Abstract
We have previously observed that HIV-1 replication is suppressed in uninflamed lung and increased during tuberculosis. In vitro THP-1 cell–derived macrophages inhibited HIV-1 replication after infection with Mycobacterium tuberculosis. Suppression of HIV-1 replication was associated with inhibition of the HIV-1 long terminal repeat (LTR) and induction of ISGF-3, a type I interferon (IFN)–specific transcription factor. Repression of the HIV-1 LTR required intact CCAAT/enhancer binding protein (C/EBP) sites. THP-1 cell–derived macrophages infected with M. tuberculosis, lipopolysaccharide, or IFN-β induced the 16-kD inhibitory C/EBPβ isoform and coincidentally repressed HIV-1 LTR transcription. C/EBPβ was the predominant C/EBP family member produced in THP-1 macrophages during HIV-1 LTR repression. In vivo, alveolar macrophages from uninflamed lung strongly expressed inhibitory 16-kD C/EBPβ, but pulmonary tuberculosis abolished inhibitory C/EBPβ expression and induced a novel C/EBP DNA binding protein. Therefore, in vitro, proinflammatory stimulation produces an IFN response inhibiting viral replication by induction of a C/EBPβ transcriptional repressor. THP-1 cell–derived macrophages stimulated with type I IFN are similar to alveolar macrophages in the uninflamed lung in vivo. In contrast, the cellular immune response in active pulmonary tuberculosis disrupts this innate immunity, switching C/EBP expression and allowing high level viral replication.
Keywords: interferon β, CCAAT/enhancer binding protein β, HIV-1 long terminal repeat, tuberculosis, repression
AIDS patients without pneumonia have very low levels of HIV-1 provirus or viral particles in the lung. Only 1 in 105 alveolar macrophages is infected (1), and AIDS patients with clear chest x rays have no detectable virus in bronchoalveolar lavage (BAL)1 fluid (2). The situation is remarkably different in patients with pulmonary infiltrates: the number of lung cells infected and the level of virus produced are increased up to 1,000-fold (2–5). The alveolar macrophage is an important source of HIV-1 production in patients with tuberculosis (6). The high levels of virus produced during infections such as tuberculosis and the increased viral mutation produced by enhanced viral replication (2) may contribute to the accelerated course of AIDS after opportunistic infection (7).
Inflammatory stimuli such as TNF-α and LPS suppress viral replication in macrophages (8–12). The negative regulatory element (NRE) is the DNA element responsible for suppressing the HIV-1 LTR after LPS stimulation (8). The transcription repressors induced by LPS stimulation of macrophages have not been identified. The suppression of HIV-1 replication seen after LPS stimulation closely resembles the suppression seen after addition of IFNs (9). Type I IFN is required for the suppressive effects of LPS and TNF-α on viral replication (11, 12). Macrophages derived from knockout mice deficient in the IFN receptor α/β p100, also known as IFNR-1, do not repress viral replication after stimulation with LPS or TNF-α (12, 13). In macrophages with intact type I IFN receptors, antibodies to IFN-β but not IFN-α will block the suppressive effects of proinflammatory stimuli (11). IFN-β is also the most potent inhibitor of HIV-1 replication in macrophages, producing a 99% inhibition of p24 production after treatment with 1 U/ml (9).
IFN-α and IFN-β are the major type I IFNs, which constitute an essential early arm of the innate immune response (14). Type I IFNs are expressed at low levels in normal tissue macrophages (15). Knockout mice deficient in type I IFN receptors have reduced levels of IFN responsive enzymes in macrophages compared with normal mice, supporting the concept that macrophages are primed by low levels of type I IFN in the absence of viral infection (13). Signal transduction in the type I IFN system is mediated by a high-affinity transmembrane receptor composed of two subunits (16). Knockout mice deficient in the p100 subunit are particularly susceptible to viral infection (13, 17). Upon binding of type I IFNs to their receptor, specific protein tyrosine kinases are activated, and signal transducer and activator of transcription (Stat) proteins are phosphorylated. IFN-stimulated gene factor 3 (ISGF-3) is then rapidly assembled without the need for protein synthesis. ISGF-3 is a heterotrimer of Stat-1, Stat-2, and p48 that rapidly translocates to the nucleus, binds promoter sequences named IFN-stimulated response elements (ISRE), and activates gene expression (18).
The HIV-1 LTR NRE has three CCAAT/enhancer binding protein (C/EBP) sites that bind C/EBPβ (19). The C/EBPβ gene has no introns, but two different proteins are produced from the same mRNA. Large 30–37-kD isoforms stimulate transcription while small (∼16–20-kD) isoforms repress transcription (20, 21). The small inhibitory C/EBPβ is likely produced when the ribosome starts protein translation from an internal ribosome entry site in the C/EBPβ mRNA. Calame and colleagues transfected constructs expressing inhibitory 16-kD C/EBPβ isoform and demonstrated inhibition of HIV-1 replication and LTR promoter function (22, 23). Further, intact C/EBP sites are required for HIV-1 replication in macrophages but not lymphocytes (24). C/EBP sites in the LTR of Rous sarcoma virus can mediate either stimulation or repression of promoter function depending on which C/EBP isoform is expressed (25). These data suggest that transcription factors binding to the C/EBP sites in the LTR are important enhancers and repressors of replication in monocytes. Cytokine genes such as TNF-α also have C/EBP binding sites, and transfection of inhibitory C/EBPβ represses TNF-α promoter activity (26).
In this investigation, we demonstrate that Mycobacterium tuberculosis infection of THP-1 macrophages suppresses HIV-1 replication and induces a type I IFN response. This is accompanied by repressed LTR transcription that depends on intact C/EBP sites in the LTR. An inhibitory 16-kD C/EBPβ transcription factor is induced by M. tuberculosis or LPS, and C/EBPβ accounts for >95% of the LTR NRE binding. Treatment of macrophages with IFN-β also represses LTR transcription and induces the inhibitory C/EBPβ. Resting alveolar macrophages strongly express the inhibitory C/EBPβ but downregulate it in pulmonary tuberculosis and switch expression to another C/EBP site binding factor.
Materials and Methods
Study Population.
We evaluated seven patients with active pulmonary tuberculosis with sputum or lung tissue culture positive for M. tuberculosis. Tuberculosis patients had received antimycobacterial therapy for 1–7 d before fiberoptic bronchoscopy. HIV-1 testing on all patients was done with ELISA and confirmed by Western blot; five out of seven were HIV-1 positive. None were on antiretroviral therapy during the acute illness. All patients had segmental infiltrates; radiographically uninvolved lobes were identified, and a separate BAL was performed and processed from this segment. Three out of the seven were smokers; all seven were male. Their mean age ± SD was 40 ± 11 yr. Two normal volunteers were lavaged; both were HIV-1 negative, and both had a normal chest radiograph and spirometry. BAL was approved by the Human Subjects Review Committees of New York University Medical Center and Bellevue Hospital Center.
BAL Cells.
BAL was performed with a flexible fiberoptic bronchoscope with local Xylocaine anesthesia. We instilled six 50-ml aliquots (total 300 ml) of normal saline and suctioned sequentially from radiographically uninvolved and involved segments. The lavage fluid was filtered through sterile gauze to remove mucous, and total cells were counted in a hemacytometer. Cell differentials were performed on cytospin slides stained with Wright-Giemsa. For purification of alveolar macrophages, BAL cells were spun and resuspended in RPMI 1640 and allowed to adhere to plastic plates for 3 h. Cells were recovered by gentle scraping with a rubber policeman. Alveolar macrophages were 95% pure by morphology and nonspecific esterase staining.
HIV-1–Mycobacteria Coinfection.
THP-1 cells (104 cells/well in 96-well plates) were incubated overnight with 10 ng/ml PMA. Cells were infected with HIV-1 strains BaL or NL4-3 (1 ng p24/ ml) and M. tuberculosis H37Ra at various multiplicities of infection (moi) for 2 h at 37°C. They were washed twice and incubated with RPMI 1640/10% FCS containing PMA. p24 was assayed by ELISA.
Stable LTR Reporter Constructs.
BF-24 cells, obtained from the AIDS Reference Reagent Program (#1296; Rockville, MD), are THP-1 cells with an integrated HIV-1 LTR.CAT construct. pHIV-CAT LTR reporter construct was obtained from the AIDS Reference Reagent Program (#2619). The HindIII-XhoI LTR fragment was transferred to the HindIII-XhoI site of pGL 3 (Stratagene, Inc., La Jolla, CA). The SalI-NotI pGL 3 LTR luciferase fragment was then transferred to a modified pCR3 plasmid carrying a NotI site (Invitrogen Corp., Carlsbad, CA). Double-stranded PCR mutagenesis was performed according to the manufacturer's instruction (Stratagene, Inc.). Three oligonucleotides and their reverse complements were used to mutate the C/ EBP domains 1, 2, and 3 of the HIV-1 LTR (see Fig. 3 for position of C/EBP domains and mutations introduced). Mutants were selected for addition of PstI or PvuII sites and were confirmed by complete sequencing of the LTR. 2 × 106 THP-1 cells were transfected with 2 μg of mutant or wild-type plasmid mixed with 10 μl of lipofectamine (GIBCO BRL, Gaithersburg, MD) for 5 h; transfected cells were cultured in RPMI/10% FCS at 105 cells/well in 24-well plates. After 24 h, 700 μg/ml of G418 was added. Resistant clones were expanded and screened for luciferase production.
