Abstract
Transcription factors of the nuclear factor of activated T cells (NFAT) family play a key role in antigen receptor–mediated responses in lymphocytes by controlling induction of a wide variety of cytokine genes. The GTPases Ras and Rac-1 have essential functions in regulation of NFAT transcriptional activity in the mast cell system, where Fcε receptor type 1 (FcεR1) ligation results in induction of multiple NFAT target genes. This report examines the precise biochemical basis for the Rac-1 dependency of FcεR1 activation of NFAT in mast cells. We are able to place Rac-1 in two positions in the signaling network that regulates the assembly and activation of NFAT transcriptional complexes in lymphocytes. First, we show that activity of Rac-1 is required for FcεR1-mediated NFATC1 dephosphorylation and nuclear import. Regulation of NFAT localization by the FcεR1 is a Rac-dependent but Ras-independent process. This novel signaling role for Rac-1 is distinct from its established regulation of the actin cytoskeleton. Our data also reveal a second GTPase signaling pathway regulating NFAT transcriptional activity, in which Rac-1 mediates a Ras signal. These data illustrate that the GTPase Rac-1 should now be considered as a component of the therapeutically important pathways controlling NFATC1 subcellular localization. They also reveal that GTPases may serve multiple functions in cellular responses to antigen receptor ligation.
Keywords: Ras, nuclear factor of activated T cells, Rac-1, Fcε receptor type 1, mast cells
Antigenic cross-linking of the high-affinity receptor for IgE, FcεR1, on mast cells results in secretion of allergic mediators and induction of the expression of genes encoding multiple cytokines and chemokines (1, 2). Transcription factors of the nuclear factor of activated T cells (NFAT)1 family play a key role in these antigen receptor– mediated responses by controlling induction of a wide variety of cytokine genes, including those for IL-2, IL-4, GM-CSF, and the Fas ligand and CD40 ligand molecules (3). FcεR1 activation of NFAT in mast cells is mediated by calcium/calcineurin (CN)-controlled signaling pathways acting in synergy with signal transduction pathways regulated by GTPases of the Ras superfamily, Ras and Rac-1 (4). GTPases cycle between GDP- (inactive) and GTP-bound (active) conformations. Exchange of GDP for GTP and, hence, transition to the activated state is promoted by a class of guanine nucleotide exchange factors (GEFs; references 5 and 6). Regulation of the Ras GEF Sos and activation of a Rac GEF Vav are immediate consequences of FcεR1 ligation (7–9). Ras and Rac GTPases are then able to regulate diverse cellular processes in lymphocytes, including NFAT activation, by virtue of their coupling to multiple biochemical effector signaling pathways (4, 10).
A simple model explaining the cooperation between calcium and Ras/Rac signaling pathways for FcεR1 activation of NFAT has been proposed: NFAT transcription factors are cytosolic in quiescent cells and are imported into the nucleus in response to calcium/CN signals triggered by ligation of antigen receptors (3, 11). In the nucleus, NFAT proteins form complexes with activator protein (AP)-1 family proteins and in this context are able to transcriptionally activate cytokine genes (10, 12). Ras/Rac-mediated signals couple antigen receptors to the activation of AP-1 complexes, and signaling by these GTPases is therefore required for antigen receptor–mediated NFAT responses (10). The translocation of NFAT from cytosol to nucleus, where it can contact DNA, is a critical commitment step for the induction of NFAT/AP-1 transcriptional activity by immunoreceptors. The subcellular localization of NFAT is tightly regulated by a phosphorylation cycle: phospho-NFAT is retained in the cytosol and upon antigen receptor activation is dephosphorylated by CN at several sites of constitutive serine phosphorylation (11). The latter pathway is the focus of much analysis because of its therapeutic importance as the target of the macrolide immunosuppressants Cyclosporin A (CsA) and FK506 (13). CsA–cyclophilin and FK506–immunophilin complexes bind to and inactivate CN. This is the basis of the immunosuppressive nature of these compounds; CN-dependent transcription of multiple NFAT-regulated cytokine genes such as IL-2, IL-4, and TNF-α is blocked in CsA/FK506-treated cells.
The role of calcium/CN signals in the control of NFAT subcellular localization is well documented (11, 14, 15), as is the role of Ras/Rac GTPases in control of AP-1 complexes (16, 17). However, two recent lines of research question the current simple “two signal” NFAT activation model. First, the regulation of NFAT subcellular localization may be a more complicated process than originally proposed; there are protein kinase pathways that can promote NFAT nuclear export and antagonize the action of calcium/CN-regulated signals (18, 19). Second, it is clear that the Ras and Rac GTPases have functions that extend beyond regulation of AP-1 complexes. For example, regulation of the actin cytoskeleton by both Ras and Rac is well documented (20). The role of the actin cytoskeleton in NFAT translocation has not been examined, although induction of certain NFAT-regulated cytokine genes is sensitive to Cytochalasin D, which inhibits actin polymerization. The relocation of a large pool of protein across the nuclear membrane may require morphological changes dependent on actin, and may encompass a role for GTPases of the Ras family. In addition to these candidate roles for Ras family GTPases in NFAT/AP-1 activation, there are defined roles for at least two GTPases in the regulation of the nuclear import process (21, 22). Ran is an integral part of the nuclear import machinery that functions constitutively to transport proteins and other cargo across the nuclear membrane. There is also biochemical evidence that a GTPase other than Ran is important for nuclear import. It remains to be seen whether this represents another GTPase activity which is part of the basic nuclear transport machinery, or is an indicator of a GTPase signaling requirement which is potentially a target for regulation by transmembrane receptors.
In this study, an assay to monitor NFAT subcellular localization in cells transiently transfected with dominant inhibitory Ras and Rac mutants was established in the RBL2H3 mast cell line. The role of these GTPases in FcεR1 regulation of NFAT nuclear import was then examined. Function of the Rac-1 GTPase, but not Ras, was absolutely required for FcεR1 induction of NFATC1 nuclear import. This novel role for Rac-1 was distinct from its function in regulation of the actin cytoskeleton. Rather, Rac-1 regulates the phosphorylation status of NFATC1 in mast cells. We also observed that Rac-1 regulates NFAT transcriptional complexes in distinct Ras-dependent and -independent pathways. The latter, Ras-independent, role for Rac-1 in regulation of NFAT nuclear import identifies Rac-1 as a significant new player in this therapeutically important pathway.
Materials and Methods
Plasmids.
The full-length NFATC1 cDNA was subcloned into the pEGFP-C1 vector (Clontech, Palo Alto, CA) to give NFATC1–green fluorescent protein (GFP). Constructs were verified by sequencing. The pSRα-CNM (activated CN; reference 17), pEF-N17-Rac-1 (dominant inhibitory Rac-1), pEF-V12Rac-1 (activated Rac-1), and pRSV-N17 Ras (dominant inhibitory Ras) have all been described previously (10, 23). All constructs were purified by CsCl density gradient centrifugation before use in transfection. The amount of plasmid used in transient transfection was 8 μg/107 cells NFATC1-GFP. Cotransfections included 30 μg/107 cells pEF-N17Rac-1, pEF-V12Rac-1, or pEF-N17 Ras, or 20 μg/107 cells pSRα-CNM. IL-4 NFAT/AP-1 chloramphenicol acetyl transferase (CAT) comprised a trimerized NFAT/ AP-1 site derived from Purine box B of the murine IL-4 promoter (24). The NFAT/AP-1 binding site oligonucleotide used in the construction of this reporter was GATCCTGAGTTTACATTGGAAAATTTTATAGAGCGAGTTG (5′–3′).
Confirmation of Transfection.
In cotransfection experiments, an excess of regulator (i.e., GTPase) plasmid over reporter (NFATC1-GFP) plasmid was used. Coexpression of Rac-1 was confirmed by immunostaining for the myc-epitope tag at the single cell level and by Western analysis of whole cell lysate. In both cases, the anti-myc mAb 9E10 was used (Hybridoma Development Unit, Imperial Cancer Research Fund).
