl of complete medium and different concentrations of ABCD-1 purified either by immunoaffinity or HighTrap Heparin fast-performance liquid chromatography (FPLC; Pharmacia Biotech AB, Uppsala, Sweden), and incubated at 37°C in 10% CO2. After 3 h, the upper chamber was removed, and the cells in the lower chamber were counted using a Neubauer hemocytometer. The phenotype of the migrated cells was determined by FACS® analysis. Results are expressed as percentage of migrated cells to total cells used in the assay.
Selection of cDNAs.
Total RNA of in vitro–stimulated cells was isolated with the RNAeasy system (QIAGEN GmbH, Hilden, Germany), and poly-A+ mRNA was selected using the Qiaex system according to the instructions of the manufacturer (QIAGEN GmbH). The quality of the isolated RNA was evaluated by Northern blot analysis using formaldehyde gels.
Suppression subtractive hybridization was performed using the PCR-Select kit (Clontech, Palo Alto, CA) essentially as described by Diatchenko et al. (see reference 50). In brief, complementary DNA was synthesized from 4 μg of mRNA either from R2BFL cells growing on stromal cells in the presence of IL-7, or from R2BFL cells stimulated with anti-CD40 mAb plus IL-4 in the absence of IL-7. The cDNAs were digested with RsaI, and two types of adapters, provided by the manufacturer, were independently ligated. First and second hybridizations were performed, and the resulting annealed material was used as PCR template. Routinely, 2 μl of 1:1,000, 1:300, 1:100, and 1:30 diluted hybridization mixture was amplified in 30 primary PCR cycles, with the primers provided by the manufacturer. 15–17 cycles of secondary PCR were done using 2 μl of 1:20, 1:10, and 1:5 diluted primary PCR as template. PCR reactions were performed with the Advantage PCR system (Clontech). PCR products were ligated in the pT7Blue vector (Novagen, Inc., Madison, WI), and sublibraries of potentially subtracted cDNAs were established. This led to the establishment of ∼500 cDNA clones. 120 randomly chosen clones were sequenced.
Sequencing Analyses.
The nucleotide sequence of the isolated cDNA clones was determined using the dye terminator cycle sequencing kit (Perkin-Elmer Corp., Foster City, CA) and running the samples in an automatic sequencer (Applied Biosystems Inc., Foster City, CA), or using super coiled plasmids and conventional sequencing protocols (46). The sequences were analyzed using Genetics Computer Group software (Madison, WI) and search launcher software available on the Internet (National Center for Biotechnical Information, http://www.ncbi.nlm.nih.gov). Expression of potentially interesting cDNA clones was studied in Northern blot analyses and/or by semiquantitative reverse transcription (RT)-PCR using specific primers.
Semiquantitative RT-PCR and Northern Blotting.
Total RNA was extracted using RNAzol B (Biotecx Laboratories, Inc., Houston, TX) according to the manufacturer's instructions. Maximally 1 μg of total RNA was reverse transcribed for 1 h at 37°C. The RT mix contained 50 mM Tris (pH 8.3), 3 mM MgCl2, 75 mM KCl, 10 mM dithiothreitol, 1 mM each dATP, dGTP, dCTP, and dTTP (Pharmacia Biotech AB), 0.2 U pd(N)6 sodium salt (Pharmacia Biotech AB), 40 U RNAse Block Ribonuclease Inhibitor (Stratagene Inc., La Jolla, CA), and 50 U M-MLV reverse transcriptase (GIBCO BRL). The PCR mix consisted of 10 mM Tris (pH 8.3), 1.5 mM MgCl2, 50 mM KCl, 10 mM 2-ME, 0.01% gelatin, 0.2 mM each dATP, dGTP, dCTP, and dTTP (Pharmacia Biotech AB), and 0.5 U Amplitaq DNA polymerase (Roche Diagnostics Systems, Basel, Switzerland).
Primers used for PCR were 5′ and 3′ ABCD-1 (5′-GCCCTCTGGTCATTAGACACCTG-3′ and 5′-TCGTTGGCAAGGCTCTTGCTG-3′), resulting in a 413-bp fragment; 5′ and 3′ RANTES (5′-TCTGCAGCTGCCCTCACCATCATC-3′ and 5′-TGCCCATTTTCCCAGGACCGAGTG-3′), amplifying a 335-bp product; 5′ and 3′ MIP-1α (5′-ATCATGAAGGTCTCCACCAC-3′ and 5′-TCTCAGGCAATCAGTTCCAG-3′), giving rise to a 284-bp fragment; 5′ and 3′ MCP-1 (5′-AAGCCAGCTCTCTCTTCCTC-3′ and 5′-CCTCTCTCTTGAGCTTGGTG-3′), resulting in a 247-bp product; and 5′ and 3′ MDC/ STCP-1 (5′-CTGCACTCCTGGTTGTCCTC-3′ and 5′-ACGGTCATCAGAGTAGGCTC-3′), producing a 297-bp fragment. PCR was performed on a thermal cycler (OmniGene; Hybaid Ltd., Hampton Hill, Middlesex, UK). Serially diluted cDNA (1:5 serial dilutions) was amplified by PCR using the following conditions: 35 cycles of 30 s at 94°C, 20 s at 58°C, and 30 s at 72°C followed by 45 s at 94°C, 90 s at 58°C, and 2 min at 72°C. Parallel PCR reactions were set using primers specific for β-actin in the mouse studies, 5′ and 3′ β-actin (5′-GTGGGAATTCGTCAGAAGGACTCCTATGTG-3′ and 5′-GAAGTCTAGAGCAACATAGCACAGCTTCTC-3′), amplifying a 537-bp product; and G3PDH in the human studies, 5′ and 3′ G3PDH (5′-ACCACAGTCCATGCCATCAC-3′ and 5′-TCCACCACCCTGTTGCTGTA-3′), amplifying a 451-bp product, to normalize for possible differences in the amount of cDNA synthesized. PCR products were separated in 1.2% TAE-agarose gels. The amount of specific mRNA relative to β-actin in the mouse or to G3PDH in the human cDNA, respectively, was estimated in serial 1:5 dilutions of the cDNA analyzed on gels stained with ethidium bromide. The results of this semiquantitative RT-PCR analysis are expressed in Results as relative quantities.
The probe encoding ABCD-1 sequences used for Northern hybridization was a 457-bp fragment containing bases 10–466 of the ABCD-1 cDNA (Fig. 1) labeled with [α-32P]dCTP (Amersham International, Little Chalfont, Bucks, UK) by random priming, using the NEBlot kit (New England Biolabs, Inc., Beverly, MA). Approximately 30 μg of total RNA of tissues or 4 μg of nonstimulated or anti-CD40 plus IL-4–induced R2BFL cells was fractionated on a 1.5% agarose formaldehyde gel, transferred to GeneScreen-plus nylon membrane (DuPont-NEN, Boston, MA), prehybridized for 4 h at 65°C in 0.5 M Na2HPO4/ NaH2PO4 (pH 7.2), 7% SDS, 1 mM EDTA, and hybridized overnight in the same solution with labeled probe. Membranes were washed twice with 40 mM Na2HPO4/NaH2PO4 (pH 7.2), 1% SDS for 30 min at 65°C. Autoradiographs were exposed at −70°C for 6 h for ABCD-1, or for 2 h for β-actin.
Figure 1.

Structure of ABCD-1 mRNA. The putative leader sequence (double-underlined), the critical cysteine residues (bold), the potential heparin binding site at the COOH terminus (underlined), and the two polyadenylation sites that are used in ABCD-1–encoding cDNAs (bold and underlined) are shown. Triangles, Positions where the coding regions are split by introns. The cDNA sequence of ABCD-1 can be found in EMBL/GenBank/DDBJ under accession no. AF052505.
Full-length cDNA Clones.
A cDNA library was constructed using total RNA from stimulated R2BFL cells. Complementary DNA was synthesized with the PCR-based system CAP-finder (Clontech) according to the instructions of the manufacturer and was then subcloned into a λ-ZAP Express vector (Stratagene Inc.). Ligated cDNAs were packaged into λ-phages using Gigapack III Gold packaging extracts (Stratagene Inc.). The library was screened using the D2-2 cDNA fragment, which was shown to contain coding sequences of ABCD-1.
Isolation of Genomic Clones.
To obtain ABCD-1 genomic DNA clones, the 129SVJ mouse genomic library (Stratagene Inc.) was screened using a full-length cDNA according to the instructions of the manufacturer. Positive phage clones were isolated and mapped as described previously (47).
Production of Recombinant ABCD-1.
A 457-bp fragment containing the complete coding region of the ABCD-1 cDNA clone (Fig. 1, bases 10–466) was cloned into the BamHI-SalI site of the expression vector pRmHa-3 (48). 3 μg of plasmid together with 0.3 μg of pBS-hs.PURO plasmid (K. Karjalainen, unpublished data) were cotransfected into SL-3 cells (49). In brief, the two plasmids were dissolved in 100 μl SF 900 II SF medium (GIBCO BRL) supplemented with 0.35 g/liter NaHCO3 (complete insect medium), and then incubated with 100 μl complete insect medium and 10 μl lipofectin (GIBCO BRL) at room temperature for 10 min. 90% confluent layer of SL-3 cells in 1.8 ml complete insect medium was incubated with the 200 ml plasmids/lipofectin mixture at 27°C for 5 h. The medium was then carefully removed and exchanged for 5 ml complete insect medium, 1% FCS (GIBCO BRL). 2 d later, cells were subjected to selection pressure in complete insect medium, 1% FCS, and 10 μg/ml puromycin (Calbiochem Corp., La Jolla, CA). ABCD-1 expression was induced in the transfected SL-3 cells by the addition of 1 mM CuSO4. 4 d later, cells were checked for ABCD-1 expression by RT-PCR using the primer pair FCC1 (5′-CTGGGATCCATGTCTGCTTGGTAGTGA-3′) and UCC1 (5′-CATGTCGACGAAGAATAGGGCTTGCTG-3′), and the conditions described above. Positive cell clones were expanded in complete insect medium, 0.2–1% FCS, and 10 μg/ml puromycin. ABCD-1 was isolated from 5-d–induced culture supernatant by passage through a 0.45-μm pore filter and over a HighTrap Heparin FPLC column (Pharmacia Biotech AB) at 0.5 ml/min. The column was washed with 20 mM Tris (pH 8), 200 mM NaCl and eluted with a linear gradient of 20 mM Tris (pH 8) and increasing 20 mM/min NaCl. Alternatively, ABCD-1 was purified from induced culture supernatant by immunoaffinity using ABCD-1 mAb (see below for production of specific mAb) coupled to CNBr-activated Sepharose (Pharmacia Biotech AB) according to the manufacturer's instructions. Eluted fractions were checked for ABCD-1 by ELISA and fractionated on an 18% acrylamide SDS-PAGE gel using a Mini Trans-Blot Cell (Bio-Rad Laboratories, Hercules, CA). The gel was either silver stained or electroblotted onto a nitrocellulose membrane followed by immunodetection of ABCD-1 by incubation with anti–ABCD-1 mAb (10 μg/ml) overnight and ECL (Amersham International) detection, or onto a polyvinylidene fluoride membrane (Bio-Rad Laboratories) for protein sequencing. The fractionated 7.8-kD protein, transferred to polyvinylidene fluoride membrane, was NH2-terminally sequenced on a protein sequencer (model 494; Applied Biosystems Inc.).