Figure 3.
Nucleotide sequence of the HIV-1 LTR. C/EBP binding sites determined by DNA footprinting are shown in boxes (from reference 19). The mutations introduced to abolish C/EBP binding are shown below the wild-type sequence. The NF-κB sites are underlined. The transcription start site is marked (arrow).
Cell Stimulation and Nuclear Protein Isolation.
Cells were incubated with PMA at 20 ng/ml and live M. tuberculosis at an moi of 0.1–10 or PMA and LPS at 10 μg/ml. When used, IFN-β was added at 1 U/ml after pretreatment for 24 h with PMA at 20 ng/ ml. Cells were harvested at various time points and were washed in PBS twice. Nuclei were isolated by NP-40 lysis and high salt extraction as described (27). Whole cells were lysed and extracted with RIPA (PBS, 1% NP-40, 0.5% deoxycholate) with 30 μl/ml aprotinin and 1 mM sodium orthovanadate for 30 min, and disrupted by passage through a 21 gauge needle; 1 mM PMSF was then added. Protein extracts for chloramphenicol acetyltransferase (CAT) ELISA or luciferase assays (both from Boehringer Mannheim) were made according to the manufacturer's instructions. Reporter activity was corrected for cell viability as measured by trypan blue exclusion.
Immunoblots.
100 μg of protein was added to a 10–20% linear gradient SDS-polyacrylamide gel. After electrophoresis, protein was electrotransferred to a nylon membrane and probed with 3 μg of polyclonal C/EBPβ antibody (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) in 50 ml blotto and visualized with 1.2 μg anti–rabbit horseradish peroxidase and ECL (Amersham Pharmacia Biotech, Piscataway, NJ). Blots were then reprobed with antibody to nuclear factor (NF)-κB p65 Rel (Santa Cruz Biotechnology, Inc.).
Electromobility Shift Assay.
DNA probe was labeled with [α- 32P]dCTP using Klenow DNA polymerase in a fill-in reaction. The overlapping oligonucleotides used for wild-type NRE probe were −184 5′AGCAAACTAGCATTTCATC3′-166 and −160 5′CCATGTGATGAAATGC3′-175, with bold letters denoting the C/EBP binding domain. The mutant sequences were −184 5′AGCAAACTAGCAGGGG3′-163 and −160 5′CCATGTCCCCTGCAGC3′-175, with bold letters denoting mutation from wild-type. Full-length 25-bp reaction products were isolated on a 20% acrylamide gel, and 2 × 105 cpm labeled DNA was mixed with 10 μg of protein extract, 2.5 μg poly dI/dC, and gel mobility shift buffer. The ISRE probe was 5′CTCGGGAAAGGGAAACCGAAACTGAAGCC3′, with the ISRE homology in bold (28). The nonspecific competitor on the ISRE electromobility shift assay (EMSA) was 5′CTCTCTGCAAGGGTCATCAGTAC3′. For supershift experiments, 1 μg of antibody was added to the reaction (C/EBP antibodies from Santa Cruz Biotechnology, Inc.; Stat-1 and Stat-2 antibodies were a gift from Chris Schindler). The DNA–protein complexes were electrophoresed on a 6% polyacrylamide gel at 4°C and analyzed by autoradiography. Films of immunoblots and EMSA were scanned with a Personal densitometer (Molecular Dynamics, Sunnyvale, CA). Results are presented as the mean ± SEM.
Results
M. tuberculosis Infection Suppresses HIV-1 Replication and LTR Promoter Function in Macrophages.
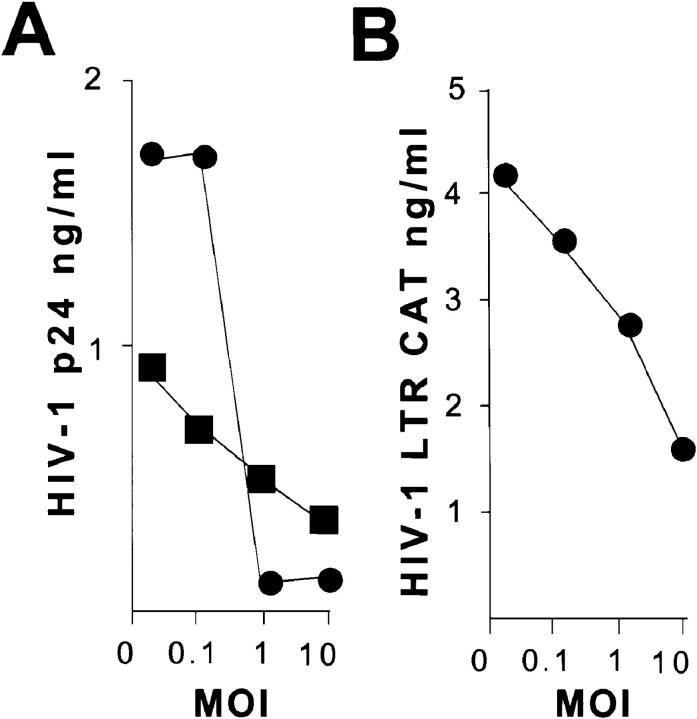
THP-1 cell–derived macrophages support high levels of HIV-1 replication, producing between 1 and 3 ng/ml of p24 antigen 7 d after infection (Fig. 1, A, moi 0). M. tuberculosis suppressed HIV-1 replication in a dose-dependent manner. At an moi of 1, there was at least a 50% reduction of HIV-1 p24 in both viral strains without reduced cell viability. At an moi of 10, there was a 70–90% reduction in viral production. M. tuberculosis suppressed replication of HIV-1 BaL (macrophage trophic) and HIV-1 NL4-3 (lymphocyte trophic). Similar results were obtained from blood monocytes differentiated with M-CSF and infected with Mycobacterium bovis (data not shown).
Figure 1.
HIV-1 replication and LTR function after infection with M. tuberculosis. (A) HIV-1 replication is suppressed by M. tuberculosis infection. Monocytic THP-1 cells differentiated to macrophages by PMA were infected with HIV-1 BaL (•) or HIV-1 NL4-3 (▪). HIV-1 replication was measured by p24 production at day 7 after infection. (B) HIV-1 LTR is repressed by M. tuberculosis infection. BF-24 cells containing a stable integrated HIV-1 LTR CAT construct were differentiated with PMA and infected with M. tuberculosis at various moi. CAT activity was measured 48 h later.
To test if the reduction in viral replication was due to reduced LTR-driven transcription, these experiments were repeated on BF-24 cells (THP-1 cells with a stable HIV-1 LTR reporter construct). M. tuberculosis suppressed HIV-1 LTR–driven CAT production in PMA-differentiated BF-24 cells (Fig. 1 B). Similar to the decrease in HIV-1 p24, CAT concentration declined by 33% at an moi of 1 and 66% at an moi of 10.
M. tuberculosis Infection of Macrophages Induces an IFN Response.
Since other proinflammatory stimuli such as LPS inhibit viral replication by a type 1 interferon response (12), we tested if M. tuberculosis infection also induced an interferon response. We assayed cell extracts for activation of a type I IFN specific transcription factor in cells infected with M. tuberculosis. Extracts of differentiated THP-1 cells were used in EMSA with an ISRE-containing DNA probe.
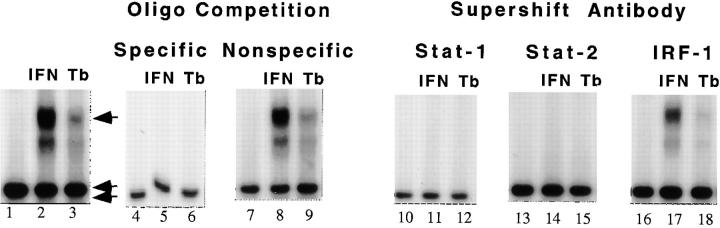
Stimulation of THP-1 macrophages with IFN-α or infection with M. tuberculosis at an moi of 1 induced an ISRE–protein complex that was not present in unstimulated THP-1 macrophages (Fig. 2, lanes 1 and 2). Both stimuli induced the same complex, although M. tuberculosis produced less ISRE–protein complex than 500 U/ml of IFN-α (Fig. 2, lane 3). The complex specifically bound ISRE sequences and contained Stat-1 and Stat-2 but not IFN regulatory factor 1 (IRF-1). Excess unlabeled ISRE oligonucleotide disrupted the DNA–protein complex (Fig. 2, lanes 4–6), but excess nonspecific oligonucleotide did not (Fig. 2, lanes 7–9). Antibodies against Stat-1 (Fig. 2, lanes 10–12) and Stat-2 (Fig. 2, lanes 13–15) disrupted the DNA–protein complex, whereas antibodies to IRF-1 did not (Fig. 2, lanes 16–18). Therefore, the DNA–protein complex induced by infection with M. tuberculosis had the characteristics of ISGF-3, a type I IFN–specific transcription factor.
Figure 2.