Transient Transfection and Cell Imaging.
RBL2H3 mast cells were cultured as described previously and electroporated using a Gene Pulser apparatus (Beckman Instruments, Inc., Fullerton, CA) at 107 cells/0.5 ml DMEM, 960 μF, 310 V. After plating onto glass coverslips, cells were allowed 6 h recovery. In the case of Ionomycin stimulation, cells were incubated for the indicated times with 500 ng/ml Ionomycin (Calbiochem Corp., La Jolla, CA). Cell stimulation via the FcεR1 was carried out by 1 h incubation at 37°C with 1 μg/ml IgE anti-DNP (Sigma Chemical Co., St. Louis, MO) followed by antigenic cross-linking of bound IgE using 250 ng/ml KLH-DNP (Calbiochem Corp.) unless otherwise indicated. Cells were fixed in 4% paraformaldehyde for 30 min at room temperature. Nuclear staining using propidium iodide (PI) was carried out by permeabilization of cells in 0.1% Triton X-100 followed by sequential incubation with 500 mg/ml RNase (37°C, 10 min) and 0.1 μg/ml PI (room temperature, 10 min). Coverslips were washed in PBS after each stage and mounted in 15 μl Gelvatol (Monsanto Co., St. Louis, MO) which was allowed to set for 4 h before imaging using a TCS-NT upright confocal microscope (Leica Inc., Deerfield, IL). Images were corrected for contribution of PI emission to GFP signal and viewed using the Imaris system (Bitplane AG, Zurich, Switzerland). In scoring experiments, cells were analyzed using an Axiophot fluorescence microscope (Nikon Inc., Melville, NY). Cells were scored for NFATC1-GFP localization as blind-coded samples.
Western Blot Analysis.
Transfected RBL2H3 mast cells were primed and stimulated as described. Cells were removed from the culture dish using cell scrapers, washed once in ice-cold PBS, then lysed for 40 min at 4°C with rotation in a buffer containing 20 mM Hepes, pH 7.9, 20% (vol/vol) glycerol, 0.42 M NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 10 mM NaF, 1 mM dithiothreitol, 1 mM PMSF, 1 mM Na2VO4, and 1% NP-40. Nuclear membranes were pelleted by centrifugation at 14,000 rpm for 20 min at 4°C, and proteins in the supernatant were then acetone precipitated. Samples were resolved under reducing conditions by 7% SDS-PAGE. The resolved proteins were transferred to polyvinylidene difluoride, and the membranes were blocked in 5% nonfat milk for 1 h at room temperature. Anti-GFP Western blot analysis was performed using an affinity-purified rabbit anti-GFP, a gift of Dr. Ken Sawin, Imperial Cancer Research Fund.
CAT Reporter Gene Assay.
RBL2H3 mast cells were transiently transfected using electroporation as described above. For CAT reporter gene assays, 5 × 106 cells were lysed in 150 μl of a buffer containing 0.65% (vol/vol) NP-40, 10 mM Tris, pH 8.0, 1 mM EDTA, and 150 mM NaCl for 15 min on ice. Lysates were then transferred to a 68°C water bath for 10 min. Cell debris was pelleted, and aliquots of lysate were removed to a fresh tube in an assay volume of 100 μl, to which 40 μl of a start solution containing 0.5 mM acetyl coenzyme A, 5 mM chloramphenicol, and 0.5 M Tris, pH 8.0, and 1 μl per point of 50 μCi/ml 14C acetyl coenzyme A was added. The assay was incubated for 16 h at 37°C before chloramphenicol was extracted using 150 μl per point ethyl acetate. The amount of radioactivity in the acetylated product (100 μl top phase) and nonacetylated substrate (50 μl bottom phase) for each reaction was determined by liquid scintillation counting of organic and aqueous phases, respectively. Results are expressed as percentage of conversion of chloramphenicol to the acetylated form.
Results
Assay of NFATC1-GFP Subcellular Localization in Transiently Transfected RBL2H3 Mast Cells.
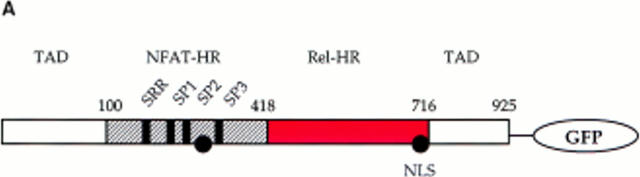
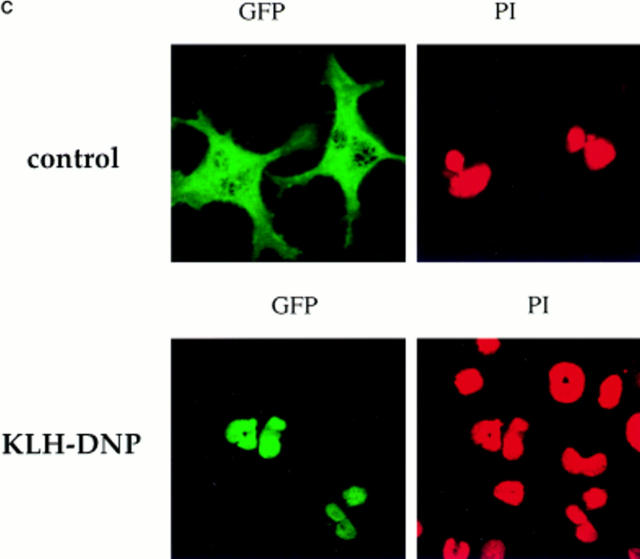
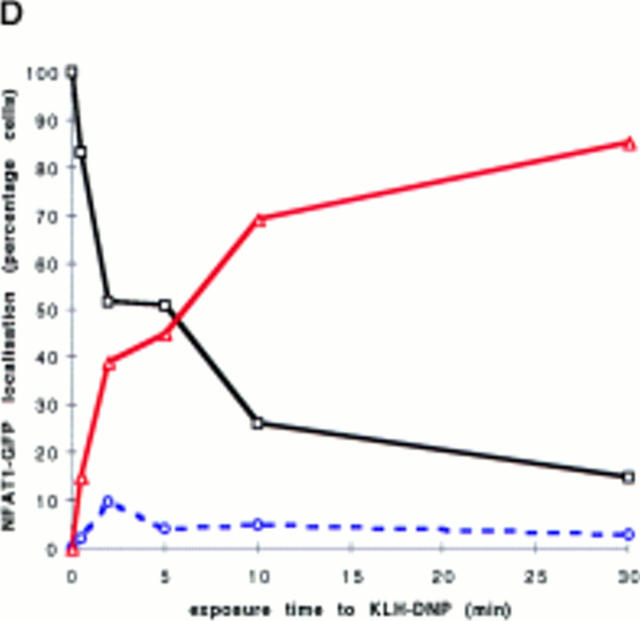
Initial experiments established a transient transfection system for the assay of NFATC1 subcellular localization in RBL2H3 mast cells. Fig. 1 A shows a schematic representation of the tagged NFATC1 molecule generated for these experiments. A fusion protein comprising NFATC1 (11) was linked to the GFP molecule of Aequorea victoria (25). The resulting expression plasmid, peGFP-NFATC1, was used in transient transfection of RBL2H3 cells. The montage of confocal microscope images in Fig. 1 B shows that NFATC1-GFP is excluded from the nucleus in quiescent RBL2H3 cells. The position of the nucleus in these cells is defined by the red fluorescence of the DNA binding dye PI. FcεR1 ligation results in expression of numerous genes controlled by NFAT transcription factor complexes. Accordingly, the location of NFATC1-GFP in resting and FcεR1-stimulated RBL2H3 cells was examined. Fig. 1 C shows that in resting RBL2H3 cells, NFATC1-GFP is predominantly localized to the cytosol and does not colocalize with the nuclear stain PI. After 30 min exposure to cross-linking antigen, NFATC1-GFP is exclusively nuclear. The kinetics of this response are shown in Fig. 1 D. In response to FcεR1 stimulation, NFATC1-GFP is exported from the cytosol, with a concomitant increase in nuclear GFP. Most nuclear import is achieved within 10 min. At 30 min stimulation, typically 80–90% cells exhibit exclusively nuclear NFATC1-GFP. Hence, FcεR1 stimulation of RBL2H3 cells results in the rapid nuclear import of NFATC1-GFP protein.