The predicted mature form of ABCD-1 was chemically synthesized at the Basel Institute for Immunology using FastMocTM chemistry on a peptide synthesizer (model 430A; Applied Biosystems Inc.). Lyophilized protein was dissolved at 5 mg/ml in PBS and stored at −70°C.
Production of ABCD-1 mAb.
Lewis rats were immunized with 100 μg of synthetic ABCD-1 emulsified with CFAs. Rats were boosted with 20 μg of chemically synthesized ABCD-1 after 5 wk and then after an additional 3 d. 1 d thereafter, the animals were killed, and the LN cells were fused with Sp2/0 myeloma cells, according to standard protocol.
Results
Development of Subtracted Pools of cDNAs.
We have used the recently described suppression subtractive hybridization method (50) to isolate genes preferentially expressed in anti-CD40 plus IL-4–stimulated cells of the IL-7–dependent pro B cell line R2BFL. This method combines normalization of cDNA abundance and subtraction in a single procedure. We have subtracted our cDNA populations as described in Materials and Methods and amplified by PCR-upregulated sequences. Southern blot analyses of the selected PCR products using the G3PDH-specific probe demonstrated a >1,000-fold elimination of sequences encoding this housekeeping gene (data not shown). The pool of cDNA fragments potentially containing cDNA sequences selectively expressed in the anti-CD40 plus IL-4– differentiated B lineage cells was subcloned into a cloning vector. Screening the sequences of the first 120 clones containing inserts >150 bp in the available databases revealed that 23 of these 120 clones were encoding parts of genes that are already known and could be expected to be upregulated in anti-CD40 and IL-4–differentiated B lineage cells (14). Hence, clones encoding the invariant chain of MHC class II (51), MHC class II molecules (52), the MHC locus gene H2-M (53), several Ig germline transcripts associated with Ig class switching (for a review, see reference 54), CD23 (55), and the IL-4 receptor (56) were found (Table 1). These results were taken as evidence for a successful enrichment of specific cDNA sequences in our subtracted sublibrary.
Table 1.
Gene Fragments Detected in B Lineage Cells Induced by CD40-specific mAb and IL-4
| Gene | No. of independent isolates (of 120 total) | |
|---|---|---|
| Invariant chain MHC class II | 7 | |
| Class II (α and β chains of I-A and I-E) | 5 | |
| H2-M | 2 | |
| Iγ1 germline transcript | 2 | |
| Iε germline transcript | 4 | |
| CD23 | 1 | |
| IL-4 receptor | 2 | |
| ABCD-1 | 10 | |
| Total | 33 |
cDNA Clone D2-2 Encodes a Novel Murine Chemokine.
A high number of cDNA fragments (10/120), including clone D2-2, showed the very same high homology to the coding sequences of the chemokine RANTES (∼59% homology at the nucleotide level in 167 bp, and 38% similarity at the potentially encoded mature protein sequence level [57–59]). This indicated that the gene(s) from which these fragments were derived could encode one or several novel chemokine(s). Using the D2-2 cDNA fragment, full-length cDNA clones were isolated from a library constructed with mRNA from anti-CD40 plus IL-4–stimulated R2BFL cells. Two types of cDNAs were deduced, differing at their 3′ ends. Sequencing analyses demonstrated that this was due to differential usage of two polyadenylation sites separated from each other by ∼300 bases (Fig. 1). The two cDNAs contained a single open reading frame encoding a 92 amino acid–long unprocessed secretory peptide. The critical cysteine residues are organized in a pattern characteristic for the CC or β chemokine subfamily. We named this novel murine CC chemokine ABCD-1.
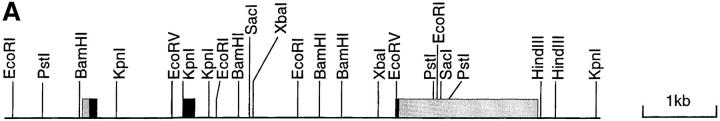
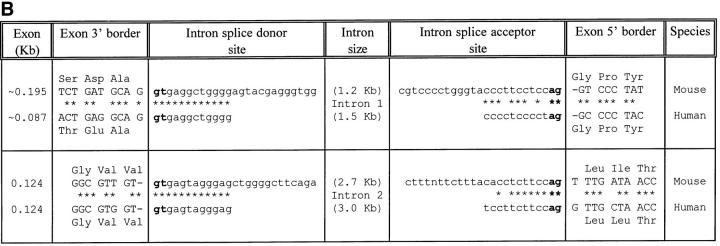
Using the cDNA clone as a probe, λ-phage genomic DNA clones were isolated that contained the entire ABCD-1 chemokine gene. Comparison of the cDNA and genomic sequences revealed that like other CC chemokine genes, three exons encoded the entire cDNA sequences (Fig. 2). The exons are separated by introns of ∼1.2 and 2.7 kb, respectively.
Figure 2.
(A) Schematic representation of the mouse ABCD-1 genetic locus. Boxes, Exons: gray boxes, 5′ and 3′ untranslated regions; black boxes, coding regions. Exon I contains the translation start site; exon II contains the critical CC residues; exon III contains the stop codon and the 3′ untranslated region. (B) Nucleotide sequences of the splice junctions and comparison with the corresponding human sequences (derived from reference 62). Lower case letters, Intron sequences. Upper case letters, Exon sequences.
Knowledge of the complete sequence of the ABCD-1 chemokine gene also made it clear that all 10 cDNA clones with close sequence homologies identified in the original subtractive library originated from the same gene.
mRNA for the Novel ABCD-1 Chemokine Is Produced Primarily by Activated B Cells and DC.
The expression pattern of ABCD-1 at the RNA level was studied by Northern blot hybridization (Fig. 3) and/or by semiquantitative RT-PCR (Fig. 4). All analyses were related to the expression of a ubiquitously expressed gene, i.e., β-actin.
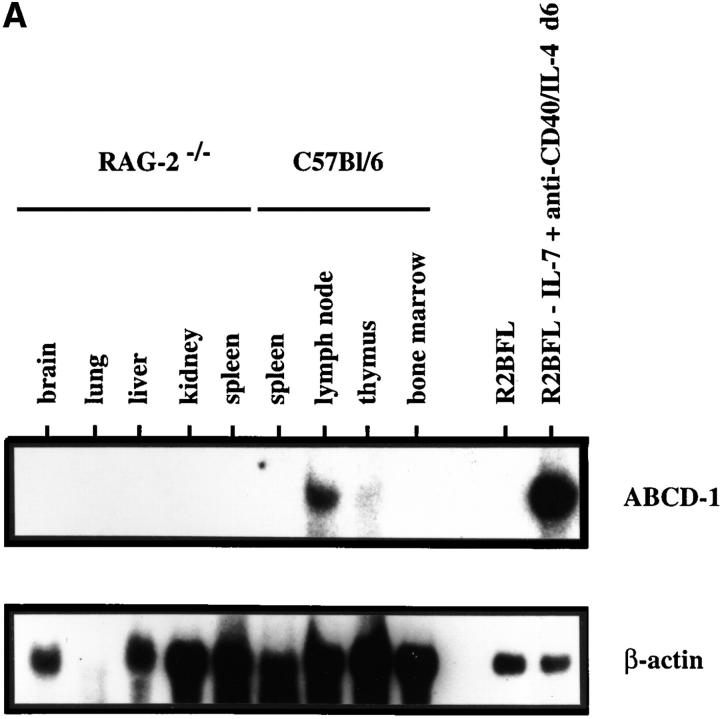
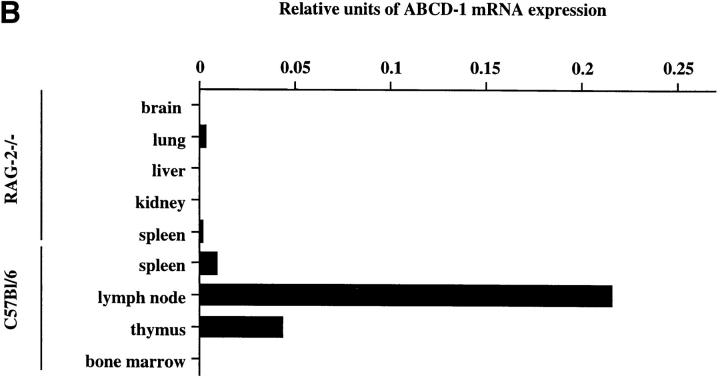
Figure 3.