ISGF-3 binding to ISRE DNA in THP-1 macrophages. Treatment with IFN-α and infection with M. tuberculosis induces ISGF-3 expression. Lane 1, Unstimulated macrophages do not express ISGF-3. Lane 2, Stimulation with IFN-α (IFN) induces an ISRE binding complex. Lane 3, Infection with M. tuberculosis (Tb) produces a comigrating complex (arrow). Lanes 4–6, The ISRE–protein complex was disrupted by excess unlabeled ISRE-containing competitor. Lanes 7–9, The ISRE– protein complex was not disrupted by excess unlabeled nonspecific oligonucleotide competitor. Lanes 10–15, The ISRE–protein complex was disrupted by antibodies to Stat-1 and Stat-2. Lanes 16–18, The ISRE– protein complex was not disrupted by antibodies to IRF-1. A nonspecific DNA–protein complex is shown at the bottom of each panel for reference (double arrow).
C/EBP Sites Are Required for HIV-1 LTR Promoter Repression in Macrophages.
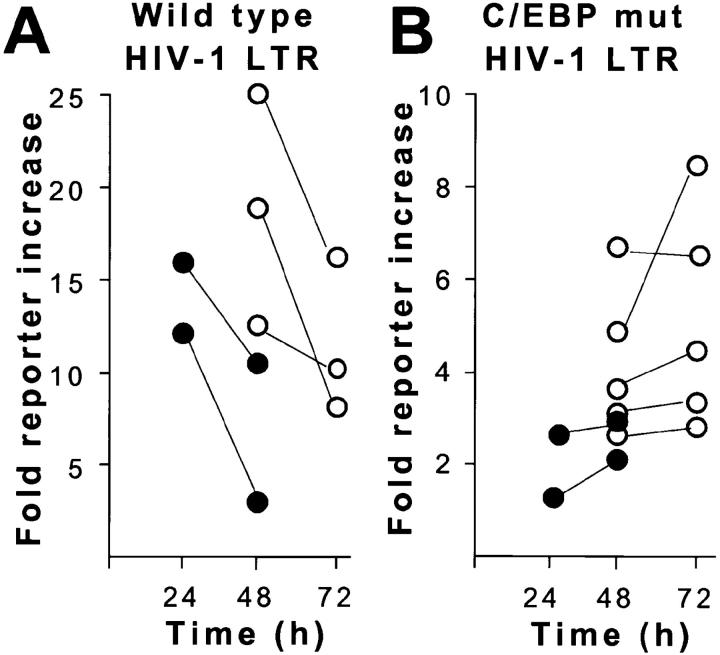
The NRE of the HIV-1 LTR is required for repression of the HIV-1 LTR after stimulation of macrophages with LPS (8), and the NRE contains C/EBP transcription factor binding sites (19). To test if C/EBP binding factors contributed to promoter repression in macrophages after infection with M. tuberculosis, HIV-1 LTR reporter constructs carrying three C/EBP binding site mutations were generated (Fig. 3). Wild-type and C/EBP mutant LTR reporter constructs were stably transfected into THP-1 cells. In time course experiments, M. tuberculosis infection inhibited wild-type LTR reporter activity 38% from 24 to 48 h after infection, and LPS stimulation inhibited wild-type LTR reporter activity by 40% from 48 to 72 h after stimulation (Fig. 4 A). HIV-1 LTR CAT and HIV-1 LTR luciferase produced similar results.
Figure 4.
Time course of HIV-1 LTR promoter activity in stably transfected THP-1 cells. (A) Wild-type HIV-1 LTR. M. tuberculosis (•) or LPS (○) produces a striking decline in reporter activity in THP-1 macrophages between 24 and 72 h. (B) HIV-1 LTR with mutated (mut) C/EBPβ sites. M. tuberculosis (•) or LPS (○) produces a moderate increase in reporter activity in THP-1 macrophages.
Mutation of the C/EBP sites in the HIV-1 LTR abolished repression of LTR reporter activity in macrophages after infection with M. tuberculosis or stimulation with LPS (Fig. 4 B). Reporter activity increased 20% after infection with M. tuberculosis and 40% after stimulation with LPS when C/EBP sites were mutated. Consistent with previous reports demonstrating the importance of C/EBP sites in transcriptional activity of the LTR (22–24), the C/EBP mutant had reduced total activity compared with the wild-type HIV-1 LTR.
An Inhibitory 16-kD C/EBPβ Isoform Is Induced by Proinflammatory Stimuli and IFN-β.
Since C/EBP binding sites are required for HIV-1 LTR–mediated transcription in macrophages (22, 23) and the C/EBPβ gene can produce a 16-kD dominant negative transcription factor (20, 21), we investigated whether changes in C/EBPβ expression could account for repression of the HIV-1 LTR. Initial experiments were conducted with M. tuberculosis infection and repeated with LPS, another proinflammatory stimulus that inhibits LTR reporter activity and HIV-1 replication in macrophages (8, 9).
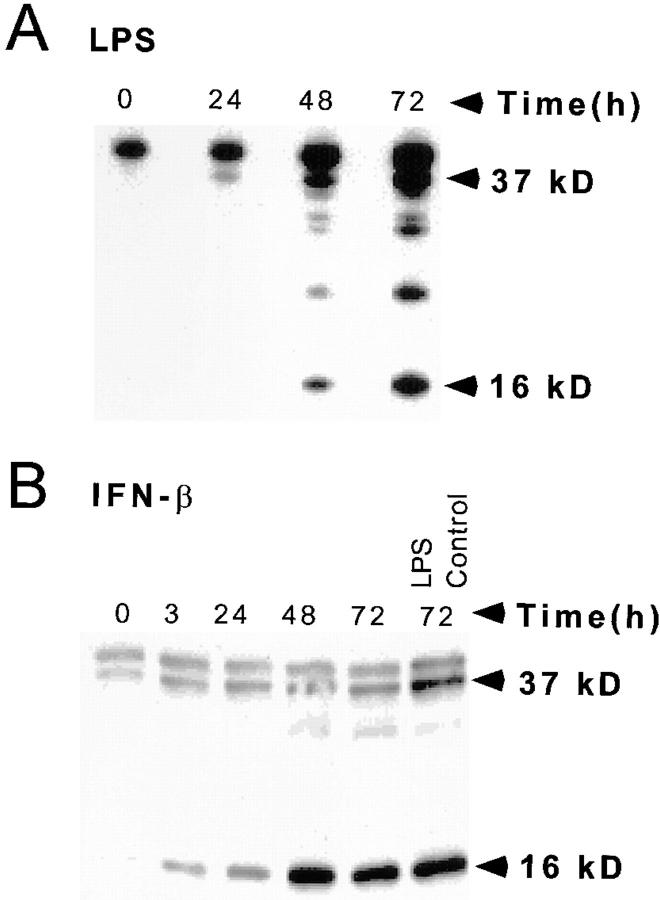
THP-1 cell–derived macrophages produced a 16-kD inhibitory C/EBPβ in nuclear extracts 48–72 h after stimulation with LPS (Fig. 5 A) or 24–48 h after infection with M. tuberculosis (data not shown). A stimulatory 37-kD C/EBPβ was also induced over the same time course. The ratio of inhibitory to stimulatory C/EBPβ was 0.8 after 72 h of LPS stimulation (Table 1). Inhibition predominates at stoichiometric ratios >0.2 (20, 21). A cross-reacting 40-kD protein was detected in the immunoblots. This band is nonspecific since it is expressed in unstimulated THP-1 cells that have negligible C/EBP DNA binding activity.
Figure 5.
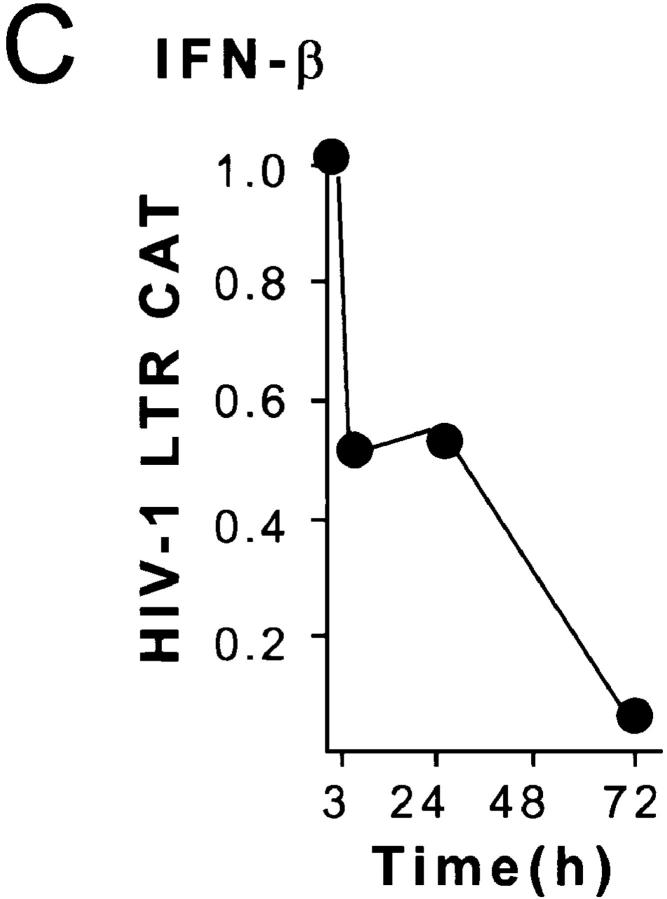
C/EBPβ expression and HIV-1 LTR reporter activity of THP-1 macrophages after LPS or IFN-β stimulation. (A) LPS induces inhibitory 16-kD C/EBPβ in the nuclear extracts between 48 and 72 h. (B) Inhibitory 16-kD C/EBPβ is increased 3 h after IFN treatment of differentiated THP-1 cells and is maximally induced after 48 h. The level of inhibitory C/EBPβ produced by LPS stimulation is the same as that produced by IFN-β. (C) The HIV-1 LTR is repressed after 1 U/ml IFN-β in differentiated BF-24 cells. There is a 50% decline in LTR activity after 3 h and a 95% decline in LTR activity at 72 h.