Figure 1.
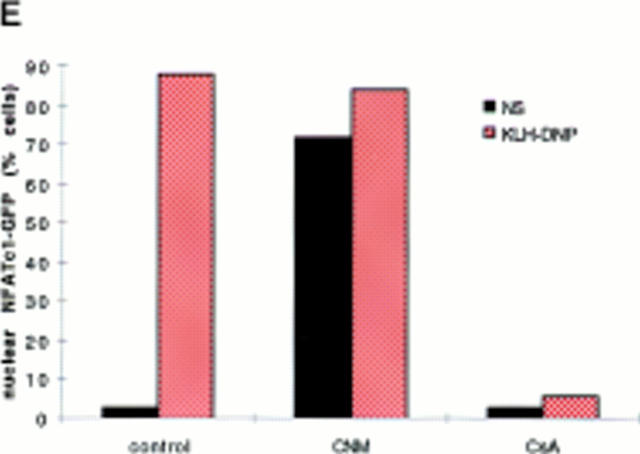
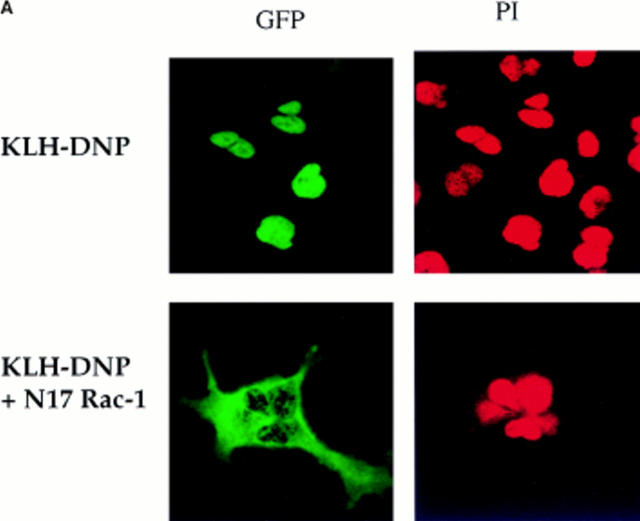
(A) NFATC1-GFP marker construct. Schematically shown here, the full-length NFATC1 molecule was fused to the GFP reporter by cloning into the pEGFP-1 vector (Clontech). TAD, Transactivation domain. HR, Homology region. SRR, Serine-rich region. SP, Ser/Pro box. (B) NFATC1-GFP is cytosolic in resting RBL2H3 mast cells. RBL2H3 cells were transfected with 8 μg NFATC1-GFP and recovered for 6 h on glass coverslips in complete medium at 37°C. Cells were costained with PI, fixed, and mounted as described. (C and D) FcεR1 cross-linking induces NFATC1-GFP nuclear import in RBL2H3 mast cells. RBL2H3 cells were transfected with 8 μg NFATC1-GFP and recovered for 6 h on glass coverslips in complete medium at 37°C. Cells were IgE primed and stimulated with 500 ng/ml KLH-DNP for 30 min (C) or for the indicated times (D). Cells were either costained with PI, fixed, and mounted as described (C), or cells were scored for predominant localization of GFP (D). (E) FcεR1 induction of NFATC1-GFP nuclear import is CN regulated and CsA sensitive. RBL2H3 cells were transfected with 8 μg NFATC1-GFP reporter alone or in combination with 20 μg activated CN plasmid (CNM) and recovered for 6 h on glass coverslips in complete medium at 37°C. Cells were primed and stimulated with 500 ng/ml KLH-DNP for 30 min in the absence or presence of 50 nM CsA. NS, Not stimulated. Cells were fixed, mounted, and scored for predominant localization of GFP as described.
Fusion to GFP is a major modification of the NFATC1 protein. Control experiments were performed to assess whether the behavior of NFATC1-GFP was comparable to that expected for endogenous NFAT. Dephosphorylation of NFAT by the CN phosphatase is permissive for nuclear import and for assembly of productive NFAT/ AP-1 complexes. CN activity is the target of the immunosuppressive drugs CsA and FK506. To be considered a good model for endogenous NFAT regulation, NFATC1-GFP nuclear import should therefore be positively regulated by CN and sensitive to inhibition by CsA. Fig. 1 E shows the results of experiments testing these criteria. In control cells singly transfected with reporter, NFATC1-GFP is predominantly nuclear after 30 min FcεR1 stimulation. In cells cotransfected with an activated mutant of CN, NFATC1-GFP is localized to the nucleus in the absence of FcεR1 stimulation. Conversely, pretreatment of RBL2H3 cells with CsA ablates FcεR1-induced NFATC1-GFP nuclear import. These data demonstrate that the subcellular localization of NFATC1-GFP is regulated by CN signaling pathways.
Rac-1– but not Ras-mediated Signals Regulate FcεR1-induced NFATC1-GFP Nuclear Import.
Previous experiments have identified that Rac-1 and Ras function is required for FcεR1 induction of NFAT transcriptional activity in mast cells (4). The effect of dominant inhibitory Rac-1 and Ras upon NFATC1-GFP nuclear import was examined. The N17 mutants of Ras and Rac-1 act to sequester GEF from endogenous pools of the GTPase, maintaining the GTPase in its GDP-bound (inactive) state. Cotransfection of these mutants prevents activation of the endogenous GTPases.
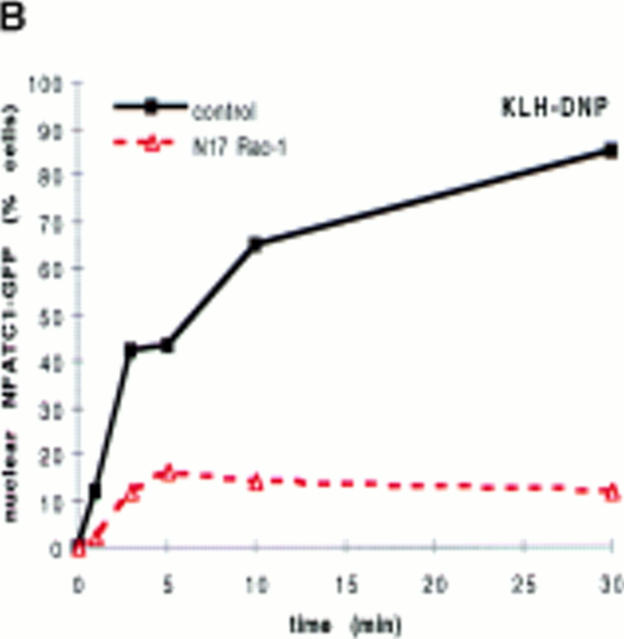
Fig. 2 A shows the subcellular localization of NFATC1-GFP in either control RBL2H3 cells or in cells cotransfected with N17Rac-1. In control cells after 30 min FcεR1 stimulation, NFATC1-GFP is exclusively localized to the nucleus. In receptor-stimulated cells expressing dominant inhibitory Rac-1, NFATC1-GFP is effectively retained in the cytosol. The data in Fig. 2 B show that across a population of cells, NFATC1-GFP nuclear accumulation is severely abrogated in N17Rac-1–expressing cells. After 30 min stimulation, only 15% of cells bearing N17Rac-1 have predominantly nuclear NFATC1-GFP, compared with 80–90% of control cells. These data suggest that Rac-1 signals are required for FcεR1-induced NFATC1-GFP nuclear import. NFAT nuclear translocation can also be induced by the pharmacological agent, Ionomycin, a calcium ionophore. The data in Fig. 2 C show the kinetics of NFATC1-GFP nuclear import in response to Ionomycin treatment in the absence or presence of N17Rac-1. In Ionomycin-treated cells, the rate of nuclear uptake of NFATC1-GFP in the presence of N17Rac-1 closely matched that of control cells. These results imply that expression of N17Rac-1 does not result in a blockade of the general nuclear import machinery in RBL2H3 cells.