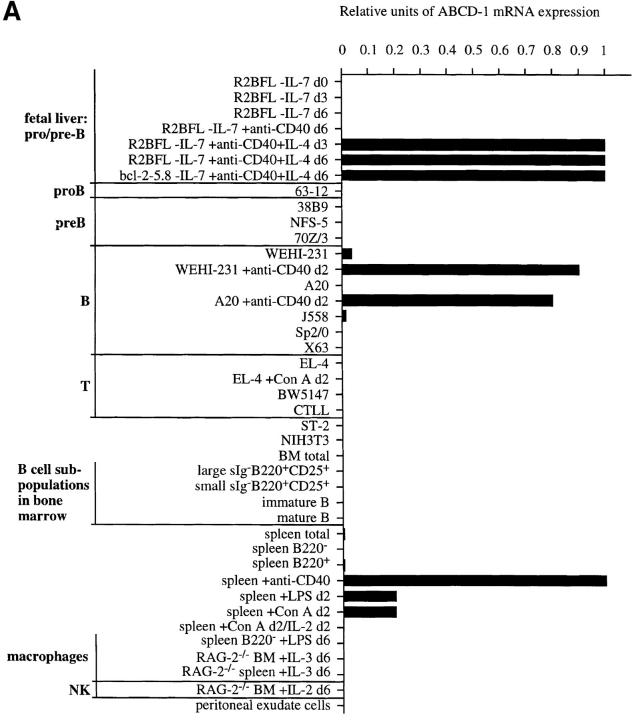
ABCD-1 expression in mouse tissues. (A) Northern blot analyses of ABCD-1 mRNA expression. Approximately 30 μg of total RNA from the indicated cell sources was electrophoresed in a 1.2% agarose formaldehyde gel. After transfer, the filter was hybridized first to a ABCD-1–specific probe and subsequently to a β-actin probe. Because of the very strong signal in RNA from induced R2BFL cells, ∼4 μg of total RNA was loaded in these lanes. (B) Semiquantitative RT-PCR of ABCD-1 mRNA levels. The same RNA samples as in A were reverse transcribed, and serial 1:5 dilutions of the cDNA samples were used as PCR template to amplify ABCD-1 and β-actin. The latter amplification was used for normalization of the results, which are given as relative units of ABCD-1 compared with β-actin mRNA expression levels.
Figure 4.
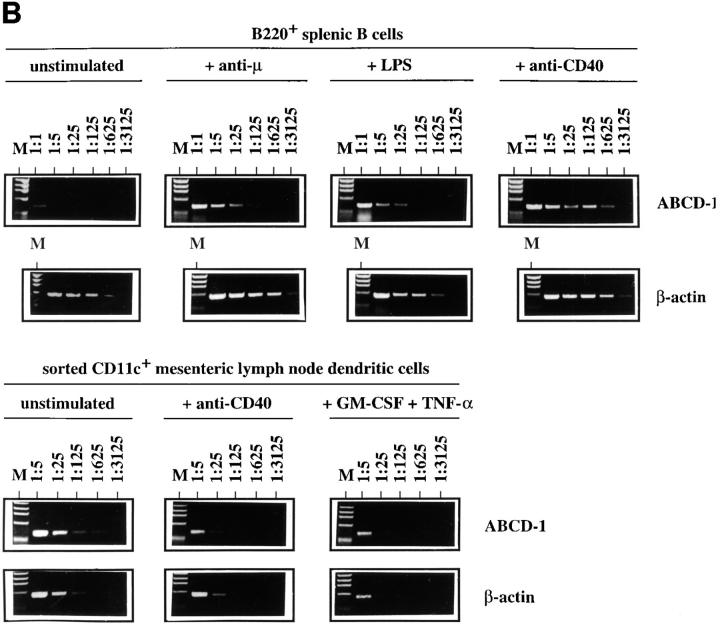
Semiquantitative RT-PCR analyses of ABCD-1 mRNA expression (A) in different cell populations and (B) in sorted splenic B cells (B220+) and purified mesenteric LN DC (CD11c+), cultured in vitro in the presence of the indicated stimuli for 2 d or 24 h, respectively. Total RNA was isolated and reverse transcribed. Serial 1:5 dilutions of cDNA prepared from the indicated cells were used as template to PCR-amplify ABCD-1 and β-actin sequences. The latter amplification was used for normalization of the results. The amount of ABCD-1 relative to β-actin mRNA is shown. BM, Bone marrow. d, Day. M, Marker.
Expression of the ABCD-1 mRNA was below levels of detections (i.e., undetectable after amplification of 50 ng of cDNA) in brain, liver, kidney (all from RAG-2−/− animals), and in total unstimulated bone marrow (Fig. 3). It was also undetectable in purified B lineage precursors of bone marrow as well as in immature or mature surface (s)IgM+ B cells from the bone marrow (Fig. 4 A). Transformed cell lines of comparable states of differentiation, i.e., the pro B cell–like 63-12 line, the pre B-I cell–like 38B9 line, the pre B-II cell–like NFS-5 and 70Z/3 lines, and the mature B cell–like A20 line, also did not express ABCD-1. By contrast, the B cell line WEHI-231 as well as the myeloma cell line J558 did express low but detectable levels. A series of transformed T cell lines (EL-4 and BW5147) and the IL-2–reactive cytotoxic T cell line CTLL, as well as NIH-3T3 fibroblasts, the ST-2 stromal cell line, and the hybridomas Sp2/0 and X63, did not express the ABCD-1 mRNA in detectable amounts. Con A–activated and IL-2– restimulated splenic T lymphocytes, IL-2–activated NK cells, and IL-3–induced macrophages also did not express detectable amounts of ABCD-1 message (Fig. 4 A).
Relatively small amounts of ABCD-1 mRNA were detectable in lung (from RAG-2–deficient mice), unstimulated spleen cells, LN cells, and thymocytes (Fig. 3).
As expected, high levels of ABCD-1 mRNA were detected in R2BFL cells stimulated with anti-CD40 plus IL-4 for either 3 or 6 d in culture (Fig. 4 A). Other pre B-I cell lines stimulated for 3–6 d with anti-CD40 plus IL-4 (like the bcl-2-5.8 line) also showed this high level of chemokine mRNA expression. Since the unstimulated pro B and pre B-I cell lines growing on stromal cells in the presence of IL-7 or differentiating in the absence of IL-7, or upon stimulation with either IL-4 or anti-CD40 alone in the absence of IL-7, did not express ABCD-1 mRNA, expression appears to be induced by the removal of IL-7 and the addition of anti-CD40 plus IL-4 to the cultured cells.
ABCD-1 mRNA expression was also inducible by anti-CD40 plus IL-4, as well as anti-CD40 alone, in B220+ spleen cells (containing in majority mature B cells) and in the transformed B cell lines A20 and WEHI-231 within 2 d of anti-CD40 stimulation (Fig. 4). These results indicate that activated B cells are major producers of the chemokine.
Splenic mature B cells, purified by FACS®, could be activated not only in the CD40-dependent way (i.e., T cell– dependent) but also in other polyclonal ways, i.e., by LPS or by Ig-specific mAbs (i.e., T cell–independent) to high levels of ABCD-1 mRNA expression (Fig. 4 B).
One other likely source for production of ABCD-1 is DC. Freshly sorted, unstimulated CD11c+ mesenteric LN DC expressed similar amounts of chemokine mRNA as did anti-CD40–activated splenic B cells (Fig. 4 B). Stimulation of CD11c+ mesenteric LN DC with either anti-CD40 or GM-CSF plus TNF-α for 24 h did not change mRNA levels of the chemokine.
Differential Expression of the ABCD-1, RANTES, MIP-1α, and MCP-1 Chemokines.
mRNA from R2BFL cells stimulated with anti-CD40 plus IL-4, spleen cells stimulated with anti-CD40 or Con A, freshly isolated cells from bone marrow and thymus, and RAG-2–deficient bone marrow cells cultured in the presence of rIL-3 (primarily macrophages) were analyzed by semiquantitative RT-PCR for the presence of ABCD-1, RANTES, MIP-1α, and MCP-1 mRNA. This analysis demonstrated that anti-CD40–stimulated R2BFL cells produced primarily ABCD-1 mRNA but not the other chemokines. Similarly, spleen cells activated by anti-CD40 produced primarily ABCD-1 and very small amounts of MIP-1α. Con A–activated spleen cells produced ABCD-1 mRNA, but only ∼20% of the amount produced by anti-CD40 stimulation, and small amounts of RANTES and MIP-1α. The ABCD-1 production observed is probably an indirect effect, in the sense that the activated T cells stimulate the B cells which, in turn, produce ABCD-1 (60). RANTES was produced primarily by thymocytes, and MCP-1 by the macrophage preparation (data not shown). These results indicate that anti-CD40 signaling rather selectively upregulates ABCD-1 expression.
Production of Recombinant ABCD-1 in Insect Cells.
The sequences encompassing the open reading frame were used to produce the putative chemokine in SL-3 insect cells (49) as described in Materials and Methods. Recombinant ABCD-1 was purified from pRmHa-3/ABCD-1–transfected SL-3 cell culture supernatants by HighTrap Heparin FPLC, or, alternatively, an mAb raised against a chemically synthesized polypeptide (amino acids 25–92) was used as an affinity reagent fixed on beads to purify the chemokine, and gel fractionation. NH2-terminal sequencing of the band migrating at the expected molecular weight resulted in the sequence GPYGANVEDS, and thus confirmed the computer-predicted NH2 terminus of the mature, 68 amino acid–long, 7.8-kD chemokine.
The Target Cells for the Biological Activity of ABCD-1.