Table 1.
Densitometry of C/EBPβ Immunoblot
| THP-1 macrophages | Normal BAL | Uninvolved BAL | ||||||
|---|---|---|---|---|---|---|---|---|
| +LPS | +IFN-β | |||||||
| n = 5 | n = 4 | n = 2 | n = 6 | |||||
| Ratio | ||||||||
| 16-kD C/EBPβ/ | 0.8 ± 0.64 | 3.6 ± 2.4 | 1.8 ± 0.9 | 1.0 ± 0.8 | ||||
| 37-kD C/EBPβ | ||||||||
IFN-β also induced the inhibitory 16-kD C/EBPβ in THP-1 macrophages (Fig. 5 B). In time course experiments, induction of inhibitory C/EBPβ started 3 h after addition of IFN-β and achieved its maximum at 48 h. The amount of inhibitory C/EBPβ induced with IFN-β and LPS were the same (Fig. 5 B, last two lanes), but induction of the inhibitory transcription factor by IFN is more rapid than with LPS stimulation, and the ratio of inhibitory to stimulatory C/EBPβ was 3.6 (Table 1). In dose–response experiments, 1 U/ml of IFN-β maximally induced the 16-kD C/EBPβ (data not shown).
1 U/ml of IFN-β reduced HIV-1 LTR CAT expression by 50% within 3 h in BF-24 cells differentiated with PMA (Fig. 5 C). The reduction of LTR activity was coincident with induction of inhibitory 16-kD C/EBPβ. There was >95% inhibition of transcription by IFN-β 72 h after treatment. This was greater than the inhibition produced by LPS stimulation or infection with M. tuberculosis (compare Figs. 1 and 2 with Fig. 5 C).
C/EBPβ Is the Predominant Transcription Factor Binding the HIV-1 NRE in THP-1 Cell–derived Macrophages.
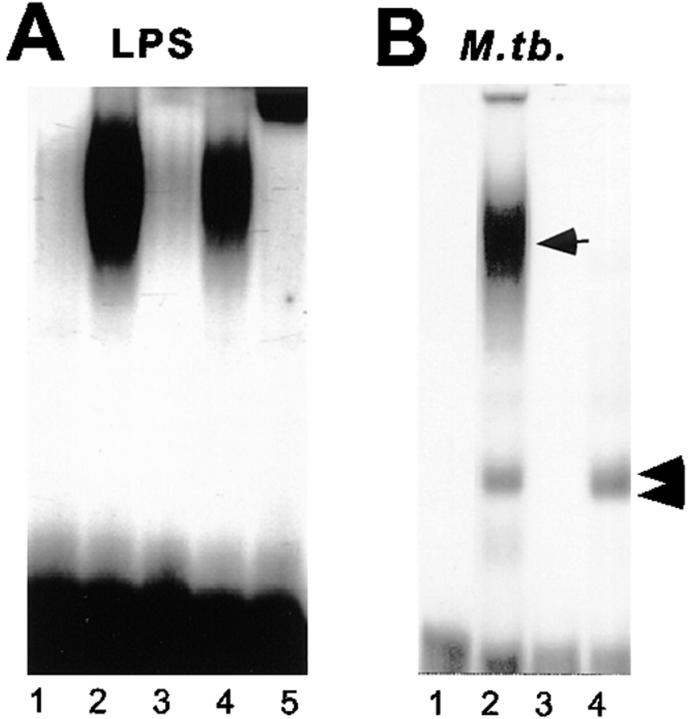
EMSA with an HIV-1 NRE probe was used to investigate which C/EBP transcription factors bound to the HIV-1 LTR. THP-1 macrophages stimulated with LPS increased NRE binding 10-fold (Fig. 6 A, lanes 1 and 2). This binding is specific to the C/EBP site of the NRE, since cold NRE competed effectively with the DNA–protein complex (Fig. 6 A, lane 3), whereas an oligonucleotide with point mutations in the C/EBP domain did not (Fig. 6 A, lane 4). Over 95% of the C/EBP-specific NRE–protein complex was supershifted with C/EBPβ antibody (Fig. 6 A, lane 5).
Figure 6.
C/EBPβ binding to the HIV-1 LTR NRE in THP-1 macrophages. (A) Cells predominantly express C/EBPβ after LPS stimulation. Lane 1, Unstimulated THP-1 cells with wild-type NRE probe. Lane 2, Wild-type NRE binding is increased 10-fold after stimulation with PMA and LPS. Lane 3, Competition with 100-fold excess unlabeled wild-type NRE probe disrupts the NRE–protein complex. Lane 4, 100-fold excess unlabeled NRE mutated in the C/EBP site does not disrupt the NRE–protein complex. Lane 5, Antibody to C/EBPβ supershifts >95% of the NRE–protein complex. (B) THP-1 macrophages infected with M. tuberculosis (M. tb.) express C/EBP-specific and C/EBP-nonspecific NRE–protein complexes. Lane 1, Wild-type NRE probe only. Lane 2, Wild-type NRE binding in THP-1 macrophage extracts after M. tuberculosis infection produces a band similar to LPS stimulation (arrow) and another more rapidly migrating NRE–protein complex (double arrow). Lane 3, C/EBP mutant NRE probe only. Lane 4, C/EBP-mutated NRE probe produces a single complex with the same mobility as the rapidly migrating wild-type NRE–protein complex (double arrow) with extracts of M. tuberculosis–infected THP-1 macrophages.
The major NRE–protein complex observed with LPS stimulation is also induced by infection of THP-1 macrophages with M. tuberculosis (Fig. 6 B, lane 2, arrow). This NRE–protein complex competed with cold wild-type NRE oligonucleotide, but not C/EBP mutant oligonucleotide and supershifted with C/EBPβ antibody (data not shown). In these extracts, another rapidly migrating band was observed (Fig. 6 B, double arrow). This band bound to non-C/EBP sequences of the NRE, since EMSA using the C/EBP-mutated NRE as a probe produced a single comigrating band (Fig. 6 B, lane 4). Wild-type or C/EBP- mutated NRE probes by themselves did not produce these bands (Fig. 6 B, lanes 1 and 3). The non-C/EBP NRE– protein complex was heat labile, whereas the NRE–C/ EBPβ complex reformed after heating extracts at 95°C for 5 min (data not shown).
Inhibitory C/EBPβ Is Strongly Expressed in Alveolar Macrophages from Lung Segments with Low Viral Replication and Is Downregulated in Inflamed Lung Segments with High Viral Replication.
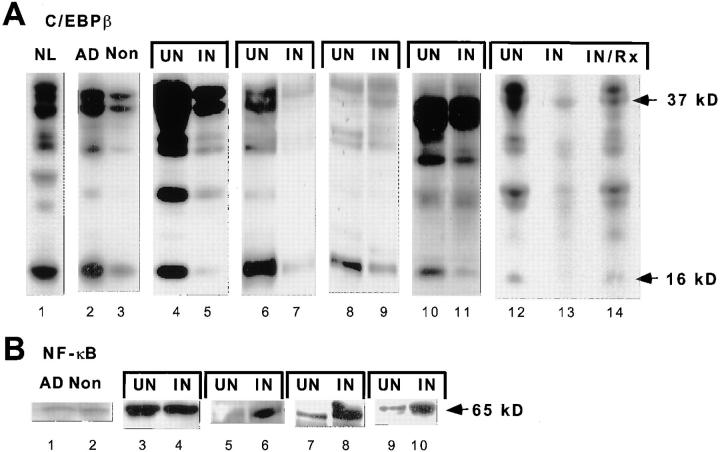
We investigated C/EBPβ expression in the lung to test if cells that suppress HIV-1 replication in vivo also express the inhibitory 16-kD C/EBPβ. The C/EBPβ immunoblot pattern of BAL cells from a normal control (Fig. 7 A, lane 1) was similar to the pattern in macrophages stimulated with LPS. Expression of the 16-kD C/EBPβ was striking. The ratio of inhibitory to stimulatory C/EBPβ was 1.8 (Table 1). To define which cell population expressed the C/EBPβ, adherence was used to separate macrophages from lymphocytes in the BAL. This procedure works well in BAL cells from uninvolved lung; adherent cells were 90–95% alveolar macrophages by morphology and nonspecific esterase staining. The nonadherent cells were 80–90% lymphocytes by morphology. Protein extracts were made from adherent (AD) and nonadherent (Non) BAL cells from an uninvolved lobe of an AIDS patient with pulmonary tuberculosis. The adherent fraction expressed at least sixfold more 16-kD C/EBPβ (Fig. 7 A, lanes 2 and 3), demonstrating that the 16-kD C/EBPβ is strongly expressed in the alveolar macrophages.