Figure 2.
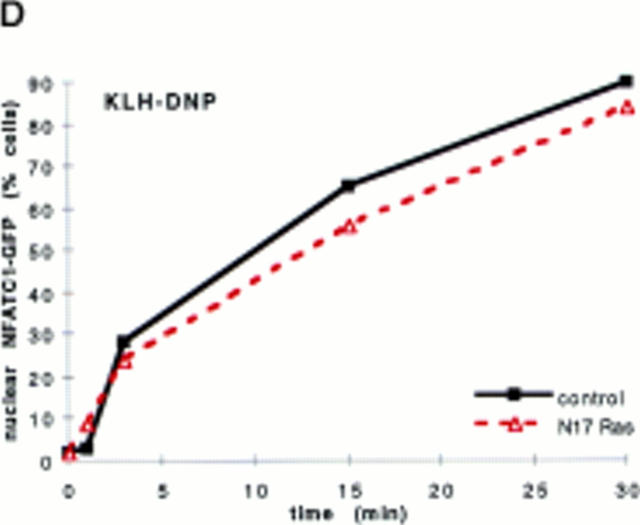
(A and B) N17Rac-1 prevents FcεR1-induced NFATC1-GFP nuclear import. RBL2H3 mast cells were transfected with 8 μg NFATC1-GFP reporter alone or in combination with 20 μg pEF-N17Rac-1. Cells were recovered for 6 h on glass coverslips before IgE priming and stimulation with 500 ng/ml KLH-DNP for 30 min (A) or for the indicated times (B). Cells were fixed and mounted, and localization of GFP was visualized as described in Materials and Methods (A) or scored for predominant localization of NFATC1-GFP (B). (C) Ionomycin induction of NFATC1-GFP nuclear accumulation is unaffected by N17Rac-1. RBL2H3 cells were transfected with 8 μg NFATC1-GFP alone or in combination with 20 μg pEF-N17Rac-1. Cells were seeded onto glass coverslips, recovered for 6 h, and stimulated for the indicated times using 500 ng/ml Ionomycin or vehicle control. Cells were fixed, mounted, and scored for predominant localization of GFP as described. (D) Dominant inhibition of Ras does not affect NFATC1-GFP nuclear import. RBL2H3 cells were transfected with 8 μg NFATC1-GFP alone or in combination with 20 μg pRSV-N17 Ras. Cells were seeded onto glass coverslips, recovered for 6 h, and stimulated for the indicated times using 500 ng/ml KLH-DNP. Cells were fixed, mounted, and scored for predominant localization of GFP as described.
Our previous work on NFAT transcriptional activation has shown that there is clearly a role for both Rac-1– and Ras-derived signals (4). The data in Fig. 2 D assess the effect of N17Ras upon FcεR1-regulated NFATC1-GFP nuclear import. The data show that N17Ras expression does not affect the ability of the FcεR1 to drive NFATC1-GFP translocation. In parallel control experiments, N17 Ras markedly inhibited the FcεR1 activation of an NFAT/AP-1 CAT reporter gene in RBL2H3 cells (data not shown). Therefore, the lack of effect upon NFATC1-GFP translocation does not reflect nonexpression of the N17Ras protein. These data suggest that Rac-1, but not Ras, has an important role in FcεR1 regulation of NFAT nuclear localization.
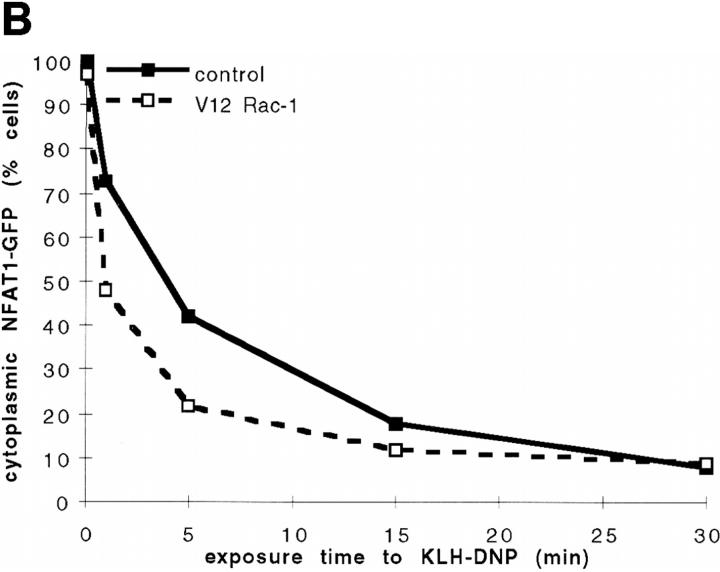
The data presented so far show that inhibition of Rac-1 signaling antagonizes FcεR1-induced NFATC1-GFP nuclear uptake. This suggests that one basis of the requirement for Rac-1 in FcεR1 activation of NFAT transcriptional activity may lie in Rac-1 regulation of NFAT nuclear accumulation. Given that activity of the Rac-1 GTPase is required for FcεR1 regulation of NFATC1-GFP subcellular localization, we asked how constitutive activation of Rac-1 would affect this phenomenon. The data in Fig. 3, A and B, show the effect of cotransfection of a constitutively active V12 mutant of Rac-1 (V12Rac-1) upon FcεR1-stimulated NFATC1-GFP nuclear localization. Cotransfection with V12Rac-1 potentiates the antigen dose response for FcεR1-stimulated NFATC1-GFP nuclear accumulation (Fig. 3 A). Moreover, Fig. 3 B shows that V12Rac-1 causes a slight but reproducible acceleration in the kinetics of FcεR1-stimulated NFATC1-GFP nuclear uptake. Activated alleles of GTPases may in theory acquire function that does not reflect that of the endogenous GTP-loaded protein; activated GTPases may interact with new regulators or effectors by virtue of overexpression or expression in an unphysiological cellular compartment. However, our combined data using inhibitory and activated alleles of the Rac-1 GTPase strongly support the conclusion that FcεR1 regulation of NFAT subcellular localization is Rac-1 dependent.
Figure 3.
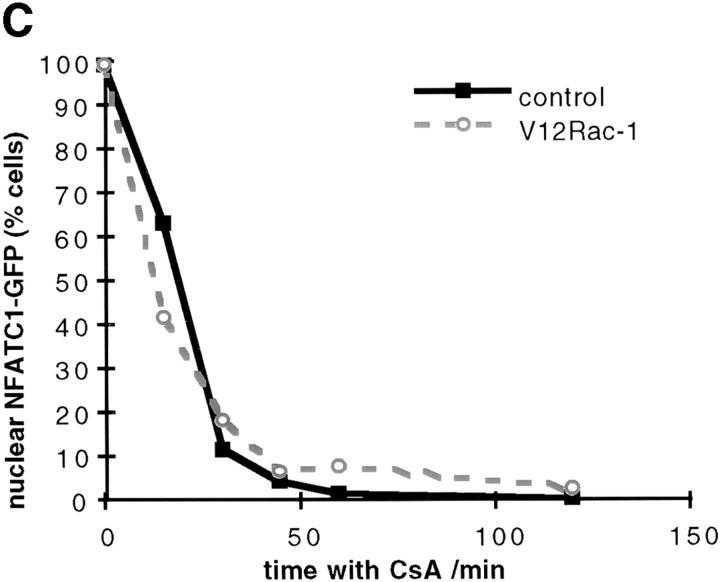
(A and B) Effect of V12Rac-1 upon NFATC1-GFP nuclear accumulation in RBL2H3 mast cells. RBL2H3 cells were transfected with 8 μg NFATC1-GFP reporter alone or in combination with 20 μg pEF-V12Rac-1. Cells were recovered for 6 h on glass coverslips before IgE priming and stimulation for 30 min with the indicated concentrations of KLH-DNP (A) or stimulation with 500 ng/ml KLH-DNP for the indicated times (B). Cells were fixed, mounted, and scored for predominant localization of GFP as described. (C) Effect of V12Rac-1 on NFATC1-GFP nuclear export in RBL2H3 mast cells. RBL2H3 cells were transfected with 8 μg NFATC1-GFP reporter alone or in combination with 20 μg pEF-V12Rac-1. Cells were recovered for 6 h on glass coverslips before stimulation for 30 min with 500 ng/ml Ionomycin to induce nuclear uptake of NFATC1-GFP. The medium was then exchanged twice with warm DMEM/10% FCS before addition of 50 nM CsA to prevent further import. At the indicated time points after CsA addition, cells were fixed, mounted, and scored for predominant localization of GFP as described.