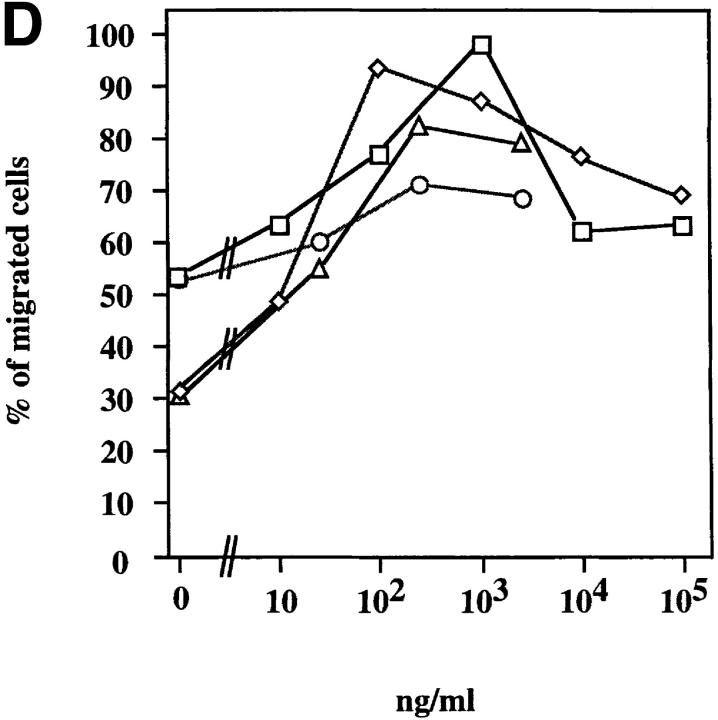
We used a microchamber migration assay to study the chemotactic properties of purified recombinant ABCD-1 on different cell populations. No significant increase in the migratory activity was observed with primary spleen cells, thymocytes, LN cells, or bone marrow cells exposed to ABCD-1 at concentrations ranging from 1 ng/ml to 10 μg/ml (Fig. 5 A). Although LPS-activated splenic B cells, which we showed to produce ABCD-1 mRNA, were not chemoattracted by ABCD-1 (data not shown), Con A–activated IL-2–restimulated splenic T lymphoblasts were (Fig. 5, A and B). Migration was concentration dependent, but inhibited at high doses. FACS® analysis of chemoattracted T cell blasts showed that CD4+ and CD8+ T lymphoblasts were attracted equally (data not shown). IL-2–activated NK1.1+, IL-2Rα−, IL-2Rβ+ NK cells from RAG-2−/− mice did not show any significant migratory response to doses up to 10 μg/ml (Fig. 5 B). ABCD-1–containing culture supernatants from either cDNA-transfected insect cells or R2BFL cells stimulated with anti-CD40 plus IL-4 were also able to induce migration of activated T lymphoblasts (Fig. 5 C).
Figure 5.
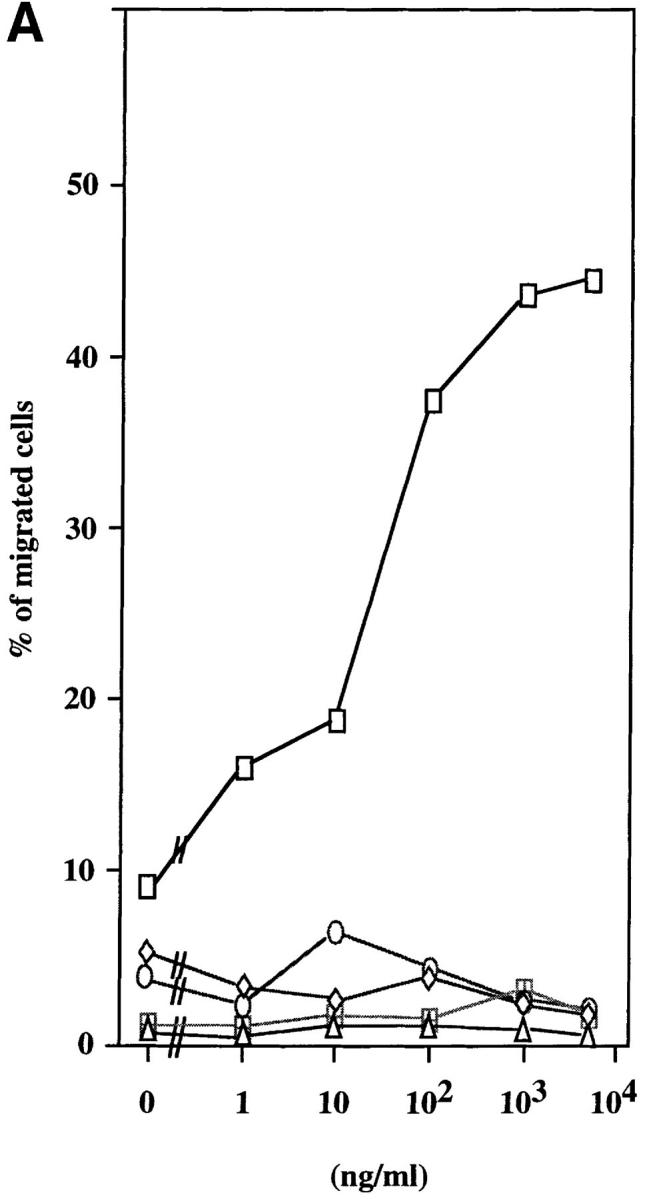
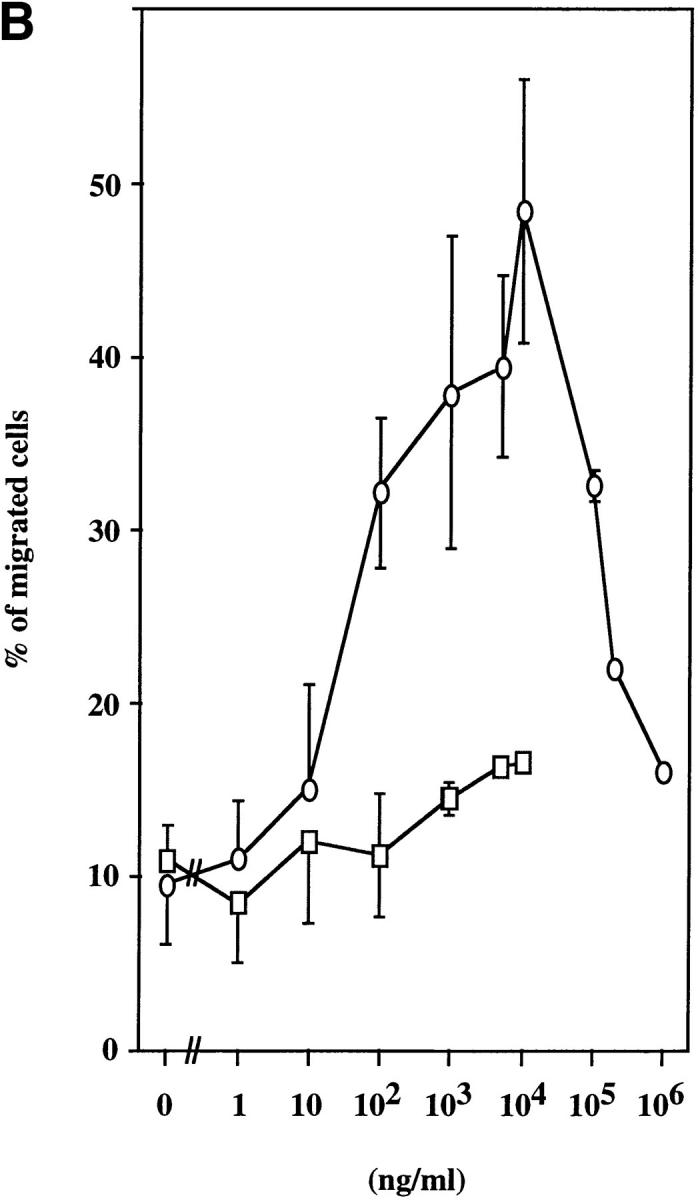
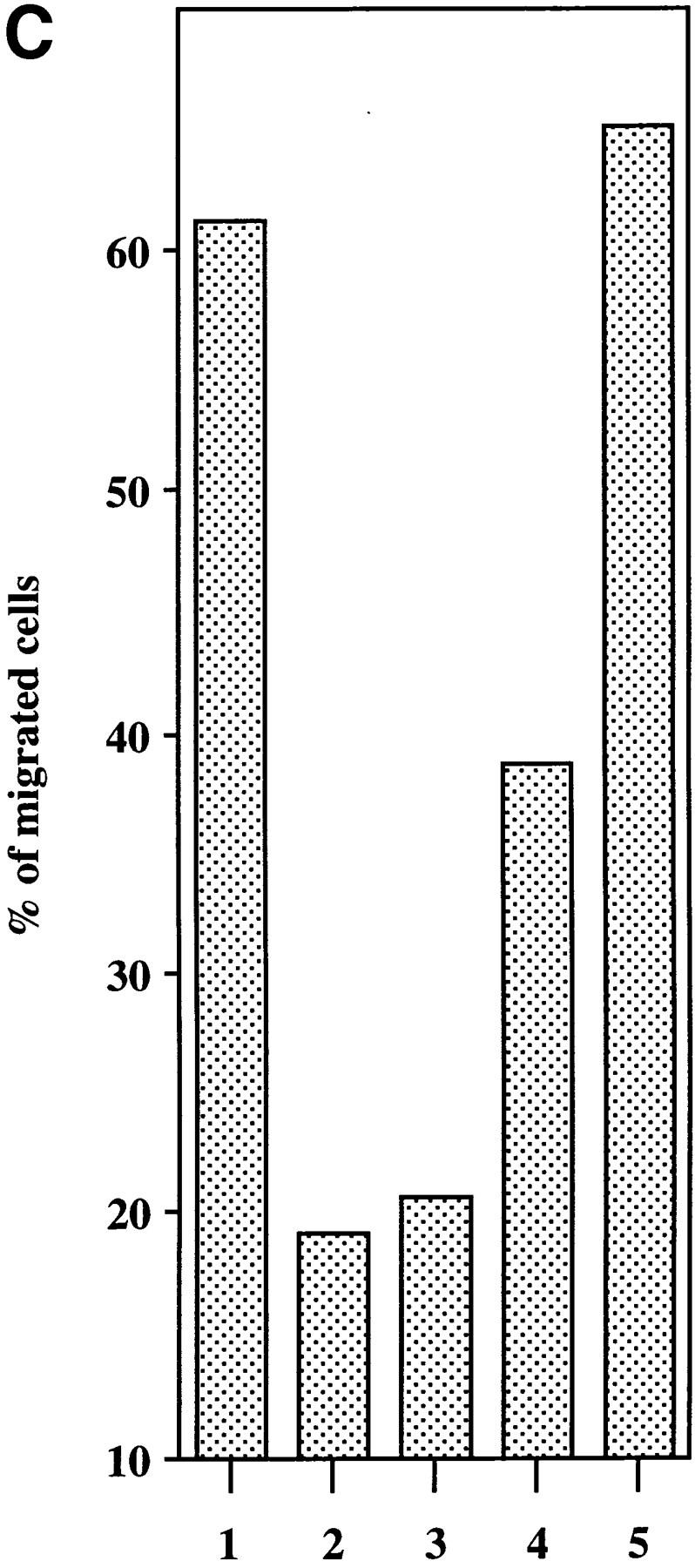
Chemotaxis of different cell populations induced by ABCD-1. (A) Migratory responsiveness to ABCD-1 of freshly isolated cells from spleen (diamonds), LN (ellipses), bone marrow (triangles), and thymus (windows), as well as Con A–activated/IL-2–restimulated T lymphocytes (rectangles). A single experiment is shown. (B) ABCD-1 induced migration of Con A–activated/IL-2–restimulated T lymphocytes (ellipses) and in vitro–generated NK1.1+, IL-2Rα−, and IL-2Rβ+ NK cells (rectangles) derived by culture of RAG-2–deficient bone marrow cells in the presence of IL-2. The migratory response of IL-2–induced NK cells was considered as nonsignificant. The mean of three to eight individual experiments is shown. (C) Chemotaxis of Con A–activated/ IL-2–restimulated T lymphocytes induced by supernatant of induced ABCD-1–transfected Schneider cells, diluted 1:50 (lane 1); supernatant of induced mock-transfected Schneider cells, diluted 1:50 (lane 2); medium (lane 3); supernatant of R2BFL induced with anti-CD40 plus IL-4 for 6 d, diluted 1:100 (lane 4) and 1:1 (lane 5). Concentration of purified recombinant ABCD-1 was determined by spectrometer measurements at a wavelength of 280 nm. OD280 of 1 was set equal to a protein concentration of 1 mg/ml.