Figure 7.
Transcription factor expression in BAL cells from a normal control and six patients with pulmonary tuberculosis. (A) Inhibitory 16-kD C/EBPβ is strongly expressed in BAL cells from uninflamed lung and is downregulated in pulmonary tuberculosis. Lane 1, The inhibitory 16-kD C/EBPβ is strongly expressed in a normal control (NL). Lane 2, Adherent BAL cells (Ad) from an uninvolved lobe of an AIDS patient with tuberculosis strongly express inhibitory 16-kD C/EBPβ. Adherent BAL cells are >95% alveolar macrophages. Lane 3, Nonadherent cells (Non) which are 80–90% lymphocytes have little inhibitory 16-kD C/EBPβ expression. Lanes 2, 4, 6, 8, 10, and 12 show that the inhibitory 16-kD C/EBPβ is strongly expressed in uninvolved lobes (UN) of six patients with tuberculosis. Four are HIV-1 infected and two are HIV-1 negative. Lanes 5, 7, 9, 11, and 13 show that the inhibitory 16-kD C/EBPβ is markedly downregulated in the involved lobes (IN) of these tuberculosis patients. Lane 14, There is an increase in 16-kD C/EBPβ in the involved lobe after 2 wk of antituberculous chemotherapy (IN/Rx). (B) NF-κB expression in BAL cells. Lanes 1–10 are the same blots as in A, lanes 2–11. Lanes 1 and 2, The level of NF-κB expression is similar in adherent (Ad) and nonadherent (Non) cells. Lanes 3–10, There is equal or increased NF-κB expression in the involved lobes (IN) compared with the uninvolved lobes (UN).
BAL cells from radiographically uninvolved lung segments of six patients with pulmonary tuberculosis were similar to the immunoblots of the normal control (Fig. 7 A, lanes 2, 4, 6, 8, 10, and 12). The ratio of inhibitory to stimulatory C/ EBPβ was 1.0 (Table 1). Lung segments involved with tuberculosis had expressed 14 ± 13% the level of 16-kD inhibitory C/EBPβ observed in uninvolved segments (Fig. 7 A, lanes 5, 7, 9, 11, and 13). Immunocompetent patients had the same expression pattern as AIDS patients. Expression of 16-kD C/ EBPβ increased in the involved lung segment after antituberculous chemotherapy (Fig. 6 B, lanes 13 and 14).
Reprobing of the blots with antibody to NF-κB p65 Rel A demonstrated that adherent and nonadherent fractions had similar amounts of NF-κB (Fig. 7 B, lanes 1 and 2). NF-κB was equally or more strongly expressed in the involved lung segment than in the uninvolved lung segment (Fig. 7 B, lanes 3–10). This demonstrated that changes in C/EBPβ expression were not due to nonspecific protein degradation or unequal loading of samples.
The BAL cellular differentials are shown in Table 2. These results are similar to larger series of BAL differentials in tuberculosis patients and normal controls (29). The lung segment involved with tuberculosis has increased percentages of lymphocytes and neutrophils compared with uninvolved lung segments or normal controls. Involved lung segments have a bimodal distribution of inflammatory cells, with three of the patients having <2% neutrophils, while the other patients had 34 ± 21% neutrophils. The 1.5-fold enrichment of alveolar macrophages in the uninvolved lobes does not account for the 6.6-fold enrichment of total C/EBPβ observed in the uninvolved BAL (Table 3).
Table 2.
BAL Cellular Differential
| Pulmonary tuberculosis | ||||||
|---|---|---|---|---|---|---|
| Normal | Uninvolved | Involved | ||||
| % | % | % | ||||
| Macrophages | 91 ± 1 | 80 ± 16 | 52 ± 17 | |||
| Lymphocytes | 9 ± 1 | 17 ± 15 | 27 ± 18 | |||
| Neutrophils | 0 | 1 ± 0.9 | 20 ± 22 | |||
Table 3.
Densitometry of BAL NRE–Protein Complexes
| C/EBPβ supershift | Nonspecific band | |||
|---|---|---|---|---|
| Ratio | ||||
| uninvolved/involved | 6.6 ± 3.7 | 1.1 ± 0.3 |
Pulmonary Tuberculosis Switches C/EBP Expression in BAL Cells.
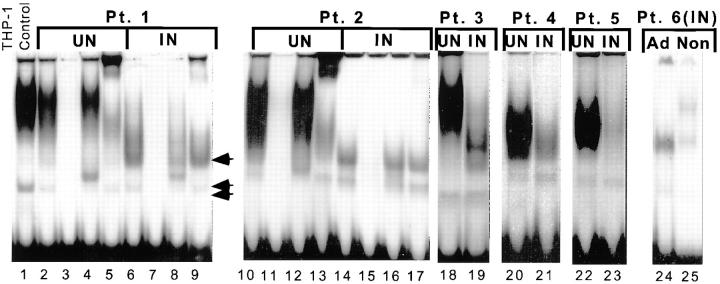
BAL cell extracts from uninvolved lobes have the same EMSA pattern as THP-1 macrophages infected with M. tuberculosis (Fig. 8, lanes 1 and 2). The uninvolved lobe from five patients with pulmonary tuberculosis strongly binds the HIV-1 LTR NRE (Fig. 8, lanes 2, 10, 18, 20, and 22). This binding is specific for the C/EBP site, since binding could be competed away by excess wild-type oligonucleotide (Fig. 8, lanes 3 and 11) but not by mutant NRE (Fig. 8, lanes 4 and 12). Over 90% of the NRE–protein complexes supershifted with antibody to C/EBPβ (Fig. 8, lanes 5 and 13). The non-C/EBP NRE–protein (nonspecific) complex observed in THP-1 macrophages infected with M. tuberculosis was also observed in BAL cell extracts from uninvolved and involved lung segments (Fig. 8, double arrow).
Figure 8.
C/EBPβ binding to the HIV-1 LTR NRE in BAL cells from six patients with pulmonary tuberculosis. Extracts from uninvolved lung of patients with pulmonary tuberculosis predominantly express C/EBPβ, whereas BAL cells from involved lung downregulate C/EBPβ and switch expression to another C/EBP family member. Lane 1, NRE binding of THP-1 macrophages infected with M. tuberculosis. Lanes 2–4, 10–13, 18, 20, and 22, NRE–protein complex in uninvolved lobes (UN) of HIV-1–infected and HIV-1–negative patients with pulmonary tuberculosis. Lanes 2, 10, 18, 20, and 22, BAL cell extract strongly binds the NRE. A nonspecific band is also expressed in BAL extracts (double arrow). Lanes 3 and 11, 100-fold excess unlabeled NRE probe disrupts both the specific and nonspecific complex. Lanes 4 and 12, 100-fold excess unlabeled mutant probe does not affect the specific complex. Lanes 5 and 13, Anti-C/EBPβ antibody supershifts >90% of the complex. Lanes 6, 14, 19, 21, 23, and 24, Tuberculosis-involved lobes (IN) lose the predominant band expressed in uninvolved lobes and switch expression to a faster migrating specific NRE–protein complex (arrow). Lanes 7 and 15, 100-fold excess unlabeled NRE disrupts the complex. Lanes 8 and 16, 100-fold excess unlabeled mutant probe does not disrupt the complex. Lanes 9 and 17, Anti-C/EBPβ antibody does not supershift the complex. Lane 24, Adherent cells (Ad) from an involved lobe strongly express specific and nonspecific NRE– protein complexes. Lane 25, Nonadherent cells (Non) from the same involved lobe have markedly reduced expression of the specific and nonspecific NRE–protein complexes.
EMSA confirmed the immunoblot observations. Uninvolved lung segments had a marked increase in NRE binding compared with involved lobes from the same patient (in Fig. 8, compare lanes 2, 10, 18, 20, and 22 with lanes 6, 14, 19, 21, and 23). Supershift with antibody to C/EBPβ demonstrated 6.6-fold greater C/EBPβ expression in the uninvolved lung segment (Table 3). In contrast, expression of the nonspecific complex was increased 1.1-fold in the uninvolved lung segment (Table 3). This nonspecific band is an internal standard for DNA binding activity in uninvolved and involved BAL extracts. The similar level of expression of the nonspecific NRE–protein complex in extracts from different sources demonstrated that changes in C/EBPβ expression were not due to protein degradation in the involved segment or to unequal loading of samples.
A new rapidly migrating band was observed in extracts of BAL cells from the involved segment of five patients (Fig. 8, lanes 5–8, arrow, 14–17, 19, 21, and 24). The induced NRE–protein complex is specific for the C/EBP site, since binding could be competed by excess wild-type oligonucleotide (Fig. 8, lanes 7 and 15) but not by mutant NRE (Fig. 8, lanes 8 and 16). Full data are shown from two patients, but similar results were obtained from all five. The rapidly migrating C/EBP-specific band expressed in involved lung segments was observed in patients with and without neutrophil-predominant BAL differentials. In addition, it did not supershift with antibody to C/EBPβ (Fig. 8, lanes 9 and 17) or with antibodies to C/EBPα, C/EBPδ, C/EBPε, or C/EBP-homologous protein (CHOP) (data not shown). Adherence was used to separate macrophages from other cells in the involved BAL. Both the C/EBP-specific and C/EBP-nonspecific NRE binding activity were enriched in the adherent cell fraction (Fig. 8, lanes 24 and 25). Another slowly migrating NRE–protein complex was expressed in nonadherent BAL cells. This complex did not supershift with C/EBPβ antibodies (data not shown).