The amount of NFAT protein present in the nucleus at any given time is controlled by a balance of the rates of nuclear import and export. The data presented above show that V12Rac-1 enhances the FcεR1-driven nuclear accumulation of NFATC1-GFP, but only slightly modifies the initial kinetics of nuclear import. Therefore, we examined whether expression of V12Rac-1 could influence NFATC1-GFP nuclear export. Protocols for the study of NFAT nuclear export have been established (11, 18). We induced nuclear accumulation of NFATC1-GFP using the calcium ionophore, Ionomycin. Ionomycin was washed out, and cells were treated with CsA to prevent continued nuclear import of NFATC1-GFP. NFATC1-GFP nuclear export kinetics were then monitored. Fig. 3 C shows the results of this experiment in control cells or those expressing V12Rac-1. The data show that relocalization of NFATC1-GFP from nucleus to cytosol in RBL2H3 cells occurs rapidly in response to CsA. Full export is achieved in 45 min exposure to CsA. It is clear that the kinetics or degree of NFATC1-GFP export from the nucleus are both unaffected by the presence of V12Rac-1.
Rac-1 Control of NFATC1-GFP Subcellular Localization Is Not Connected to Rac-1 Regulation of the Actin Cytoskeleton.
There is an established body of work on the role of the Rac GTPase in orchestration of actin cytoskeleton rearrangements (26, 27). FcεR1 stimulation of RBL2H3 mast cells results in marked rearrangements of the actin cytoskeleton. Stimulated RBL2H3 cells form actin plaques/focal complexes at their adherent surface and membrane ruffles at the top of the cell (28, 29). FcεR1-mediated NFATC1-GFP nuclear import involves the translocation of a considerable pool of protein from the cytosol to the nucleus. It is possible that this type of event requires a contribution from the actin cytoskeleton in the form of morphological changes in the RBL2H3 cells. Since there is evidence that these changes can be under the control of Rac-1, they are candidates for the basis of the role for Rac-1 in regulation of NFATC1 subcellular localization. Accordingly, the role of actin cytoskeleton rearrangements in NFATC1-GFP nuclear import was examined.
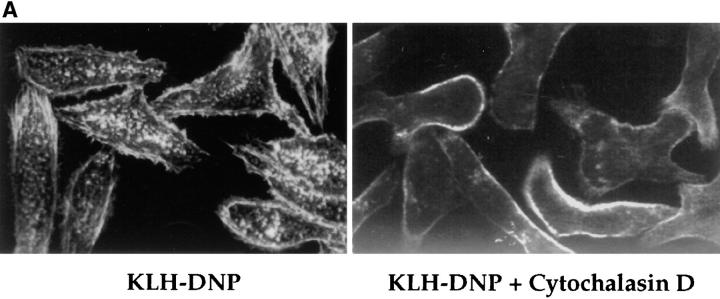
FcεR1 stimulation of RBL2H3 cells causes the formation of actin structures such as stress fibers, focal complexes, and membrane ruffles (Fig. 4 A, left), which are visualized using Rhodamine-Phalloidin staining for polymerized actin. Cytochalasin D is an inhibitor of actin polymerization. This compound prevents FcεR1-induced cytoskeletal changes such as the induction of focal complex formation and stress fibers. Fig. 4 A, left, shows FcεR1-stimulated RBL2H3 cells stained for polymerized actin. Marked membrane ruffles and punctate structures are observed in stimulated cells. In cells pretreated with Cytochalasin D for 30 min before FcεR1 cross-linking, complete disruption of the cytoskeleton was observed (Fig. 4 A, right). These data confirm the efficacy of Cytochalasin D.
Figure 4.
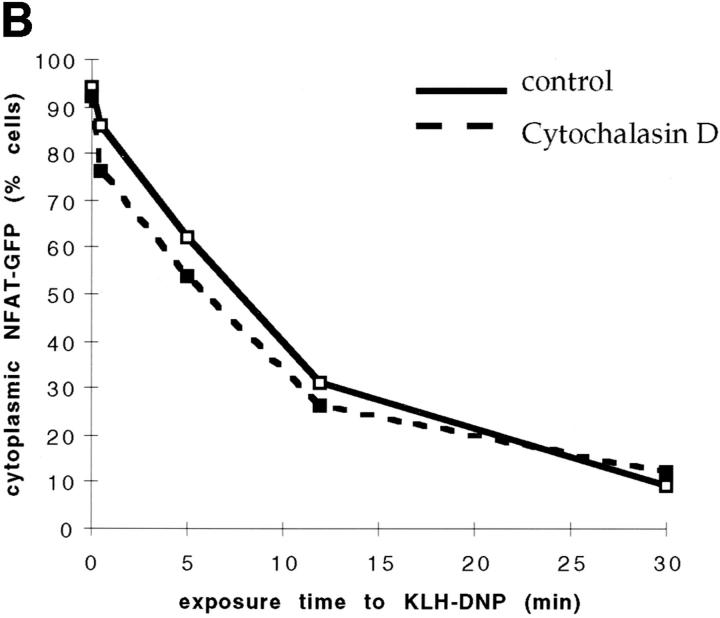
Cytochalasin D does not affect NFATC1-GFP nuclear import. (A) RBL2H3 mast cells were seeded onto glass coverslips. Cells were IgE primed and stimulated with 500 ng/ml KLH-DNP for 30 min in the absence (left) or presence (right) of 500 nM Cytochalasin D. Cells were fixed and permeabilized with Triton X-100, and polymerized actin was visualized using 0.25 μg/ml Rhodamine-Phalloidin. (B) RBL2H3 cells were transfected with 8 μg NFATC1-GFP reporter. Cells were recovered for 6 h on glass coverslips before IgE priming and stimulation for the indicated times with 500 ng/ml KLH-DNP in the absence or presence of 500 nM Cytochalasin D.
The effect of disruption of the actin cytoskeleton on NFATC1-GFP nuclear accumulation was then assessed. RBL2H3 cells were transfected with NFATC1-GFP reporter and incubated either with vehicle or with Cytochalasin D for 30 min before stimulation via IgE/FcεR1. The data in Fig. 4 B show that application of Cytochalasin D does not affect FcεR1-induced NFATC1-GFP nuclear translocation. In both control and Cytochalasin D–treated cells, FcεR1 stimulation leads to the rapid export of NFATC1-GFP from the cytosol and its concomitant accumulation in the nucleus. These data suggest that rearrangements of the actin cytoskeleton are in fact not required for FcεR1 regulation of NFAT nuclear import. Therefore, the basis of Rac-1 involvement in the regulation of NFATC1 subcellular localization is distinct from the published roles for Rac-1 in the regulation of the actin cytoskeleton.
In summary of the data presented so far, it is clear that a novel role for Rac-1 has been described. Rac-1 activity is a previously unrecognized player in the regulation of NFAT subcellular localization. Rac-1 is apparently a selective regulator of NFAT, functioning as a component of the FcεR1 signaling pathways regulating this transcription factor. This novel role for Rac-1 cannot be ascribed to a function for the GTPase at the level of general nuclear transport. Moreover, the mechanism by which Rac-1 accomplishes this function is separate from the established Rac-1 regulation of the actin cytoskeleton.