ABCD-1 Is Most Likely the Murine Orthologue of the Human Chemokine MDC/STCP-1.
During the progress of our studies, a structurally and functionally similar chemokine was reported in two independent studies, named macrophage-derived chemokine (MDC [61]) and stimulated T cell chemotactic protein 1 (STCP-1 [62]), respectively. ABCD-1 and MDC/STCP-1 share 64% identity and 83% similarity at the amino acid level (Fig. 6 A). The human chemokine is composed of 93 amino acids, but the mouse chemokine contains 92 amino acids, the last Q93 residue being absent.
Figure 6.
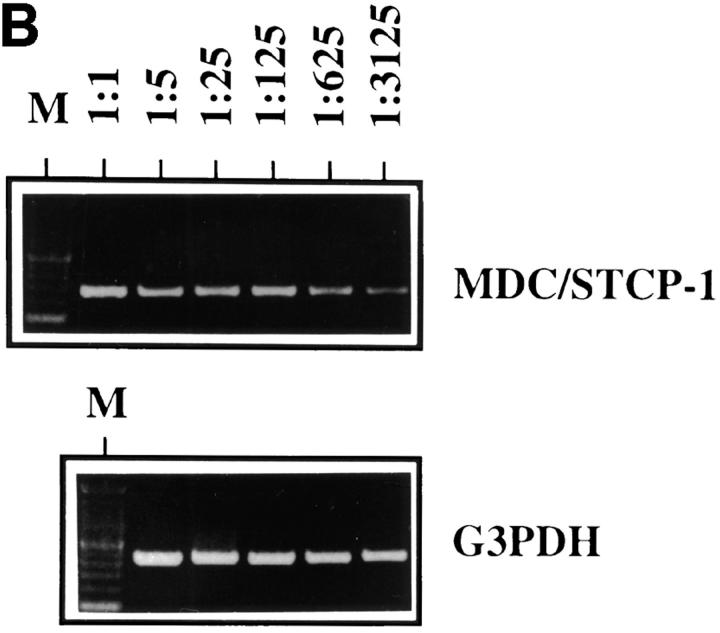
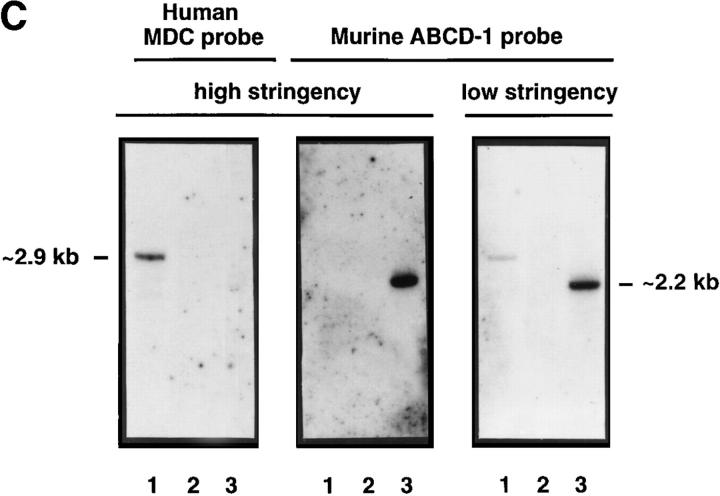
ABCD-1 is the most likely orthologue of MDC/STCP-1. (A) Alignment of amino acid sequences of mouse ABCD-1 and human MDC/ STCP-1 chemokines (references 61 and 62). Vertical lines, Identities; semicolons, similarities. The overall identity is 64.8%, and the similarity 82.4%. (B) Semiquantitative RT-PCR of MDC/STCP-1 mRNA expression in anti-CD40 plus IL-4–activated human tonsil B cells. Total RNA was extracted and reverse transcribed. MDC/STCP-1 and G3PDH were PCR amplified using serial 1:5 dilutions of cDNA as template. M, Marker. (C) 300 ng of cDNA prepared using the CAP-finder system (see Materials and Methods) from anti-CD40 plus IL-4–stimulated human tonsil B cells (lane 1), nonstimulated (lane 2), or 3-d–induced (lane 3) R2BFL cells were separated in 1.2% TAE-agarose gels. After transfer, replica filters were hybridized at high stringency with the MDC-specific probe or at either high or low stringency with the ABCD-1– specific probe. At high stringency, each probe recognized sequences of its corresponding species. However, at low stringency, the ABCD-1 probe, in addition to the ABCD-1 signal in the lane with the activated mouse R2BFL cDNA (lane 3), recognized a human species of identical size as that of MDC (lane 1). (D) Chemotactic responsiveness to ABCD-1 of human IL-12/anti–IL-4–activated (squares), or IL-4/anti–IL-12–activated (diamonds) CD4+ T cells. Migratory response to human TARC (PeproTech Ltd., London, UK) of human IL-12/anti–IL-4–activated (circles) or IL-4/anti–IL-12–activated (triangles) CD4+ T cells was used as positive control in this experiment.
The high amino acid sequence homology between MDC and ABCD-1 prompted us to investigate whether these chemokines represent murine and human orthologues. Neither of the human studies had investigated the expression of MDC/STCP-1 by activated B cells. Hence, we have stimulated B cells from human tonsils with anti-CD40 plus IL-4 and found them to produce high levels of MDC/STCP-1–specific mRNA (Fig. 6 B), indicating that MDC/STCP-1 and ABCD-1 are expressed in the same type of human and mouse cells. Moreover, we synthesized cDNA from human or mouse B cells activated with anti-CD40 plus IL-4 using the PCR-based system Cap-finder (Clontech) and hybridized it with an ABCD-1– or MDC-specific probe, respectively, at high stringency. Under these conditions, the ABCD-1 probe recognized a species of cDNAs of ∼2.2 kb in the mouse cDNA, whereas the MDC probe recognized a species of cDNA of ∼2.9 kb in the human cDNA (Fig. 6 C). No cross-reaction between the species was observed. However, when the ABCD-1 probe was used under low stringency conditions, it recognized the ABCD-1–specific sequences in the mouse cDNA and, in addition, recognized a species of cDNAs in the human sample that comigrated with the signal detected using the MDC probe. This indicates that the species of human cDNAs with the closest homology to ABCD-1 are comigrating with those detected by the MDC/STCP-1 probe, a finding consistent with the notion that ABCD-1 is the murine orthologue of MDC/STCP-1.
The chemotactic activity of recombinant ABCD-1 is not restricted to activated mouse T cells, since ABCD-1 was also found to be a potent chemoattractant for human CD4+ T cells stimulated by anti-CD3/anti-CD28 and either IL-12 and anti–IL-4 or IL-4 and anti–IL-12 (Fig. 6 D). IL-4/anti–IL-12–stimulated T cells migrated optimally at lower concentrations of chemokine than did IL-12/anti– IL-4–stimulated cells.
All of the above findings are consistent with the notion that ABCD-1 is the most likely murine orthologue of MDC/STCP-1.
Discussion
We have generated a sublibrary of cDNA clones that appears to be highly enriched for sequences which are specifically upregulated when IL-7–dependent pro/pre B-I cells are induced by the removal of IL-7 and the addition of CD40-specific mAb and IL-4 to differentiate to B cells with a mature phenotype and Sμ-Sε–switched genotype (14). Because of the very broad developmental changes that occur during such a stimulation, a very heterogeneous group of genes are expected to be found in the subtracted sublibrary. Genes involved in the changes from pro/pre B-I to mature B cell, genes induced by either the anti-CD40 or the IL-4 signal transduction pathways, genes controlling Ig V-gene hypermutation, and genes involved in Sμ-Sε switching are likely candidates. Indeed, several genes already known to be upregulated when pro/pre B cells develop to mature B cells were found among the selected clones. They are genes critical for the capacity of developing B cells to present antigen, namely those encoding MHC class II proteins (52), the invariant chain of MHC class II (51), and the MHC locus–linked H-2M gene (53); genes related to IL-4 signaling, such as the IL-4 receptor (56); and, finally, genes related to the anti-CD40 plus IL-4 class-switching process, such as the Iγ1 and Iε germline transcripts (for a review, see reference 54). In fact, their frequency within the total collection was one in six cDNA sequences in the sublibrary.