Discussion
M. tuberculosis and other opportunistic infections including Pneumocystis carinii, cryptococcus, CMV, and bacterial infections enhance HIV-1 replication in vivo (2, 3, 30–34). We previously reported significantly increased HIV-1 RNA and p24 levels in radiographically involved lobes in tuberculosis/HIV-1 coinfected patients compared with uninvolved lobes or HIV-1–infected patients with no lung disease (2). We also demonstrated that M. tuberculosis stimulated HIV-1 replication in vitro as measured by increased p24 in cell supernatants of undifferentiated U937 and THP-1 cells over 4 d (30). We correlated these increases with transcriptional activation at the tandem NF-κB sites and the C/EBP sites on the HIV-1 LTR using CAT reporter constructs (30). Studies with the promonocyte U937 and monocytic THP-1 cell lines have shown that LPS and TNF-α also enhance HIV-1 replication through activation of NF-κB (35–37). Surprisingly, when monocytes are differentiated to macrophages, LPS or other proinflammatory stimuli suppress HIV-1 replication (8, 9). Inhibition of viral replication in macrophages after proinflammatory stimuli is a type I IFN effect (11, 12). We now demonstrate a novel mechanism of transcriptional control by type I IFNs that alter expression of C/EBPβ, producing a dominant negative transcription factor and repressing the HIV-1 LTR. In vivo, alveolar macrophages from uninflamed lung behave like IFN-β–treated macrophages, expressing the inhibitory 16-kDa C/EBPβ and inhibiting HIV-1 replication. This may contribute to viral latency in the uninflamed lung.
We showed that M. tuberculosis suppressed HIV-1 replication, repressed LTR activity, and induced ISGF-3, a type I IFN–specific transcription factor, in THP-1 macrophages. Two separate strains of HIV-1 were suppressed, demonstrating that the effect was not strain specific. The formation of a type I IFN–specific transcription factor complex is early in the IFN signaling cascade, induced by receptor- directed protein phosphorylation. Since type I IFN mediates strong antiviral activity, it is likely that the reduction of viral replication after M. tuberculosis infection is an IFN effect. The suppression of HIV-1 replication correlated with repression of LTR activity, suggesting that transcriptional downregulation is one mechanism of inhibition of viral replication in this system. The HIV-1 LTR has an NRE with three C/EBP sites located 5′ to a tandem NF-κB site (19, 38, 39). The NRE is the sequence that mediates HIV-1 LTR repression after an inflammatory stimulus (8).
Productive infection of macrophages by HIV-1 occurs predominantly in alveolar macrophages and macrophage-lineage cells, e.g., in the brain, although monocyte-derived macrophages are highly susceptible (6, 30, 40, 41). Differentiated monocytic U1 cells suppress HIV-1 expression after treatment with IFN-γ, IL-6, or GM-CSF (42). These cytokines act transcriptionally on the stably integrated HIV-1 LTR in U1 cells. Macrophage differentiation upregulates NF-IL6 (C/EBPβ), and NF-IL6 is responsible for regulating gene expression in mature macrophages (43). M. tuberculosis stimulates production of macrophage cytokines IL-1β, IL-6, and TNF-α, and C/EBP binding sites are involved in the transcriptional regulation of these genes (44, 45). HIV-1 transcriptional regulation occurred independent of NF-κB after U1 cells were treated with proinflammatory cytokines (39). Positive regulation can also occur through interactions of C/EBP transcription factors with the adjacent NF-κB (30). C/EBPβ–NF-κB heterodimers are more potent activators of the HIV-1 LTR than p50– p65 complexes (46, 47). HIV-1 replication requires intact C/EBP sites in monocytes but not lymphocytes, demonstrating a requirement for C/EBP activators by HIV-1 only in monocytes and macrophages (24). Thus, we evaluated the C/EBP family of transcription factors to clarify the mechanism of activation and suppression of the HIV-1 LTR in differentiated and activated macrophages.
Point mutations in the C/EBP sites of the HIV-1 LTR NRE abolished promoter repression after stimulation with LPS or infection with M. tuberculosis. The continued increase in C/EBP-mutated HIV-1 LTR reporter activity over the course of the experiments demonstrates that these macrophages could sustain high levels of transcription after inflammatory stimuli. Therefore, the decline in the wild-type HIV-1 LTR after M. tuberculosis or LPS was not due to a nonspecific toxicity produced by the inflammatory stimuli. These results were obtained with stably integrated LTR CAT and LTR luciferase reporter constructs, demonstrating that the level of stimulation is not critically dependent on the site of integration or the reporter used. Repression of the wild-type HIV-1 LTR was caused by a specific interaction of a transcriptional repressor(s) with the C/EBP sites in the HIV-1 LTR.
Descombes and Schibler described two rat C/EBPβ proteins which either activated the albumin promoter (liver-activating protein [LAP]) or repressed the albumin promoter (liver-enriched inhibitory protein [LIP]) (20). Both were transcribed from the same mRNA by using in-frame AUGs. Both proteins share the 145 COOH-terminal amino acids that contain the basic DNA-binding domain and the leucine zipper dimerization helix. LAP uses the first and second AUG start site and LIP the third, resulting in only LAP having the transcriptional activation domain (20). Consequently, LAP is a longer protein (∼39 kD) than LIP (∼16 kD). However, LIP has higher binding affinity for its DNA cognate sequences, resulting in its ability to attenuate the transcriptional stimulation of LAP in substoichiometric amounts. Small changes in the LIP to LAP ratio resulted in large differences in transcription, with promoter repression occurring at ratios >0.2 (20, 21). LIP is also induced in mouse liver by LPS (48). Our data suggest that Kupffer cells may contribute to the increase in LIP observed in the liver after LPS treatment.
In vitro, expression of the inhibitory 16-kD C/EBPβ represses promoters containing C/EBP sites in monocytes and macrophages. When the 16-kD C/EBPβ is cotransfected with the HIV-1 LTR, transcription is strongly inhibited (22), and cell lines with engineered expression of the C/EBPβ dominant negative isoform suppress induction of provirus and HIV-1 replication (23). Cytokine genes such as TNF-α also have C/EBP binding sites, and transfection of inhibitory C/EBPβ represses TNF-α promoter activity (26). The relevance of these in vitro studies is undefined, since endogenous expression of inhibitory 16-kD C/EBPβ in monocytes and macrophages has not been studied.
We demonstrate here that macrophages induce a dominant negative 16-kD C/EBPβ transcription factor after stimulation with LPS or infection with M. tuberculosis. C/EBPβ is the predominant C/EBP family member in these cells. EMSA demonstrates that C/EBPβ is in >95% of NRE– protein complexes induced by these inflammatory stimuli. The induction of this transcriptional repressor in our model is coincident with repression of the HIV-1 LTR. Both stimulatory and inhibitory C/EBPβ isoforms are expressed, but repression would be expected because the 16-kD C/EBPβ is dominant negative, inhibiting transcription when expressed at 20% the level of the 37-kD isoform (20, 21). All cells that inhibited HIV-1 replication or LTR promoter function had a higher ratio of inhibitory to stimulatory isoforms (Table 1). Since C/EBPβ is the predominant transcription factor induced by inflammatory stimuli and intact C/EBP sites are required for LTR repression, induction of endogenous inhibitory 16-kD C/EBPβ is most likely responsible for LTR repression after inflammatory stimulation. This conclusion is strongly supported by the observation that engineered expression of 16-kD C/EBPβ represses the LTR and inhibits HIV-1 replication (22, 23).
Treatment of macrophages with IFN-β induces a dominant negative C/EBPβ transcription factor and represses HIV-1 LTR promoter within 3 h. Given the rapid effect of IFN-β on 16-kD C/EBPβ induction and LTR repression, it is likely that these are direct IFN effects. These effects occur at low doses. 1 U/ml of IFN-β maximally induces inhibitory C/EBPβ and maximally represses the HIV-1 LTR. Therefore, one mechanism of inhibition of viral replication by type I IFN is transcriptional repression. The ratio of inhibitory to stimulatory C/EBPβ is 3.6 after treatment with IFN-β compared with a ratio of 0.8 after treatment with LPS. This may account for the greater LTR repression produced by IFN-β. Taken together with the induction of ISGF-3 by M. tuberculosis, these data suggest that M. tuberculosis and LPS repress the LTR by producing an IFN response in macrophages.