Rac-1 Regulates NFATC1 Phosphorylation.
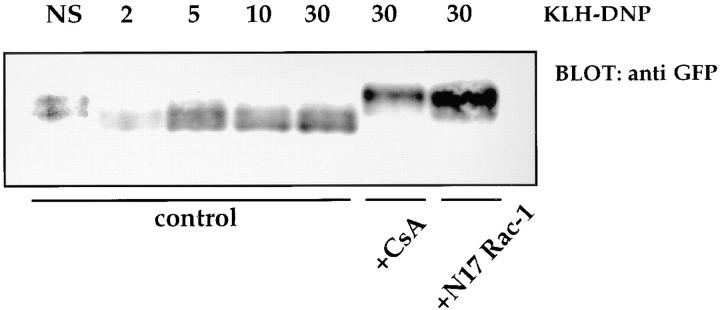
NFAT proteins are constitutively serine phosphorylated at multiple phosphoacceptor sites in quiescent cells. Upon antigen receptor ligation, calcium-dependent activation of the CN phosphatase results in the dephosphorylation of a number of these sites and permits nuclear import. The effect of N17Rac-1 upon FcεR1 regulation of NFATC1-GFP phosphorylation status was examined. RBL2H3 cells were transfected with NFATC1-GFP alone or in combination with pEF-N17Rac-1. Phosphorylated and dephosphorylated NFAT proteins have distinct electrophoretic mobilities in SDS-PAGE. The data in Fig. 5 show that antigenic cross-linking of the FcεR1 results in increased mobility of NFATC1-GFP, reflecting dephosphorylation by CN. Dephosphorylation is evident within 2 min exposure to cross-linking antigen and is sustained for 30 min. Pretreatment of RBL2H3 cells with CsA prevents FcεR1-mediated dephosphorylation of NFATC1-GFP; in this case, NFATC1-GFP is retained in the lower mobility, phosphorylated form. Fig. 5 also shows the effect of N17Rac-1 expression on the FcεR1-stimulated NFATC1-GFP mobility shift. In cells bearing N17Rac-1, NFATC1-GFP is effectively retained in the lower electrophoretic mobility form. Hence, dominant inhibition of Rac-1 function blocks NFATC1-GFP dephosphorylation.
Figure 5.
FcεR1-mediated NFATC1-GFP dephosphorylation is inhibited by N17Rac-1 or CsA. 107 RBL2H3 cells per point were transfected with 10 μg NFATC1-GFP alone or in combination with 20 μg pEF-N17Rac-1 and recovered for 6 h in the absence or presence of 50 nM CsA, before IgE priming and stimulation for the indicated times (in minutes) with 500 ng/ml KLH-DNP. NS, Not stimulated. Cells were harvested and lysed for Western blot analysis in a buffer containing 1% NP-40, 50 mM Hepes, pH 7.4, 10 mM iodoacetamide, 1 mM PMSF, and 0.42 M NaCl. Proteins were acetone precipitated and resolved by 6% SDS-PAGE before anti-GFP Western blot analysis using 1 μg/ml anti-GFP.
Ras-dependent and -independent Rac-1 Signals Target the NFAT–AP-1 Transcription Factor Complex.
The requirement for Rac- but not Ras-mediated signals for NFATC1 nuclear import indicates that these GTPases can function in independent signaling pathways to control NFAT function. There are also examples where Rac mediates Ras-induced cellular responses. Ras regulates fibroblast transformation via activation of a Rac-1 signaling pathway (30, 31). Moreover, Ras-mediated activation of NFAT in T lymphocytes is dependent on Rac function (10). Therefore, we explored the possibility that Rac might have a second function in mast cells and mediate Ras regulation of NFAT. The relationship between Ras and Rac-1 in mast cells was examined more closely using an assay for NFAT transcriptional activity. The NFAT/AP-1 reporter gene consisted of a CAT reporter controlled by an NFAT/AP-1 site derived from the murine IL-4 promoter (IL-4 NFAT/AP-1 CAT). These experiments tested the ability of activated mutants of Ras and Rac to rescue the effects of the inhibitory Ras and Rac-1 mutants on FcεR1 induction of IL-4 NFAT/AP-1 CAT.
The data in Fig. 6 A show that N17Ras cotransfection causes a marked inhibition of NFAT/AP-1 transcriptional activity induced in response to antigenic cross-linking of the FcεR1. However, if N17Ras and the activated V12 mutant of Rac-1 (V12Rac-1) are doubly cotransfected, the inhibition observed with N17Ras alone is alleviated. Hence, V12Rac-1 rescues N17Ras inhibition of FcεR1-induced NFAT/AP-1 transcriptional activity. These data suggest that there is a linear Ras/Rac-1 pathway regulating the activity of the NFAT/AP-1 complex in mast cells. The data in Fig. 6 B show that conversely, an activated mutant of Ras cannot rescue the N17Rac-1 inhibition of NFAT induction.
Figure 6.
Activated Rac-1 can rescue the dominant inhibition of NFAT/AP-1 transcriptional activity by N17Ras. 107 RBL2H3 cells per condition were transfected with 15 μg IL-4 NFAT/AP-1 CAT reporter alone or in combination with the indicated regulatory plasmids (A) 15 μg pEF-N17Rac-1, 15 μg RSV-N17 Ras; (B) 15 μg pEF-V12Rac-1, and 15 μg pEF-V12 Ras. Cells were recovered for 6 h, then stimulated using the indicated doses of KLH-DNP for 16 h. Cells were harvested and assayed for CAT activity as described. Results are representative of three experiments.
Discussion
Regulation of NFAT transcription factor complexes by antigen receptors such as the FcεR1 is crucial to the initiation and maintenance of immune responses. This report describes that the GTPase Rac-1 has a critical function in FcεR1 control of the subcellular localization of NFATC1 and a separate function in the regulation of the transcriptional activity of nuclear localized NFAT molecules. Accordingly, activity of Rac-1 is required for FcεR1-mediated NFATC1 dephosphorylation and nuclear import. Disruption of actin polymerization and, hence, cytoskeletal structure has no impact on the regulated nuclear import of NFAT. Thus, the role of Rac-1 in control of NFAT subcellular localization is independent of the established role of this GTPase in control of the actin cytoskeleton. These results also show that in FcεR1-activated cells, NFAT complexes are nuclear localized in the absence of endogenous Ras function but transcriptionally inactive. However, NFAT transcriptional function can be restored by activation of Rac signaling pathways. Thus, we are able to place Rac-1 in two positions in the signaling network that regulates the assembly and activation of NFAT/AP-1 transcriptional complexes in lymphocytes, as follows.
Dephosphorylation of NFAT molecules permits their subsequent nuclear import. In quiescent cells, NFAT is phosphorylated upon multiple sites within the NH2-terminal regulatory region. Upon antigen receptor stimulation, dephosphorylation of NFAT is rapid and sustained, and it is proposed that dephosphorylation results in a conformational change that exposes previously buried nuclear localization sequences (NLS). Thus, NFAT subcellular localization is controlled by the phosphorylation state of this molecule. The effect of inhibition of Rac-1 signaling in RBL2H3 cells is to maintain NFATC1 in a phosphorylated state and, hence, in a “closed” conformation where NLS remain buried, causing cytosolic retention of NFATC1. Protein phosphorylation levels are determined by a balance of regulatory kinase and phosphatase activities. The calcium-dependent phosphatase CN is responsible for NFAT dephosphorylation, whereas NFAT kinases have a fundamental role in regulating NFAT subcellular localization. In nonlymphoid cells, the nuclear localization of NFAT4 is prevented when this NFAT isoform is phosphorylated by the MAP kinase c-Jun NH2-terminal kinase (JUN; reference 19). It has also been described that NFATC1 is a substrate for glycogen synthase kinase (GSK)-3β and that phosphorylation of NFATC1 by GSK-3β promotes its nuclear export (18). In a variety of cell types, GSK-3β is inactivated in response to stimulation of cells via protein kinase C or protein kinase B (32, 33). A failure to inactivate GSK-3β would be assumed to promote NFAT phosphorylation and cytoplasmic retention. For the interpretation of our current data, two clear possibilities exist. First, Rac-1 function may regulate the activity of the kinase(s) responsible for NFAT phosphorylation and nuclear export. Second, Rac-1 may regulate the activity of the NFAT phosphatases that control nuclear import. These results show that dominant activation of Rac signaling pathways did not prevent NFAT nuclear export. These data suggest that Rac-1 is exerting its effects through regulation of NFAT import into the nucleus. Therefore, in terms of the current simple models for the regulation of NFAT subcellular localization, it seems most likely that Rac-1 regulates NFAT nuclear import by regulating the activity and/or localization of the NFAT phosphatase CN.