One sequence of the remaining clones of the library— and the structure of the corresponding gene—is described in this paper. It was present in surprisingly high frequency among the enriched clones (10 of 120). It was found to encode a novel murine member of the CC or β chemokine family, which we name ABCD-1. We show that this gene is expressed rather selectively by DC and activated B cells, but not by resting B cells, T cells, IL-3–induced macrophages, or IL-2–stimulated NK cells.
While our characterization of the ABCD-1 gene and its protein was in progress, two groups independently described a human chemokine molecule that is structurally and functionally related to ABCD-1 (MDC [61] and STCP-1 [62]). The overall amino acid sequence identity between the mouse ABCD-1 and the human protein MDC/STCP-1 is ∼64% (similarity 84%). In addition, the genomic organization of the ABCD-1 and STCP-1 genes is very similar; particularly striking is the identity of sequences at the intron–exon boundaries of the mouse and human gene (Fig. 2 B). All of these findings are strong support for the notion that the mouse ABCD-1 gene is the orthologue of the human MDC/STCP-1 gene.
A number of characteristics are the same, or comparable, for the mouse ABCD-1 and human MDC/STCP-1 chemo-kines (references 61 and 62, and this report). All three studies reported that they are expressed at low levels in freshly isolated cells from thymus, LNs, spleen, and lung. In our studies, high levels of ABCD-1 mRNA expression were only seen in DC and activated B lymphocytes, and we have shown that MDC/STCP-1 is expressed by activated human B cells. Hence, the patterns of expression are consistent in mouse and human cells. Further evidence for the close relationship between mouse ABCD-1 and human MDC comes from hybridization experiments with mouse and human probes under low or high stringency, suggesting that in human anti-CD40 plus IL-4–stimulated B cells, the sequences most similar to mouse ABCD-1 are those encoded by MDC/STCP-1 (Fig. 6 D).
Finally, the chemotactic properties are similar. Activated mouse T cells of the CD8+ as well as CD4+ phenotype are the best chemoattracted cell populations. Activated human T cells are also chemoattracted by the mouse chemokine (Fig. 6 D). It is generally accepted that stimulation of anti-CD3/anti-CD28–activated CD4+ human T cells by IL-12 plus anti–IL-4 generates Th1-like cells, whereas stimulation with IL-4 plus anti–IL-12 develops Th2-like cells. Our results show that the murine chemokine ABCD-1 acts on both types of Th cells. These results suggest a close relationship between the mouse and the human chemokine. Its role appears to be to attract activated T cells to DC and activated B cells, but not to polarize them or select helper or killer T cell subpopulations.
Other than a low level production of the MIP-1α and -3α chemokines in some malignant B cell lines (63, 64), expression of chemokines by normal B cells has not previously been described. Our discovery that ABCD-1 and MDC/STCP-1 are produced by mouse and human activated B cells, respectively, provides a missing link in a scenario that attempts to describe the T cell–dependent activation of B cells and the formation of germinal centers. In such a scenario, foreign antigen is first taken up and processed by immature DC, e.g., Langerhans cells in the epidermis. Inflammatory stimuli lead to activation and differentiation of these immature DC to mature professional antigen-presenting DC (65), which migrate into the spleen and into regional LNs. Antigen-presenting mature DC could do three things. First, they could attract naive T cells by the production of the DC-CK-1 chemokine (25). Second, they could present antigen in the context of MHC molecules and thus activate the T cells. Third, the production of ABCD-1 by the DC would then keep the T cells attracted to the DC and continuously activated. This is all likely to occur in or near the T cell–rich regions, e.g., in or near the periarteriolar lymphocyte sheath of the spleen. As a result, a focus of antigen-activated T cells could be formed around the ABCD-1–producing antigen-presenting DC.
B cells in B cell–rich follicular regions also have to be activated. Our studies show that T cell–independent as well as T cell–dependent stimulation activates B cells to ABCD-1 production. Once activated, B cells become attractive for Th cells, which migrate from the periarteriolar lymphocyte sheath into the follicular areas of the spleen and begin to interact with activated B cells.
At the same time, the B cells are attracted into the follicular regions by the CXC chemokine BLC (23), alternatively known as BCA-1 (24), which is most likely produced by FDC. Thereafter, Th cells follow the activated B cells through chemoattraction via ABCD-1 into the follicle, where T and B cells begin with the formation of a germinal center, i.e., with extensive proliferation, Ig V-region hypermutation, class switching, and antibody secretion.
Such a scenario makes a number of predictions. First, all attracted cells should express chemokine-specific receptors. Hence, activated but not necessarily resting T cells are expected to express ABCD-1 receptors. In humans, MDC and TARC bind to CCR4 and attract cells transfected with CCR4 (66, 67). On the other side, CCR4 is expressed selectively on Th2 cells (68, 69). However, we show that Th1 and Th2 cells are attracted by ABCD-1. Hence, ABCD-1 is suspected to have a second receptor expressed on Th1 cells, and perhaps also on Th2 cells. Future desensitization experiments with the CCR4-specific TARC and MDC/STCP-1 chemokines (61, 62, 70) and with ABCD-1 might clarify how many chemokine receptors function with the different chemokines on the different T cells.
The second prediction is that targeted disruption of the ABCD-1 gene might impede the initiation of T cell– dependent B cell stimulation and the formation of germinal centers, like the targeted disruption of BLR-1, the BLC/ BCA receptor (16). The targeted disruption of the ABCD-1 gene is under way.
An exciting new development came from the discovery that certain chemokines can function as HIV-suppressive factors. RANTES, MIP-1α, and MIP-1β were shown to be potent inhibitors of viral infection by monocyte/macrophage-tropic HIV strains (71). Interestingly, Pal and co-workers (72) have recently identified an HIV-inhibitory activity in the supernatants of some immortalized CD8+ cell lines. When this activity was purified and the protein sequence was determined, the molecule was identical to the MDC/STCP-1 chemokine. Hence, we would expect that ABCD-1, which we show here to chemoattract human T cells, might also act as an HIV-suppressive factor. Such experiments are now in progress. Because of this suggested suppressive effect of a B cell–produced chemokine, administration to HIV-infected individuals of soluble CD40 ligand or anti-CD40 mAb could be an additional way of inhibiting viral spread.
Acknowledgments
The authors thank Dr. R. Jessberger for help with ABCD-1 purification by HighTrap Heparin FPLC from ABCD-1–transfected Schneider cell supernatants; Mr. D. Avila for synthesizing chemical ABCD-1 and for protein sequencing; and Dr. M. Colonna for critical reading of the manuscript.
Abbreviations used in this paper
- BCA-1
B cell–attracting chemokine 1
- BLC
B lymphocyte chemoattractant
- BLR
Burkitt's lymphoma receptor
- DC
dendritic cell(s)
- FDC
follicular dendritic cell(s)
- FPLC
fast-performance liquid chromatography
- MCP
monocyte chemotactic protein
- MDC
macrophage-derived chemokine
- MIP
macrophage-inflammatory protein
- RAG
recombination activating gene
- RANTES
regulated upon activation, normal T cell expressed and secreted
- RT
reverse transcription
- STCP-1
stimulated T cell chemotactic protein 1
- TARC
thymus and activation–regulated chemokine
Footnotes
The Basel Institute for Immunology was founded and is supported by F. Hoffmann-La Roche Ltd., Basel, Switzerland. P. Sideras, E. Pardali, and M. Speletas were supported by the Swedish Medical Research Council (MFR), the Swedish Cancer Foundation “Cancerfonden,” and the “Petrus and Augusta Hedlund”-Stiftelsen.