Induction of a dominant negative C/EBPβ transcription factor by type I IFN reveals a highly unusual form of regulation. The C/EBPβ gene has no introns, so the change in coding potential of the mRNA is regulated by a ribosome scanning mechanism (20, 48). After IFN-β stimulation, the ribosome selects an internal ribosome entry site in the C/EBPβ mRNA, producing dominant negative transcription factors 16–20-kD in size. In the absence of IFN stimulation, the ribosome selects the 5′ translation start sites, producing stimulatory transcription factors 35–37-kD in size. IFN stimulation can produce a rapid switch from transcriptional stimulation to transcriptional repression without changes in levels of C/EBPβ mRNA. It is possible that phosphorylation of translation initiation factors by double-stranded protein kinase alters ribosome function, producing altered translation start site usage. IFN-β–treated macrophages are the first in vitro system to regulate inhibitory C/EBPβ production, and this model will be useful to investigate translation start site switching in the C/EBPβ mRNA.
Alveolar macrophages from normal lung are primed by type I IFN even in the absence of viral infection (13). Expression of innate immunity in the uninflamed lung may contribute to viral latency. In vivo, resting alveolar macrophages from AIDS patients with no detectable viral replication strongly express the inhibitory 16-kD C/EBPβ. The ratio of inhibitory to stimulatory C/EBPβ is 1.0 in these lung segments. The inhibition of viral replication in uninflamed lung is similar to IFN-β–treated macrophages in vitro (9). Therefore, IFN-treated macrophages are a good model for resting alveolar macrophages. AIDS does not alter regulation of C/EBPβ, since the ratio of inhibitory to stimulatory C/EBPβ is similar in HIV-1–infected and HIV-1–negative patients. HIV-1 latency in alveolar macrophages in uninflamed lung is encouraged by increased expression of this C/EBPβ 16-kD inhibitory isoform, and our in vitro experiments with differentiated and stimulated macrophages support this novel concept.
Our data demonstrate that M. tuberculosis infection in vivo suppresses C/EBPβ proteins, particularly the 16-kD inhibitory isoform. There is 6.6-fold greater C/EBPβ– mediated NRE binding in the uninvolved lobes than in the involved lobes (Table 3). NF-κB expression is equal or higher in the involved segment on the immunoblot. Furthermore, nonspecific NRE binding activity is similar in uninvolved and involved lung segments (only 1.1-fold increased) in EMSA from five patients. Therefore, it is unlikely that the diminished C/EBPβ expression in involved lobes is due to nonspecific protein degradation. We propose that loss of the inhibitory C/EBPβ expression observed in pulmonary tuberculosis derepresses the HIV-1 LTR. Derepression may be one mechanism for enhanced HIV-1 replication observed in pulmonary tuberculosis.
A C/EBP DNA binding protein is induced in BAL cells during pulmonary tuberculosis. This NRE–protein complex is not supershifted by antibodies to C/EBPα, C/EBPβ, C/EBPδ, C/EBPε, or CHOP, raising the possibility that the NRE binding protein is a novel member of the C/EBP transcription factor family. This NRE binding activity is enriched in adherent BAL cells, suggesting that it is expressed in alveolar macrophages. Since C/EBP binding sites of the LTR are required for HIV-1 replication in macrophages (24), it is possible that the novel C/EBP binding protein is important in the high level of replication observed in pulmonary tuberculosis (2, 6). This transcription factor is a candidate to stimulate C/EBP-containing promoters during pulmonary tuberculosis and possibly other diseases associated with macrophage activation. Further experiments defining the modulation of these nuclear proteins may improve the understanding of HIV-1 latency in tissue macrophages and provide novel approaches to pursuing elimination of this clever pathogen.
Acknowledgments
This work was supported by National Institutes of Health grants MO1 RR00096, HL-51470, and HL-51494, and AI-37877, by the American Lung Association, and by Fred Friedman.
Abbreviations used in this paper
- BAL
bronchoalveolar lavage
- CAT
chloramphenicol acetyltransferase
- C/EBP
CCAAT/enhancer binding protein
- EMSA
electromobility shift assay
- IRF-1
IFN regulatory factor 1
- ISGF-3
IFN-stimulated gene factor 3
- ISRE
IFN-stimulated response element(s)
- LAP
liver-activating protein
- LIP
liver-enriched inhibitory protein
- moi
multiplicity of infection
- NF
nuclear factor
- NRE
negative regulatory element
- Stat
signal transducer and activator of transcription
Footnotes
Y. Honda's current address is Department of Medicine, Sendai Kosei Hospital, Sendai 980, Japan.
References
- 1.Nakata K, Weiden M, Harkin T, Ho D, Rom WN. Low copy number and limited variability of proviral DNA in alveolar macrophages from HIV-1 infected patients: evidence for genetic differences in HIV-1 between lung and blood macrophage populations. Mol Med. 1995;1:744–757. [PMC free article] [PubMed] [Google Scholar]
- 2.Nakata K, Rom WN, Honda Y, Condos R, Kanegasaki S, Cao Y, Weiden M. M. tuberculosisenhances human immunodeficiency virus-1 replication in the lung. Am J Respir Crit Care Med. 1997;155:996–1003. doi: 10.1164/ajrccm.155.3.9117038. [DOI] [PubMed] [Google Scholar]
- 3.Itescu S, Simonelli PF, Winchester RJ, Ginsberg HS. Human immunodeficiency virus type 1 strains in the lungs of infected individuals evolve independently from those in the peripheral blood and are highly conserved in the C-terminal region of the envelope V3 loop. Proc Natl Acad Sci USA. 1994;91:11378–11382. doi: 10.1073/pnas.91.24.11378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Israel-Biet D, Cadranel J, Beldjord K, Andrieu J, Jeffrey A, Even P. Tumor necrosis factor production in HIV-seropositive subjects: relationship with lung opportunistic infections and HIV expression in alveolar macrophages. J Immunol. 1991;147:490–494. [PubMed] [Google Scholar]
- 5.Sierra-Madero J, Toossi Z, Hom D, Finegan C, Hoenig E, Rich E. Relationship between load of virus in alveolar macrophages from human immunodeficiency virus type 1-infected persons, production of cytokines, and clinical status. J Infect Dis. 1994;169:18–27. doi: 10.1093/infdis/169.1.18. [DOI] [PubMed] [Google Scholar]
- 6.Orenstein JM, Fox C, Whal SM. Macrophages as a source of HIV during opportunistic infections. Science. 1997;276:1857–1861. doi: 10.1126/science.276.5320.1857. [DOI] [PubMed] [Google Scholar]
- 7.Whalen C, Horsburgh CR, Hom D, Lahart C, Simberkoff M, Ellner J. Accelerated course of human immunodeficiency virus infection after tuberculosis. Am J Respir Crit Care Med. 1995;151:129–135. doi: 10.1164/ajrccm.151.1.7812542. [DOI] [PubMed] [Google Scholar]
- 8.Bernstein MS, Tong-Starksen SE, Locksley RM. Activation of human monocyte-derived macrophages with lipopolysaccharide decreases human immunodeficiency virus replication at the level of gene expression. J Clin Invest. 1991;88:540–545. doi: 10.1172/JCI115337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kornbluth RS, Oh PS, Munis JR, Cleveland PH, Richman DD. Interferons and bacterial lipopolysaccharide protect macrophages from productive infection by human immunodeficiency virus in vitro. J Exp Med. 1989;169:1137–1151. doi: 10.1084/jem.169.3.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herbein G, Gordon S. 55- and 75-kilodalton tumor necrosis factor receptors mediate distinct actions in regard to human immunodeficiency virus type 1 replication in primary human macrophages. J Virol. 1997;71:4150–4156. doi: 10.1128/jvi.71.5.4150-4156.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gessani S, Testa U, Varano B, Di Marzio P, Borghi P, Conti L, Barberi T, Tritarelli E, Martucci R, Seripa D, et al. Enhanced production of LPS-induced cytokines during differentiation of human monocytes to macrophages: role of LPS receptors. J Immunol. 1993;151:3758–3766. [PubMed] [Google Scholar]
- 12.Hamilton JA, Genevieve WA, Kola I, Hertzog PJ. Endogenous Interferon-α/β suppresses colony-stimulating factor (CSF-1) stimulated macrophage DNA synthesis and mediates inhibitory effects of LPS and TNF-α. J Immunol. 1996;156:2553–2557. [PubMed] [Google Scholar]
- 13.Hwang S, Herzog PJ, Holland K, Sumarsono S, Tymms MJ, Hamilton JA, Whitty G, Bertoncello I, Kola I. A null mutation in the gene encoding a type I interferon receptor component eliminates antiproliferative and antiviral responses to interferon α and β and alters macrophage responses. Proc Natl Acad Sci USA. 1995;92:11284–11288. doi: 10.1073/pnas.92.24.11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van der Broek M, Muller U, Huang S, Zinkernagel R, Aguet M. Immune defense in mice lacking type I and/or type II interferon receptors. Immunol Rev. 1995;148:5–18. doi: 10.1111/j.1600-065x.1995.tb00090.x. [DOI] [PubMed] [Google Scholar]