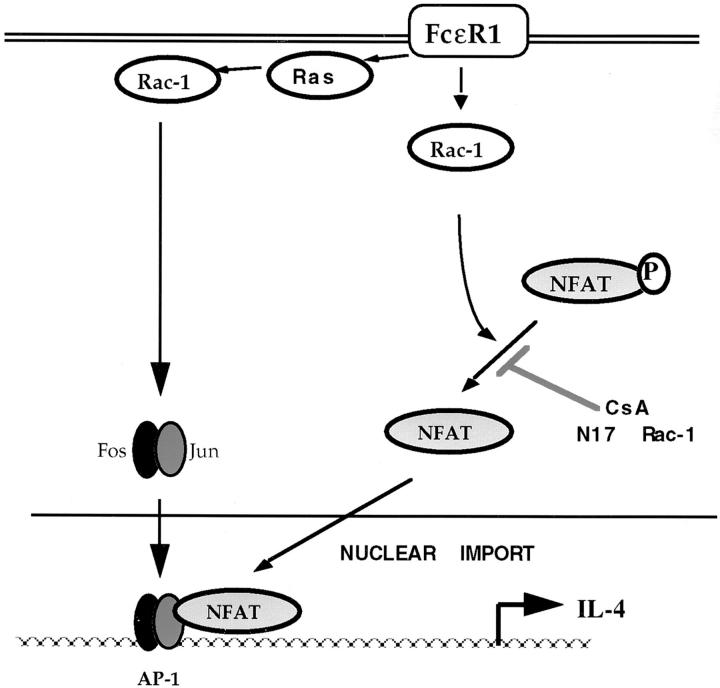
Rac-1 signals that affect NFATC1 nuclear import are independent of the activity of the Ras GTPase. However, Ras signals are necessary for FcεR1 induction of the transcriptional activity of nuclear localized NFAT. Therefore, a simple model would show parallel Ras/Rac FcεR1 signaling pathways regulating NFAT. In this model, a Rac-1–mediated pathway controls NFATC1 subcellular localization, whereas Ras regulates the transcriptional activity of the complex without regulating NFATC1 distribution. However, this simple model is not adequate to explain our previous observations that Ras regulates NFAT transcriptional activity in a Rac-1–dependent fashion. Moreover, the data presented here show that activation of Rac-1 signaling pathways can restore transcriptional activity of nuclear localized NFAT in cells lacking endogenous Ras function. Thus, there is a component of NFAT regulation in mast cells that is both Ras and Rac-1 dependent; Rac-1 signals can clearly regulate the transcriptional activity of NFAT complexes as well as regulating their subcellular location. To reconcile these data, a model is proposed in Fig. 7.
Figure 7.
Model of NFAT/AP-1 complex regulation by Ras family GTPases in mast cells. Ras and Rac-1 are required for transcriptional activity of the NFAT/AP-1 complex. Ras signals do not regulate the subcellular localization of NFAT protein. Rac-1 regulates NFAT phosphorylation status and, hence, nuclear import. The effect of dominant inhibition of Ras upon NFAT/AP-1 transcriptional activity can be rescued by activated Rac-1, indicating that there is a Ras-dependent Rac-1 signaling pathway acting upon the NFAT/AP-1 complex in addition to the Ras-independent effect of Rac-1 upon NFAT subcellular localization. This latter pathway is postulated to target the AP-1 binding partner of NFAT.
In Fig. 7, two pools of Rac-1 are shown to affect distinct processes contributing to NFAT transcriptional activity. First, Rac-1 is postulated to couple Ras to NFAT complexes. Second, a Ras-independent Rac-1 signal regulates the phosphorylation status and, hence, subcellular localization of NFAT protein. NFAT transcription factors function in the context of a dimer of AP-1 family proteins. In the T cell system, Ras and Rac-1 functions have clearly defined roles in AP-1 regulation and are required for AP-1 transcriptional activity. Therefore, the role of these GTPases in the induction of transcriptional activity of NFAT complexes could reflect their contribution to AP-1 activity. Nevertheless, the data presented here show that the role of Rac-1 in NFAT induction cannot be considered solely in the context of AP-1 regulation; Rac-1 also directly influences the subcellular localization of NFAT protein.
In summary, the Rac-1 GTPase has a previously unsuspected role as a regulator of NFATC1 phosphorylation status and subcellular localization. Rac-1 also functions to control the transcriptional activity of nuclear localized NFAT complexes. These experiments on the role of Rac-1 in NFAT regulation illustrate several important aspects of signal transduction by Rac GTPases. First, it is clear that not all functions of Rac GTPases can be attributed to the role of the GTPase in regulation of the actin cytoskeleton. Second, our data show that functionally distinct pools of a GTPase may coexist within the same cell and regulate disparate targets in response to similar upstream signals. Numerous Rac effectors have been identified, including the Pak family of serine/threonine kinases and mixed lineage kinases (MLK)2,3, the tyrosine kinase p120Ack, partner of Rac-1 (POR1), p67phox, MEKs, and the phosphatidylinositol 4-phosphate 5-kinase (PI4-P 5-kinase). Rac GTPases are able to regulate diverse cellular responses because they can couple to these multiple effector molecules. Hence, Rac-1 regulation of NFAT cellular localization or NFAT transcriptional activity may reflect the activation of distinct effector cascades.
Acknowledgments
The authors thank Alex Stokes and Peter Jordan (Confocal Imaging Laboratory, Imperial Cancer Research Fund) for excellent advice on microscopy, and Dr. Paul Brennan and Steve Cleverley (Lymphocyte Activation Laboratory, Imperial Cancer Research Fund) for discussion of and help with the manuscript. Rac-1 constructs were generously provided by Dr. Alan Hall (University College, London, UK).
Abbreviations used in this paper
- AP-1
activator protein 1
- CAT
chloramphenicol acetyl transferase
- CN
calcineurin
- CsA
Cyclosporin A
- GEF
guanine nucleotide exchange factor
- GFP
green fluorescent protein
- GSK
glycogen synthase kinase
- PI
propidium iodide
- NFAT
nuclear factor of activated T cells
- NLS
nuclear localization sequences
Footnotes
H. Turner was supported by Glaxo-Wellcome and Imperial Cancer Research Fund.
H. Turner's present address is Laboratory of Allergy and Immunology, Beth Israel Hospital, Harvard Medical School, 99 Brookline Ave., Boston, MA 02215.