C. Schaniel and E. Pardali contributed equally to this work.
References
- 1.Gray D, Skarvall H. B-cell memory is short-lived in the absence of antigen. Nature. 1988;336:70–73. doi: 10.1038/336070a0. [DOI] [PubMed] [Google Scholar]
- 2.Jacob J, Kelsoe G. In situ studies of the primary immune response to (4-hydroxy-3-nitrophenyl)acetyl. II. A common clonal origin for periarteriolar lymphoid sheath–associated foci and germinal centers. J Exp Med. 1992;176:679–687. doi: 10.1084/jem.176.3.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.MacLennan IC, Gulbranson-Judge A, Toellner KM, Casamayor-Palleja M, Chan E, Sze DM, Luther SA, Orbea HA. The changing preference of T and B cells for partners as T-dependent antibody responses develop. Immunol Rev. 1997;156:53–66. doi: 10.1111/j.1600-065x.1997.tb00958.x. [DOI] [PubMed] [Google Scholar]
- 4.Steinman RM, Pack M, Inaba K. Dendritic cells in the T-cell areas of lymphoid organs. Immunol Rev. 1997;156:25–37. doi: 10.1111/j.1600-065x.1997.tb00956.x. [DOI] [PubMed] [Google Scholar]
- 5.Liu YJ, Arpin C. Germinal center development. Immunol Rev. 1997;156:111–126. doi: 10.1111/j.1600-065x.1997.tb00963.x. [DOI] [PubMed] [Google Scholar]
- 6.Liu YJ, Zhang J, Lane PJ, Chan EY, MacLennan IC. Sites of specific B cell activation in primary and secondary responses to T cell-dependent and T cell-independent antigens. Eur J Immunol. 1991;21:2951–2962. doi: 10.1002/eji.1830211209. [DOI] [PubMed] [Google Scholar]
- 7.Coico RF, Bhogal BS, Thorbecke GJ. Relationship of germinal centers in lymphoid tissue to immunologic memory. VI. Transfer of B cell memory with lymph node cells fractionated according to their receptors for peanut agglutinin. J Immunol. 1983;131:2254–2257. [PubMed] [Google Scholar]
- 8.Klaus GG, Humphrey JH, Kunkl A, Dongworth DW. The follicular dendritic cell: its role in antigen presentation in the generation of immunological memory. Immunol Rev. 1980;53:3–28. doi: 10.1111/j.1600-065x.1980.tb01038.x. [DOI] [PubMed] [Google Scholar]
- 9.Berek C, Berger A, Apel M. Maturation of the immune response in germinal centers. Cell. 1991;67:1121–1129. doi: 10.1016/0092-8674(91)90289-b. [DOI] [PubMed] [Google Scholar]
- 10.Kawabe T, Naka T, Yoshida K, Tanaka T, Fujiwara H, Suematsu S, Yoshida N, Kishimoto T, Kikutani H. The immune responses in CD40-deficient mice: impaired immunoglobulin class switching and germinal center formation. Immunity. 1994;1:167–178. doi: 10.1016/1074-7613(94)90095-7. [DOI] [PubMed] [Google Scholar]
- 11.Xu J, Foy TM, Laman JD, Elliott EA, Dunn JJ, Waldschmidt TJ, Elsemore J, Noelle RJ, Flavell RA. Mice deficient for the CD40 ligand. Immunity. 1994;1:423–431. doi: 10.1016/1074-7613(94)90073-6. [DOI] [PubMed] [Google Scholar]
- 12.Lederman S, Yellin MJ, Covey LR, Cleary AM, Callard R, Chess L. Non-antigen signals for B-cell growth and differentiation to antibody secretion. Curr Opin Immunol. 1993;5:439–444. doi: 10.1016/0952-7915(93)90066-2. [DOI] [PubMed] [Google Scholar]
- 13.Snapper CM, Mond JJ. Towards a comprehensive view of immunoglobulin class switching. Immunol Today. 1993;14:15–17. doi: 10.1016/0167-5699(93)90318-F. [DOI] [PubMed] [Google Scholar]
- 14.Rolink A, Melchers F, Andersson J. The SCID but not the RAG-2 gene product is required for S mu-S epsilon heavy chain class switching. Immunity. 1996;5:319–330. doi: 10.1016/s1074-7613(00)80258-7. [DOI] [PubMed] [Google Scholar]
- 15.Rolink A, Kudo A, Karasuyama H, Kikuchi Y, Melchers F. Long-term proliferating early pre B cell lines and clones with the potential to develop to surface Ig-positive, mitogen reactive B cells in vitro and in vivo. EMBO (Eur Mol Biol Organ) J. 1991;10:327–336. doi: 10.1002/j.1460-2075.1991.tb07953.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forster R, Mattis AE, Kremmer E, Wolf E, Brem G, Lipp M. A putative chemokine receptor, BLR1, directs B cell migration to defined lymphoid organs and specific anatomic compartments of the spleen. Cell. 1996;87:1037–1047. doi: 10.1016/s0092-8674(00)81798-5. [DOI] [PubMed] [Google Scholar]
- 17.Baggiolini M, Dahinden CA. CC chemokines in allergic inflammation. Immunol Today. 1994;15:127–133. doi: 10.1016/0167-5699(94)90156-2. [DOI] [PubMed] [Google Scholar]
- 18.Hedrick JA, Zlotnik A. Chemokines and lymphocyte biology. Curr Opin Immunol. 1996;8:343–347. doi: 10.1016/s0952-7915(96)80123-3. [DOI] [PubMed] [Google Scholar]
- 19.Schall TJ, Bacon KB. Chemokines, leukocyte trafficking, and inflammation. Curr Opin Immunol. 1994;6:865–873. doi: 10.1016/0952-7915(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 20.Kelner GS, Kennedy J, Bacon KB, Kleyensteuber S, Largaespada DA, Jenkins NA, Copeland NG, Bazan JF, Moore KW, Schall TJ, et al. Lymphotactin: a cytokine that represents a new class of chemokine. Science. 1994;266:1395–1399. doi: 10.1126/science.7973732. [DOI] [PubMed] [Google Scholar]
- 21.Bazan JF, Bacon KB, Hardiman G, Wang W, Soo K, Rossi D, Greaves DR, Zlotnik A, Schall TJ. A new class of membrane-bound chemokine with a CX3C motif. Nature. 1997;385:640–644. doi: 10.1038/385640a0. [DOI] [PubMed] [Google Scholar]
- 22.Murphy PM. The molecular biology of leukocyte chemoattractant receptors. Annu Rev Immunol. 1994;12:593–633. doi: 10.1146/annurev.iy.12.040194.003113. [DOI] [PubMed] [Google Scholar]
- 23.Gunn MD, Ngo VN, Ansel KM, Ekland EH, Cyster JG, Williams LT. A B-cell-homing chemokine made in lymphoid follicles activates Burkitt's lymphoma receptor-1. Science. 1998;391:799–803. doi: 10.1038/35876. [DOI] [PubMed] [Google Scholar]
- 24.Legler DF, Loetscher M, Ross RS, Clark-Lewis I, Baggiolini M, Moser B. B cell–attracting chemokine 1, a human CXC chemokine expressed in lymphoid tissues, selectively attracts B lymphocytes via BLR1/CXCR5. J Exp Med. 1998;187:655–660. doi: 10.1084/jem.187.4.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adema GJ, Hartgers F, Verstraten R, de Vries E, Marland G, Menon S, Foster J, Xu Y, Nooyen P, McClanahan T, et al. A dendritic-cell-derived C-C chemokine that preferentially attracts naive T cells. Nature. 1997;387:713–717. doi: 10.1038/42716. [DOI] [PubMed] [Google Scholar]
- 26.Grawunder U, Rolink A, Melchers F. Induction of sterile transcription from the kappa L chain gene locus in V(D)J recombinase-deficient progenitor B cells. Int Immunol. 1995;7:1915–1925. doi: 10.1093/intimm/7.12.1915. [DOI] [PubMed] [Google Scholar]
- 27.Shinkai Y, Rathbun G, Lam KP, Oltz EM, Stewart V, Mendelsohn M, Charron J, Datta M, Young F, Stall AM, et al. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell. 1992;68:855–867. doi: 10.1016/0092-8674(92)90029-c. [DOI] [PubMed] [Google Scholar]
- 28.Alt F, Rosenberg N, Lewis S, Thomas E, Baltimore D. Organization and reorganization of immunoglobulin genes in A-MULV-transformed cells: rearrangement of heavy but not light chain genes. Cell. 1981;27:381–390. doi: 10.1016/0092-8674(81)90421-9. [DOI] [PubMed] [Google Scholar]
- 29.Alt FW, Yancopoulos GD, Blackwell TK, Wood C, Thomas E, Boss M, Coffman R, Rosenberg N, Tonegawa S, Baltimore D. Ordered rearrangement of immunoglobulin heavy chain variable region segments. EMBO (Eur Mol Biol Organ) J. 1984;3:1209–1219. doi: 10.1002/j.1460-2075.1984.tb01955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davidson WF, Fredrickson TN, Rudikoff EK, Coffman RL, Hartley JW, Morse HC., III A unique series of lymphomas related to the Ly-1+ lineage of B lymphocyte differentiation. J Immunol. 1984;133:744–753. [PubMed] [Google Scholar]
- 31.Paige CJ, Kincade PW, Ralph P. Murine B cell leukemia line with inducible surface immunoglobulin expression. J Immunol. 1978;121:641–647. [PubMed] [Google Scholar]
- 32.Kim KJ, Kanellopoulos-Langevin C, Merwin RM, Sachs DH, Asofsky R. Establishment and characterization of BALB/c lymphoma lines with B cell properties. J Immunol. 1979;122:549–554. [PubMed] [Google Scholar]
- 33.Warner NL, Daley MJ, Richey J, Spellman C. Flow cytometry analysis of murine B cell lymphoma differentiation. Immunol Rev. 1979;48:197–243. doi: 10.1111/j.1600-065x.1979.tb00304.x. [DOI] [PubMed] [Google Scholar]
- 34.Weigert MG, Cesari IM, Yonkovich SJ, Cohn M. Variability in the lambda light chain sequences of mouse antibody. Nature. 1970;228:1045–1047. doi: 10.1038/2281045a0. [DOI] [PubMed] [Google Scholar]
- 35.Kearney JF, Radbruch A, Liesegang B, Rajewsky K. A new mouse myeloma cell line that has lost immunoglobulin expression but permits the construction of antibody-secreting hybrid cell lines. J Immunol. 1979;123:1548–1550. [PubMed] [Google Scholar]
- 36.Shulman M, Wilde CD, Kohler G. A better cell line for making hybridomas secreting specific antibodies. Nature. 1978;276:269–270. doi: 10.1038/276269a0. [DOI] [PubMed] [Google Scholar]
- 37.Ralph P. Retention of lymphocyte characteristics by myelomas and theta+ -lymphomas: sensitivity to cortisol and phytohemagglutinin. J Immunol. 1973;110:1470–1475. [PubMed] [Google Scholar]
- 38.Hyman R, Lacorbiere M, Stavarek S, Nicolson G. Derivation of lymphoma variants with reduced sensitivity to plant lectins. J Natl Cancer Inst. 1974;52:963–969. doi: 10.1093/jnci/52.3.963. [DOI] [PubMed] [Google Scholar]