- 15.Khan N, Pulford A, Farquharson M, Howatson A, Stewart C, Jackson R, McNicol A, Foulis A. The distribution of immunoreactive interferon-alpha in normal human tissues. Immunology. 1989;66:201–206. [PMC free article] [PubMed] [Google Scholar]
- 16.Novic D, Cohen B, Rubenstein M. The human interferon α/β receptor: characterization and molecular cloning. Cell. 1994;77:391–400. doi: 10.1016/0092-8674(94)90154-6. [DOI] [PubMed] [Google Scholar]
- 17.Muller U, Steinhoff U, Reis L, Hemmi S, Povlovic J, Zinkernagel R, Aguet M. Functional role of type I and type II interferons in antiviral defense. Science. 1994;264:1918–1921. doi: 10.1126/science.8009221. [DOI] [PubMed] [Google Scholar]
- 18.Bluyssen H, Durbin J, Levy D. ISGF3γ p48, a specificity switch for interferon-activated transcription factors. Cytokine Growth Factor Rev. 1996;7:11–17. doi: 10.1016/1359-6101(96)00005-6. [DOI] [PubMed] [Google Scholar]
- 19.Tesmer VM, Rajadhyaksha A, Babin J, Bina M. NF-IL6-mediated transcriptional activation of the long terminal repeat of the human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1993;90:7298–7303. doi: 10.1073/pnas.90.15.7298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Descombes P, Schibler U. A liver-enriched transcriptional activator protein, LAP, and a transcriptional inhibitory protein, LIP, are translated from the same mRNA. Cell. 1991;67:569–579. doi: 10.1016/0092-8674(91)90531-3. [DOI] [PubMed] [Google Scholar]
- 21.Ossipow V, Descombes P, Schibler U. CCAAT/enhancer-binding protein mRNA is translated into multiple proteins with different transcription activation potentials. Proc Natl Acad Sci USA. 1993;90:8219–8223. doi: 10.1073/pnas.90.17.8219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henderson AJ, Zou X, Calame K. C/EBP proteins activate transcription from human immunodeficiency virus type 1 long terminal repeat in macrophages/monocytes. J Virol. 1995;69:5337–5344. doi: 10.1128/jvi.69.9.5337-5344.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henderson AJ, Connor RI, Calame KL. C/EBP activators are required for HIV-1 replication and proviral induction in monocytic cell lines. Immunity. 1996;5:91–101. doi: 10.1016/s1074-7613(00)80313-1. [DOI] [PubMed] [Google Scholar]
- 24.Henderson AJ, Calame KL. CCAAT/enhancer binding protein (C/EBP) sites are required for HIV-1 replication in primary macrophages but not CD4+ T cells. Proc Natl Acad Sci USA. 1997;94:8714–8719. doi: 10.1073/pnas.94.16.8714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sears R, Sealy L. Multiple forms of C/EBPβ bind the EFII enhancer sequence in Rous sarcoma virus long terminal repeat. Mol Cell Biol. 1994;14:4855–4871. doi: 10.1128/mcb.14.7.4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pope RM, Lentz A, Ness SA. C/EBPβ regulation of the tumor necrosis factor α gene. J Clin Invest. 1995;94:1449–1455. doi: 10.1172/JCI117482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dyer RB, Herzog NK. Isolation of intact nuclei for nuclear extract preparation from a fragile B-lymphocyte cell line. Biotechniques. 1995;19:192–195. [PubMed] [Google Scholar]
- 28.Pine R. Convergence of TNFα and IFNγ signaling pathways through synergistic induction of IRF-1/ISGF-1 is mediated by a composite GAS/κB promoter element. Nucleic Acids Res. 1997;25:4346–4354. doi: 10.1093/nar/25.21.4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Law KF, Jagirdar J, Weiden M, Bodkin M, Rom WN. Tuberculosis in HIV-positive patients: cellular response and immune activation in the lung. Am J Respir Crit Care Med. 1996;153:1377–1384. doi: 10.1164/ajrccm.153.4.8616569. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Y, Nakata K, Weiden M, Rom WN. Mycobacterium tuberculosisenhances HIV-1 replication by transcriptional activation at the long terminal repeat. J Clin Invest. 1995;95:2324–2331. doi: 10.1172/JCI117924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Orendi JM, Nottet HSLM, Visser MR, Verheul AFM, Snippe H, Verhoef J. Enhancement of HIV-1 replication in peripheral blood mononuclear cells by Cryptococcus neoformansis monocyte-dependent but tumor necrosis factor-independent. AIDS (Lond) 1994;8:423–429. doi: 10.1097/00002030-199404000-00003. [DOI] [PubMed] [Google Scholar]
- 32.Peterson PK, Gekker G, Chao CC, Hu S, Edelman C, Balfour HH, Verhoef J. Human cytomegalovirus–stimulated peripheral blood mononuclear cells induce HIV-1 replication via a tumor necrosis factor-α–mediated mechanism. J Clin Invest. 1992;89:574–580. doi: 10.1172/JCI115623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Donovan RM, Bush CE, Markowitz NP, Baxa DM, Saravolatz LD. Changes in virus load markers during AIDS-associated opportunistic diseases in human immunodeficiency virus-infected persons. J Infect Dis. 1996;174:401–403. doi: 10.1093/infdis/174.2.401. [DOI] [PubMed] [Google Scholar]
- 34.Goletti D, Weissman D, Jackson RW, Graham NMH, Vlahov D, Klein RS, Munsiff SS, Ortona L, Cauda R, Fauci AS. Effect of Mycobacterium tuberculosison HIV replication. Role of immune activation. J Immunol. 1996;157:1271–1278. [PubMed] [Google Scholar]
- 35.Poli G, Kinter AL, Vicenzi E, Fauci AS. Interleukin-1 induces expression of the human immunodeficiency virus alone and in synergy with interleukin-6 in chronically infected U1 cells: inhibition of inductive effects by the interleukin-1 receptor antagonist. Proc Natl Acad Sci USA. 1994;91:108–112. doi: 10.1073/pnas.91.1.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pomerantz RJ, Feinberg MB, Trono D, Baltimore D. Lipopolysaccharide is a potent monocyte/macrophage-specific stimulator of human immunodeficient virus type 1 expression. J Exp Med. 1990;172:253–261. doi: 10.1084/jem.172.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tadmori W, Mondal D, Tadmori I, Prakash O. Transactivation of human immunodeficiency virus type 1 long terminal repeats by cell surface tumor necrosis factor α. J Virol. 1991;65:6425–6429. doi: 10.1128/jvi.65.12.6425-6429.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu Y, Stenzel M, Sodroski J, Haseltine W. Effects of long terminal repeat mutations on human immunodeficiency virus type 1 replication. J Virol. 1989;63:4115–4119. doi: 10.1128/jvi.63.9.4115-4119.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu Y, Touzjian N, Stenzel M, Dorfman T, Sodroski J, Haseltine W. Identification of cis-acting repressive sequences within the negative regulatory element of HIV-1. J Virol. 1990;64:5226–5229. doi: 10.1128/jvi.64.10.5226-5229.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rich E, Chen I, Zack J, Leonard M, O'Brien W. Increased susceptibility of differentiated mononuclear phagocytes to productive infection with human immunodeficiency virus-1. J Clin Invest. 1992;89:176–183. doi: 10.1172/JCI115559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schuitemaker H, Kootstra NA, Koppelman MHGM, Bruistein SM, Huisman HG, Tersmette M, Miedema F. Proliferation-dependent HIV-1 infection of monocytes occurs during differentiation into macrophages. J Clin Invest. 1992;89:1154–1160. doi: 10.1172/JCI115697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goletti D, Kinter A, Biswas P, Bende S, Poli G, Fauci A. Effects of cellular differentiation on cytokine- induced expression of human immunodeficiency virus in chronically infected promonocytic cells: dissociation of cellular differentiation and viral expression. J Virol. 1995;69:2540–2546. doi: 10.1128/jvi.69.4.2540-2546.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Natsuka S, Akira S. Macrophage differentiation-specific expression of NF-IL6, a transcription factor for interleukin 6. Blood. 1992;79:460–466. [PubMed] [Google Scholar]
- 44.Zhang Y, Broser M, Rom WN. Activation of the interleukin-6 gene by Mycobacterium tuberculosisor lipopolysaccharide is mediated by NF-IL6 and NF-κB. Proc Natl Acad Sci USA. 1994;91:2225–2229. doi: 10.1073/pnas.91.6.2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Y, Rom WN. Regulation of the interleukin-1β gene by mycobacterial components and lipopolysaccharide is mediated by two NF-IL6-like motifs. Mol Cell Biol. 1993;13:3831–3837. doi: 10.1128/mcb.13.6.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vietor I, Oliveira IC, Vilcek J. CCAAT box enhancer binding protein α (C/EBP-α) stimulates κB element-mediated transcription in transfected cells. J Biol Chem. 1996;271:5595–5602. doi: 10.1074/jbc.271.10.5595. [DOI] [PubMed] [Google Scholar]
- 47.Ruocco MR, Chen X, Ambrosino C, Dragonetti E, Liu W, Mallardo M, De Falco G, Palmieri C, Franzoso G, Quinto I, et al. Regulation of HIV-1 long terminal repeats by interaction of C/EBP (NF-IL6) and NF-κB/rel transcription factors. J Biol Chem. 1996;247:22479–22486. doi: 10.1074/jbc.271.37.22479. [DOI] [PubMed] [Google Scholar]
- 48.An MR, Hsieh CC, Reisner PD, Rabek JP, Scott SG, Kuninger DT, Papaconstantinou J. Evidence for posttranscriptional regulation of C/EBPα and C/EBPβ isoform expression during lipopolysaccharide-mediated acute-phase response. Mol Cell Biol. 1996;16:2295–2306. doi: 10.1128/mcb.16.5.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]