References
- 1.Daeron M. Fc receptor biology. Annu Rev Immunol. 1997;15:203–234. doi: 10.1146/annurev.immunol.15.1.203. [DOI] [PubMed] [Google Scholar]
- 2.Jouvin MH, Numerof RP, Kinet JP. Signal transduction through the conserved motifs of the high affinity IgE receptor Fc epsilon RI. Semin Immunol. 1995;7:29–35. doi: 10.1016/1044-5323(95)90005-5. [DOI] [PubMed] [Google Scholar]
- 3.Rao A, Luo C, Hogan P. Transcription factors of the NFAT family: regulation and function. Annu Rev Immunol. 1997;15:707–748. doi: 10.1146/annurev.immunol.15.1.707. [DOI] [PubMed] [Google Scholar]
- 4.Turner H, Cantrell DA. Distinct Ras effector pathways are involved in FcεR1 regulation of the transcriptional activity of Elk-1 and NFAT in mast cells. J Exp Med. 1997;185:43–57. doi: 10.1084/jem.185.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boguski MS, McCormick F. Proteins regulating Ras and its relatives. Nature. 1993;366:643–654. doi: 10.1038/366643a0. [DOI] [PubMed] [Google Scholar]
- 6.Downward J. Control of ras activation. Cancer Surv. 1996;27:87–100. [PubMed] [Google Scholar]
- 7.Turner H, Reif K, Rivera J, Cantrell DA. Regulation of the adapter molecule Grb2 by the FcεR1 in the mast cell line RBL2H3. J Biol Chem. 1995;270:9500–9506. doi: 10.1074/jbc.270.16.9500. [DOI] [PubMed] [Google Scholar]
- 8.Crespo P, Schuebel KE, Ostrom AA, Gutkind JS, Bustelo XR. Phosphotyrosine-dependent activation of Rac-1 GDP/GTP exchange by the vavproto-oncogene product. Nature. 1997;385:169–172. doi: 10.1038/385169a0. [DOI] [PubMed] [Google Scholar]
- 9.Margolis B, Hu P, Katzav S, Li W, Oliver JM, Ullrich A, Weiss A, Schlessinger J. Tyrosine phosphorylation of vav proto-oncogene product containing SH2 domain and transcription factor motifs. Nature. 1992;356:71–74. doi: 10.1038/356071a0. [DOI] [PubMed] [Google Scholar]
- 10.Genot E, Cleverley S, Henning S, Cantrell DA. Multiple p21ras effector pathways regulate nuclear factor of activated T cells. EMBO (Eur Mol Biol Organ) J. 1996;15:3923–3933. [PMC free article] [PubMed] [Google Scholar]
- 11.Beals CR, Clipstone NA, Ho SN, Crabtree GR. Nuclear localization of NF-ATc by a calcineurin-dependent, cyclosporin A-sensitive intramolecular interaction. Genes Dev. 1997;11:824–834. doi: 10.1101/gad.11.7.824. [DOI] [PubMed] [Google Scholar]
- 12.Jain J, McCaffrey PG, Miner Z, Kerppola TK, Lambert JN, Verdine GL, Curran T, Rao A. The T-cell transcription factor NFATp is a substrate for calcineurin and interacts with Fos and Jun. Nature. 1993;365:352–355. doi: 10.1038/365352a0. [DOI] [PubMed] [Google Scholar]
- 13.Schreiber SL, Crabtree GR. The mechanism of action of cyclosporin A and FK506. Immunol Today. 1992;13:136–142. doi: 10.1016/0167-5699(92)90111-J. [DOI] [PubMed] [Google Scholar]
- 14.Loh C, Shaw KT, Carew J, Viola JP, Luo C, Perrino BA, Rao A. Calcineurin binds the transcription factor NFAT1 and reversibly regulates its activity. J Biol Chem. 1996;271:10884–10891. doi: 10.1074/jbc.271.18.10884. [DOI] [PubMed] [Google Scholar]
- 15.Shibasaki F, Price ER, Milan D, McKeon F. Role of kinases and the phosphatase calcineurin in the nuclear shuttling of transcription factor NF-AT4. Nature. 1996;382:370–373. doi: 10.1038/382370a0. [DOI] [PubMed] [Google Scholar]
- 16.Genot EM, Parker PJ, Cantrell DA. Analysis of the role of protein kinase Cα, -ε, and -ζ in T cell activation. J Biol Chem. 1995;270:9833–9839. doi: 10.1074/jbc.270.17.9833. [DOI] [PubMed] [Google Scholar]
- 17.Woodrow M, Clipstone NA, Cantrell D. p21ras and calcineurin synergize to regulate the nuclear factor of activated T cells. J Exp Med. 1993;178:1517–1522. doi: 10.1084/jem.178.5.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beals CR, Sheridan CM, Turck CW, Gardner P, Crabtree GR. Nuclear export of NF-ATc enhanced by glycogen synthase kinase-3. Science. 1997;275:1930–1933. doi: 10.1126/science.275.5308.1930. [DOI] [PubMed] [Google Scholar]
- 19.Chow CW, Rincon M, Cavanagh J, Dickens M, Davis RJ. Nuclear accumulation of NFAT4 opposed by the JNK signal transduction pathway. Science. 1997;278:1638–1641. doi: 10.1126/science.278.5343.1638. [DOI] [PubMed] [Google Scholar]
- 20.Ridley AJ, Paterson HF, Johnston CL, Diekmann D, Hall A. The small GTP binding protein rac regulates growth factor-induced membrane ruffling. Cell. 1992;70:401–410. doi: 10.1016/0092-8674(92)90164-8. [DOI] [PubMed] [Google Scholar]
- 21.Nigg EA. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature. 1997;386:779–786. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- 22.Sweet DJ, Gerace L. A GTPase distinct from Ran is involved in nuclear protein import. J Cell Biol. 1996;133:971–983. doi: 10.1083/jcb.133.5.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Izquierdo M, Leevers SJ, Marshall C, Cantrell DA. p21ras couples the T cell antigen receptor to extracellular signal–regulated kinase 2 in T lymphocytes. J Exp Med. 1993;178:1199–1208. doi: 10.1084/jem.178.4.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chuvpilo S, Schomberg C, Gerwig R, Heinfling A, Reeves R, Grummt F, Serfling E. Multiple closely linked NFAT/octamer and HMG I(Y) binding sites are part of the interleukin-4 promoter. Nucleic Acids Res. 1993;21:5694–5704. doi: 10.1093/nar/21.24.5694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heim R, Tsien RY. Engineering green fluorescent protein for improved brightness, longer wavelengths and fluorescence resonance energy transfer. Curr Biol. 1996;6:178–182. doi: 10.1016/s0960-9822(02)00450-5. [DOI] [PubMed] [Google Scholar]
- 26.Nobes C, Hall A. Rho, Rac and Cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 1995;81:53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- 27.Tapon N, Hall A. Rho, Rac and Cdc42 GTPases regulate the organization of the actin cytoskeleton. Curr Opin Cell Biol. 1997;9:86–92. doi: 10.1016/s0955-0674(97)80156-1. [DOI] [PubMed] [Google Scholar]
- 28.Norman JC, Price LS, Ridley A, Hall A, Koffer A. Actin filament organization on activated mast cells is regulated by heterotrimeric and small GTP-binding proteins. J Cell Biol. 1994;126:1005–1015. doi: 10.1083/jcb.126.4.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pfeiffer JR, Oliver JM. Tyrosine kinase-dependent assembly of actin plaques linking Fc epsilon R1 cross-linking to increased cell substrate adhesion in RBL-2H3 tumor mast cells. J Immunol. 1994;152:270–279. [PubMed] [Google Scholar]
- 30.Khosravi-Far R, Solski PA, Clark GJ, Kinch MS, Der CJ. Activation of Rac1, RhoA, and mitogen-activated protein kinases is required for Ras transformation. Mol Cell Biol. 1995;15:6443–6453. doi: 10.1128/mcb.15.11.6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qiu RG, Chen J, Kirn D, McCormick F, Symons M. An essential role for Rac in Ras transformation. Nature. 1995;374:457–459. doi: 10.1038/374457a0. [DOI] [PubMed] [Google Scholar]
- 32.Cross DAE, Alessi DR, Cohen P, Andjelkovich M, Hemmings BA. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature. 1995;378:785–789. doi: 10.1038/378785a0. [DOI] [PubMed] [Google Scholar]
- 33.Welsh G, Miyamoto S, Price N, Safer B, Proud C. T cell activation leads to rapid stimulation of eIF2B and inactivation of glycogen synthase kinase 3. J Biol Chem. 1996;271:11410–11413. doi: 10.1074/jbc.271.19.11410. [DOI] [PubMed] [Google Scholar]