- 39.Ogawa M, Nishikawa S, Ikuta K, Yamamura F, Naito M, Takahashi K, Nishikawa S. B cell ontogeny in murine embryo studied by a culture system with the monolayer of a stromal cell clone, ST2: B cell progenitor develops first in the embryonal body rather than in the yolk sac. EMBO (Eur Mol Biol Organ) J. 1988;7:1337–1343. doi: 10.1002/j.1460-2075.1988.tb02949.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gillis S, Smith KA. Long term culture of tumour-specific cytotoxic T cells. Nature. 1977;268:154–156. doi: 10.1038/268154a0. [DOI] [PubMed] [Google Scholar]
- 41.Karasuyama H, Melchers F. Establishment of mouse cell lines which constitutively secrete large quantities of interleukin 2, 3, 4 or 5, using modified cDNA expression vectors. Eur J Immunol. 1988;18:97–104. doi: 10.1002/eji.1830180115. [DOI] [PubMed] [Google Scholar]
- 42.Leptin M, Potash MJ, Grutzmann R, Heusser C, Shulman M, Kohler G, Melchers F. Monoclonal antibodies specific for murine IgM. I. Characterization of antigenic determinants on the four constant domains of the mu heavy chain. Eur J Immunol. 1984;14:534–542. doi: 10.1002/eji.1830140610. [DOI] [PubMed] [Google Scholar]
- 43.Rolink A, ten Boekel E, Melchers F, Fearon DT, Krop I, Andersson J. A subpopulation of B220+ cells in murine bone marrow does not express CD19 and contains natural killer cell progenitors. J Exp Med. 1996;183:187–194. doi: 10.1084/jem.183.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ruedl C, Hubele S. Maturation of Peyer's patch dendritic cells in vitro upon stimulation via cytokines or CD40 triggering. Eur J Immunol. 1997;27:1325–1330. doi: 10.1002/eji.1830270605. [DOI] [PubMed] [Google Scholar]
- 45.Lanzavecchia A, Scheidegger D. The use of hybrid hybridomas to target human cytotoxic T lymphocytes. Eur J Immunol. 1987;17:105–111. doi: 10.1002/eji.1830170118. [DOI] [PubMed] [Google Scholar]
- 46.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sideras P, Muller S, Shiels H, Jin H, Khan WN, Nilsson L, Parkinson E, Thomas JD, Branden L, Larsson I, et al. Genomic organization of mouse and human Bruton's agammaglobulinemia tyrosine kinase (Btk) loci. J Immunol. 1994;153:5607–5617. [PubMed] [Google Scholar]
- 48.Bunch TA, Grinblat Y, Goldstein LS. Characterization and use of the Drosophila metallothionein promoter in cultured Drosophila melanogastercells. Nucleic Acids Res. 1988;16:1043–1061. doi: 10.1093/nar/16.3.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schneider I. Cell lines derived from late embryonic stages of Drosophila melanogaster. . J Embryol Exp Morphol. 1972;27:353–365. [PubMed] [Google Scholar]
- 50.Diatchenko L, Lau YF, Campbell AP, Chenchik A, Moqadam F, Huang B, Lukyanov S, Lukyanov K, Gurskaya N, Sverdlov ED, Siebert PD. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc Natl Acad Sci USA. 1996;93:6025–6030. doi: 10.1073/pnas.93.12.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Koch N, Lauer W, Habicht J, Dobberstein B. Primary structure of the gene for the murine Ia antigen-associated invariant chains (Ii). An alternatively spliced exon encodes a cysteine-rich domain highly homologous to a repetitive sequence of thyroglobulin. EMBO (Eur Mol Biol Organ) J. 1987;6:1677–1683. doi: 10.1002/j.1460-2075.1987.tb02417.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cardell S, Merkenschlager M, Bodmer H, Chan S, Cosgrove D, Benoist C, Mathis D. The immune system of mice lacking conventional MHC class II molecules. Adv Immunol. 1994;55:423–440. doi: 10.1016/s0065-2776(08)60515-5. [DOI] [PubMed] [Google Scholar]
- 53.Le Bouteiller P. HLA class I chromosomal region, genes, and products: facts and questions. Crit Rev Immunol. 1994;14:89–129. doi: 10.1615/critrevimmunol.v14.i2.10. [DOI] [PubMed] [Google Scholar]
- 54.Esser C, Radbruch A. Immunoglobulin class switching: molecular and cellular analysis. Annu Rev Immunol. 1990;8:717–735. doi: 10.1146/annurev.iy.08.040190.003441. [DOI] [PubMed] [Google Scholar]
- 55.Bettler B, Hofstetter H, Rao M, Yokoyama WM, Kilchherr F, Conrad DH. Molecular structure and expression of the murine lymphocyte low-affinity receptor for IgE (Fc epsilon RII) Proc Natl Acad Sci USA. 1989;86:7566–7570. doi: 10.1073/pnas.86.19.7566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mosley B, Beckmann MP, March CJ, Idzerda RL, Gimpel SD, VandenBos T, Friend D, Alpert A, Anderson D, Jackson J, et al. The murine interleukin-4 receptor: molecular cloning and characterization of secreted and membrane bound forms. Cell. 1989;59:335–348. doi: 10.1016/0092-8674(89)90295-x. [DOI] [PubMed] [Google Scholar]
- 57.Schall TJ, Simpson NJ, Mak JY. Molecular cloning and expression of the murine RANTES cytokine: structural and functional conservation between mouse and man. Eur J Immunol. 1992;22:1477–1481. doi: 10.1002/eji.1830220621. [DOI] [PubMed] [Google Scholar]
- 58.Heeger P, Wolf G, Meyers C, Sun MJ, O'Farrell SC, Krensky AM, Neilson EG. Isolation and characterization of cDNA from renal tubular epithelium encoding murine Rantes. Kidney Int. 1992;41:220–225. doi: 10.1038/ki.1992.31. [DOI] [PubMed] [Google Scholar]
- 59.Danoff TM, Lalley PA, Chang YS, Heeger PS, Neilson EG. Cloning, genomic organization, and chromosomal localization of the Scya5 gene encoding the murine chemokine RANTES. J Immunol. 1994;152:1182–1189. [PubMed] [Google Scholar]
- 60.Andersson J, Melchers F. Induction of immunoglobulin M synthesis and secretion in bone-marrow-derived lymphocytes by locally concentrated concanavalin A. Proc Natl Acad Sci USA. 1973;70:416–420. doi: 10.1073/pnas.70.2.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Godiska R, Chantry D, Raport CJ, Sozzani S, Allavena P, Leviten D, Mantovani A, Gray PW. Human macrophage-derived chemokine (MDC), a novel chemoattractant for monocytes, monocyte-derived dendritic cells, and natural killer cells. J Exp Med. 1997;185:1595–1604. doi: 10.1084/jem.185.9.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chang M, McNinch J, Elias C, III, Manthey CL, Grosshans D, Meng T, Boone T, Andrew DP. Molecular cloning and functional characterization of a novel CC chemokine, stimulated T cell chemotactic protein (STCP-1), that specifically acts on activated T lymphocytes. J Biol Chem. 1997;272:25229–25237. doi: 10.1074/jbc.272.40.25229. [DOI] [PubMed] [Google Scholar]
- 63.Sharma V, Walper D, Deckert R. Modulation of macrophage inflammatory protein-1alpha and its receptors in human B-cell lines derived from patients with acquired immunodeficiency syndrome and Burkitt's lymphoma. Biochem Biophys Res Commun. 1997;235:576–581. doi: 10.1006/bbrc.1997.6828. [DOI] [PubMed] [Google Scholar]
- 64.Rossi DL, Vicari AP, Franz-Bacon K, McClanahan TK, Zlotnik A. Identification through bioinformatics of two new macrophage proinflammatory human chemokines: MIP-3alpha and MIP-3beta. J Immunol. 1997;158:1033–1036. [PubMed] [Google Scholar]
- 65.Cella M, Sallusto F, Lanzavecchia A. Origin, maturation and antigen presenting function of dendritic cells. Curr Opin Immunol. 1997;9:10–16. doi: 10.1016/s0952-7915(97)80153-7. [DOI] [PubMed] [Google Scholar]
- 66.Imai T, Chantry D, Raport CJ, Wood CL, Nishimura M, Godiska R, Yoshie O, Gray PW. Macrophage-derived chemokine is a functional ligand for the CC chemokine receptor 4. J Biol Chem. 1998;273:1764–1768. doi: 10.1074/jbc.273.3.1764. [DOI] [PubMed] [Google Scholar]
- 67.Imai T, Baba M, Nishimura M, Kakizaki M, Takagi S, Yoshie O. The T cell-directed CC chemokine TARC is a highly specific biological ligand for CC chemokine receptor 4. J Biol Chem. 1997;272:15036–15042. doi: 10.1074/jbc.272.23.15036. [DOI] [PubMed] [Google Scholar]
- 68.Bonecchi R, Bianchi G, Bordignon PP, D'Ambrosio D, Lang R, Borsatti A, Sozzani S, Allavena P, Gray PA, Mantovani A, Sinigaglia F. Differential expression of chemokine receptors and chemotactic responsiveness of type 1 T helper cells (Th1s) and Th2s. J Exp Med. 1998;187:129–134. doi: 10.1084/jem.187.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sallusto F, Lenig D, MacKay CR, Lanzavecchia A. Flexible programs of chemokine receptor expression on human polarized T helper 1 and 2 lymphocytes. J Exp Med. 1998;187:875–883. doi: 10.1084/jem.187.6.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Imai T, Yoshida T, Baba M, Nishimura M, Kakizaki M, Yoshie O. Molecular cloning of a novel T cell- directed CC chemokine expressed in thymus by signal sequence trap using Epstein-Barr virus vector. J Biol Chem. 1996;271:21514–21521. doi: 10.1074/jbc.271.35.21514. [DOI] [PubMed] [Google Scholar]
- 71.Cocchi F, DeVico AL, Garzino-Demo A, Arya SK, Gallo RC, Lusso P. Identification of RANTES, MIP-1 alpha, and MIP-1 beta as the major HIV-suppressive factors produced by CD8+ T cells. Science. 1995;270:1811–1815. doi: 10.1126/science.270.5243.1811. [DOI] [PubMed] [Google Scholar]
- 72.Pal R, Garzino-Demo A, Markham PD, Burns J, Brown M, Gallo RC, DeVico AL. Inhibition of HIV-1 infection by the beta-chemokine MDC. Science. 1997;278:695–698. doi: 10.1126/science.278.5338.695. [DOI] [PubMed] [Google Scholar]